Abstract
Background
The purpose of our study is to analyze the microbiological and clinical characteristics of carbapenem‐resistant hypervirulent Klebsiella pneumoniae (CR‐hvKP) that causes nosocomial infection.
Methods
We collected the carbapenem‐resistant K. pneumoniae (CRKP) strains that caused nosocomial infection in a hospital in China and collected the relevant clinical data. We characterized these strains for their antimicrobial and virulence‐associated phenotype and genotype and analyzed the clonal relatedness. We screened hypervirulent strains and compared them with non‐hypervirulent strains.
Results
We retrospectively analyzed 62 CRKP strains that caused nosocomial infection in a tertiary hospital within 1 year, of which 41 (41/62, 66.1%) CRKP were considered as CR‐hvKP. All CR‐hvKP strains were multi‐drug resistance (MDR) and the vast majority of isolates (39/41, 95.1%) were ST11 KPC‐2‐producing strains. Two hypermucoviscous isolates and 4 capsular types were found in 41 CR‐hvKP. Twenty‐nine isolates (29/41, 70.7%) showed hypervirulence in Galleria mellonella infection model. PFGE showed that ST11‐KL47 CR‐hvKP and ST11‐KL64 CR‐hvKP exhibited a high degree of clonality, while non‐hypervirulent strains were not significant. CR‐hvKP had higher positive rates of bla KPC‐2 and bla CTX‐M‐65 and higher levofloxacin resistance (p < 0.001, p = 0.005 and p = 0.046, respectively) when compared to the non‐hypervirulent strains. There was no significant difference between the two groups in terms of in‐hospital mortality (7/41, 17.1% vs 5/21, 23.8%, p = 0.743).
Conclusion
Our research finds that ST11 KPC‐2‐producing CR‐hvKP is the main type of CRKP that caused nosocomial infection, and clonal spread has occurred. We provide more information about CR‐hvKP in health care.
Keywords: carbapenem resistance, hypervirulence, Klebsiella pneumoniae, nosocomial infection
Our research finds that ST11 KPC‐2‐producing CR‐hvKP is the main type of CRKP that caused nosocomial infection, which poses a new threat to people's health. And we provide more information about CR‐hvKP in health care.

1. INTRODUCTION
Klebsiella pneumoniae is an important opportunistic pathogen associated with bacterial infections in hospitals. Carbapenem‐resistant K. pneumoniae (CRKP), which can resist almost all clinically used antibiotics and cause serious infections, poses a huge threat to global public health. 1 , 2 K. pneumoniae sequence type (ST) 258 has contributed significantly to the dissemination of CRKP in the United States and in European countries, but ST11 is predominant in China, 3 and the genomic characteristics of ST11 K. pneumoniae strains are significantly different from that of other countries. 4 The most predominant carbapenemase genes of ST11 CRKP in China are bla KPC‐2, bla NDM‐1, bla OXA‐48, and ST11 KPC‐2‐producing K. pneumoniae is spreading at an alarming speed. 5 , 6 , 7 ST11 CRKP is emerging in almost all Chinese provinces, and its prevalence in China forces us to conduct better tracing and control of such a pathogen.
Hypervirulent K. pneumoniae (hvKP) is a more virulent variant of K. pneumoniae, known for causing invasive infections among young and relatively healthy individuals. This phenomenon is first observed in community infections and mainly occurs in the Asian Pacific Rim. 8 However, infections caused by hvKP are increasingly being reported worldwide in recent years. 9 , 10 Researches find that the genetic determinants of hypervirulence of hvKP are generally on a typical ~200 kb virulence plasmid, as well as integrative conjugal elements. 8 , 11 Alarmingly, hvKP can continuously increase antibiotic resistance by acquiring resistance elements. 12 Convergence of hypervirulent and resistance make hvKP a global pathogen of current concern. 13
In the past years, reports on carbapenem‐resistant hypervirulent K. pneumoniae (CR‐hvKP) infection were increasing, even a fatal outbreak caused by CR‐hvKP in a Chinese hospital had been observed. 14 Worryingly, ST11 CRKP with both hypervirulence and high drug resistance demonstrated limited adaptive cost and enhanced environmental survival, 10 , 13 , 15 which might cause CR‐hvKP to spread more quickly and widely, particularly in the hospital environment.
However, our knowledge of CR‐hvKP is still very limited so far, and more monitoring and research results are urgently needed to fight this pathogen, particularly the pheno‐ and genotypic characteristics of prevalent strains from continually survey in hospitals. This study analyzed the presence of CR‐hvKP among CRKP which caused nosocomial infection in hospitalized patients, and studied the phenotypes and genotypes of the strains, as well as clinical characteristics of the associated patients, which provided more information on understanding CR‐hvKP.
2. METHOD
2.1. Study design and definition
A one‐year retrospective study was conducted in Hunan Provincial People's Hospital, the First Affiliated Hospital of Hunan Normal University. This study was approved by the ethics committee of Hunan Provincial People's Hospital. The hospital is located in Changsha, Hunan Province, China. It is a large‐scale comprehensive tertiary hospital with nearly 4000 beds. We used VITEK 2 Compact system (bioMérieux, Marcy‐l’Étoile, France) conducting identification and antimicrobial susceptibility test of all strains isolated in Clinical Microbiology Laboratory from January to December 2018. The results of the antimicrobial susceptibility test were interpreted according to the rules provided by the CLSI guidelines. 16 Strains identified as K. pneumoniae and resistant to imipenem and/or meropenem (defined as CRKP in this study) were collected. Strains isolated from the same part of the same patient were excluded. Identification results were reconfirmed through VITEK MS (bioMérieux, Marcy‐l’Étoile, France). For strains resistant to imipenem or meropenem, the E test method was used to verify their susceptibility. The antimicrobial susceptibility tests of tigecycline and colistin were carried out by the micro‐broth dilution method. The interpretation of the results was carried out according to the FDA guidelines and EUCAST guidelines, respectively. 17 , 18 Multi‐drug resistance (MDR) was defined as resistance to three or more different antimicrobial classes. Disease‐related data of all patients with CRKP infection were collected from medical records, including demographic characteristics, underlying diseases, invasive treatments, infection sites, length of stay, and infection outcome. Nosocomial infection was defined as a new infection which developed 48 h after patient admission. Only patients with a complete hospitalization history and who developed CRKP nosocomial infection were included in the study. CR‐hvKP was defined by the presence of virulence gene iucA, iroN, prmpA, or prmpA2 in CRKP. 14
2.2. Carbapenem‐resistant phenotype and hypermucoviscous phenotype detection
The carbapenem‐resistant phenotype of strains was detected by CLSI recommended modified carbapenem inactivation method (mCIM). 19 And, the hypermucoviscous phenotype was detected by string test. 14
2.3. Resistance genes, virulence genes, and capsular type genes dection
We detected resistance genes by PCR amplification, including genes encoding carbapenemases (bla KPC, bla NDM, bla IMP, bla VIM, and bla OXA‐48) and genes encoding other β‐lactamases (bla CTX‐M, bla SHV, and bla TEM). 20 We detected virulence genes by PCR amplification, including siderophores (entB, irp‐1, and irp‐2), fimbriae biosynthesis (fimH and mrkD), lipopolysaccharide biosynthesis (wabG), fucose synthesis (wcaG), allantoin metabolism (allS), and putative transporter (peg‐344). 21 , 22 , 23 , 24 The positive amplification products after agarose gel electrophoresis were sequenced and compared on the NCBI BLAST website (https://blast.ncbi.nlm.nih.gov/blast.cgi). We genotyped the capsular polysaccharide synthesis (cps) gene of strains by amplifying and sequencing the wzi gene, as previously described. 25 All primers used in this study are provided in Appendix S1.
