Abstract
A gram-positive bacterium with antagonistic activity towards soilborne fungal pathogens has been isolated from the mycorrhizosphere of Sorghum bicolor inoculated with Glomus mosseae. It has been identified as Paenibacillus sp. strain B2 based on its analytical profile index and on 16S ribosomal DNA analysis. Besides having antagonistic activity, this bacterium stimulates mycorrhization.
In recent years, several types of microorganisms have been reported to be associated with the rhizospheres of different host plants colonized by arbuscular mycorrhizal (AM) fungi. These have been identified as associative N2-fixing bacteria (13), plant growth-promoting rhizobacteria (17), phosphate-solubilizing bacteria (15), and antagonists of plant pathogens (4). AM fungi are a ubiquitous component of most agroecosystems and play an important role in key rhizosphere processes (14), including plant protection against soilborne diseases (1, 5). Associated microorganisms may complement mycorrhizal activities, particularly in biological control in agricultural systems. However, few data on the compatibility between these microorganisms and AM fungi are available. There are reports that biocontrol agents, like gram-negative Pseudomonas strains, do not have inhibitory effects on AM formation (2, 11). We report the isolation and identification of a gram-positive bacterium from the mycorrhizosphere of sorghum and describe its activity towards root pathogens and AM fungi in vitro and in vivo.
Eight morphologically different bacteria (B1 to B8) were isolated from the mycorrhizosphere of sorghum plants (Sorghum bicolor L. var. Esquirol) inoculated with the AM fungus Glomus mosseae (Nicol et Gerd) Gerdemann et Trappe (BEG 12) (obtained from the Banque Européenne des Glomales [1a]). Seeds of sorghum were surface sterilized (30 min in 7% calcium hypochlorite with a few drops of Tween 20, followed by 30 min in 4% chloramine-T), and sporocarps of G. mosseae were surface sterilized as previously described (3). Plants were grown in a sterilized clay loam soil-calcined clay (Oil Dry Type III; OIL · DRI, Limited, Wisbech, United Kingdom) mixture (1:1) in sterile Sunbags (catalog no. 7026; Sigma) under constant conditions (22 to 24°C, 16-h photoperiod, 300 μmol of photons/m2/s, and 70% relative humidity) for 12 weeks. All the components (soil-clay mixture sporocarps, seeds, and water) were extensively checked for the absence of contaminants by incubation in malt agar medium. Bacteria were isolated from the growth substrate of G. mosseae-inoculated plants according to the method described by Zuberer (18) and were selected by colony characteristics: shape, size, edge morphology, surface, and pigment. No bacteria were obtained from the growth substrate of plants not inoculated with G. mosseae sporocarps. Bacteria were screened in vitro for their antagonistic activity towards Phytophthora parasitica isolate 204 by measuring the radial colony growth of the fungus and the zoosporangium production in the absence and presence of the bacteria according to the methods of Dal-Soo et al. (6). Zoospore germination on Millipore filters left unsaturated or saturated with the filtered supernatant of a B2 culture was evaluated, and the in vivo antagonism of the bacteria in terms of plant growth and the percentage of necrosed roots, in the presence or absence of G. mosseae, was assessed as described by Cordier et al. (5). AM development was determined according to the method of Trouvelot et al. (16). In vitro experiments were performed with five replicates for each experiment and were repeated at least twice. A randomized block design was used for in vivo growth room experiments, with seven replicates per treatment. Percentages were arcsine transformed prior to analysis. All data were analyzed by one-way analysis of variance and the Newman-Keuls test at a P value of <0.05. Results are given for one representative experiment.
Only one strain, B2, out of the eight isolated showed a significant antagonistic activity towards P. parasitica in vitro (Table 1). The antagonistic effect was also obtained with partially purified (by ion-exchange chromatography after heat treatment) media from a B2 bacterial culture, indicating that the strain secretes an antifungal factor (data not shown). The effect of antagonistic B2 bacteria was also tested in vivo on tomato plants grown in the presence or absence of G. mosseae and/or the bacterial strain B2 and subsequently inoculated with P. parasitica. Root necrosis caused by P. parasitica was reduced by 32, 53, and 63% when plants were inoculated with B2, G. mosseae, and B2 and G. mosseae, respectively (Table 2). The presence of B2 increased the root and shoot fresh weights of the mycorrhizal tomato plants, and together with G. mosseae it abolished the negative growth effect of P. parasitica. Inoculation with the bacteria also stimulated root colonization by G. mosseae (Table 2).
