Significance
Scramblases translocate lipids across the lipid bilayer without consumption of ATP, thereby regulating lipid distributions in cellular membranes. Cytosol-to-lumen translocation across the endoplasmic reticulum (ER) membrane is a common process among lipid glycoconjugates involved in posttranslational protein modifications in eukaryotes. These translocations are thought to be mediated by specific ER-resident scramblases, but the identity of these proteins and the underlying molecular mechanisms have been elusive. Here, we show that CLPTM1L, an integral membrane protein with eight putative transmembrane domains, is the major lipid scramblase involved in efficient glycosylphosphatidylinositol biosynthesis in the ER membrane. Our results validate the long-standing hypothesis that lipid scramblases ensure the efficient translocations of lipid glycoconjugates across the ER membrane for protein glycosylation pathways.
Keywords: endoplasmic reticulum, scramblase, glycosylphosphatidylinositol, glycobiology, CLPTM1L
Abstract
Glycosylphosphatidylinositols (GPIs) are complex glycolipids that act as membrane anchors of many eukaryotic cell surface proteins. Biosynthesis of GPIs is initiated at the cytosolic face of the endoplasmic reticulum (ER) by generation of N-acetylglucosaminyl-phosphatidylinositol (GlcNAc-PI). The second intermediate, glucosaminyl-phosphatidylinositol (GlcN-PI), is translocated across the membrane to the luminal face for later biosynthetic steps and attachment to proteins. The mechanism of the luminal translocation of GlcN-PI is unclear. Here, we report a genome-wide CRISPR knockout screen of genes required for rescuing GPI-anchored protein expression after addition of chemically synthesized GlcNAc-PI to PIGA-knockout cells that cannot synthesize GlcNAc-PI. We identified CLPTM1L (cleft lip and palate transmembrane protein 1-like), an ER-resident multipass membrane protein, as a GlcN-PI scramblase required for efficient biosynthesis of GPIs. Knockout of CLPTM1L in PIGA-knockout cells impaired the efficient utilization of chemically synthesized GlcNAc-PI and GlcN-PI for GPI biosynthesis. Purified CLPTM1L scrambled GlcN-PI, GlcNAc-PI, PI, and several other phospholipids in vitro. CLPTM1L, a member of the PQ-loop family of proteins, represents a type of lipid scramblase having no structural similarity to known lipid scramblases. Knockout of CLPTM1L in various wild-type mammalian cultured cells partially decreased the level of GPI-anchored proteins. These results suggest that CLPTM1L is the major lipid scramblase involved in cytosol-to-lumen translocation of GlcN-PI across the ER membrane for efficient GPI biosynthesis.
Glycosylphosphatidylinositol (GPI) anchoring is a posttranslational modification of many eukaryotic cell surface proteins, including more than 150 human proteins (1). GPI is a highly conserved complex glycolipid in eukaryotic organisms, with a core structure comprising EtNP-6Manα−2Manα−6Manα−4GlcNα−6Inositol−PL (where EtNP, Man, GlcN, and PL are ethanolamine phosphate, mannose, glucosamine, and phospholipid, respectively) (SI Appendix, Fig. S1A). Defects in the GPI biosynthetic pathway can result in rare acquired diseases, such as paroxysmal nocturnal hemoglobinuria and a group of congenital disorders of glycosylation, known as inherited GPI deficiencies (IGD) (2, 3). Biosynthesis of GPI starting from phosphatidylinositol (PI) is a stepwise sequence of at least 10 reactions occurring on the endoplasmic reticulum (ER) membrane (Fig. 1A). The first biosynthetic step is transfer of N-acetylglucosamine (GlcNAc) from uridine-diphosphate (UDP)-GlcNAc to PI to generate GlcNAc-PI, which is then de-N-acetylated to generate GlcN-PI. These two enzymatic reactions occur on the cytosolic face (4, 5) and the resulting GlcN-PI is translocated across the ER membrane into the luminal face, where later steps and attachment to proteins occur (Fig. 1A). Whereas enzymes that catalyze transfers of monosaccharides and other components to growing GPI have been molecularly cloned and characterized, the molecular basis of cytosol-to-lumen transmembrane (TM) translocation of GlcN-PI is unknown.
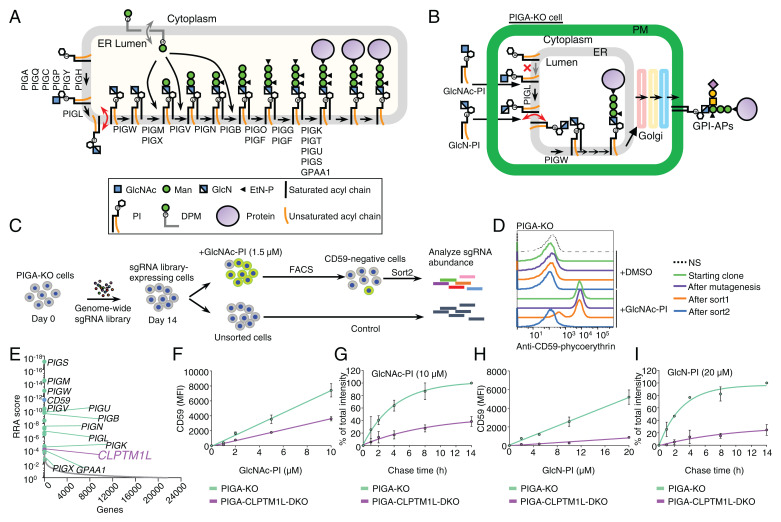
Fig. 1.
A genetic screen for components of the GPI biosynthetic pathway identified CLPTM1L. (A) Schematic of the GPI biosynthetic pathway in the ER. The GlcN-PI scrambling step is indicated in red and DPM scrambling is indicated in gray. Genes essential for GPI biosynthesis and GPI anchoring to protein are listed. (B) Schematic of the restoration of GPI biosynthesis in PIGA-KO cells in the presence of chemically synthesized GlcNAc-PI or GlcN-PI. (C) Scheme depicting a FACS-based genome-wide CRISPR screen for genes involved in efficient GlcNAc-PI utilization for GPI biosynthesis using PIGA-KO HEK293 cells. (D) Loss of an ability to restore GPI-AP using exogenous GlcNAc-PI by PIGA-KO cells after the FACS-based genome-wide CRISPR screen. Starting clone, cells after mutagenesis, sort1 cells, and sort2 cells incubated with DMSO or 2 µM GlcNAc-PI overnight were stained with anti-CD59 mAb and analyzed by flow cytometry. NS, cells not stained with first antibody. Representative of two biological repeats. (E) Gene scores in unsorted versus sorted PIGA-KO cells. Known GPI biosynthetic pathway genes are shown in green with some of their names, CD59 is shown in blue, and CLPTM1L is shown in purple. See also Datasets S2 and S3. (F and H) Restoration of CD59 expression in PIGA-KO and CLPTM1L-PIGA-DKO cells in the presence of various concentrations of GlcNAc-PI (F) or GlcN-PI (H) for 24 h. MFI: mean fluorescence intensity. (G and I) Time course of CD59 expression in PIGA-KO and CLPTM1L-PIGA-DKO cells preincubated with GlcNAc-PI (G) or GlcN-PI (I) for 2 h. Data are mean ± SEM in F–I of two independent biological repeats.
Other lipid glycoconjugates involved in protein modifications—including dolichol-pyrophosphate-heptasaccharide, dolichol-phosphate-mannose (DPM), and dolichol-phosphate-glucose (DPG)—are also generated on the cytoplasmic face of the ER and then are translocated to the luminal side. It is a long-standing hypothesis that specific scramblases mediate the TM translocation of these lipid glycoconjugates involved in posttranslational modification of proteins in the ER lumen, and while the activity of these scramblases has been measured (6, 7), their molecular identity has been elusive (6, 8–10). Many genes required for GPI biosynthesis have been identified by establishing GPI-deficient mutants from various cultured cell lines. Given that the entry of GlcN-PI into the ER lumen is an essential step for GPI assembly, it is surprising that previous intensive forward genetic screens all failed to identify GlcN-PI scramblase. It seems possible that more than one protein works redundantly to scramble GlcN-PI or that the scramblase is a moonlighting protein essential for cell survival in its other role.
In this study, we used PIGA-knockout (KO) human embryonic kidney 293 (HEK293) cells that cannot synthesize GlcNAc-PI. Performing a new CRISPR-based genome-wide screen for genes that are needed by these cells to utilize exogenously supplied, synthetic GlcNAc-PI for GPI biosynthesis, we identified CLPTM1L (cleft lip and palate transmembrane protein 1-like). We show that CLPTM1L is an ER-resident lipid scramblase that mediates translocation of GlcN-PI across the ER membrane for efficient GPI biosynthesis in the ER lumen. As biosynthesis of GPI at steady state is partially impaired in CLPTM1L-KO cells, our results further suggest that another protein may act redundantly with CLPTM1L to scramble GlcN-PI.
Results
A Genetic Screen for Components of the GPI Biosynthetic Pathway Identified CLPTM1L.
Aiming to identify new genes, including the GlcN-PI scramblase, that are involved in biosynthesis and trafficking of GPI-anchored proteins (GPI-APs), we took advantage of our recent finding that chemically synthesized GlcNAc-PI or GlcN-PI (SI Appendix, Fig. S1B) is able to rescue GPI-AP generation in PIGA-KO cells that do not make GlcNAc-PI due to a defect in the first step in GPI biosynthesis (Fig. 1B) (11). Previous studies suggest that exogenous PI can be flipped into the inner leaflet of the plasma membrane (12), and then reach other organelles, including the ER (13), where it can be used for GPI biosynthesis (14). Probably through the same transport route, exogenous GlcNAc-PI is translocated to the ER membrane. After de-N-acetylation by PIGL to generate GlcN-PI at the cytoplasmic face of the ER, the translocation of GlcN-PI across the ER membrane is necessary for efficient utilization of GlcN-PI for GPI biosynthesis (Fig. 1B).
