Abstract
Background:
The World Health Organization (WHO) recommends the introduction of rotavirus vaccine in the immunization program of all countries. In the Central African Republic (CAR), sentinel surveillance for rotavirus gastroenteritis was established in 2011 by the Ministry of Health, with the support of the Surveillance en Afrique Centrale Project (SURVAC). The purpose of this study was to assess the burden of rotavirus gastroenteritis and to identify rotavirus strains circulating in CAR before the introduction of rotavirus vaccine planned for this year, 2014.
Methods:
One sentinel site and one laboratory at the national level were designated by the CAR Ministry of Health to participate in this surveillance system. Stool samples were collected from children who met the WHO rotavirus gastroenteritis case definition (WHO, 2006). The samples were first screened for group A rotavirus antigen by enzyme immunoassay (EIA), and genotyping assays performed using a multiplex reverse transcriptase PCR (RT-PCR) technique.
Results:
Between October 2011 and September 2013, 438 stool samples were collected and analyzed for detection of rotavirus antigen; 206 (47%) were positive. Among the 160 (78%) that could be genotyped, G2P[6] was the predominant strain (47%) followed by G1P[8] (25%) and G2P[4] (13%).
Conclusions:
Almost half of stool samples obtained from children hospitalized with gastroenteritis were positive for rotavirus. These baseline rotavirus surveillance data will be useful to health authorities considering rotavirus vaccine introduction and for evaluating the efficacy of rotavirus vaccine once it is introduced into the routine immunization system.
Keywords: Pediatrics, Rotavirus, Surveillance, Gastroenteritis, Central African Republic
Group A rotavirus is a major cause of severe gastroenteritis in young children, causing an estimated 453,000 deaths in 2008, mostly in Africa and South Asia (WHO, 2012). In 2009, WHO recommended the introduction of rotavirus vaccine in all countries (WHO, 2009).
Rotavirus belongs to the family of Reoviridae. The virus genome consists of 11 segments of double-stranded RNA which encodes 12 proteins. The protease sensitive outer capsid protein VP4 defines the P genotypes, and the surface glycoprotein VP7 defines the G genotypes. Rotavirus strain genotyping is commonly based on a dual nomenclature using both VP4 and VP7 genes (Hoshino et al., 1985). Studies in African countries have shown a diversity of rotavirus strains, including some less common human rotavirus types such as G12, G9, G8 and P[6] (Mwenda et al., 2010). The most prevalent rotavirus strains found in Africa are G1P[8], G2P[6], G8P[6], G3P[8] (Todd et al., 2010). There are two licensed rotavirus vaccines ; (1) Rotarix which is based on a monovalent human G1P[8] strain, and (2) RotaTeq, a pentavalent vaccine consist of 5 human-bovine reassortant strains. The genetic evolution or changes in circulating virus population may represent a problem for vaccines efficacy. For this reason, it is important to collect genotype information before the introduction of the rotavirus vaccine. These data will help monitor the impact of the vaccine in the future.
CAR is a developing country located in Central African region with a population estimated at 5,166,510 in July 2013 (CIA, 2013). Very limited data on rotavirus were obtained in CAR during the 1980s (Georges-Courbot et al., 1988a,b). Since then, only few data on rotavirus epidemiology in CAR in 2008 have indicated that the most common genotypes found were G1P[8] followed by G1P[6] and G2P[4] (Gouandjika-Vasilache et al., 2014). In July 2011, the Ministry of Health of CAR established sentinel surveillance for rotavirus gastroenteritis, with support of the SURVAC project (The_Laboratory_Working_Group_for_SURVAC, 2011), The goal of the SURVAC project was to strengthen disease surveillance and response in selected countries in Central Africa. Rotavirus surveillance was established in CAR using the generic protocol provided by the World Health Organization (WHO) (WHO, 2006).
This study aimed to describe the prevalence of rotavirus gastroenteritis among children less than 5 years of age and identify rotavirus strains circulating in CAR. The findings reported here describe the co-circulation of different rotavirus strains in CAR.