TABLE 1.
Effect of B2 bacteria on in vitro growth and zoosporangium production of mycelial cultures of P. parasitica and effect of filtered culture supernatant of B2 on zoospore germinationa
| Status of B2 | Radial mycelium growth (cm) after 6 days | No. of sporangia/culture after 6 days | % Zoospore germination after 6 h |
|---|---|---|---|
| Absent | 5.3a | 1,040a | 62.5a |
| Present | 2.1 (75.0)b | 0 (100)b | 25.9 (58.6)b |
Values in a column followed by the same letter do not differ significantly from each other at a P value of <0.05. Values in parentheses are the percents inhibition.
TABLE 2.
Effect of B2 bacteria on mycorrhizal colonization by G. mosseae and on damage of tomato roots by P. parasiticaa
| Content of inoculum (n = 7) | Fresh wt (g)
|
Root colonization by G. mosseae
|
% Necrosis by P. parasitica | ||
|---|---|---|---|---|---|
| Shoot | Root | % Colonized | AA (%)b | ||
| None | 16.07 bc | 7.12 c | |||
| G. mosseae | 16.44 b | 8.08 b | 32.4 b | 22.6 a | |
| B2 | 15.89 bc | 7.90 b | |||
| G. mosseae + B2 | 17.57 a | 9.06 a | 43.7 a | 28.4 a | |
| P. parasitica | 15.30 c | 5.42 d | 15.1 a | ||
| G. mosseae + P. parasitica | 16.20 bc | 7.18 c | 34.2 b | 22.7 a | 7.2 c |
| B2 + P. parasitica | 15.49 bc | 7.10 c | 10.3 b | ||
| G. mosseae + B2 + P. parasitica | 17.27 a | 8.88 a | 45.7 a | 31.0 a | 5.6 d |
Values in a column followed by the same letter do not differ significantly at a P value of <0.05.
AA, arbuscule abundancy.
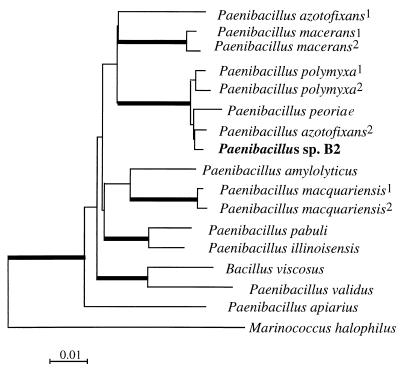
The bacterial strain B2 (gram positive) was characterized by using the analytical profile index (API) system (BioMérieux, Marcy l’Etoile, France) with API 50 CHB galleries and by sequence analysis (sequencing by Genome Express, Grenoble, France) of the small ribosomal subunit (16S ribosomal DNA [rDNA]) after PCR amplification with the eubacterial primers 27f (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492r (5′-TA CGG YTA CCT TGT TAC GAC TT-3′) and cloning. Of the 1,510 nucleotide bases of the DNA sequence obtained, 1,410 were aligned with those of the most closely related isolates by the Clustalx program. Phylogenetic distances were calculated according to the neighbor-joining method with Kimura parameters (7). A consensus tree obtained from 1,000 bootstrap replicates was drawn with Treeview (10). The API of strain B2 differed from that of Paenibacillus polymyxa only by the use of 3 carbohydrates (methyl-d-glucoside, melezitose, and d-tagatose) out of 49. Phylogenetic comparison of the 16S rDNA sequence with those of other Paenibacillus isolates (Fig. 1) confirmed the high similarity to P. polymyxa. Strain B2 was grouped with P. polymyxa, Paenibacillus peoriae, and Paenibacillus azotofixans in 100% of the trees obtained after bootstrap analysis, with its highest similarity being to the last species. However, the reference strain P. azotofixans P3 L-5 (DSM 5976T) did not show any antagonistic activity towards P. parasitica (Fig. 2).
FIG. 1.