We established a genome-wide CRISPR-Cas9 KO screen using PIGA-KO HEK293 cells as the parent cells. PIGA-KO cells were transduced with a genome-wide single-guide RNA (sgRNA) library and maintained for 2 wk, followed by culturing with GlcNAc-PI at a low concentration overnight (15). Mutant cells defective in restoring CD59, a GPI-AP marker, were collected by cell sorting and enriched sgRNA sequences were determined (Fig. 1 C and D). As expected, most previously known genes involved in the GPI biosynthetic pathway were highly enriched in the sorted CD59-deficient cells, supporting the validity of this screening strategy. Notably, CLPTM1L of unknown function was one of the top-ranking genes from this screen (Fig. 1E). To confirm the screening result, CLPTM1L-PIGA-double-KO (DKO) HEK293 cells were generated (SI Appendix, Fig. S1 C and D) and their ability to restore GPI-APs after incubation with GlcNAc-PI and GlcN-PI was assessed. GlcNAc-PI was 2-fold less effective in promoting CD59 synthesis in DKO vs. PIGA-KO cells when measured at a single time point (Fig. 1F), and 2.6-fold less effective when assessed in a pulse-chase experiment (Fig. 1G). Restoration efficiency with GlcN-PI was even more strongly affected by CLPTM1L KO, being 5.1-fold and 8.1-fold less efficient in the two types of assays used (Fig. 1 H and I). Therefore, CLPTM1L is required for efficient generation of GPI-APs from GlcN-PI.
CLPTM1L Is a Widely Expressed ER-Resident PQ-Loop Family Protein with Eight TM Domains.
CLPTM1L, also known as cisplatin resistance-related protein 9, was originally identified as a gene associated with cisplatin-induced apoptosis of carcinoma cells and is a member of the CLPTM1 family (16). Consistent with involvement in GPI biosynthesis, CLPTM1L is ubiquitously expressed in human tissues and organs similar to PIGA (SI Appendix, Fig. S2 A and B). Both plasma membrane and intracellular localizations of CLPTM1L were reported in pancreatic adenocarcinoma cells and antibody-producing plasma cells, which express high levels of this protein (17–19). In human HEK293 cells, a noncarcinoma cell line, we used antibodies against endogenous CLPTM1L protein (the specificity of the antibodies was verified by using CLPTM1L-KO HEK293 cells) (SI Appendix, Fig. S1C), and observed colocalization with an ER membrane protein UBE2J1 (Fig. 2A). The N-glycans of CLPTM1L were sensitive to endoglycosidase H treatment, consistent with ER localization of the protein (Fig. 2B). ER localization was also observed for overexpressed monomeric enhanced green fluorescent protein (mEGFP)–tagged CLPTM1L (CLPTM1L-mEGFP) (SI Appendix, Fig. S2C). Whereas in carcinoma cells, a fraction of CLPTM1L might exit the ER when it is expressed at high levels, our results indicate that CLPTM1L acts at the level of the ER in regulating GlcN(Ac)-PI–mediated restoration of GPI biosynthesis in PIGA-KO HEK293 cells.
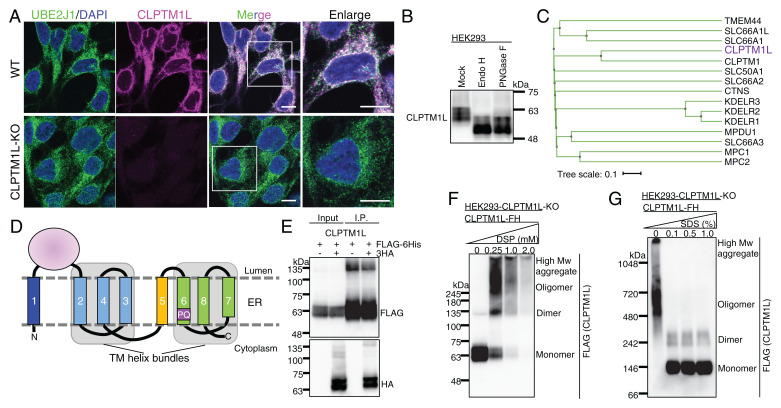
Fig. 2.
CLPTM1L is an ER-resident PQ-loop family protein with eight TM domains. (A) IF detection of endogenous CLPTM1L (magenta) and the ER membrane protein marker UBE2J1 (green) in wild-type (WT) and CLPTM1L-KO HEK293 cells. (Scale bars, 10 μm.) (B) Western blotting of endogenous CLPTM1L. Lysates of HEK293 cells treated with or without EndoH or PNGase F were analyzed by Western blotting. (C) The human PQ-loop family tree. Protein sequences taken from UniProt were aligned by the MAFFT to generate the tree file and illustrated using iTOL (65, 66). Gene names are shown, and CLPTM1L is shown in purple. (D) Topology model of human CLPTM1L in the ER membrane. TM5 is the inversion linker TM helix between two TM helix bundles. The TM domains are indicated by numbers. The PQ motif within TM6 is indicated. (E) Empty vector or CLPTM1L-3HA was coexpressed with CLPTM1L-FLAG-6His (CLPTM1L-FH) in CLPTM1L-KO HEK293 cells. Immunoprecipitates with anti-FLAG antibody resins were separated by SDS/PAGE and analyzed by Western blotting with anti-FLAG and anti-HA antibodies. (F) CLPTM1L-KO HEK293 cells expressing CLPTM1L-FH were incubated with various concentrations of a chemical cross-linker dithiobis (succinimidyl propionate) (DSP). Cell lysates were analyzed by Western blotting. (G) Lysates from CLPTM1L-KO HEK293 cells expressing CLPTM1L-FH were treated with various concentrations of SDS. Proteins were separated by blue-native gel electrophoresis and were analyzed by Western blotting. Experiments in A, B, and E–G are representative of two biological repeats.
Various algorithms predict five to eight TM domains in human CLPTM1L (SI Appendix, Fig. S3A). Since the predicted number of TM domains of CLPTM1L from other species are very different, the number of TM domains suggested by secondary-structure predictions may be inaccurate. We therefore generated a structure model for human CLPTM1L by trRosetta (20), which detected two SWEET/PQ-loop (PF04193) family members from Arabidopsis thaliana, atSWEET13 and atSWEET2, as the homologous templates (SI Appendix, Fig. S3 B and C). SWEETs are bidirectional sugar transporters (21), which might also transport other metabolites, like gibberellins (22). The PQ-loop family is a functionally diverse family of membrane proteins including CLPTM1L (Fig. 2C) (23). The PQ-loop family proteins usually have seven TM domains in a 1–3–2–4–5–7–6 helix arrangement (SI Appendix, Fig. S3D) (24); the CLPTM1L structure model contains eight TM domains (SI Appendix, Fig. S3C). Despite lack of significant sequence similarity between CLPTM1L and atSWEET13 (SI Appendix, Fig. S3B), the TM arrangement of TM2 to TM8 from the predicted CLPTM1L structure has similarity to atSWEET13 (SI Appendix, Fig. S3E). To verify the membrane topology of CLPTM1L, we determined the orientation of the hydrophilic region between TM1 and TM2, and the C terminus in HEK293 cells. The region between TM1 and TM2 was detected by a region-specific antibody when both the plasma membrane and ER membrane were permeabilized by Triton X-100 but not when only the plasma membrane was permeabilized by digitonin (the latter treatment allowed detection of the cytoplasmic region of a tail-anchored ER membrane protein, UBE2J1) (SI Appendix, Fig. S4A). CLPTM1L fused with N-terminal HA tag and C-terminal mEGFP was stained with monoclonal antibodies (mAbs) after permeabilization by Triton X-100 or digitonin (SI Appendix, Fig. S4B). Based on these results, we propose that CLPTM1L contains eight TM domains and that the N and C termini are cytosolically oriented, and the region between TM1 and TM2 is luminally oriented (Fig. 2D).
CLPTM1L-Mediated Efficient Utilization of GlcN-PI Is Crucial for the Downstream GPI Biosynthetic Reactions in the ER Lumen.
Although no PQ-loop protein has been demonstrated to have lipid scramblase activity, ANY1, a PQ-loop protein antagonizing yeast PL flippase, was predicted as a lipid scramblase candidate (25). The PQ-loop family belongs to the transporter-opsin-G protein–coupled receptor (TOP) superfamily. Some TOP family proteins, such as the rhodopsin-like class-A G protein–coupled receptors, have lipid scramblase activity (26, 27). Furthermore, CLPTM1L, similar to SWEET proteins and many scramblases (24, 28), appeared to make homodimers or bigger oligomers on analysis by coimmunoprecipitation (Fig. 2E), chemical cross-linking, and blue-native PAGE (Fig. 2 F and G), supporting the idea that CLPTM1L may function as a GlcN-PI scramblase. The size of CLPTM1L estimated by blue-native PAGE was approximately twofold bigger than the size estimated by SDS/PAGE. A similar observation was reported for various membrane proteins (29).