The surveillance was conducted at one sentinel hospital, the Complexe Pédiatrique de Bangui Hospital, located in Bangui, the capital of CAR. This hospital is the largest pediatric hospital in the country. From October 2011 to September 2013, stool specimens were collected as described in WHO manual (WHO, 2006), within 48 h of hospital admission, from all children less than 5 years old who met the WHO case definition of gastroenteritis: the occurrence of at least 3 looser than normal or watery stools in a 24 h period and/or two or more episodes of vomiting unexplained by other reasons (WHO, 2006).
Stool samples were first screened for group A Rotavirus antigen by EIA at the Complexe Pediatrique de Bangui Hospital laboratory. Aliquots were then stored at −20 °C before being transported to the Institut Pasteur de Bangui, where results were confirmed by EIA and genotyping assays were performed using a multiplex reverse transcription polymerase chain reaction (RT-PCR) technique as previously described (Pukuta et al., 2014). Samples subjected to genotyping were subsequently confirmed using the same method at the Centers for Disease Control and Prevention (CDC), Atlanta, USA, for quality control.
From October 2011 through September 2013, a total of 438 children <5 years old with diarrhea were enrolled at the Complex Pédiatrique sentinel site in Bangui, CAR. For each of these 438 children, a stool sample was tested for group A rotavirus antigen by EIA, and rotavirus was identified in 206 (47%), ranging from 37% in 2012 to 52% in 2013. This rotavirus prevalence is consistent with that reported in a 2008 Bangui study, where rotavirus was responsible for 40% of hospitalized diarrhea cases (Gouandjika-Vasilache et al., 2014) and with studies conducted in a neighboring country, Cameroon, where rotavirus was found in 42.8% of diarrhea cases (Ndze