Phylogenetic consensus tree based on the alignment of 1,410 bases from the small ribosomal subunit of Paenibacillus sp. strain B2 and 15 strains showing the highest nucleotide sequence similarities. Accession no. are as follows: Paenibacillus macerans, X57306; P. macerans2, D78319; P. polymyxa1, X60632; P. polymyxa2, D16276; P. peoriae, D78476; P. azotofixans1, X60608; P. azotofixans2, D78318; Paenibacillus amylolyticus, X60606; Paenibacillus macquariensis1, X60625; P. macquariensis2, X57305; Paenibacillus pabuli, X60630; Paenibacillus illinoisensis, D85397; Bacillus viscosus, X77792; Paenibacillus validus DSM 3037, D78320; Paenibacillus apiarius NRRL NRS-1438, U49247; and Marinococcus halophilus, X90835 (superscripts indicate different strains). M. halophilus was used as an out-group. Bootstrap values higher than 99% are in heavy lines.
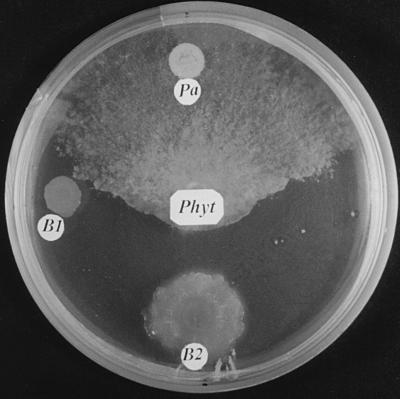
FIG. 2.
Antagonistic activity of Paenibacillus sp. strain B2 (B2) against P. parasitica (Phyt) compared with those of another bacterial isolate (B1) and the reference strain P. azotofixans P3 L-5 (DSM 5976T) (Pa). The organisms were grown on malt agar medium at 25°C for 2 weeks.
The occurrence of unidentified antagonistic bacteria in pot cultures of G. mosseae on strawberry has recently been reported (4). In the present study, such antagonistic bacteria were found in pot cultures of S. bicolor inoculated with sporocarps of G. mosseae, one strain of which (B2) strongly inhibited the in vitro and in vivo development of P. parasitica. Characterization of this bacterium with the API system and sequence analysis of the 16S rDNA indicated a close relationship to P. polymyxa and P. azotofixans. However, it differed from these species in carbohydrate use and antagonistic activity, respectively. These data indicate that the B2 isolate represents a new strain of Paenibacillus, and we propose that this bacterium be called Paenibacillus sp. strain B2.
Paenibacillus sp. strain B2 and its metabolites not only suppress the in vitro mycelial growth of P. parasitica but also inhibit sporangium production, zoospore germination, and germ tube elongation by the pathogen. This is very important since such effects can hinder the completion of the life cycle of the plant pathogen in vivo. In addition to inhibiting the in vitro growth of P. parasitica, Paenibacillus sp. strain B2 also reduced the hyphal growth of other pathogenic fungi: Fusarium oxysporum Foeu1 (FPSS) (reduced by 35.0%), Fusarium culmorum Fcul 1 (35.7%), Aphanomyces euteiches 502 (50.0%), Chalara elegans 84.1 (82.6%), Pythium sp. strain 0P 4 (36.0%), and Rhizoctonia solani AG3 (53.0%). This suggests that this bacterium has a broad spectrum of antagonistic activity. Interestingly, Paenibacillus sp. strain B2 stimulates AM fungal root colonization and also appears to be compatible with the germination and hyphal growth of G. mosseae in vitro (results not shown). Similar data have been reported by Barea et al. (2) for Pseudomonas strains.
There are several reports about the potential use of AM fungi as biological control agents against soilborne diseases (1, 5, 8, 9). The discovery of a Paenibacillus strain that can act as a biological control agent against soilborne fungal diseases while improving AM formation opens the possibility of using dual bacterial-fungal inoculation for ensuring the production of high-value plants in systems compatible with the environment.
Nucleotide sequence accession number.
The nucleotide sequence for isolate B2 was deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under the accession no. AJ011687.
Acknowledgments
We are grateful to Bachar Blal (BIORIZE) for supplying sorghum seeds and to V. Gianinazzi-Pearson for critically reading the manuscript. Origins of fungal isolates were as follows: P. parasitica, INRA—Antibes; F. oxysporum, F. culmorum, R. solani, and a Pythium sp., INRA—Dijon; A. euteiches and C. elegans, C. Richard (Agriculture Canada, Quebec, Canada); and the Tobacco Institute (Bergerac, France).
This work was partially supported by the INCO-DC EU, project no. ERBIC18 CT 970180, and Center Grant Program, Directorate General of Higher Education, Republic of Indonesia (grant to S.W.B.).