2.4. Galleria mellonella infection model
Further virulence assessment for CR‐hvKP was conducted by in vivo G. mellonella infection model. 14 Healthy G. mellonella larvae weighing 250–350 mg were obtained from Huiyude Biotech Company (Huiyude Biotech Company, Tianjin, China). Setting up each strain as an experimental group, each experimental group contained 10 larvae. The mid‐log phase culture was diluted with PBS into a bacterial suspension with a concentration of 106 cfu/ml, and 10 μl of the bacterial suspension was injected with a microsyringe into the left forefoot of each larva. After injection, the larvae were kept at 37°C in dark to observe the 72‐h mortality. Strain which the pathogenicity was similar or stronger than the positive control was assessed as hypervirulence in vivo. NTUH‐K2044, ZR2 (a non‐hypervirulent K. pneumoniae strain identified in our previous study), and PBS were used as positive control, negative control, and blank control, respectively.
2.5. Multilocus sequence typing (MLST) and pulsed field gel electrophoresis (PFGE)
We conducted MLST by amplifying and sequencing 7 housekeeping genes (gapA, infB, mdh, pgI, phoE, rpoB, and tonB). 26 The allele types and STs were confirmed based on the database provided on the Institut Pasteur website (https://bigsdb.pasteur.fr/). Genetic correlations among the CRKP strains were subsequently analyzed by PFGE. 27 The PFGE banding pattern was clustered by BioNumerics software version 5.1 (Applied Maths, Kortrijk, Belgium). Isolates with 100% similarity were considered the same PFGE cluster.
2.6. Statistic analysis
Statistical analysis was performed using SPSS 19.0 software (IBM Corporation, Armonk, NY, USA). Continuous variables were assessed by Student's t tests. The comparison of the categorical variables was calculated by Chi‐square test or Fisher's exact test. p value <0.05 was considered to be statistically significant.
3. RESULTS
3.1. Strain and patient information
In this study, a total of 1743 K. pneumoniae strains from Hunan Provincial People's Hospital were collected between January 1 and December 31, 2018, by routine monitoring, among which 125 (7.17%) CRKP were identified (Figure 1). By consulting medical records, 62 CRKP causing nosocomial infections from 62 patients were confirmed, including 36 (58.1%) in surgical department, 19 (30.6%) in ICU, 5 (8.1%) in internal medicine department, and 2 (3.2%) in pediatric department. CRKP strains were isolated from a variety of sample types. Samples of respiratory tract and bile accounted for the largest proportion, with 25 (40.3%) cases and 11 (17.7%) cases, respectively. The others were 9 (14.5%) ascites cases, 8 (12.9%) blood cases, 4 (6.5%) wound cases, 3 (4.9%) pus cases, 1 (1.6%) reproductive tract case, and 1(1.6%) urine case. No pyogenic liver abscess or metastatic infection was reported in the 62 CRKP‐infected patients.
FIGURE 1.
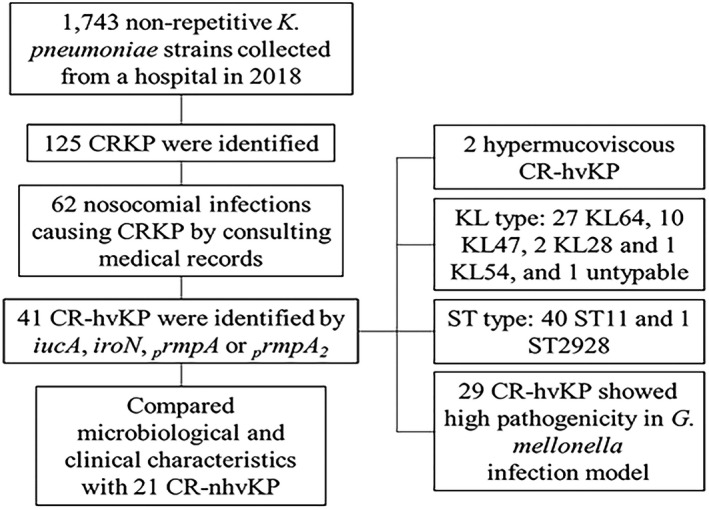
Routine monitoring of K. pneumoniae and research process of this study
More than half of the 62 (41/62, 66.1%) CRKP were CR‐hvKP. The positive rates of iucA, iroN, prmpA, and prmpA2 in 41 CR‐hvKP were 40/41 (97.6%), 30/41 (73.2%), 3/41 (7.3%), and 19/41 (46.3%), respectively, among which 3 isolates had all four virulence genes (Figure 2). CR‐hvKP mainly came from respiratory tract specimens (14/41, 34.2%). Males were mostly infected (30/41, 73.2%). Surgery department and ICU were the main epidemic departments (25/41, 61.0% and 12/41, 29.3%, respectively). Only 7 cases of CR‐hvKP infection caused fatal outcomes (7/41, 17.1% mortality rate) (Table 1). We compared the clinical features between CR‐hvKP and carbapenem‐resistant non‐hypervirulent K. pneumoniae (CR‐nhvKP). Through our analysis, there were no significant differences between the two groups in terms of age, gender, underlying diseases, invasive treatments, infection sites, length of hospital stay, and in‐hospital mortality (Table 2).