Results with CLPTM1L-PIGA-DKO HEK293 cells indicated that CLPTM1L is necessary for efficient utilization of exogenous GlcNAc-PI and GlcN-PI for GPI biosynthesis in the ER (Fig. 1 F–I). Next, we sought to determine whether CLPTM1L is also critical for endogenous GPI biosynthesis by generating CLPTM1L-KO HEK293 cells. CLPTM1L-KO caused approximately a 30% reduction in cell surface CD59 at steady state, which was rescued by expression of CLPTM1L-mEGFP (Fig. 3A and SI Appendix, Fig. S5A). These results are consistent with a role for CLPTM1L as a lipid scramblase candidate involved in GPI biosynthesis in HEK293 cells, although the partial decrease in CD59 level in CLPTM1L-KO cells suggests that there may be additional scramblases functioning redundantly alongside CLPTM1L.
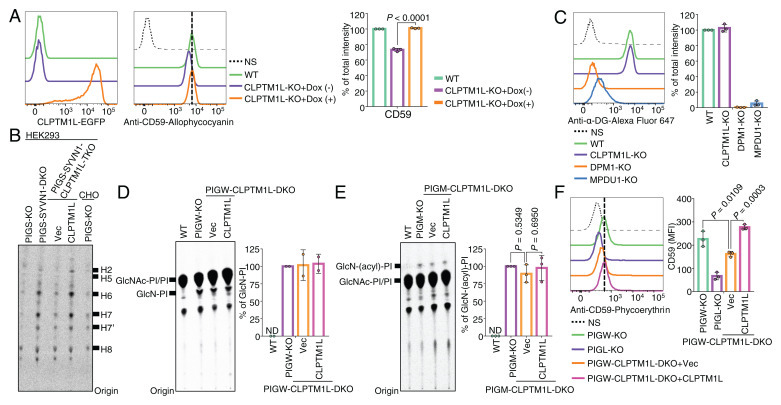
Fig. 3.
CLPTM1L is crucial for efficient utilization of GlcN-PI by the GPI biosynthetic pathway in the ER lumen. (A) Flow cytometry analysis of WT and CLPTM1L-KO HEK293 cells stably expressing CLPTM1L-mEGFP controlled by a doxycycline (Dox)-inducible Tet-On system (Left) stained with anti-CD59 mAb (Center). CLPTM1L-mEGFP expression is induced by 1 µg/mL of Dox. (B) HPTLC analysis of GPI intermediates from cells metabolically labeled with [3H]Man for 1 h. Representative of two biological repeats. (C) Flow cytometry analysis of WT, DPM1-KO, MPDU1-KO, and CLPTM1L-KO HEK293 cells stained with anti–α-dystroglycan (α-DG) mAb. DPM1 and MPDU1 are essential for biosynthesis and utilization of DPM, respectively. (D and E) HPTLC analysis of GlcN-PI (D) or GlcN-(acyl)-PI (E) from cells metabolically labeled with [3H]inositol for 10 h. (F) Flow cytometry analysis of PIGW-KO, PIGL-KO, and PIGW-CLPTM1L-DKO HEK293 cells stably expressing empty vector (Vec) or CLPTM1L stained with anti-CD59 mAb. Data shown in Right panels of A, C, and D–F are mean ± SD of three (A, C, E, and F) or two (D) independent biological repeats. In A, P value is from t test (unpaired and two-tailed). In E and F, P values are from one-way ANOVA followed by Dunnett’s test for multiple comparisons.
To further characterize GPI biosynthesis in the absence of CLPTM1L, we analyzed Man-containing GPI intermediates after metabolic labeling of cells with [3H]Man. After cellular uptake, Man is converted in several steps to DPM, which is the direct donor of Man for GPI synthesis (Fig. 1A). For this experiment, we used PIGS-KO and PIGS-SYVN1-DKO HEK293 cells that are defective in GPI-transamidase required for attachment of preassembled GPIs to proteins. In these cells, synthesized GPIs remain unlinked to proteins, undergo structural maturation similar to protein-linked GPI, and appear as free GPIs on the cell surface (30). We reported that GPI biosynthesis in PIGS-KO cells is enhanced several times after inactivation of the SYVN1-dependent ER-associated protein degradation pathway (31), enabling GPI intermediates to be more easily determined after [3H]Man-labeling (Fig. 3B). To study the effects of CLPTM1L-defect on GPI biosynthetic intermediates, we generated PIGS-SYVN1-CLPTM1L-triple KO (TKO) HEK293 cells and those rescued by CLPTM1L cDNA (SI Appendix, Fig. S5B). KO of CLPTM1L caused clear but partial decrease of all the detected Man-containing GPI intermediates (H2 to H6) and mature forms of GPIs (H7 and H8) that were restored by re-expression of CLPTM1L (Fig. 3B and SI Appendix, Fig. S5B). No new and unusual GPI species were generated. Because the reduction of Man-labeled GPI intermediates could be caused by inefficient usage of DPM, we addressed this possibility. Since DPM is required not only for GPI biosynthesis but also for protein O-mannosylation, N-glycosylation (SI Appendix, Fig. S5C), and protein C-mannosylation, we assessed the status of α-dystroglycan, an O-mannosylation marker, and found that its level was not affected by KO of CLPTM1L (Fig. 3C). As expected, the α-dystroglycan levels were greatly reduced by KO of DPM1 and MPDU1 required for synthesis and utilization of DPM, respectively (Fig. 3C). Together, these results indicate that the reduction of Man-containing GPI intermediates was indeed caused by limited availability of GlcN-PI rather than DPM and support the conclusion that CLPTM1L-mediated GlcN-PI utilization is a key step for the downstream stepwise reactions of GPI biosynthesis in the ER lumen.
To test whether CLPTM1L is involved in the N-glycosylation pathway, in which scrambling of dolichol-pyrophosphate-heptasaccharide and DPM is involved (SI Appendix, Fig. S5C), we analyzed sizes of N-glycosylated proteins LAMP1 and CD59 by SDS/PAGE. In DPM1-KO and MPDU1-KO cells, in which N-glycan precursor and GPI biosynthesis are abnormal, the smaller size of LAMP1 and a loss of CD59 expression were detected as expected (SI Appendix, Fig. S5 D and E). In contrast, their sizes were normal in CLPTM1L-KO cells (SI Appendix, Fig. S5E), albeit a lower amount of CD59 was observed similar to flow cytometry data shown in Fig. 3A. These results suggest that CLPTM1L is not involved in scrambling the N-glycan precursors dolichol-pyrophosphate-heptasaccharide and DPM.
We next analyzed GPI intermediates prior to mannosylation steps. Since GlcN-PI and GlcN-(acyl)-PI have very short lifetimes when the GPI biosynthetic pathway is ongoing, their detection by metabolic [3H]inositol labeling can be achieved in practice only by using cells defective in either inositol acylation (PIGW-KO cells) or the first mannosylation step (PIGM-KO cells). Therefore, we made PIGW-CLPTM1L-DKO cells and PIGM-CLPTM1L-DKO cells (SI Appendix, Fig. S5 F and G). The levels of GlcN-PI in PIGW-KO HEK293 cells was not appreciably affected by CLPTM1L KO (Fig. 3D), nor did we observe any significant decrease of GlcN-(acyl)-PI in PIGM-CLPTM1L-DKO cells compared to PIGM-KO cells (Fig. 3E). Considering that detection of GlcN-(acyl)-PI by inositol labeling takes several hours of labeling, sufficient GlcN-PI is scrambled on this time frame even in the absence of CLPTM1L, explaining why there is no significant change in the total amount of GlcN-PI or GlcN-(acyl)-PI in the PIGW-CLPTM1L-DKO and PIGM-CLPTM1L-DKO cells, respectively. Compared to wild-type cells, GPI-AP expression in PIGW-KO cells is 20-fold lower due to inefficient downstream reactions in the absence of inositol acylation (32). Indeed, efficient utilization of GlcN-PI mediated by CLPTM1L is necessary for GPI biosynthesis independent of the inositol acylation step, because the low level of CD59 expression in PIGW-KO cells made directly from GlcN-PI was still CLPTM1L-dependent (Fig. 3F).
CLPTM1L Scrambles GlcN-PI, GlcNAc-PI, and Various PLs In Vitro.
To test directly whether CLPTM1L can scramble GlcN-PI (Fig. 4A), we used an in vitro assay. Bacterial PI-specific phospholipase C (PI-PLC) has been previously used as a membrane-impermeable topological probe for PI, GlcNAc-PI, and GlcN-PI (12, 33, 34), and a scramblase assay using PI-PLC has been described to test activities of other scramblases, including opsin and nhTMEM16, toward [3H]PI in reconstituted vesicles (35). In this assay system, as PI-PLC only accesses the outer leaflet of liposomes and because spontaneous translocation of [3H]GlcN-PI across the membrane bilayer is expected to be negligible in a short time, about 50% of [3H]GlcN-PI will be cleaved on adding PI-PLC. If the vesicles possess a GlcN-PI scramblase, then greater than 50% of [3H]GlcN-PI will be hydrolyzed, because molecules from the inner leaflet of the vesicles will be translocated across the bilayer in a short period of time (Fig. 4B). To test whether CLPTM1L can translocate GlcN-PI, we purified CLPTM1L-FLAG-6His protein (Fig. 4C) and [3H]GlcN-PI (Fig. 4D), and reconstituted them into liposomes consisting of nine parts of egg phosphatidylcholine (PC) and one part of bovine brain phosphatidylserine (PS) (27, 36). In some experiments, [3H]PI or [3H]GlcNAc-PI were included instead of [3H]GlcN-PI. In an initial time-course study, we observed ∼80% hydrolysis of [3H]GlcN-PI in CLPTM1L-proteioliposomes and ∼50% hydrolysis of [3H]GlcN-PI in protein-free liposomes within 10 min (Fig. 4E). Similar results were obtained using [3H]PI (Fig. 4F), indicating that CLPTM1L facilitates transbilayer movement of both lipids.