REFERENCES
- 1.Azcon-Aguilar C, Barea J M. Arbuscular mycorrhizas and biological control of soil borne pathogens—an overview of the mechanisms involved. Mycorrhiza. 1996;6:457–464. [Google Scholar]
- 1a.Banque Européenne des Glomales. [Online.] University of Kent, Canterbury, United Kingdom. http://wwwbio.ukc.ac.uk/beg/. [19 September 1999, last date accessed.]
- 2.Barea J M, Andrade G, Bianciotto V, Dowling D, Lohrke S, Bonfante P, O’Gara F, Azcon-Aguilar C. Impact on arbuscular mycorrhiza formation of Pseudomonas strains used as inoculants for biocontrol of soil-borne fungal plant pathogens. Appl Environ Microbiol. 1998;64:2304–2307. doi: 10.1128/aem.64.6.2304-2307.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budi S W, Blal B, Gianinazzi S. Surface-sterilization of sporacarps of Glomus mosseae for studying endomycorrhization in vitro. Mycorrhiza, 1999;9:65–68. [Google Scholar]
- 4.Citernesi A S, Fortuna P, Filippi C, Bagnoli G, Giovannetti M. The occurrence of antagonistic bacteria in Glomus mosseae pot cultures. Agronomie. 1996;16:671–677. [Google Scholar]
- 5.Cordier C, Pozo M J, Barea J M, Gianinazzi S, Gianinazzi-Pearson V. Cell defense responses associated with localized and systemic resistance to Phytophthora parasitica induced in tomato by an arbuscular mycorrhizal fungus. Mol Plant-Microbe Interact. 1998;11:1017–1028. [Google Scholar]
- 6.Dal-Soo K, Cook J R, Weller D M. Bacillus sp. L324-92 for biological control of three root diseases of wheat grown with reduced tillage. Phytopathology. 1997;87:551–558. doi: 10.1094/PHYTO.1997.87.5.551. [DOI] [PubMed] [Google Scholar]
- 7.Kimura M. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 8.Linderman R G. Mycorrhizal interactions with the rhizosphere microflora: the mycorrhizosphere effect. Phytopathology. 1988;78:366–371. [Google Scholar]
- 9.Liu R J. Effect of vesicular-arbuscular mycorrhizal fungi on Verticillium wilt of cotton. Mycorrhiza. 1995;5:293–297. [Google Scholar]
- 10.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 11.Paulitz T C, Linderman R G. Interactions between fluorescent pseudomonads and VA mycorrhizal fungi. New Phytol. 1989;113:37–45. [Google Scholar]
- 12.Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Secilia J, Bagyaraj D J. Bacteria and actinomycetes associated with pot cultures of vesicular-arbuscular mycorrhizas. Can J Microbiol. 1987;33:1069–1073. [Google Scholar]
- 14.Smith S E, Read D J. Mycorrhizal symbiosis. London, England: Academic Press; 1997. [Google Scholar]
- 15.Toro M, Nedialkova K, Azcon R, Barea J M. Establishment of two rock phosphate solubilizing bacteria in the rhizosphere of mycorrhizal onion plants and their effect on plant growth in a microcosm. In: Azcon-Aguilar C, Barea J M, editors. Mycorrhizas in integrated systems: from genes to plant development (EUR 16728). Luxembourg, Luxembourg: European Commission; 1996. pp. 665–668. [Google Scholar]
- 16.Trouvelot A, Kough J L, Gianinazzi-Pearson V. Mesure du taux de mycorhization VA d’un système radiculaire. Recherche de méthodes d’estimation ayant une signification fonctionnelle. In: Gianinazzi-Pearson V, Gianinazzi S, editors. Mycorrhizae: physiology and genetics. Paris, France: INRA Press; 1986. pp. 217–221. [Google Scholar]
- 17.von Alten H, Lindemann A, Schonbeck F. Stimulation of vesicular-arbuscular mycorrhiza by fungicides or rhizosphere bacteria. Mycorrhiza. 1993;2:167–173. [Google Scholar]
- 18.Zuberer D A. Recovery and enumeration of viable bacteria. In: Weaver R W, et al., editors. Method of soil analysis, part 2. Microbiological and biochemical properties. Madison, Wis: Soil Science Society of America; 1994. pp. 119–144. [Google Scholar]