FIGURE 2.
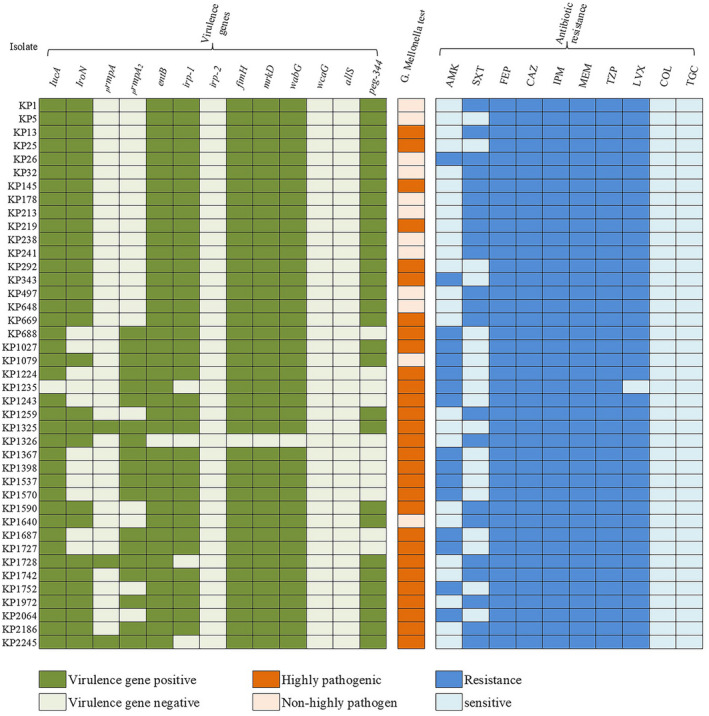
Carriage of virulence genes, in vivo virulence and antimicrobial resistance features of 41 CR‐hvKP strains. AMK, amikacin; SXT, trimethoprim/sulfamethoxazole; FEP, cefepime; CAZ, ceftazidime; IPM, imipenem; MEM, meropenem; TZP, piperacillin/Tazobactam; LVX, levofloxacin; COL, colistin; TGC, tigecycline
TABLE 1.
Clinical characteristics of 41 CR‐hvKP strains
| Strain ID | Source | Department | Collection date | Outcome | mCIM | Antibiotic resistance genes | String test | Capsular type | ST |
|---|---|---|---|---|---|---|---|---|---|
| KP1 | Reproductive tract | SSD | 2018/3/7 | Survived | + | bla KPC‐2, bla SHV‐11, bla TEM‐1C | − | KL64(wzi64) | 11 |
| KP5 | Blood | GSD | 2018/3/1 | Survived | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP13 | Sputum | RD | 2018/3/12 | Death | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP25 | Ascites | GSD | 2018/1/5 | Survived | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP26 | Wound | SSD | 2018/1/6 | Survived | + | bla KPC‐2, bla SHV‐11, bla TEM‐1 | − | KL64(wzi64) | 11 |
| KP32 | Blood | GSD | 2018/3/7 | Survived | + | bla KPC‐2, bla SHV‐12 | − | KL64(wzi64) | 11 |
| KP145 | Bile | GSD | 2018/3/31 | Survived | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP178 | Blood | ICU | 2018/5/14 | Death | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP213 | Blood | ICU | 2018/4/17 | Death | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP219 | Blood | GSD | 2018/4/8 | Survived | + | bla KPC‐2, bla SHV‐11 | + | KL64(wzi64) | 11 |
| KP238 | Ascites | HSD | 2018/4/17 | Survived | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP241 | Sputum | ICU | 2018/4/19 | Survived | + | bla KPC‐2, bla SHV‐12 | − | KL64(wzi64) | 11 |
| KP292 | Ascites | GSD | 2018/4/25 | Survived | + | bla KPC‐2, bla SHV‐12 | − | KL64(wzi64) | 11 |
| KP343 | Ascites | HSD | 2018/5/8 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11, bla TEM‐1 | − | KL64(wzi64) | 11 |
| KP497 | Sputum | GSD | 2018/6/11 | Survived | + | bla KPC‐2, bla SHV‐12 | − | Untypable | 11 |
| KP648 | Urine | ICU | 2018/6/26 | Survived | + | bla KPC‐2, bla SHV‐12 | − | KL64(wzi64) | 11 |
| KP669 | Sputum | ICU | 2018/6/26 | Survived | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP688 | Bile | HSD | 2018/6/29 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL47(wzi209) | 11 |
| KP1027 | Blood | HSD | 2018/8/2 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL47(wzi209) | 11 |
| KP1079 | Bile | HSD | 2018/8/4 | Survived | + | bla KPC‐2, bla SHV‐12, bla TEM‐1 | − | KL64(wzi64) | 11 |
| KP1224 | Sputum | ICU | 2018/8/14 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL47(wzi209) | 11 |
| KP1235 | Sputum | RD | 2018/8/15 | Survived | + | bla IMP‐4, bla CTX‐M‐3, bla SHV‐187, bla TEM‐1 | − | KL54(wzi115) | 2928 |
| KP1243 | Sputum | ICU | 2018/8/15 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL47(wzi209) | 11 |
| KP1259 | Blood | HSD | 2018/8/19 | Death | + | bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP1325 | Ascites | HSD | 2018/8/25 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐12 | − | KL64(wzi64) | 11 |
| KP1326 | Sputum | HSD | 2018/8/26 | Survived | + | bla KPC‐2, bla TEM‐1 | − | KL64(wzi64) | 11 |
| KP1367 | Pus | GSD | 2018/8/29 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL47(wzi209) | 11 |
| KP1398 | Sputum | NSD | 2018/8/14 | Death | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL47(wzi209) | 11 |
| KP1537 | Wound | GSD | 2018/9/5 | Death | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL47(wzi209) | 11 |
| KP1570 | Ascites | HSD | 2018/9/6 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL47(wzi209) | 11 |
| KP1590 | Pus | HSD | 2018/9/9 | Survived | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP1640 | Sputum | ICU | 2018/9/13 | Survived | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP1687 | Sputum | NSD | 2018/9/16 | Death | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL47(wzi209) | 11 |
| KP1727 | Wound | ICU | 2018/9/20 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL47(wzi209) | 11 |
| KP1728 | Sputum | ICU | 2018/9/21 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL28(wzi84) | 11 |
| KP1742 | Ascites | HSD | 2018/9/23 | Survived | + | bla KPC‐2, bla SHV‐12 | − | KL64(wzi64) | 11 |
| KP1752 | Ascites | HSD | 2018/9/25 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐12, bla TEM‐1 | − | KL64(wzi64) | 11 |
| KP1972 | Sputum | ND | 2018/10/15 | Survived | + | bla KPC‐2, bla SHV‐12 | + | KL64(wzi64) | 11 |
| KP2064 | Bile | ICU | 2018/11/5 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11, bla TEM‐1 | − | KL64(wzi64) | 11 |
| KP2186 | Bile | HSD | 2018/12/6 | Survived | + | bla KPC‐2, bla SHV‐11 | − | KL64(wzi64) | 11 |
| KP2245 | Sputum | ICU | 2018/12/23 | Survived | + | bla KPC‐2, bla CTX‐M‐65, bla SHV‐11 | − | KL28(wzi84) | 11 |
Abbreviations: SSD, Spinal Surgery Department; GSD, General Surgery Department; RD, Respiratory Department; ICU, Intensive Care Unit; HSD, Hepatobiliary Surgery Department; NSD, Neurological Surgery Department; ND, Nephrology Department.