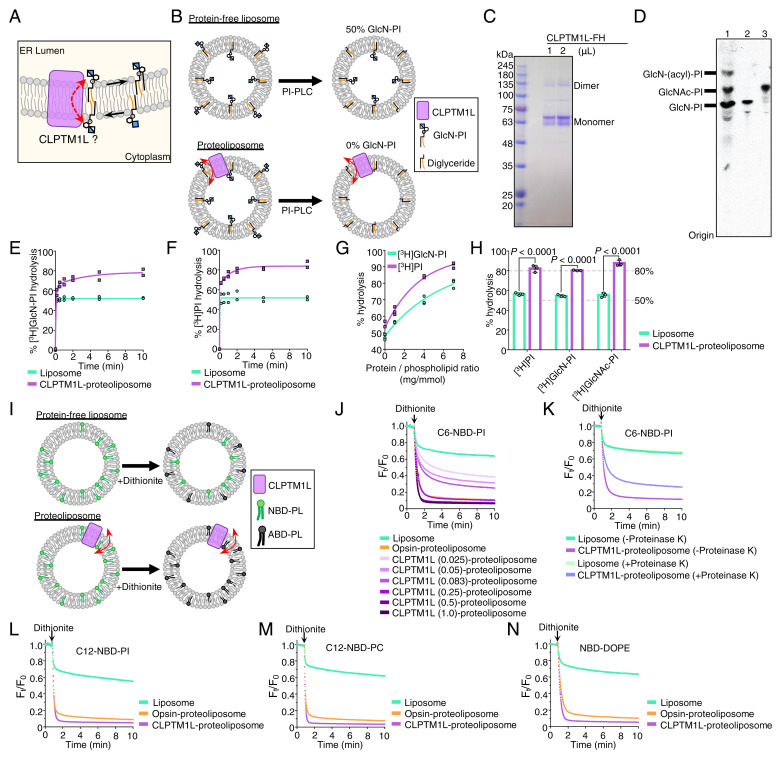
Fig. 4.
CLPTM1L scrambles GlcN-PI, GlcNAc-PI, and various PL in vitro. (A) Schematic of CLPTM1L-mediated ER luminal translocation of GlcN-PI. (B) Schematic of PI-PLC–based scramblase assay. PI-PLC hydrolyzes [3H]GlcN-PI in the outer leaflet of liposomes. (C) SDS/PAGE analysis of purified human CLPTM1L-FLAG-6His. (D) HPTLC analysis of a mixture of early GPI precursors (lane 1), purified [3H]GlcN-PI (lane 2), and purified [3H]GlcNAc-PI. (E and F) Time course of [3H]GlcN-PI (E) or [3H]PI (F) hydrolysis by PI-PLC in CLPTM1L-proteoliposomes using protein to PPR of ∼7 mg/mmol. Protein-free liposomes were used as controls. (G) PI-PLC–mediated hydrolysis of [3H]GlcN-PI or [3H]PI in CLPTM1L-proteoliposomes for 10 min at different PPRs. (H) PI-PLC–mediated hydrolysis of [3H]GlcNAc-PI in CLPTM1L-proteoliposomes (PPR ∼7 mg/mmol) for 10 min. Data are from two (E and F) or three (G and H) independent measurements. In H, P values are from t test (unpaired and two-tailed). (I) Schematic of dithionite-based scramblase assay. NBD-PLs in the outer leaflet of liposomes are bleached to nonfluorescent 7-amino-2,1,3-benzoxadiazol-4-yl (ABD)-PLs by dithionite. (J) Scrambling of acyl-C6-NBD-PI in CLPTM1L-proteoliposomes at different PPRs and bovine opsin-proteoliposomes using PPR of ∼0.8 mg/mmol. The PPR value (mg/mmol) of CLPTM1L-proteoliposome is indicated. Ft/F0 represents the fluorescence intensity at each time point (Ft) divided by F0 at time 0. (K) Scrambling of acyl-C6-NBD-PI in proteinase K or mock-treated CLPTM1L-proteoliposomes using PPR of ∼0.25 mg/mmol. (L–N) Substrate specificity of CLPTM1L toward acyl-C12-NBD-PI (L), acyl-C12-NBD-PC (M), and N-NBD-DOPE (N). Scrambling of NBD-PLs in CLPTM1L-proteoliposomes using PPR of ∼1 mg/mmol and opsin-proteoliposomes using PPR of ∼0.8 mg/mmol. Experiments in J–N are representative of two independent measurements.
Consistent with a previous study using nhTMEM16 and opsin (35), hydrolysis of GlcN-PI or PI by PI-PLC in CLPTM1L-containing proteoliposomes was a biphasic process (Fig. 4 E and F), with an initial rapid burst corresponding to hydrolysis of substrate in the outer leaflet followed by a slower phase in which molecules from the inner leaflet cross the membrane to the outer leaflet. To test whether scrambling of GlcN-PI and PI is CLPTM1L dose dependent, we measured end-point (10 min) scramblase activity of CLPTM1L-proteoliposomes prepared with different protein to PL ratios (PPRs). Hydrolysis of [3H]GlcN-PI and [3H]PI increased from ∼50% to >80%, with increased PPRs from 0 to 7 mg/mmol (Fig. 4G), similar to the previous study using nhTMEM16 and opsin (35). CLPTM1L also scrambled [3H]GlcNAc-PI prepared by acetylation of [3H]GlcN-PI (Fig. 4D). Approximately 80% of [3H]GlcNAc-PI was hydrolyzed by PI-PLC in CLPTM1L-proteoliposomes within 10 min (Fig. 4H), similar to the results for [3H]GlcN-PI and [3H]PI.
To address whether CLPTM1L can translocate other kinds of PLs, we tested nitrobenzoxadiazole (NBD)–labeled PLs. The membrane-impermeant reductant dithionite bleaches the fluorescence of NBD-PLs in the outer leaflet of the liposomes (26, 27), thus ∼50% bleaching is expected on adding dithionite to protein-free liposomes and nearly complete reduction of fluorescence is expected if all the vesicles contain a scramblase (Fig. 4I). We used NBD acyl-labeled PI (C6-NBD-PI and C12-NBD-PI) as positive control lipids (SI Appendix, Fig. S6) (35) and purified bovine opsin as a positive control scramblase. Dithionite reduced the fluorescence of NBD-PI from bovine opsin-proteoliposomes by ∼90% (Fig. 4J) (26, 35). As seen by the greater loss of fluorescence in dithionite-treated CLPTM1L-proteoliposomes versus protein-free liposomes, CLPTM1L scrambled C6-NBD-PI in a dose-dependent manner (Fig. 4J). The reduction in NBD fluorescence upon adding dithionite to CLPTM1L-proteoliposomes treated with proteinase K was less than that seen for the untreated sample (Fig. 4K). The partial loss of activity is most likely caused by incomplete proteolysis of membrane-embedded proteins, as has been observed previously (37). We next tested the ability of CLPTM1L to scramble other PLs. Similar to opsin-proteoliposomes, we observed nearly complete fluorescence reduction on dithionite addition to CLPTM1L-proteoliposomes containing C12-NBD-PI (Fig. 4L), NBD acyl-labeled PC (C12-NBD-PC) (Fig. 4M), and NBD head group-labeled dioleoylphosphatidylethanolamine (NBD-DOPE) (Fig. 4N) as transport reporters. Taking these data together, we find that CLPTM1L exhibits scramblase activity toward both anionic and zwitterionic PLs in vitro. We conclude that CLPTM1L mediates scrambling of GlcN-PI in a nonspecific manner.
Other Scramblases Might Contribute to Scrambling of GlcN-PI in the Absence of CLPTM1L.
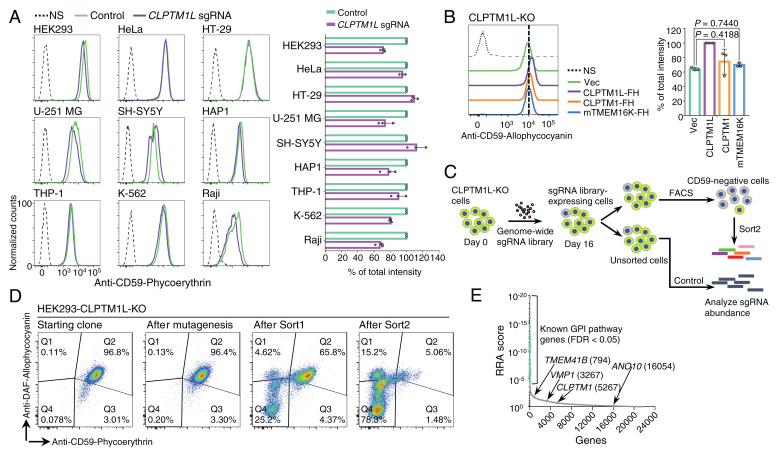
While CLPTM1L appears to be the major lipid scramblase required for GPI biosynthesis in HEK293 cells, results with CLPTM1L-PIGA-DKO or CLPTM1L-KO cells indicated that cytosol-to-lumen translocation of GlcN-PI is mediated redundantly by more than one scramblase in HEK293 cells. As in CLPTM1L-KO HEK293 cells, the partial deficiency (10 to 30% reduction) of CD59 at steady state was also observed in most but not all CLPTM1L-KO human cells derived from various cell lines (Fig. 5A and SI Appendix, Fig. S7A). Therefore, it is likely that GlcN-PI scrambling in various cell types is mediated by redundantly acting scramblases, among which CLPTM1L plays a dominant role in many cells. Overexpression of TMEM16K, a Ca2+-activated ER scramblase (38), failed to rescue GPI deficiency in CLPTM1L-KO cells (Fig. 5B and SI Appendix, Fig. S7B). Although the other CLPTM1L homologous protein, CLPTM1, is likely a lipid scramblase, deficiency of GPI biosynthesis in CLPTM1L-KO cells was not effectively rescued by overexpression of CLPTM1 (Fig. 5B and SI Appendix, Fig. S7 B and C). Therefore, CLPTM1 might have no functional overlap with CLPTM1L (39, 40).