et al., 2012), as well as with an overall prevalence of rotavirus infection estimated at 40% in the African region (Mwenda et al., 2010). During the study period, rotavirus infection occurred year-round but was more common from November through March, with a high peak in January (2012) and March (2013), and from June to September with a smaller peak in August (2012) and June (2013) (Fig. 1). These biannual peaks correspond to the large and small dry seasons respectively in the country. This pattern is similar to that seen in Namibia in 1999 where rotavirus infection also showed biannual peaks (Page et al., 2010).
Fig. 1.
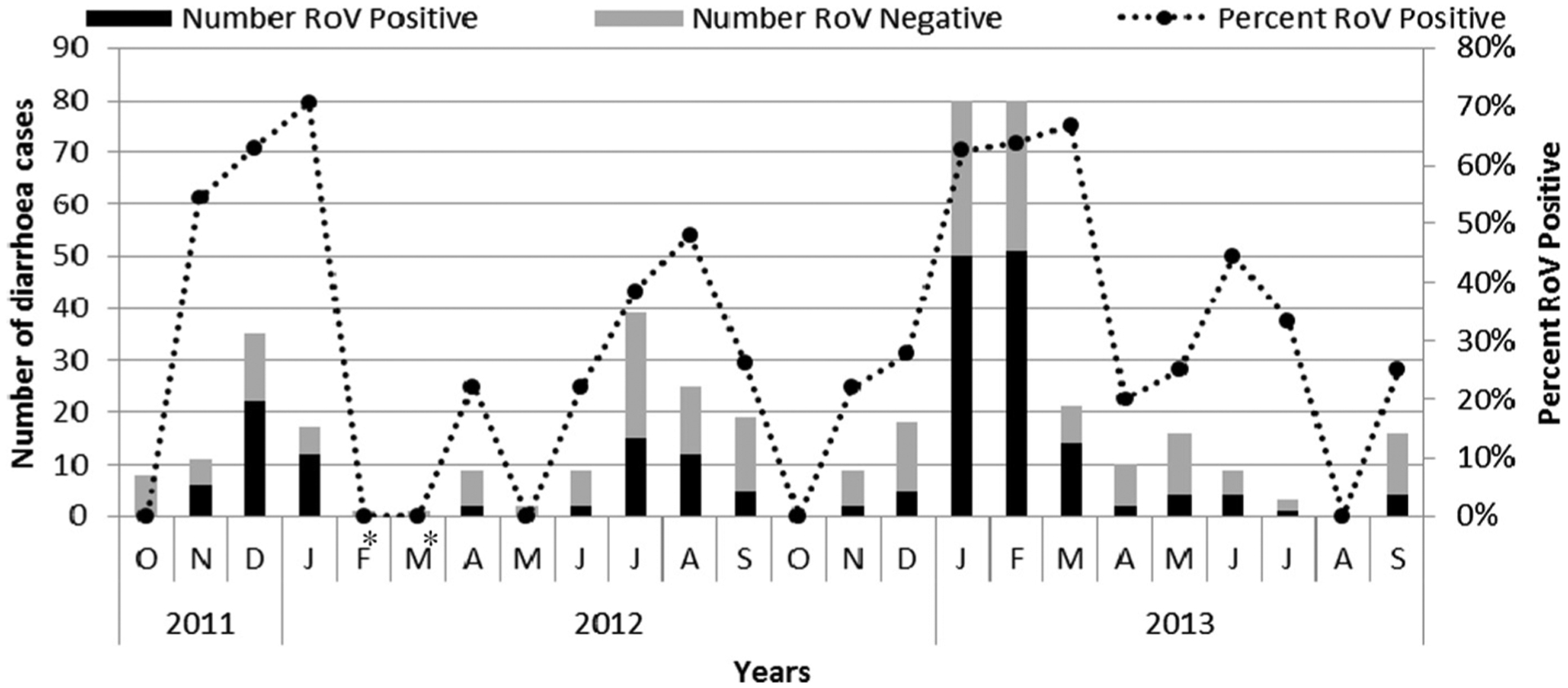
Seasonality of rotavirus diarrhea CAR, from October 2011–September 2013. Number of rotavirus-positives (shaded dark bars), rotavirus-negatives (shaded gray bars) and percentage of rotavirus-positives (dotted line) are indicated. *Enrollment during these months declined as a result of staffing issues at the sentinel site.
The age distribution of diarrhea cases showed that the highest rotavirus detection rate was among children less than 5 months old (56%), followed by children 6–11 (49%), 12–23 (34%) and 24–59 (17%) months of age (data not shown). The cumulative rotavirus positivity rate showed that 180 of 206 (87.4%) positive specimens were from children in the 0–11 month age group, including 38% from the 0–5 months age group (Fig. 2). Only 5% of positive cases were observed in the 24–59 month age group. These findings are consistent with those from other Central African countries such as the Democratic Republic of the Congo, where in 2003–2005 the majority of the patients affected by rotavirus infection were less than 12 months old (Kabue et al., 2010). Similar results were observed in other African countries, including Ghana, Kenya, Ethiopia (Mwenda et al., 2010).
Fig. 2.