TABLE 2.
Comparison of clinical features between CR‐hvKP strains and CR‐nhvKP strains
| Variables | Total (n = 62) | CR‐hvKP (n=41) | CR‐nhvKP (n=21) | χ2 | p value |
|---|---|---|---|---|---|
| Demographics | |||||
| Age(mean ± SD) | 56.9 ± 19.9 | 57.07 ± 18.14 | 56.86 ± 23.50 | NA a | 0.923 |
| Age (<1) | 2 (3.2%) | 0 (0%) | 2 (9.5%) | 0.698 b | 0.403 |
| Age (1–49) | 17 (27.4%) | 14 (34.1%) | 3 (14.3%) | ||
| Age (≥50) | 43 (69.4%) | 27 (65.9%) | 16 (76.2%) | ||
| Male | 41 (66.1%) | 30 (73.2%) | 11 (52.4%) | 2.680 | 0.102 |
| Underlying diseases | |||||
| Diabetes mellitus | 8 (12.9%) | 7 (17.1%) | 1 (4.8%) | NA | 0.247 |
| Chronic cardiovascular system diseases | 6 (9.7%) | 2 (4.9%) | 4 (19.0%) | NA | 0.167 |
| Chronic respiratory system diseases | 4 (6.5%) | 1 (2.4%) | 3 (14.3%) | NA | 0.108 |
| Tumor | 7 (11.3%) | 3 (7.3%) | 4 (19.0%) | NA | 0.214 |
| Chronic kidney diseases | 1 (1.6%) | 1 (2.4%) | 0 (0%) | NA | 1.000 |
| Chronic Hepatitis/cirrhosis | 4 (6.5%) | 3 (7.3%) | 1 (4.8%) | NA | 1.000 |
| Invasive treatments | |||||
| Puncture or venous catheterization | 30 (48.4%) | 19 (46.3%) | 11 (52.4%) | 0.203 | 0.652 |
| Mechanical Ventilation | 18 (29.0%) | 13 (31.7%) | 5 (23.8%) | 0.420 | 0.517 |
| History of surgery | 38 (61.3%) | 27 (65.9%) | 11 (52.4%) | 1.062 | 0.303 |
| Infection sites | |||||
| Respiratory tract | 25 (40.3%) | 14 (34.2%) | 11 (52.4%) | 1.919 | 0.166 |
| Biliary tract | 11 (17.7%) | 5 (12.2%) | 6 (28.6%) | NA | 0.160 |
| Abdomen | 9 (14.5%) | 8 (19.5%) | 1 (4.8%) | NA | 0.150 |
| Blood | 8 (12.9%) | 7 (17.1%) | 1 (4.8%) | NA | 0.247 |
| Wound | 4 (6.5%) | 3 (7.3%) | 1 (4.8%) | NA | 1.000 |
| Pus | 3 (4.9%) | 2 (4.9%) | 1 (4.8%) | NA | 1.000 |
| Urinary tract | 1 (1.6%) | 1 (2.4%) | 0 (0%) | NA | 1.000 |
| Reproductive tract | 1 (1.6%) | 1 (2.4%) | 0 (0%) | NA | 1.000 |
| Hospitalization | |||||
| <30 days | 51 (82.3%) | 31 (75.6%) | 20 (95.2%) | 1.052 | 0.305 |
| ≥30 days | 11 (17.7%) | 10 (24.4%) | 1 (4.8%) | NA | 0.080 |
| Outcome | |||||
| In‐hospital mortality | 12 (19.4%) | 7 (17.1%) | 5 (23.8%) | 0.404 | 0.525 |
NA: Data not applicable.
Assessed by Student's t tests.
For the number of patients younger than 1 year old is too small, it is combined with "Age (1–49)" in statistical analysis.
3.2. Antibiotic resistance characteristics of CR‐hvKP compared to CR‐nhvKP in nosocomial infection
All 62 CRKP were MDR. The 62 CRKP had high drug resistance rates for most of the antibacterial drugs that were tested, except for colistin and tigecycline. For CR‐hvKP, the resistance levels for cefepime, ceftazidime, imipenem, meropenem, and piperacillin/tazobactam were all 100%, followed by levofloxacin (97.6%), trimethoprim/sulfamethoxazole (51.2%), amikacin (41.5%), colistin (0%), and tigecycline (0%) (Figure 2, Table 3). In terms of resistance rate for levofloxacin from the CR‐hvKP group, it was significantly higher than those from the CR‐nhvKP group (97.6% vs 52.4%, p < 0.001) (Table 3).
TABLE 3.