Fig. 5.
Other scramblases might contribute to translocation of GlcN-PI in the absence of CLPTM1L. (A) Flow cytometry analysis of various human cultured cell lines with CLPTM1L deficiency. Cells stably expressing lentiCRISPR v2 with CLPTM1L-targeting sgRNAs were stained with anti-CD59 mAb. (B) Flow cytometry analysis of CLPTM1L-KO HEK293 cells stained with anti-CD59 mAb. Overexpression of CLPTM1L-FH, CLPTM1-FH, and mTMEM16K-FH controlled by the Tet-On system was induced by 1 µg/mL Dox. In A and B, data on the right are mean ± SD of three independent biological repeats. In B, P values are from one-way ANOVA followed by Dunnett’s test for multiple comparisons. (C) Scheme depicting a FACS-based genome-wide CRISPR screen for genes involved in GPI biosynthesis using CLPTM1L-KO HEK293 cells. (D) Flow cytometry analysis of CLPTM1L-KO HEK293 cells during the FACS-based genome-wide CRISPR screen. Starting clone, cells after mutagenesis, sort1 cells, and sort2 cells were stained with anti-CD59 and anti-CD55/DAF antibodies. (E) Gene scores in unsorted versus sorted CLPTM1L-KO cells. Known GPI biosynthetic pathway genes are shown in green. CLPTM1 and reported ER scramblase genes, including TMEM41B, VMP1, and TMEM16K/ANO10, are shown in pink. The ranking numbers based on the gene scores are shown in parentheses after gene names. See also Datasets S4 and S5.
To search for other scramblases, we performed a conventional genome-wide CRISPR-Cas9 KO screen using CLPTM1L-KO HEK293 cells (Fig. 5 C and D). If only one scramblase is capable of compensating for GlcN-PI scrambling in the absence of CLPTM1L, this approach would identify the other scramblase candidate. However, top-ranked genes were mainly previously known genes required for GPI-AP generation, and CLPTM1 and reported ER scramblases—including TMEM16K, TMEM41B, and VMP1 (38, 41–44)—were not highly ranked (Fig. 5E). No new scramblase candidates were identified (Fig. 5E). This could be due to a possibility that only a slight decrease in GPI-AP expression occurs by KO of the second scramblase gene. These results suggest that multiple redundant scramblases might contribute to GlcN-PI translocation in the absence of CLPTM1L.
Discussion
Taking these data together, we conclude that CLPTM1L is a long-sought scramblase that mediates luminal translocation of GlcN-PI for biosynthesis of GPI in the ER. Because the level of GPI-APs is reduced significantly but not eliminated in CLPTM1L-KO cells, CLPTM1L should have a major role in GPI biosynthesis and there must be at least one more protein capable of luminal translocation of GlcN-PI, accounting for the remaining GPI biosynthesis in these cells. For GPI biosynthesis, DPM and PE are used as Man- and EtNP-donors, respectively (45, 46), and both donor lipids must be translocated into the ER lumen after their synthesis. We determined that CLPTM1L is not required for DPM usage in the ER lumen. Recently, Vps13-like lipid transfer protein, yeast Csf1, and its mammalian homolog KIAA1109/TWEEK were shown to supply PE from the mitochondria to the ER for GPI biosynthesis (47). Since CLPTM1L scrambled PE in vitro, CLPTM1L might also be involved in luminal translocation of PE for EtNP transfer. However, inefficient rescue of CD59 in PIGA-CLPTM1L-DKO cells after addition of low concentrations of GlcNAc-PI or GlcN-PI must be caused by inefficient luminal translocation of GlcN-PI rather than that of PE. GlcNAc-PI is short-lived and immediately de-N-acetylated to form GlcN-PI by PIGL in normal cells, and theoretically translocation of GlcNAc-PI across the ER membrane is not required for GPI biosynthesis. As accumulation of GlcNAc-PI was observed in cells carrying mutations in PIGL (48), it would be interesting to learn whether CLPTM1L is capable of mediating the luminal translocation of GlcNAc-PI in cells from patients carrying mutations in PIGL (49).
No deficiency of N-glycosylation is observed in CLPTM1L-KO cells, so CLPTM1L is not the major scramblase involved in luminal translocation of dolichol-pyrophosphate-heptasaccharide. Although complex lipid glycoconjugates are not the authentic substrates for opsin under physiological conditions, it was shown that opsin was capable of scrambling mature GPI but not dolichol-pyrophosphate-heptasaccharide in vitro (27). It is possible that CLPTM1L is active to more complex GPI precursors in vitro; however, its major in vivo role in scrambling GlcN-PI might be due to its distribution within the ER that limits its contribution to the early steps of GPI biosynthesis. In Trypanosoma brucei, more complex GPI intermediates, such as those with acylated inositol, and more than one Man are thought to scramble across the ER membrane (34, 50). Although a similar phenomenon has not been shown in mammalian cells, it should be clarified in future studies if the T. brucei CLPTM1L homolog (SI Appendix, Fig. S8A) scrambles those complex GPI intermediates as well as the early intermediates.
Clptm1l-KO mice showed very low embryonic and neonatal survival, but the survivors allowed further phenotypic characterization (17). The phenotypic data of Clptm1l-KO mice from the International Mouse Phenotyping Consortium website (https://www.mousephenotype.org) suggest that the survivors have neurological, skeletal, and eye abnormalities (51). The early embryonic lethality was observed in mice with complete deficiency of GPI biosynthesis (52). The Pigv341E knockin mice with partial deficiency of GPI biosynthesis were viable but had shortened life time and provided a good model of IGD characterized by neurological deficits (53). Clptm1l-KO mice might be an alternative IGD model. The partial deficiency of GPI biosynthesis should account for the embryonic and neonatal death of the Clptm1l-KO mice, and the overlapping disease phenotypes of Pigv mutation knockin mice, suggesting a compensation by the second mechanism of luminal translocation of GlcN-PI might be insufficient in some tissues.
Although CLPTM1L is highly conserved in eukaryotes, it is notable that CLPTM1L homologs exist in only a few yeasts, but not Saccharomyces, suggesting the existence of other scramblases for GlcN-PI in Saccharomyces spp. (SI Appendix, Fig. S8A). This functional redundancy explains previous failures in identifying the enzyme for scrambling GlcN-PI. It remains to be seen whether the GlcN-PI scramblases in Saccharomyces are homologous to the mammalian scramblases. Our genetic screen using CLPTM1L-KO cells failed to identify the additional scramblases, suggesting the existence of more than one protein that is capable of GlcN-PI translocation. Such a protein might be discovered by another strategy based on using GlcNAc-PI for GPI biosynthesis in CLPTM1L-PIGA-DKO cells. Nevertheless, our conclusion that multiple human ER scramblases are capable of GlcN-PI scrambling suggest that lipid scrambling is a fundamental event in the ER membrane, and that redundancy is necessary to ensure that this system is infallible. It is interesting to consider that high levels of signal peptides generated by signal peptidase activity on translocated proteins in the ER membrane might contribute some scrambling activity in the ER. However, these peptides are quickly eliminated from the membrane (54), and very low scramblase activity from such peptides was observed from previous in vitro assays (55, 56). Therefore, the major scrambling activity in the ER would be contributed by scramblases rather than these peptides.
Our discovery of CLPTM1L as a lipid scramblase further expands the functional scope of the PQ-loop family of proteins. Although CLPTM1 has been implicated in efficient GPI-inositol deacylation and limiting forward trafficking of GABAA receptors (39, 40), it is possible that CLPTM1 might function as a scramblase in the ER because its sequence similarity to CLPTM1L is high (37.7% identity in 467 residues overlap) (SI Appendix, Fig. S8B). The PQ-loop family proteins usually share very little sequence identity, and whether other PQ-loop family members, such as MPDU1 (Mannose-P-dolichol utilization defect 1 protein), yeast ANY1 and its homolog SLC66A2, are scramblases is unclear. Probably due to the confusing phenotypes observed from MPDU1-deficient cells, efficient utilization of DPM and DPG is thought to be mediated by MPDU1 through an unknown mechanism (10). Although yeast ANY1 has been predicted as a lipid scramblase in the Golgi (25), a recent study suggested yeast ANY1 might not function as a scramblase due to its interaction with a PL flippase (57). Further characterization of their scramblase activity in vitro is needed to conclude whether these PQ-loop proteins are also scramblases.
Very recently, TMEM16K (ANO10), VMP1, and TMEM41B have been reported as ER scramblases (38, 41–44). TMEM16K was active as a scramblase only in the presence of calcium (38, 44), and further study suggested that TMEM16K functions at ER–endosome contact sites to regulate endosomal sorting (58). Both TMEM41B and VMP1 have been implicated in autophagosome biogenesis, lipoprotein formation, and lipid droplet formation. It has been proposed that scramblase TMEM41B or VMP1 is required for mediating the bulk lipid supply to generate these membrane structures or lipid-rich particles (41–43). Although these scramblases exhibit activity toward various PLs in vitro, they might function as ER scramblases locally to contribute to distinct pathways. It will be interesting to further investigate how these scramblases are organized and regulated in the ER membrane.