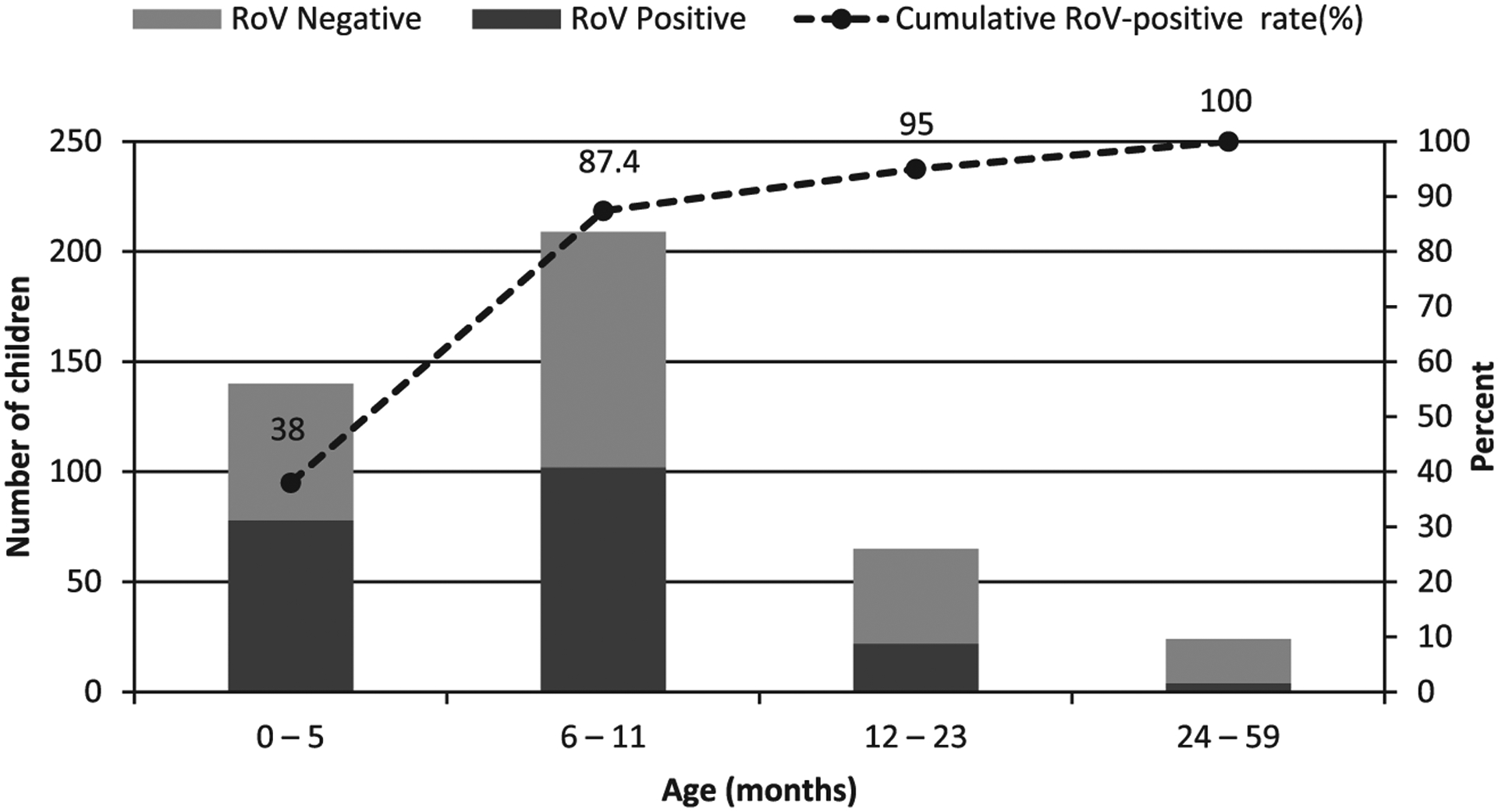
Distribution of diarrhea cases by age group of children less than 5 years of age admitted with gastroenteritis to the Complex Pediatrique hospital in Central African Republic, October 2011 to September 2013. Number of rotavirus-positives (shaded dark bars), rotavirus negatives (shaded gray bars) and Cumulative RoV positivity rate (dotted line). RoV = rotavirus.