Comparison of resistance features between CR‐hvKP and CR‐nhvKP
| Variables | Total (n = 62) | CR‐hvKP (n = 41) | CR‐nhvKP (n = 21) | χ2 | P value |
|---|---|---|---|---|---|
| Antibiotic resistance rates | |||||
| AMK | 24 (38.7%) | 17 (41.5%) | 7 (33.3%) | 0.387 | 0.534 |
| SXT | 34 (54.8%) | 21 (51.2%) | 13 (61.9%) | 0.640 | 0.424 |
| FEP | 62 (100.0%) | 41 (100.0%) | 21 (100.0%) | NA | NA |
| CAZ | 62 (100.0%) | 41 (100.0%) | 21 (100.0%) | NA | NA |
| IPM | 62 (100.0%) | 41 (100.0%) | 21 (100.0%) | NA | NA |
| MEM | 62 (100.0%) | 41 (100.0%) | 21 (100.0%) | NA | NA |
| TZP | 61 (98.4%) | 41 (100.0%) | 20 (95.2%) | NA | 0.339 |
| LVX | 52 (83.9%) | 40 (97.6%) | 11 (52.4%) | NA | <0.001 |
| COL | 0 (0%) | 0 (0%) | 0 (0%) | NA | NA |
| TGC | 0 (0%) | 0 (0%) | 0 (0%) | NA | NA |
| Antibiotic‐resistant genes | |||||
| bla KPC‐2 | 49 (79.0%) | 39 (95.1%) | 10 (47.6%) | NA | <0.001 |
| bla IMP‐4 | 1 (1.6%) | 1 (2.4%) | 0 (0%) | NA | 1.000 |
| bla CTX‐M‐3 | 3 (4.8%) | 1 (2.4%) | 2 (9.5%) | NA | 0.263 |
| bla CTX‐M‐14 | 1 (1.6%) | 0 (0%) | 1 (4.8%) | NA | 0.339 |
| bla CTX‐M‐15 | 1 (1.6%) | 0 (0%) | 1 (4.8%) | NA | 0.339 |
| bla CTX‐M‐65 | 19 (30.6%) | 16 (39.0%) | 3 (14.3%) | 3.999 | 0.046 |
| bla SHV‐1 | 2 (3.2%) | 0 (0%) | 2 (9.5%) | NA | 0.111 |
| bla SHV‐11 | 40 (64.5%) | 29 (70.7%) | 11 (52.4%) | 2.043 | 0.153 |
| bla SHV‐12 | 11 (17.7%) | 10 (24.4%) | 1 (4.8%) | NA | 0.080 |
| bla SHV‐28 | 1 (1.6%) | 0 (0%) | 1 (4.8%) | NA | 0.339 |
| bla SHV‐187 | 1 (1.6%) | 1 (2.4%) | 0 (0%) | NA | 1.000 |
| bla TEM‐1 | 17 (27.4%) | 8 (19.5%) | 9 (42.9%) | 3.803 | 0.051 |
Abbreviations: AMK, amikacin; SXT, trimethoprim/sulfamethoxazole; FEP, cefepime; CAZ, ceftazidime; IPM, imipenem; MEM, meropenem; TZP, piperacillin/tazobactam; LVX, levofloxacin; COL, colistin; TGC, tigecycline; NA, Data not applicable.
The mCIM tests of all 62 CRKP strains were positive. Two families of carbapenemase genes were identified from 62 CRKP strains: bla KPC‐2 (49/62, 79.0%) and bla IMP‐4 (1/62, 1.6%). Most (39/41, 95.1%) CR‐hvKP harbored bla KPC‐2, and 1 (1/41, 2.4%) harbored bla IMP‐4. 1 CR‐hvKP isolate did not carry bla KPC, bla NDM, bla IMP, bla VIM, or bla OXA‐48, but the mCIM phenotype test was positive, indicating that it might harbor other carbapenem resistance genes. Other β‐lactamase genes were detected in CR‐hvKP strains, including bla CTX‐M‐3, bla CTX‐M‐15, bla CTX‐M‐65, bla SHV‐11, bla SHV‐12, and bla TEM‐1. The positive rates of bla KPC‐2 and bla CTX‐M‐65 in CR‐hvKP group were both higher than that of CR‐nhvKP group (p < 0.001 and p = 0.046, respectively) (Table 3).
3.3. Virulence‐related characteristics of CR‐hvKP compared with CR‐nhvKP in nosocomial infection
By string test, only 2/62 (3.2%) CRKP strains were hypermucoviscous (both were CR‐hvKP). wzi sequencing revealed 11 capsular types in 62 CRKP (28 KL64, 17 KL47, 2 KL28, 1 KL54, 1 KL136, 1 KL38, 1 KL19, 1 KL17, 1 KL123, 1 KL23, and 1 KL25, with 7 untypable). The vast majority of CR‐hvKP belonged to KL64 and KL47, and KL64 was the dominant capsular type (27/41 KL64, 10/41 KL47, 2/41 KL28, and 1/41 KL54, with 1 untypable), while in the CR‐nhvKP group more diverse capsular types were found. Capsule type KL64 (27/41 vs 1/21, p < 0.001) was significantly associated with the CR‐hvKP group when compared to the CR‐nhvKP group (Table 4). For virulence background genes in 62 CRKP strains, the positive rates of fimH, wabG, entB, mrkD, irp‐1, peg‐344, irp‐2, wcaG, and allS were 96.8%, 95.2%, 93.5%, 93.5%, 77.4%, 58.1%, 0%, 0%, and 0%, respectively. Virulence genes irp‐1 (37/41 vs 11/21, p = 0.003) and peg‐344 (30/41 vs 6/21, p=0.001) were significantly associated with the CR‐hvKP group when compared to the CR‐nhvKP group (Table 4). By G. mellonella infection model, we found that 29/41 (70.7%) CR‐hvKP strains showed hypervirulence in vivo (Figure 2).
TABLE 4.