Materials and Methods
Antibodies and Chemicals.
GlcNAc-PI and GlcN-PI used in this study were chemically synthesized as recently described (11); the PI portion is 1-stearoyl-2-oleoyl-sn-glycero-3-phospho-1D-myo-inositol (SI Appendix, Fig. S1B). Acyl-NBD-PIs including 16:0–6:0 NBD-PI (C6-NBD-PI) and 16:0–12:0 NBD-PI (C12-NBD-PI) with palmitic acid (C16:0) at the sn1 position were used in this study, N-hexanoyl (C6:0)-NBD or N-dodecanoyl (C12:0)-NBD is the chain at the sn2 position. C6-NBD-PI was synthesized as previously described (35, 59); the synthesis of C12-NBD-PI is described in SI Appendix. Primary antibodies, including anti-CD55 (clone IA10, BD Biosciences) and anti-CD59 (clone 5HA) mAbs (1:100 for FACS; 1:1,000 for Western blot, or WB), were described previously (60). Phycoerythrin-labeled mouse mAb against human CD59 (1:50 for FACS) was from BioLegend. Mouse mAb against DYKDDDDK (FLAG) (1:1,000 for WB) was from Wako. Mouse mAb against GAPDH (1:1,000 for WB) was from Thermo Fisher Scientific. Rabbit Ab against CLPTM1L (1:1,000 for WB; 1:500 for immunofluorescence, or IF) was from Atlas Antibodies. Mouse mAbs against UBE2J1 (1:250 for IF) was from Santa Cruz Biotechnology. Mouse mAbs against α-tubulin (1:1,000 for WB) were from Sigma-Aldrich. Rabbit mAb against HA (1:1,000 for WB; 1:500 for IF) and LAMP1 (1:1,000 for WB) were from Cell Signaling Technology. Goat Ab against GFP (1:1,000 for WB; 1:500 for IF) was from Abcam. Mouse mAb against α-dystroglycan (1:100 for FACS) was from Merck Millipore.
Cells and Culture.
HEK293 cells (ATCC CRL-1573), HT-29 cells (ATCC HTB-38), U-251 MG cells (CVCL_0021), SH-SY5Y cells (ATCC CRL-2266), HeLa cells (ATCC CCL-2), and their KO derivatives were cultured in Dulbecco's Modified Eagle Medium (DMEM) with high glucose (Nacalai Tesque). Raji cells (ATCC CCL-86), THP-1 cells (ATCC TIB-202), K-562 cells (ATCC CCL-243), and their derivatives were cultured in RPMI medium 1640 (Nacalai Tesque). HAP1 cells (kindly provided by Thijn Brummelkamp, The Netherlands Cancer Institute, Amsterdam, The Netherlands) and their KO derivatives were cultured in Iscove's Modified Dulbecco's Medium (Nacalai Tesque). CHO (Chinese Hamster Ovary) K1 cells (ATCC CCL-61) and their derivatives were maintained in DMEM/F-12 (Nacalai Tesque). All cell culture medium contained 10% heat-inactivated fetal bovine serum (FBS) (172012-500ML, Sigma). All cells were maintained in an incubator at 37 °C and 5% CO2.
Recombinant DNA.
Human GeCKOv2 CRISPR KO pooled library (Pooled Library #1000000048), pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene plasmid #42230), and lentiCRISPR v2 (Addgene plasmid #52961) were gifts from Feng Zhang, Broad Institute of MIT and Harvard, Cambridge, MA (15, 61). pX330-EGFP plasmid was generated from pX330-U6-Chimeric_BB-CBh-hSpCas9 (62). The plasmids of the PiggyBac Tet-On expression system including pPB-tetIRB-tetON-BSD and pPB-tetON-BSD-tightFF were gifts from Yusuke Maeda, Osaka University, Osaka, Japan. The hyperactive PB transposase expression vector (pCMV-hyPBase) was a gift from Kosuke Yusa, Wellcome Trust Sanger Institute, Cambridge, United Kingdom (63). Mouse Ano10/Tmem16k cDNA was a gift from Toyoshi Fujimoto, Juntendo University, Tokyo, Japan (44). Retroviral vector pLIB2-BSD-hCLPTM1 was previously described (39). Human CLPTM1L cDNA was amplified by PCR from a human cDNA library and cloned into the vector pLIB2-BSD to construct pLIB2-BSD-hCLPTM1L. Plasmid pLIB2-BSD-hCLPTM1L-FLAG-6His was constructed by subcloning the EcoRI-MluI fragment of hCLPTM1L from pLIB2-BSD-hCLPTM1L into the same sites of pLIB2-BSD-hB3GALT4-FLAG-6His (31). To construct pLIB2-Hyg-hCLPTM1L-3HA, an EcoRI-MluI fragment of hCLPTM1L was cloned into pME-3HA, and the EcoRI-MluI fragment of hCLPTM1L-3HA was cloned into pLIB2-Hyg. To construct pPB-tetON-hCLPTM1L-FLAG-6His-BSD-tightFF plasmid, hCLPTM1L-FLAG-6His was amplified by PCR and cloned into pPB-tetON-BSD-tightFF plasmid by In-fusion cloning (Clontech). To construct pPB-tetON-Tmem16k-FLAG-6His-BSD-tightFF plasmid, mouse Ano10/Tmem16k cDNA was amplified by PCR from pMXs-IRES-Puro-mTMEM16K-FLAG, then cloned into pPB-tetON-hCLPTM1L-FLAG-6His-BSD-tightFF plasmid by In-fusion cloning. Other plasmids including pPB-tetON-hCLPTM1-FLAG-6His-BSD-tightFF, pPB-tetIRB-tetON-CLPTM1L-mEGFP-BSD, and pPB-tetIRB-tetON-HA-CLPTM1L-mEGFP-BSD were also constructed by in-fusion cloning. Oligos for generation of the plasmids used in this study are shown in Dataset S1.
Gene KO.
To generate HEK293 KO clones, sgRNAs targeting to the exon regions of each gene was cloned into pX330-EGFP plasmids. CLPTM1L-KO HEK293 clones—including CLPTM1L-KO, PIGS-SYVN1-CLPTM1L-TKO, PIGW-CLPTM1L-DKO, and PIGM-CLPTM1L-DKO—were validated by PCR to confirm a large deletion of the amplicon from CLPTM1L gene and immunoblotting to confirm the loss of endogenous CLPTM1L protein. To generate CLPTM1L-PIGA-DKO cells, PIGA-KO from CLPTM1L-KO cells were confirmed by FACS analysis to confirm complete loss of CD59 expression. DPM1-KO and MPDU1-KO cells were confirmed by FACS analysis of cell surface CD59 and α-dystroglycan expression. To knockout CLPTM1L in various human cell lines, sgRNAs targeting to the exon regions of CLPTM1L were cloned into lentiCRISPR v2 vector. Cells were transduced with lentiCRISPR v2 lentivirus and maintained in their medium containing 1 to 2 μg/mL puromycin. The polyclonal cell populations with CLPTM1L deficiency were validated by Western blotting to confirm the loss of CLPTM1L protein in each cell line. Other KO HEK293 cells, including PIGA-KO, PIGW-KO, PIGL-KO, and PIGM-KO cells were recently described (11). sgRNA sequences used in this study are in Dataset S1.
Retrovirus-Mediated Gene Overexpression.
PLAT-GP retroviral packaging cells were transfected with pLIB2-BSD or pLIB2-Hyg plasmid bearing cDNA of human CLPTM1L or CLPTM1 by using PEI “Max” transfection reagent (Polyscience) and cultured for 12 h. Cells were incubated in medium containing 10 mM sodium butyrate (Sigma-Aldrich) overnight. Cells were incubated in fresh medium at 32 °C for 1 d. Viral medium was added to HEK293 cells, followed by centrifugation at 1,100 × g at 32 °C for 4 h. Cells were maintained at 32 °C overnight, cultured in DMEM at 37 °C for another 2 d, then maintained in the medium with 10 μg/mL blasticidin (InvivoGen) or 400 μg/mL hygromycin B (Sigma-Aldrich) for at least 2 wk to achieve complete antibiotic selection before using for other experiments.
Tet-On Tetracycline-Inducible Gene Overexpression.
Expression of CLPTM1L-mEGFP, HA-CLPTM1L-mEGFP, CLPTM1L-FLAG-6His, CLPTM1-FLAG-6His, or mouse Ano10/Tmem16k-FLAG-6His in HEK293 cells or Expi293F cells was achieved through the PiggyBac Tet-On expression system. Cells were transfected with pCMV-hyPBase, and pPB-tetIRB-tetON-BSD plasmid bearing CLPTM1L-mEGFP or pPB-tetON-BSD-tightFF plasmid bearing CLPTM1L-FLAG-6His using X-tremeGENE 9 DNA transfection reagent (Roche), and incubated at 37 °C for 2 d. Cells were then maintained in the medium containing 10 μg/mL blasticidin for 2 wk, achieving complete antibiotic selection before using for other experiments.
Flow Cytometry.