All rotavirus positive samples (206) and 22 rotavirus negative samples (228 total) were subjected to RT-PCR for rotavirus genotyping and genotypes could be assigned to 160 strains (i.e., 160 samples). Rotavirus strains bearing the G2 genotype were found in 105 of 160 (66%) samples and were the most common found during the study period, followed by G1, found in 45 (28%) samples, and 5 (3%) each of G9 and G12 genotypes (Table 1). The most common P types associated with rotavirus infections in Bangui were P[6] found in 83 (52%) specimens, followed by P[8] in 56 (35%), and P[4] in 21 (13%). The most common G and P combinations were G2P[6] (47%), followed by G1P[8] (25%) and G2P[4] (13%). Unusual combinations, such as G12P[6], were also detected (2%). These findings are consistent with 2008 data from Bangui, which also showed that G1P[8] was the most common rotavirus strain detected (Gouandjika-Vasilache et al., 2014). Genotype G1P[8] is recognized as the most prevalent in Africa (Audu et al., 2002; Steele et al., 1998) and worldwide (Gentsch et al., 2005). Among the P types, genotype P[6], found in 52% of strains, was the most common. Our findings confirm the high prevalence of P[6], which previously was documented to be 18–75% in African countries (Gentsch et al., 2005); recent studies have reported the rapid expansion of P[6] prevalence in Africa (Seheri et al., 2014; Steele and Ivanoff, 2003). During the two years of this evaluation, G2 accounted for 66% of G types and were found in combination with P[6] (71%), P[4] (20%) and P[8] (9%). Previous studies of rotavirus infection in CAR in the 1980’s reported G2 strains in only 15.4% (Georges-Courbot et al., 1988a). More recently, in 2008, G2 strains were reported to account for 10% of G types (Gouandjika-Vasilache et al., 2014). The disparities among these studies are most likely due to the fact that neither of the two first studies was sentinel site based surveillance and the population targeted was probably different; however, they may also reflect natural temporal changes in circulating strains. It is noteworthy that G12 was detected for the first time in CAR in association with P[6] and P[8]. G12 strains have been reported previously to circulate in Cameroon and in Ethiopia (Mwenda et al., 2010), in South Africa (Page et al., 2009), in India (Das et al., 2003) and worldwide (Rahman et al., 2007).
Table 1.
Rotavirus G and P types detected from sentinel site surveillance, Bangui, Central African Republic, October 2011 to September 2013.
| P type | No. of strains, by G type | ||||
|---|---|---|---|---|---|
| G1 | G2 | G9 | G12 | Total | |
| P[4] | 0(0) | 21(13) | 0 | 0 | 21(13) |
| P[6] | 5(3) | 75(47) | 0 | 3(2) | 83(52) |
| P[8] | 40(25) | 9(6) | 5(3) | 2(1) | 56(35) |
| Total | 45(28) | 105(66) | 5(3) | 5(3) | 160(100) |
The percentage of each type is indicated in bracket (%).
As the emergence of new genotypes and the changes in circulating genotypes represents potential problem for vaccines protection, knowledge of circulating rotavirus strains will help measure the efficacy of vaccine. Both rotavirus vaccines (Rotarix and Rotateq) have been shown to be efficacious against the common strains causing disease but protection against rare or novel strains is yet to be documented. Hence, rotavirus strain surveillance after vaccine introduction will be critical for a better understanding of vaccine impact in CAR and will help detect genotype replacement after vaccine introduction.
There are some limitations in this study related to the fact that this surveillance was inpatient hospital-based surveillance of children with diarrhea, in only one sentinel site in Bangui, and therefore, these data may not be representative of the entire country. Additional sentinel sites for rotavirus surveillance in other cities would likely capture a more complete picture of rotavirus burden in the country. In addition, enrollment at the sentinel site in February and March 2012 was affected by staffing issues and the collection of stool samples was likely incomplete during that time period. Also, political instability and internal conflicts have affected the country for more than a decade, and more recently the coup d’état in March 2013 impacted study enrollment and laboratory testing, as a shortage of laboratory reagents permitted only a portion of samples to be genotyped.
The data reported here represent baseline data on sentinel site surveillance of rotavirus infection in CAR in the absence of a rotavirus vaccination program. These data on rotavirus genotypes provide information that will be helpful in monitoring the impact of the vaccine on circulating strains once it is introduced.