Comparison of bacterial characteristics between CR‐hvKP and CR‐nhvKP
| Variables | Total (n = 62) | CR‐hvKP (n = 41) | CR‐nhvKP (n = 21) | χ2 | P value |
|---|---|---|---|---|---|
| Hypermucoviscosity | 2 (3.2%) | 2 (4.9%) | 0 (0%) | NA | 0.545 |
| Capsular types | |||||
| KL64 | 28 (45.2%) | 27 (65.9%) | 1 (4.8%) | 20.928 | <0.001 |
| KL47 | 17 (27.4%) | 10 (24.4%) | 7 (33.3%) | 0.558 | 0.455 |
| KL28 | 2 (3.2%) | 2 (4.9%) | 0 (0%) | NA | 0.545 |
| KL54 | 1 (1.6%) | 1 (2.4%) | 0 (0%) | NA | 1.000 |
| Other capsular type | 14 (22.6%) | 1 (2.4%) | 13 (61.9%) | NA | <0.001 |
| MLST | |||||
| ST11 | 49 (79.0%) | 40 (97.6%) | 9 (42.9%) | NA | <0.001 |
| ST2928 | 1 (1.6%) | 1 (2.4%) | 0 (0%) | NA | 1.000 |
| Other ST | 12 (19.4%) | 0 (0%) | 12 (57.1%) | NA | <0.001 |
| Siderophores | |||||
| entB | 58 (93.5%) | 40 (97.6%) | 18 (85.7%) | NA | 0.108 |
| irp‐1 | 48 (77.4%) | 37 (90.2%) | 11 (52.4%) | NA | 0.003 |
| irp‐2 | 0 (0%) | 0 (0%) | 0 (0%) | NA | NA |
| Adhesin | |||||
| fimH | 60 (96.8%) | 40 (97.6%) | 20 (95.2%) | NA | 1.000 |
| mrkD | 58 (93.5%) | 40 (97.6%) | 18 (85.7%) | NA | 0.108 |
| Lipopolysaccharide biosynthesis | |||||
| wabG | 59 (95.2%) | 40 (97.6%) | 19 (90.5%) | NA | 0.263 |
| Fucose synthesis | |||||
| wcaG | 0 (0%) | 0 (0%) | 0 (0%) | NA | NA |
| Allantoin metabolism | |||||
| allS | 0 (0%) | 0 (0%) | 0 (0%) | NA | NA |
| Putative transporter | |||||
| peg‐344 | 36 (58.1%) | 30 (73.2%) | 6 (28.6%) | 11.344 | 0.001 |
NA, Data not applicable.
3.4. Phylogenetic characteristics of CR‐hvKP isolates
A total of 13 STs among 62 CRKP were identified in the study. ST11 was the most prevalent ST (49/62), followed by ST571 (2/62), ST15 (1/62), ST37 (1/62), ST70 (1/62), ST101 (1/62), ST414 (1/62), ST515 (1/62), ST1040 (1/62), ST1779 (1/62,), ST1933 (1/62), ST2928 (1/62), and new ST (1/62). The majority of CR‐hvKP belonged to ST11 (40/41, 97.6%) and 1 belonged to ST2928 (Table 1). More scattered clones were observed in the CR‐nhvKP group. ST11 (40/41 vs 9/21, p < 0.001) was strongly associated with CR‐hvKP when compared to CR‐nhvKP (Table 4).
In the PFGE test, 1 CRKP strain had no complete bands despite several repeats. 61 CRKP strains displayed 36 PFGE clusters (Appendix S2). Clusters 10, 15, and 18 were the three main clusters, which all consisting of CR‐hvKP strains. Cluster 10 included 7 strains that all belonged to ST11‐KL47, cluster 15 included 11 strains that all belonged to ST11‐KL64, and cluster 18 included 8 strains that all belonged to ST11, among which 7 were KL64. Notably, all 7 CR‐hvKP strains in cluster 10 shared the same virulence genes and resistance genes, as well as high lethality in the G. mellonella infection model and resistance pattern. The 7 strains were all isolated between June and September 2018, suggesting clonal transmission. For CR‐nhvKP, the homology of isolates was more diverse; only one cluster included 2 strains with 100% similarity was found.
4. DISCUSSION
Hypervirulent K. pneumoniae is originally known for community infections and has become a global clinical problem. This study analyzed the CR‐hvKP that caused nosocomial infection during a specific period in a Chinese hospital, and we provided more medical institution related data about the “superbug.”
By detecting virulence genes highly related to hypervirulence, we found 66.1% CR‐hvKP from 62 CRKP strains. We further investigated the virulence genetic background of these strains, and we found 70.7% CR‐hvKP showed hypervirulence in G. mellonella infection model. Our data showed that the hypervirulent K. pneumoniae with carbapenem resistance was the main member of the CRKP that caused nosocomial infection in hospitalized patients and clonal spread of this pathogen had occurred. To date, reports on nosocomial infection CR‐hvKP are very limited. Liu et al 28 found 45.3% hypervirulent K. pneumoniae strains from 117 cases of nosocomial K. pneumoniae infection, reporting a substantial change in the epidemiology of hypervirulent K. pneumoniae. Yang et al. found that CRKP strains harbored a virulence‐encoding plasmid accounted for 58% of 784 bla KPC‐2‐bearing CRKP strains in a recently published study in China. 29 Our data partially supported this trend.
These CR‐hvKP were mainly isolated from respiratory tract specimens, and most of the cases occurred in surgical department and ICU. These findings were partly consistent with the results of previous studies. 10 Zhao et al 30 found that hypervirulent K. pneumoniae strains were prevalent in surgical site infections, with an infection rate as high as 46.0%, suggesting surgical patients should be considered to take more stringent infection prevention measures. In previous studies on hypervirulent K. pneumoniae infection, it was found that males were more commonly infected, and those in the fifth and sixth decades of life were of the highest risk. 8 In our study, we did not find similar results. It is worth noting that we had observed CRKP infections in hospitalized infants. Although the strains were non‐hypervirulent, it releases a danger signal, as Wang et al. 31 reported. Metastatic spreading or pyogenic liver abscess are typical clinical features of hypervirulent K. pneumoniae infection, but in our research, we did not find such performances.
No significant differences were found in age, gender, underlying diseases, invasive treatments, infection sites, hospitalization time, and mortality rates between hypervirulent group and non‐hypervirulent group. From these existing data, it is difficult to find the serious impact of hypervirulence. These results could be explained from the following two aspects. Firstly, patients were in the period of medical observation when the infection occurred. Due to timely intervention, the infections were prevented from worsening. Secondly, these hypervirulent strains might lose some virulence factors, which reduced their pathogenicity to host.
All CR‐hvKP strains were MDR in this study. Of all carbapenemase genes detected, gene bla KPC‐2 showed the highest detection rate, which is the most common type of carbapenemase gene carried by ST11 CR‐hvKP in China. 10 , 14 , 32 And, we found that bla KPC‐2 and bla CTX‐M‐65 tended to exist in hypervirulent strains. Hypervirulent strains were more resistant to levofloxacin, which may result from the co‐transfer of bla KPC and quinolone resistance factors. For bla KPC is usually carried by plasmid, the plasmid often simultaneously carries other drug resistance genes. 11 In addition, all strains in this research showed a high level of susceptibility to colistin and tigecycline. Continuous resistance screening is still necessary.