Cells stained with biotin-labeled anti-CD59 mAb (clone 5H8), biotin-labeled anti-CD55/DAF mAb (clone IA10), or anti–α-dystroglycan (clone IIH6C4) in FACS buffer (phosphate-buffered saline containing 1% BSA and 0.1% NaN3) were incubated on ice for 25 min. Cells were then washed twice in FACS buffer followed by staining with Alexa Fluor 647-conjugated goat anti-mouse IgM (Abcam) for α-dystroglycan mAb, allophycocyanin-labeled streptavidin (BioLegend) for biotin-labeled anti-CD59 or anti-CD55 mAb, or phycoerythrin-labeled anti-CD59 mAb (BioLegend) in FACS buffer. After twice washing by FACS buffer, cells were analyzed by using the BD FACSCanto II.
Structure Prediction for Human CLPTM1L Protein.
The full sequence of human CLPTM1L (UniProtKB = Q96KA5) was submitted to trRosetta for structure predictions using the default setting (20). The trRosetta succeeded in generating CLPTM1L structure models using AtSWEET13 (PDB ID code 5XPD) and AtSWEET2 (PDB ID code 5CTG) (24, 28). The top-ranking model was selected, and the figures were generated by using Open-Source PyMOL 2.3.
Proteoliposome Reconstitution for the PI-PLC–Based Scramblase Assay.
For the PI-PLC–based scramblase assay, a previously reported one-pot method was used to make liposomes and proteoliposomes (27, 36) using a mixture of lipids, including egg PC and brain PS at a molar ratio of 9:1. Briefly, 8.2 μmol egg PC, 0.92 μmol brain PS, and a trace amount of [3H]PI (∼25,000 cpm), [3H]GlcN-PI (∼12,000 cpm), or [3H]GlcNAc-PI (∼12,000 cpm) were combined in a glass screw-cap tube, dried under a stream of N2 gas, and solubilized in 1.9 mL of buffer A (10 mM Hepes-NaOH, pH 7.4,100 mM NaCl, 1% [wt/vol] Triton X-100). The lipid solution was vortexed at moderate speed for 20 min until the solution become completely transparent. The solubilized lipids were divided into two aliquots and mixed with or without purified CLPTM1L-FH in a final volume of 1 mL buffer A, and incubated at 4 °C for 1 h with end-over-end rotation. Proteoliposomes or protein-free liposomes were generated by two rounds of treatment with a total of 300 mg of wet SM-2 Bio-Beads (Bio-Rad) for 1 mL of sample. The samples were placed in 2-mL Eppendorf tubes, and end-over-end mixed with 100 mg of Bio-beads at room temperature for 3 h. A second portion of 200 mg of Bio-beads was added and the incubation was continued at 4 °C for 16 h. Finally, the samples were transferred to 1.5-mL Eppendorf tubes without disturbing the Bio-beads and used immediately for scramblase assays.
PI-PLC–Based Scramblase Activity Assay.
The PI-PLC–based scramblase assay was described previously (35). Briefly, 100 μL of liposomes or proteoliposomes containing a trace amount of [3H]PI, [3H]GlcN-PI, or [3H]GlcNAc-PI were mixed with 10 μL of buffer B (10 mM Hepes, pH7.4, 100 mM NaCl) and incubated at room temperature for 5 min. After adding 3 μL of 10 U/mL PI-PLC (P6466, Thermo Fisher Scientific) diluted in buffer B, samples were incubated at 25 °C for 0 to 10 min. The reaction was stopped by adding 13 μL of 100% ice cold trichloroacetic acid (TCA), then mixed with 5 μL of 4 mg/mL cytochrome c from horse heart (105201, Sigma-Aldrich). The mixtures were kept on ice for 1 h with occasional vortex at high speed, then centrifuged at 21,900 × g for 15 min to precipitate proteins and PLs. The resulting supernatant (130 μL) was mixed with 10 mL of Clear-sol II scintillation fluid (09136-83, Nacalai Tesque) and counted with a liquid scintillation counter (TRI-CARB 4810TR, PerkinElmer). TCA samples prepared from samples without PI-PLC were taken as background, and TCA samples taken after PI-PLC treatment for 10 min in the presence of 0.91% Triton X-100 were taken as 100% hydrolysis control.
Proteoliposome Reconstitution for the Dithionite-Based Scramblase Assay.
For sodium dithionite-based scramblase assays, protein-free liposomes and proteoliposomes were made as previously described (64). Briefly, 9.45 μmol 1-palmitoyl-2-oleoyl-sn-glycero-3-PC (POPC), 1.05 μmol Dioleoyl-PS, and 0.4 mol% NBD-labeled PLs were combined. The dried lipid film was rehydrated in 2 mL of buffer C (50 mM Hepes-NaOH, pH 7.4, 100 mM NaCl). After five freeze-thaw cycles, the liposomes were prepared by extrusion of the lipid suspensions 31 times through a polycarbonate membrane with a 400-nm pore size using the Mini Extruder (Avanti Polar Lipids). After destabilizing of 1.6 mL of liposomes by 0.41% (wt/vol) n-dodecyl-β-D-maltoside (DDM) at room temperature for 3 h, the lipids were then mixed with or without purified protein (CLPTM1L-FH or opsin-3FLAG) in a final volume of 1 mL of buffer C containing 0.36% (wt/vol) DDM, then incubated at room temperature for 1 h. Proteoliposomes or protein-free liposomes were generated by three rounds of treatment with 400 mg of wet SM-2 Bio-Beads in total for 1 mL of each sample. The samples were placed in 2-mL Eppendorf tubes, and end-over-end mixed at room temperature with 80 mg of Bio-beads for 1 h, and 160 mg of Bio-beads for 2 h. The samples were mixed with 160 mg of Bio-beads at 4 °C for 16 h. Finally, the samples were transferred to 1.5-mL Eppendorf tubes and kept at 4 °C before scramblase assays.
Dithionite-Based Scramblase Activity Assay Using NBD-PLs.
The dithionite-based scramblase assay was described previously (64). Briefly, 50 μL of NBD-PL containing liposomes were diluted into 2 mL of buffer C in a disposable fluorescence cuvette (67.754.00002, SARSTEDT) at room temperature. In some cases, the samples were incubated at 37 °C with or without proteinase K from Tritirachium album (P2308, Sigma-Aldrich) for 2 h prior to scramblase assay. The total fluorescence intensity was monitored over time using the Hitachi F-2700 fluorescence spectrophotometer (λex = 470 nm, λem = 530 nm). After obtaining a stable fluorescence signal, 40 μL of freshly prepared 1 M sodium dithionite (TCI) was added to the cuvette and the fluorescence signal change was monitored for 10 min. Data were collected and analyzed using the FL Solutions 4.1 software (Hitachi).
Statistical Analysis.
All statistical analyses were performed using GraphPad Prism7. Error bars and sample size for each experiment are indicated in figure legends. Comparisons between two individual groups were analyzed by unpaired Student’s t test, and multiple comparisons between groups were analyzed by one-way ANOVA followed by Dunnett’s post hoc test. P < 0.05 was considered statistically significant.
