Abstract
The occurrence of the acmA gene, encoding the lactococcal N-acetylmuramidase in new lactococcal isolates from raw milk cheeses, has been determined. Isolates were genotypically identified to the subspecies level with a PCR technique. On the basis of PCR amplification of the acmA gene, the presence or absence of an additional amplicon of approximately 700 bp correlated with Lactococcus lactis subspecies. L. lactis subsp. lactis exhibits both the expected 1,131-bp product and the additional amplicon, whereas L. lactis subsp. cremoris exhibits a single 1,131-bp fragment.
The development of new starter cultures for the manufacture of fermented dairy products usually involves the identification and characterization of lactic acid bacteria from raw milk cheeses manufactured without commercial cultures (3). Lactococcus lactis is the most important organism in dairy starter cultures.
Lactococcus strains with high autolytic activity are sought out because of their beneficial effect on cheese ripening. Autolysis of lactococci used as starter cultures in the manufacture of cheese results in the leakage of peptidases and other intracellular components, which play an important role in flavor development during ripening. Bacterial peptidoglycan hydrolases (autolysins) degrade the peptidoglycan of cell walls, causing cell lysis. The lactococcal gene acmA, encoding the major peptidoglycan hydrolase of L. lactis subsp. cremoris MG1363, the lactococcal N-acetylmuramidase required for cell separation during growth, was cloned, and its DNA was sequenced (2). PCR with the two sequencing primers PALA-4 and PALA-14 showed the amplification of a 1,131-bp fragment from the chromosomal DNA of the L. lactis subsp. cremoris strains AM1, HP, and MG1363, L. lactis subsp. lactis IL1403, and L. lactis subsp. lactis biovar diacetylactis 18-16S (2).
There is a discrepancy between the phenotypic identification to the subspecies level of some strains of Lactococcus and their genotypes determined with rRNA-targeted probes (4–8). A fast PCR approach to differentiate the two subspecies and the diacetylactis biovar of L. lactis, based on the mosaic structure of the L. lactis histidine biosynthesis operon, has been reported (1).
During a search for autolytic strains of lactococci with potential interest as dairy starters, new isolates were identified by PCR protocols and the occurrence of the acmA gene in L. lactis subsp. lactis and L. lactis subsp. cremoris isolates was investigated.
New isolates of lactococci were obtained on MRS agar (pH 5.7) (Biolife, Milan, Italy) plates from 1-day-old ewe’s milk cheeses and selected for their ability to grow and coagulate milk in 6 h at 30°C. L. lactis subsp. cremoris strains MG1363, 9B4, LMG2130, AM2, and ATCC 19257 and L. lactis subsp. lactis strains IL1403, CNRZ481, NCDO763, ATCC 9936, and ATCC 11454 were the reference strains. Isolates were maintained as stock cultures at −80°C, transferred to M17 broth (Biolife), and subcultured twice before use.
Differentiation of new isolates was performed by phenotypic and genotypic characterization. Phenotypic characteristics such as Gram reaction and shape by phase-contrast microscopy were determined with overnight cultures in Elliker broth (Difco Laboratories, Detroit, Mich.). Growth after 2 days at 40°C and arginine hydrolysis in Elliker broth supplemented with l-arginine monochlorhydrate at 0.3%, checked after 3 and 7 days at 30°C with Nessler reagent, were also investigated. Genomic DNA was prepared from colonies grown in APT agar (Biolife) plates for 24 h at 30°C. Colonies were suspended in 20 μl of sterile double-distilled water in microcentrifuge tubes, mixed thoroughly, and stored at −40°C. This preparation was used as the template. The PCR approach for genotypic Lactococcus lactis subspecies differentiation developed by Beimfohr et al. (1) was carried out according to the methodology described by these authors by using DNA from colonies as the template.
The presence of the lactococcal cell wall hydrolase encoded by the gene acmA was investigated by PCR amplification (2) with the primers PALA-4 and PALA-14 (LAB-Center, Madrid, Spain). The PCR mixture contained 1 μl of prepared cells, 0.5 μM each of the two primers, 250 μM (each) dATP, dGTP, dCTP, and dTTP, 1.25 U of Taq DNA polymerase (Advanced Biotechnologies, Surrey, United Kingdom), 1.5 mM MgCl2, and 5 μl of the reaction buffer supplied with the enzyme. The final volume was 50 μl. The amplification mixture was overlaid with two drops of mineral oil. The PCR amplification was performed with a thermal cycler, Gene ATAQ Controller (Pharmacia LKB, Uppsala, Sweden), according to the following conditions: initial denaturation at 94°C for 3 min followed by 25 cycles consisting of denaturation for 30 s at 92°C, primer annealing for 30 s at 45°C, and primer extension for 60 s at 72°C. Amplified products were separated by electrophoresis on 1% agarose gels in Tris-acetate-EDTA buffer at 100 V and visualized by staining with ethidium bromide.