Obviously, we reported a lower incidence of hypermucoviscosity phenotype of CR‐hvKP in this study. Two hypermucoviscous strains were from blood and sputum; both showed high lethality in the G. mellonella infection model. Gene prmpA2 was detected in hypermucoviscous strain KP1972, which is consistent with the finding that prmpA2 is responsible for hypermucoviscosity. 33 However, we observed a strain (KP219) with hypermucoviscosity but lack of prmpA and prmpA2 , as well as strains carrying prmpA and/or prmpA2 but not hypermucoviscous. These cases are less common in hypervirulent K. pneumoniae, but both have been reported. 9 , 34 This phenomenon deserves further investigation.
Through MLST and capsular type detection, we found that 41 CR‐hvKP in this study could mainly be divided into two series: 27 ST11‐KL64 isolates and 10 ST11‐KL47 isolates. All 27 ST11‐KL64 CR‐hvKP were positive for a combination of iucA‐iroN genes (with a few strains that also carrying prmpA and/or prmpA2 ), while all 10 ST11‐KL47 CR‐hvKP were positive for a combination of iucA‐ prmpA2 genes, suggesting that two different developmental series might have formed. This was confirmed by PFGE analysis. 7 ST11‐KL47 CR‐hvKP which from the PFGE cluster 10 shared the same virulence and resistance characteristics and all isolated from a short period of time, suggested that clonal spread had occurred in our hospital. Interestingly, among ST11‐KL64 CR‐hvKP in PFGE cluster 15 and 18, no consistent characteristics were observed. This might result from the rapid reshaping and diversification of the genome pool of ST11‐KL64 CR‐hvKP. 35 Moreover, we found 2 ST11‐KL28 KPC‐2‐producing CR‐hvKP and 1 ST2928‐KL54 IMP‐4‐producing CR‐hvKP. As far as we know, hvKP of these types had not been reported, suggesting the diversity of CR‐hvKP evolution.
In this study, only 3 strains with all the four virulence genes positive were detected; the three all showed high lethality in the G. mellonella infection model, suggesting complete pLVPK plasmid harboring. Interestingly, KP13, KP145, KP669, and KP1590 shared the same drug resistance type and genotype with KP178, KP213, KP238, and KP1640, but they showed opposite results in the G. mellonella infection model. This might be caused by the presence of undetected virulence factors. In addition, we also found that virulence genes irp‐1 and peg‐344 mainly existed in hypervirulent strains, indicating that these two genes may contribute to the virulence. In future research, the mechanism of hypervirulence is still worthy of attention.
This study only analyzed CRKP isolated from one hospital in China in one year. The sample size is very limited, and larger‐scale or multi‐center research results are needed to support the above results. In addition, the condition for screening hypervirulent strains in this study is the presence of one or more of virulence gene markers, rather than virulence plasmid, which may cause an overestimation of CR‐hvKP. And, we cannot provide data related to plasmid analysis. Given the excellent ability of K. pneumoniae to obtain exogenous genes encoding resistance and virulence, we need to arouse sufficient vigilance over CR‐hvKP.
5. CONCLUSION
ST11 KPC‐2‐producing CR‐hvKP is widespread in our hospital, and two significant lineages of ST11‐KL64 and ST11‐KL47 have emerged and clonal transmission occurred. It is urgent to develop novel rapid and convenient methods to accurately distinguish hypervirulent K. pneumoniae and classical K. pneumoniae in order to perform precise treatment and deal with the spread of CR‐hvKP.
AUTHOR CONTRIBUTIONS
LX and PO conceived and designed the study. PO, BJ, NP, and JW contributed to data collection and experiments implementation. PO, LC, YW, JY, YC, HY, CT, and LT contributed to data analysis and data interpretation. PO and LX contributed to writing the report and revising the report. All authors read and approved the final version of the manuscript.
CONFLICT OF INTEREST
All authors‐none to declare.
Supporting information
Appendix S1
Appendix S2
ACKNOWLEDGMENTS
This work was funded by Natural Science Foundation of Hunan Province, China (grant number: 2017JJ3173), Research Foundation of Education of Hunan Province, China (grant number: 20A292), and Renshu Foundation of Hunan Provincial People's Hospital, Changsha, China (grant number: 2016).
Ouyang P, Jiang B, Peng N, et al. Characteristics of ST11 KPC‐2‐producing carbapenem‐resistant hypervirulent Klebsiella pneumoniae causing nosocomial infection in a Chinese hospital. J Clin Lab Anal. 2022;36:e24476. doi: 10.1002/jcla.24476
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Munoz‐Price L, Poirel L, Bonomo R, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785‐796. doi: 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fraenkel‐Wandel Y, Raveh‐Brawer D, Wiener‐Well Y, et al. Mortality due to blaKPC Klebsiella pneumoniae bacteraemia. J Antimicrob Chemother. 2016;71(4):1083‐1087. doi: 10.1093/jac/dkv414 [DOI] [PubMed] [Google Scholar]
- 3. Wang Q, Wang X, Wang J, et al. Phenotypic and Genotypic Characterization of Carbapenem‐resistant Enterobacteriaceae: Data From a Longitudinal Large‐scale CRE Study in China (2012‐2016). Clin Infect Dis. 2018;67(suppl_2):S196‐S205. 10.1093/cid/ciy660 [DOI] [PubMed] [Google Scholar]
- 4. Zhao J, Liu C, Liu Y, et al. Genomic characteristics of clinically important ST11 Klebsiella pneumoniae strains worldwide. J Glob Antimicrob Resist. 2020;22:519‐526. doi: 10.1016/j.jgar.2020.03.023 [DOI] [PubMed] [Google Scholar]
- 5. Qi Y, Wei Z, Ji S, et al. ST11, the dominant clone of KPC‐producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307‐312. doi: 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 6. Qu H, Wang X, Ni Y, et al. NDM‐1‐producing Enterobacteriaceae in a teaching hospital in Shanghai, China: IncX3‐type plasmids may contribute to the dissemination of blaNDM‐1. Int J Infect Dis. 2015;34:8‐13. doi: 10.1016/j.ijid.2015.02.020 [DOI] [PubMed] [Google Scholar]