Acknowledgements
We are grateful to the staff of the Complexe Pédiatrique de Bangui, Bangui, Central African Republic, for their assistance with case enrolment and data collection. We would like to thank Mrs Kouagou Astrid, Complexe Pédiatrique de Bangui, Bangui, for technical assistance. We are grateful to Mrs Mazikoyo, Complexe Pédiatrique de Bangui, Bangui, for her help with data management. We also want to thank the WHO office, Bangui, Central African Republic, for logistical help in the country. We would like to thank Mrs Jamie Lewis, United States Centers for Disease Control and Prevention, Atlanta, USA for technical support. We are grateful to Olen Kew and Paul Rota of the Centers for Disease Control and Prevention, Atlanta, USA, for guidance, helpful discussions, and constructive criticisms leading to the completion of this work.
Funding Sources
Funding for this work was provided by the Ministry of Health of the Central African Republic, the Bill and Melinda Gates Foundation though the SURVAC Project (Grant No. 51214), the World Health Organization, the U.S. Centers for Diseases Control and Prevention, the CDC Foundation and the Pasteur Institute of Bangui.
Footnotes
Publisher's Disclaimer: Disclaimer
Publisher's Disclaimer: The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the US Department of Health and Human Services.
References
- Audu R, Omilabu SA, de Beer M, Peenze I, Steele AD, 2002. Diversity of human rotavirus VP6, VP7, and VP4 in Lagos State, Nigeria. J. Health Popul. Nutr 20, 59–64. [PubMed] [Google Scholar]
- CIA, 2013. Africa: Central African Republic. <https://www.cia.gov/library/publications/the-world-factbook/geos/ct.html>.
- Das S, Varghese V, Chaudhury S, Barman P, Mahapatra S, Kojima K, Bhattacharya SK, Krishnan T, Ratho RK, Chhotray GP, Phukan AC, Kobayashi N, Naik TN, 2003. Emergence of novel human group A rotavirus G12 strains in India. J. Clin. Microbiol 41, 2760–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O, Kirkwood CD, Fischer TK, Parashar UD, Bresee JS, Jiang B, Glass RI, 2005. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J. Infect. Dis 192 (Suppl 1), S146–S159. [DOI] [PubMed] [Google Scholar]
- Georges-Courbot MC, Beraud AM, Beards GM, Campbell AD, Gonzalez JP, Georges AJ, Flewett TH, 1988a. Subgroups, serotypes, and electrophoretypes of rotavirus isolated from children in Bangui, Central African Republic. J. Clin. Microbiol 26, 668–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges-Courbot MC, Monges J, Beraud-Cassel AM, Gouandjika I, Georges AJ, 1988b. Prospective longitudinal study of rotavirus infections in children from birth to two years of age in Central Africa. Ann. Inst. Pasteur Virol 139, 421–428. [DOI] [PubMed] [Google Scholar]
- Gouandjika-Vasilache I, Manirakiza A, Gody JC, Banga-Mingo V, Kongombe OO, Esona MD, Bowen MD, Waku-Kouomou D, 2014. Rotavirus epidemiology in bangui, central african republic, 2008(1.). Emerg. Infect. Dis 20, 1254–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y, Sereno MM, Midthun K, Flores J, Kapikian AZ, Chanock RM, 1985. Independent segregation of two antigenic specificities (VP3 and VP7) involved in neutralization of rotavirus infectivity. Proc. Natl. Acad. Sci. U.S.A 82, 8701–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabue JP, Peenze I, de Beer M, Esona MD, Lunfungula C, Biamungu M, Simba TR, Muyembe Tamfum JJ, Steele AD, 2010. Characterization of human rotavirus recovered from children with acute diarrhea in Kinshasa, Democratic Republic Of Congo. J. Infect. Dis 202 (Suppl), S193–S197. [DOI] [PubMed] [Google Scholar]