L. lactis subsp. lactis grows at 40°C and in 4% NaCl and hydrolyzes arginine. In contrast, L. lactis subsp. cremoris does not. However, some strains with the L. lactis subsp. cremoris genotype could be identified as L. lactis subsp. lactis on the basis of their ability to grow at 40°C and in 4% NaCl and to hydrolyze arginine (6, 9). Our results from phenotypic differentiation based on growth at 40°C and arginine hydrolysis and from genotypic differentiation based on PCR amplification with primers complementary to positions in the histidine biosynthesis operon (1) confirm the discrepancy between phenotyping and genotyping of L. lactis subspecies strains (Table 1). Of 57 isolates able to hydrolyze arginine, 13 were identified as L. lactis subsp. cremoris and 44 were identified as L. lactis subsp. lactis on the basis of genotypic differentiation. With regard to reference strains, all isolates exhibited the expected pattern, with the exception of the L. lactis subsp. lactis strains CNRZ481 and NCDO763, which presented with primer pair 1 the 556-bp DNA fragment and with primer pair 3 the 1,149-bp DNA fragment typical for L. lactis subsp. cremoris. L. lactis subsp. lactis CNRZ481 hydrolyzed arginine, whereas L. lactis subsp. lactis NCDO763 was negative for this characteristic. Thus, on the basis of PCR data, these reference strains should be reclassified as L. lactis subsp. cremoris. Godon et al. (4) earlier reported the misclassification of L. lactis subsp. lactis NCDO763 after DNA hybridization with DNA probes, showing the relationship of this strain with L. lactis subsp. cremoris. Taken together, these data show the variability in the expression of phenotypic traits and the relative stability of genotypic characteristics.
TABLE 1.
Phenotypic and genotypic characteristics of L. lactis reference strains and new isolates
| Strain | NH3 from arginine | Growth at 40°C | Amplification by PCR
|
|||
|---|---|---|---|---|---|---|
| Pair 1, 556 bp | Pair 2, 343 bp | Pair 3
|
||||
| 1,149 bp | 934 bp | |||||
| L. lactis subsp. cremoris MG1363 | + | − | + | − | + | − |
| L. lactis subsp. cremoris 9B4 | − | − | + | − | + | − |
| L. lactis subsp. cremoris LMG2130 | + | + | + | − | + | − |
| L. lactis subsp. cremoris ATCC 19257 | − | − | + | − | + | − |
| L. lactis subsp. cremoris AM2 | + | − | + | − | + | − |
| L. lactis subsp. cremoris (13 isolates)a | + | + (7) | + | − | + | − |
| L. lactis subsp. lactis NCDO763 | − | − | + | − | + | − |
| L. lactis subsp. lactis CNRZ481 | + | − | + | − | + | − |
| L. lactis subsp. lactis IL1403 | − | − | − | + | − | + |
| L. lactis subsp. lactis ATCC 11454 | + | + | − | + | − | + |
| L. lactis subsp. lactis ATCC 9936 | + | − | − | + | − | + |
| L. lactis subsp. lactis (44 isolates)a | + | + | − | + | − | + |
Isolated in the present work.
Results of PCR amplification on the presence of the acmA gene with the two sequencing primers of Buist et al. (2) are shown in Table 2. All L. lactis subsp. cremoris strains tested presented the same 1,131-bp DNA fragment expected, whereas all L. lactis subsp. lactis strains tested presented, in addition, a fragment of approximately 700 bp, with the exception of L. lactis subsp. lactis strains CNRZ481 and NCDO763, which exhibited only the 1,131-bp fragment amplified in all L. lactis subsp. cremoris strains. This result agrees with the characterization of these two strains as L. lactis subsp. cremoris by PCR amplification with the primer pairs of Beimfohr et al. (1). Ethidium bromide-stained agarose gels after PCR with primers PALA-4 and PALA-14 show the results obtained with reference strains and new L. lactis isolates (Fig. 1).
TABLE 2.
PCR fragments amplified with primers PALA-4 and PALA-14 for acmA gene detection in L. lactis
| Strain | 1,131-bp fragment | ∼700-bp fragment |
|---|---|---|
| L. lactis subsp. cremoris MG1363 | + | − |
| L. lactis subsp. cremoris 9B4 | + | − |
| L. lactis subsp. cremoris LMG2130 | + | − |
| L. lactis subsp. cremoris ATCC 19257 | + | − |
| L. lactis subsp. cremoris AM2 | + | − |
| L. lactis subsp. cremoris (13 isolates)a | + | − |
| L. lactis subsp. lactis NCDO763 | + | − |
| L. lactis subsp. lactis CNRZ481 | + | − |
| L. lactis subsp. lactis IL1403 | + | + |
| L. lactis subsp. lactis ATCC 11454 | + | + |
| L. lactis subsp. lactis ATCC 9936 | + | + |
| L. lactis subsp. lactis (44 isolates)a | + | + |
Isolated in the present work.