- 7. Lu M, Chen Y, Tang H, et al. Transmission and evolution of OXA‐48‐producing Klebsiella pneumoniae ST11 in a single hospital in Taiwan. J Antimicrob Chemother. 2020;75(2):318‐326. doi: 10.1093/jac/dkz431 [DOI] [PubMed] [Google Scholar]
- 8. Russo T, Marr C. Hypervirulent Klebsiella pneumoniae . Clin Microbiol Rev. 2019;32(3):e00001‐19. doi: 10.1128/CMR.00001-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cubero M, Grau I, Tubau F, et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin Microbiol Infect. 2016;22(2):154‐160. doi: 10.1016/j.cmi.2015.09.025 [DOI] [PubMed] [Google Scholar]
- 10. Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem‐resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327‐336. doi: 10.1093/jac/dkz446 [DOI] [PubMed] [Google Scholar]
- 11. Yang X, Dong N, Chan E, et al. Carbapenem Resistance‐Encoding and Virulence‐Encoding Conjugative Plasmids in Klebsiella pneumoniae . Trends Microbiol. 2021;29(1):65‐83. doi: 10.1016/j.tim.2020.04.012 [DOI] [PubMed] [Google Scholar]
- 12. Feng Y, Lu Y, Yao Z, et al. Carbapenem‐Resistant Hypervirulent Klebsiella pneumoniae of Sequence Type 36. Antimicrob Agents Chemother. 2018;62(7):e02644‐e2717. doi: 10.1128/AAC.02644-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen L, Kreiswirth B. Convergence of carbapenem‐resistance and hypervirulence in Klebsiella pneumoniae . Lancet Infect Dis. 2018;18(1):2‐3. doi: 10.1016/S1473-3099(17)30517-0 [DOI] [PubMed] [Google Scholar]
- 14. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem‐resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37‐46. doi: 10.1016/S1473-3099(17)30489-9 [DOI] [PubMed] [Google Scholar]
- 15. Zhou K, Xiao T, David S, et al. Novel Subclone of Carbapenem‐Resistant Klebsiella pneumoniae Sequence Type 11 with Enhanced Virulence and Transmissibility, China. Emerg Infect Dis. 2020;26(2):289‐297. doi: 10.3201/eid2602.190594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. CLSI . Performance standards for antimicrobial susceptibility testing, 27th ed. CLSI supplement M100, 2017.
- 17. FDA . Tygacil®(Tigecycline) for Injection, 2009. https://www.accessdata.fda.gov/drugsatfda_docs/label/2009/021821s016lbl
- 18. EUCAST . Breakpoints Tables for Interpretation of MICs and Zone Diameters, Version 6.0, 2016. http://www.eucast.org/clinical_breakpoints
- 19. CLSI . Performance standards for antimicrobial susceptibility testing, 28th ed. CLSI supplement M100, 2018.
- 20. Wang X, Xu X, Li Z, et al. An outbreak of a nosocomial NDM‐1‐producing Klebsiella pneumoniae ST147 at a teaching hospital in mainland China. Microb Drug Resist. 2014;20(2):144‐149. doi: 10.1089/mdr.2013.0100 [DOI] [PubMed] [Google Scholar]
- 21. Kim D, Park B, Choi M, et al. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J Antimicrob Chemother. 2019;74(1):190‐199. doi: 10.1093/jac/dky397 [DOI] [PubMed] [Google Scholar]
- 22. Turton J, Perry C, Elgohari S, et al. PCR characterization and typing of Klebsiella pneumoniae using capsular type‐specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59(Pt 5):541‐547. doi: 10.1099/jmm.0.015198-0 [DOI] [PubMed] [Google Scholar]
- 23. Schubert S, Cuenca S, Fischer D, et al. High‐pathogenicity island of Yersinia pestis in enterobacteriaceae isolated from blood cultures and urine samples: prevalence and functional expression. J Infect Dis. 2000;182(4):1268‐1271. doi: 10.1086/315831 [DOI] [PubMed] [Google Scholar]
- 24. Russo T, Olson R, Fang C, et al. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae . J Clin Microbiol. 2018;56(9):e00776‐e818. doi: 10.1128/JCM.00776-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brisse S, Passet V, Haugaard A, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51(12):4073‐4078. doi: 10.1128/JCM.01924-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diancourt L, Passet V, Verhoef J, et al. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178‐4182. doi: 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu B, Zhou H, Zhang X, et al. Molecular characterization of Klebsiella pneumoniae isolates from stool specimens of outpatients in sentinel hospitals Beijing, China, 2010–2015. Gut Pathog. 2017;30(9):39. doi: 10.1186/s13099-017-0188-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu C, Du P, Xiao N, et al. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare‐associated infections in Beijing, China. Virulence. 2020;11(1):1215‐1224. doi: 10.1080/21505594.2020.1809322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang X, Sun Q, Li J, et al. Molecular epidemiology of carbapenem‐resistant hypervirulent Klebsiella pneumoniae in China. Emerg Microbes Infect. 2022;11(1):841‐849. doi: 10.1080/22221751.2022.2049458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao Q, Guo L, Wang L, et al. Prevalence and characteristics of surgical site hypervirulent Klebsiella pneumoniae isolates. J Clin Lab Anal. 2020;34(9):e23364. doi: 10.1002/jcla.23364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang B, Pan F, Wang C, et al. Molecular epidemiology of Carbapenem‐resistant Klebsiella pneumoniae in a paediatric hospital in China. Int J Infect Dis. 2020;93:311‐319. doi: 10.1016/j.ijid.2020.02.009 [DOI] [PubMed] [Google Scholar]
- 32. Hao M, Shi X, Lv J, et al. In vitro Activity of Apramycin Against Carbapenem‐Resistant and Hypervirulent Klebsiella pneumoniae Isolates. Front Microbiol. 2020;11:425. doi: 10.3389/fmicb.2020.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lai Y, Peng H, Chang H. RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol. 2003;185(3):788‐800. doi: 10.1128/JB.185.3.788-800.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fang C, Chuang Y, Shun C, et al. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697‐705. doi: 10.1084/jem.20030857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang X, Ouyang J, He W, et al. Co‐occurrence of Rapid Gene Gain and Loss in an Interhospital Outbreak of Carbapenem‐Resistant Hypervirulent ST11‐K64 Klebsiella pneumoniae. Front Microbiol. 2020;12(11):579618. doi: 10.3389/fmicb.2020.579618 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Appendix S2
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