- Mwenda JM, Ntoto KM, Abebe A, Enweronu-Laryea C, Amina I, McHomvu J, Kisakye A, Mpabalwani EM, Pazvakavambwa I, Armah GE, Seheri LM, Kiulia NM, Page N, Widdowson MA, Steele AD, 2010. Burden and epidemiology of rotavirus diarrhea in selected African countries: preliminary results from the African Rotavirus Surveillance Network. J. Infect. Dis 202 (Suppl), S5–S11. [DOI] [PubMed] [Google Scholar]
- Ndze VN, Akum AE, Kamga GH, Enjema LE, Esona MD, Banyai K, Therese OA, 2012. Epidemiology of rotavirus diarrhea in children under 5 years in Northern Cameroon. Pan Afr. Med. J 11, 73. [PMC free article] [PubMed] [Google Scholar]
- Page NA, de Beer MC, Seheri LM, Dewar JB, Steele AD, 2009. The detection and molecular characterization of human G12 genotypes in South Africa. J. Med. Virol 81, 106–113. [DOI] [PubMed] [Google Scholar]
- Page N, Pager C, Steele AD, 2010. Characterization of rotavirus strains detected in Windhoek, Namibia during 1998–1999. J. Infect. Dis 202 (Suppl), S162–S167. [DOI] [PubMed] [Google Scholar]
- Pukuta ES, Esona MD, Nkongolo A, Seheri M, Makasi M, Nyembwe M, Mondonge V, Dahl BA, Mphahlele MJ, Cavallaro K, Gentsch J, Bowen MD, Waku-Kouomou D, Muyembe JJ, Group SW, 2014. Molecular surveillance of rotavirus infection in the Democratic Republic of the Congo August 2009 to June 2012. In: Pediatr. Infect. Dis. J 33, 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Matthijnssens J, Yang X, Delbeke T, Arijs I, Taniguchi K, Iturriza-Gomara M, Iftekharuddin N, Azim T, Van Ranst M, 2007. Evolutionary history and global spread of the emerging g12 human rotaviruses. J. Virol 81, 2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seheri M, Nemarude L, Peenze I, Netshifhefhe L, Nyaga MM, Ngobeni HG, Maphalala G, Maake LL, Steele AD, Mwenda JM, Mphahlele JM, 2014. Update of rotavirus strains circulating in Africa from 2007 through 2011. Pediatr. Infect. Dis. J 33 (Suppl 1), S76–S84. [DOI] [PubMed] [Google Scholar]
- Steele AD, Ivanoff B, 2003. Rotavirus strains circulating in Africa during 1996–1999: emergence of G9 strains and P[6] strains. Vaccine 21, 361–367. [DOI] [PubMed] [Google Scholar]
- Steele AD, Kasolo FC, Bos P, Peenze I, Oshitani H, Mpabalwani E, 1998. Characterization of VP6 subgroup, VP7 and VP4 genotype of rotavirus strains in Lusaka, Zambia. Ann. Trop. Paediatr 18, 111–116. [DOI] [PubMed] [Google Scholar]
- The_Laboratory_Working_Group_for_SURVAC, 2011. Subregional Training on Diagnostics for Bacterial Meningitis and Rotavirus Gastroenritis. Global Immunization News, pp. 9–11. [Google Scholar]
- Todd S, Page NA, Duncan Steele A, Peenze I, Cunliffe NA, 2010. Rotavirus strain types circulating in Africa: review of studies published during 1997–2006. J. Infect. Dis 202 (Suppl), S34–S42. [DOI] [PubMed] [Google Scholar]
- WHO, 2006. African Rotavirus Surveillance Network, Draft Standard Operating Procedure. WHO Regional office for Africa 2006. [Google Scholar]
- WHO, 2009. Meeting of the immunization strategic advisory group of experts, April 2009-conclusions and recommendations. Wkly. Epidemiol. Rec 84, pp. 220–236. [PubMed] [Google Scholar]
- WHO, 2012. Estimated rotavirus deaths for children under 5 years of age: 2008, 453000. <http://www.who.int/immunization/monitoring_surveillance/burden/estimates/rotavirus/en/>.