FIG. 1.
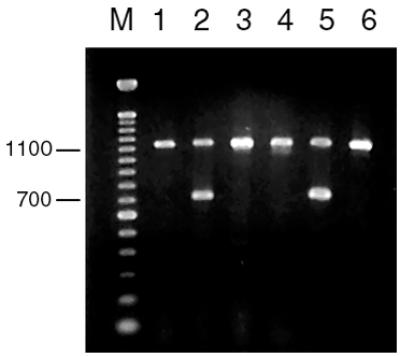
PCR products formed from total DNA of lactococcal strains with primers PALA-4 and PALA-14. L. lactis subsp. cremoris exhibits a single 1,131-bp fragment (lanes 1, 3, 4, and 6). L. lactis subsp. lactis exhibits both the 1,131-bp product and an additional amplicon of approximately 700 bp (lanes 2 and 5). Lanes: M, molecular size marker (100-bp DNA ladder; Gibco BRL); 1, L. lactis subsp. cremoris MG1363; 2, L. lactis subsp. lactis IL1403; 3, L. lactis subsp. lactis CNRZ481; 4, L. lactis subsp. lactis NCDO763; 5, L. lactis subsp. lactis CL23; 6, L. lactis subsp. cremoris CL173.
PCR products with the primers for the acmA gene from L. lactis subsp. cremoris and L. lactis subsp. lactis were purified with the GFX PCR DNA and Gel Band Purification Kit (Pharmacia Biotech) according to the supplier’s instructions. PCR-generated fragments were sequenced with an ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin- Elmer) and the Applied Biosystems model 377 automated DNA sequencer (Perkin-Elmer). Direct sequencing of the 1,131-bp PCR product from L. lactis subsp. cremoris and L. lactis subsp. lactis revealed no differences with the acmA gene of Buist et al. (2) after comparison with the BLAST programs of the National Center for Biotechnology Information. However, the fragment of approximately 700 bp resulting from amplification with primers targeting the acmA gene presented no significant similarity to the acmA gene after comparison with the BLAST 2 SEQUENCES program. This fragment present in L. lactis subsp. lactis strains showed 85% identity to the sequences of two ATPases from the Streptococcus faecalis strains H+ and F1F0.
Even though the exact nature of the product encoded by the fragment of approximately 700 bp remains unknown, the difference between L. lactis subsp. lactis and L. lactis subsp. cremoris genotypes regarding PCR amplification of the acmA gene has been proven. This characteristic may be useful to screen and group new lactococcal isolates.
Acknowledgments
This work was supported by project ALI96-2511 from the Spanish Plan Nacional de Investigación Científica y Desarrollo Tecnológico.
S.G. is the recipient of a fellowship from the Comunidad Autónoma de Madrid.
REFERENCES
- 1.Beimfohr C, Ludwig W, Schleifer K-H. Rapid genotypic differentiation of Lactococcus lactis subspecies and biovar. Syst Appl Microbiol. 1997;20:216–221. [Google Scholar]
- 2.Buist G, Kok J, Leenhouts K J, Dabrowska M, Venema G, Haandrikman A J. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cogan T M, Barbosa M, Beuvier E, Bianchi-Salvadori B, Cocconcelli P S, Fernandes I, Gomez J, Gomez R, Kalantzopoulos G, Ledda A, Medina M, Rea M, Rodriguez E. Characterization of the lactic acid bacteria in artisanal dairy products. J Dairy Res. 1997;64:409–421. [Google Scholar]
- 4.Godon J-J, Delorme C, Ehrlich S D, Renault P. Divergence of genomic sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1992;58:4045–4047. doi: 10.1128/aem.58.12.4045-4047.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klijn N, Weerkamp A H, de Vos W N. Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl Environ Microbiol. 1995;61:788–792. doi: 10.1128/aem.61.2.788-792.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salama M S, Musafija-Jeknic T, Sandine W E, Giovannoni S J. An ecological study of lactic acid bacteria: isolation of new strains of Lactococcus including Lactococcus lactis subspecies cremoris. J Dairy Sci. 1995;78:1004–1017. [Google Scholar]
- 7.Salama M S, Sandine W E, Giovannoni S J. Isolation of Lactococcus lactis subsp. cremoris from nature by colony hybridization with rRNA probes. Appl Environ Microbiol. 1993;59:3941–3945. doi: 10.1128/aem.59.11.3941-3945.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salama M, Sandine W, Giovannoni S. Development and application of oligonucleotide probes for identification of Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1991;57:1313–1318. doi: 10.1128/aem.57.5.1313-1318.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weerkamp A H, Klijn N, Neeter R, Smit G. Properties of mesophilic lactic acid bacteria from raw milk and naturally fermented milk products. Neth Milk Dairy J. 1996;50:319–332. [Google Scholar]