Additional experimental procedures are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Tamao Endo and Dr. Hiroshi Manya (Tokyo Metropolitan Institute of Gerontology) for α-dystroglycan mAb (clone IIH6C4); Dr. Toyoshi Fujimoto (Juntendo University) for expression plasmids of mouse Ano10; Indu Menon for providing purified thermostable bovine opsin; Dr. Junji Takeda, Dr. Yusuke Maeda, and Dr. Yasu Morita (University of Massachusetts Amherst) for discussion; Yuki Uchikawa and Yuko Kabumoto for assistance with cell sorting; and Keiko Kinoshita, Saori Umeshita, and Kae Imanishi for technical help. Y.W. is supported by the Immunology Frontier Research Center Kishimoto Foundation Fellowship. This work was supported by Japan Society for the Promotion of Science and Ministry of Education, Culture, Sports, Science and Technology of Japan KAKENHI Grants 21H02415 and 17H06422 (to T.K.); a grant from the Joint Research Project of the Research Institute for Microbial Diseases, Osaka University (to M.F. and T.K.); a grant from the Max Planck Society (to D.V.S. and P.H.S.); a grant from RIKEN-Max Planck Joint Research Center for Systems Chemical Biology (to P.A.G., D.V.S., and P.H.S.); and a grant from the Mizutani Foundation for Glycoscience (to Y. Murakami).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115083119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Kinoshita T., Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol. 10, 190290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeda J., et al. , Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria. Cell 73, 703–711 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Bellai-Dussault K., Nguyen T. T. M., Baratang N. V., Jimenez-Cruz D. A., Campeau P. M., Clinical variability in inherited glycosylphosphatidylinositol deficiency disorders. Clin. Genet. 95, 112–121 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Vidugiriene J., Menon A. K., Early lipid intermediates in glycosyl-phosphatidylinositol anchor assembly are synthesized in the ER and located in the cytoplasmic leaflet of the ER membrane bilayer. J. Cell Biol. 121, 987–996 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pottekat A., Menon A. K., Subcellular localization and targeting of N-acetylglucosaminyl phosphatidylinositol de-N-acetylase, the second enzyme in the glycosylphosphatidylinositol biosynthetic pathway. J. Biol. Chem. 279, 15743–15751 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Sanyal S., Menon A. K., Flipping lipids: Why an’ what’s the reason for? ACS Chem. Biol. 4, 895–909 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vishwakarma R. A., Menon A. K., Flip-flop of glycosylphosphatidylinositols (GPI’s) across the ER. Chem. Commun. (Camb.) (4), 453–455 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Helenius J., et al. , Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415, 447–450 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Frank C. G., Sanyal S., Rush J. S., Waechter C. J., Menon A. K., Does Rft1 flip an N-glycan lipid precursor? Nature 454, E3–E4, discussion E4–E5 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Anand M., et al. , Requirement of the Lec35 gene for all known classes of monosaccharide-P-dolichol-dependent glycosyltransferase reactions in mammals. Mol. Biol. Cell 12, 487–501 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerrero P. A., et al. , Rescue of glycosylphosphatidylinositol-anchored protein biosynthesis using synthetic glycosylphosphatidylinositol oligosaccharides. ACS Chem. Biol. 16, 2297–2306 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bütikofer P., Lin Z. W., Chiu D. T., Lubin B., Kuypers F. A., Transbilayer distribution and mobility of phosphatidylinositol in human red blood cells. J. Biol. Chem. 265, 16035–16038 (1990). [PubMed] [Google Scholar]
- 13.Zewe J. P., et al. , Probing the subcellular distribution of phosphatidylinositol reveals a surprising lack at the plasma membrane. J. Cell Biol. 219, e201906127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wongkajornsilp A., Rosenberry T. L., Uptake of exogenous sn-1-acyl-2-lyso-phosphatidylinositol into HeLa S3 cells. Reacylation on the cell surface and metabolism to glucosaminyl(acyl)phosphatidylinositol. J. Biol. Chem. 270, 9147–9153 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto K., Okamoto A., Isonishi S., Ochiai K., Ohtake Y., A novel gene, CRR9, which was up-regulated in CDDP-resistant ovarian tumor cell line, was associated with apoptosis. Biochem. Biophys. Res. Commun. 280, 1148–1154 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Trezise S., et al. , Mining the plasma cell transcriptome for novel cell surface proteins. Int. J. Mol. Sci. 19, 2161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia J., et al. , CLPTM1L promotes growth and enhances aneuploidy in pancreatic cancer cells. Cancer Res. 74, 2785–2795 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke W. R., Amundadottir L., James M. A., CLPTM1L/CRR9 ectodomain interaction with GRP78 at the cell surface signals for survival and chemoresistance upon ER stress in pancreatic adenocarcinoma cells. Int. J. Cancer 144, 1367–1378 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Yang J., et al. , Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U.S.A. 117, 1496–1503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L.-Q., et al. , Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanno Y., et al. , AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 7, 13245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saudek V., Cystinosin, MPDU1, SWEETs and KDELR belong to a well-defined protein family with putative function of cargo receptors involved in vesicle trafficking. PLoS One 7, e30876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao Y., et al. , Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 527, 259–263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Leeuwen J., et al. , Exploring genetic suppression interactions on a global scale. Science 354, aag0839 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goren M. A., et al. , Constitutive phospholipid scramblase activity of a G protein-coupled receptor. Nat. Commun. 5, 5115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menon I., et al. , Opsin is a phospholipid flippase. Curr. Biol. 21, 149–153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han L., et al. , Molecular mechanism of substrate recognition and transport by the AtSWEET13 sugar transporter. Proc. Natl. Acad. Sci. U.S.A. 114, 10089–10094 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heuberger E. H. M. L., Veenhoff L. M., Duurkens R. H., Friesen R. H. E., Poolman B., Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J. Mol. Biol. 317, 591–600 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., et al. , Free, unlinked glycosylphosphatidylinositols on mammalian cell surfaces revisited. J. Biol. Chem. 294, 5038–5049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., et al. , Cross-talks of glycosylphosphatidylinositol biosynthesis with glycosphingolipid biosynthesis and ER-associated degradation. Nat. Commun. 11, 860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami Y., et al. , PIG-W is critical for inositol acylation but not for flipping of glycosylphosphatidylinositol-anchor. Mol. Biol. Cell 14, 4285–4295 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higgins J. A., Hitchin B. W., Low M. G., Phosphatidylinositol-specific phospholipase C of Bacillus thuringiensis as a probe for the distribution of phosphatidylinositol in hepatocyte membranes. Biochem. J. 259, 913–916 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vidugiriene J., Menon A. K., The GPI anchor of cell-surface proteins is synthesized on the cytoplasmic face of the endoplasmic reticulum. J. Cell Biol. 127, 333–341 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L., et al. , Scrambling of natural and fluorescently tagged phosphatidylinositol by reconstituted G protein-coupled receptor and TMEM16 scramblases. J. Biol. Chem. 293, 18318–18327 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lévy D., Bluzat A., Seigneuret M., Rigaud J.-L., A systematic study of liposome and proteoliposome reconstitution involving Bio-Bead-mediated Triton X-100 removal. Biochim. Biophys. Acta 1025, 179–190 (1990). [DOI] [PubMed] [Google Scholar]
- 37.Vishwakarma R. A., et al. , New fluorescent probes reveal that flippase-mediated flip-flop of phosphatidylinositol across the endoplasmic reticulum membrane does not depend on the stereochemistry of the lipid. Org. Biomol. Chem. 3, 1275–1283 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Bushell S. R., et al. , The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K. Nat. Commun. 10, 3956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y. S., et al. , N-Glycan-dependent protein folding and endoplasmic reticulum retention regulate GPI-anchor processing. J. Cell Biol. 217, 585–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge Y., et al. , Clptm1 limits forward trafficking of GABAA receptors to scale inhibitory synaptic strength. Neuron 97, 596–610.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghanbarpour A., Valverde D. P., Melia T. J., Reinisch K. M., A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc. Natl. Acad. Sci. U.S.A. 118, e2101562118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y. E., et al. , TMEM41B and VMP1 are scramblases and regulate the distribution of cholesterol and phosphatidylserine. J. Cell Biol. 220, e202103105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang D., et al. , TMEM41B acts as an ER scramblase required for lipoprotein biogenesis and lipid homeostasis. Cell Metab. 33, 1655–1670.e8 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Tsuji T., et al. , Predominant localization of phosphatidylserine at the cytoplasmic leaflet of the ER, and its TMEM16K-dependent redistribution. Proc. Natl. Acad. Sci. U.S.A. 116, 13368–13373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menon A. K., Eppinger M., Mayor S., Schwarz R. T., Phosphatidylethanolamine is the donor of the terminal phosphoethanolamine group in trypanosome glycosylphosphatidylinositols. EMBO J. 12, 1907–1914 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Menon A. K., Mayor S., Schwarz R. T., Biosynthesis of glycosyl-phosphatidylinositol lipids in Trypanosoma brucei: Involvement of mannosyl-phosphoryldolichol as the mannose donor. EMBO J. 9, 4249–4258 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toulmay A., et al. , Vps13-like proteins provide phosphatidylethanolamine for GPI anchor synthesis in the ER. J. Cell Biol. 221, e202111095 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura N., et al. , Expression cloning of PIG-L, a candidate N-acetylglucosaminyl-phosphatidylinositol deacetylase. J. Biol. Chem. 272, 15834–15840 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Fujiwara I., et al. , Mutations in PIGL in a patient with Mabry syndrome. Am. J. Med. Genet. A. 167A, 777–785 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Güther M. L., Ferguson M. A., The role of inositol acylation and inositol deacylation in GPI biosynthesis in Trypanosoma brucei. EMBO J. 14, 3080–3093 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dickinson M. E., et al. ; International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center, High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawagoe K., et al. , Glycosylphosphatidylinositol-anchor-deficient mice: Implications for clonal dominance of mutant cells in paroxysmal nocturnal hemoglobinuria. Blood 87, 3600–3606 (1996). [PubMed] [Google Scholar]
- 53.Rodríguez de Los Santos M., et al. , A CRISPR-Cas9-engineered mouse model for GPI-anchor deficiency mirrors human phenotypes and exhibits hippocampal synaptic dysfunctions. Proc. Natl. Acad. Sci. U.S.A. 118, e2014481118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyko F., Martoglio B., Jungnickel B., Rapoport T. A., Dobberstein B., Signal sequence processing in rough microsomes. J. Biol. Chem. 270, 19873–19878 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Kol M. A., de Kroon A. I. P. M., Rijkers D. T. S., Killian J. A., de Kruijff B., Membrane-spanning peptides induce phospholipid flop: A model for phospholipid translocation across the inner membrane of E. coli. Biochemistry 40, 10500–10506 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Nakao H., Ikeda K., Ishihama Y., Nakano M., Membrane-spanning sequences in endoplasmic reticulum proteins promote phospholipid flip-flop. Biophys. J. 110, 2689–2697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takar M., Huang Y., Graham T. R., The PQ-loop protein Any1 segregates Drs2 and Neo1 functions required for viability and plasma membrane phospholipid asymmetry. J. Lipid Res. 60, 1032–1042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petkovic M., Oses-Prieto J., Burlingame A., Jan L. Y., Jan Y. N., TMEM16K is an interorganelle regulator of endosomal sorting. Nat. Commun. 11, 3298 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muraki M., Damnjanović J., Nakano H., Iwasaki Y., Salt-induced increase in the yield of enzymatically synthesized phosphatidylinositol and the underlying mechanism. J. Biosci. Bioeng. 122, 276–282 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Sugita Y., et al. , Recombinant soluble CD59 inhibits reactive haemolysis with complement. Immunology 82, 34–41 (1994). [PMC free article] [PubMed] [Google Scholar]
- 61.Cong L., et al. , Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirata T., et al. , Post-Golgi anterograde transport requires GARP-dependent endosome-to-TGN retrograde transport. Mol. Biol. Cell 26, 3071–3084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yusa K., Zhou L., Li M. A., Bradley A., Craig N. L., A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U.S.A. 108, 1531–1536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ploier B., Menon A. K., A fluorescence-based assay of phospholipid scramblase activity. J. Vis. Exp. (115), 54635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Letunic I., Bork P., Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.