Abstract
Rubella is a contagious disease caused by the rubella virus (RuV) that can lead to serious birth defects when women are infected in early pregnancy. This study aimed to describe the epidemiology and genetic diversity of rubella viruses in Cote d’Ivoire (CIV). Blood or oral fluid samples collected from suspected measles cases were first tested for the presence of measles specific IgM antibodies by enzyme-linked immunosorbent assay (ELISA). All measles IgM negative or indeterminate samples were tested for rubella IgM antibody using ELISA. Rubella-IgM–positive samples were tested by real-time reverse transcription polymerase chain reaction (RT-PCR) for the presence of rubella virus RNA. Real-time RT-PCR–positive RNA samples were used as template to amplify the 739 nt region used for rubella genotyping. PCR-positive samples were sequenced and phylogenetic analysis performed. Between 2012 and 2016, 4121 serums and 126 oral fluids were collected through the measles surveillance system. Of these, 3823 and 108 respectively were measles IgM negative or indeterminate. Subsequent testing for rubella found that 690 of 3823 (18%) serum samples and 25 of 108 (23%) oral fluid samples were rubella IgM-positive. The 739 nt segment of the E1 glycoprotein gene was amplified and sequenced for two serums and seven oral fluids samples. Phylogenetic analysis showed that the rubella viruses from CIV belonged to genotypes 1G (eight samples) and 2B (one sample). Rubella virus genotype 2B was found in CIV for the first time. These data contribute to baseline information on rubella virus strains found in CIV before the introduction of rubella vaccine.
Keywords: Africa, Cote d’Ivoire, epidemiology, genotyping, rubella infection
1 |. INTRODUCTION
Rubella is caused by rubella virus (RV), the only member of the genus Rubivirus within the Togaviridae family. RV replicates in the nose or throat of infected persons and spreads by direct contact with susceptible hosts through droplet sprays during coughing and sneezing. The infection is vaccine preventable.1 Both vaccine and natural infection are thought to result in life long immunity. If a pregnant woman is infected with rubella virus during the first trimester of pregnancy, the risk of the child developing congenital rubella syndrome (CRS) is up to 90%.2 It is estimated that approximately 100,000 children are born each year with CRS worldwide.2 Prevention of CRS is the main reason for rubella vaccination programs.3
The World Health Organization (WHO) recommends that all countries establish molecular surveillance of RV to track progress towards the goal of eliminating rubella, to help with case classification and to document transmission pathways. The WHO established a systematic RV nomenclature and an RV genome fragment of at least 739 nt within the E1 gene is required for genotype identification.4 To date, the WHO recognizes 12 RV genotypes, 1B, 1C, 1D, 1E, 1F, 1G, 1H, 1I, 1J, 2A, 2B, and 2C, and one provisional genotype, 1a.5 Among them, genotypes 1E and 2B have wide geographic distributions while four others (1D, 1F, 1I, and 2A) are considered inactive and probably extinct because they have not been reported in circulation within the past 10 years.5,6
Although rubella is vaccine preventable and an effective single dose vaccine is available, many developing countries, including Cote d’Ivoire (CIV) have not yet introduced RV vaccine in their routine immunization schedule; thus, many people remain susceptible to rubella infection. Previous studies reporting serological survies of rubella infections by detecting heamagglutination inhibiting antibodies in pregnant women in CIV, showed that antibodies were presents in 59%7 and more than 80% of the participants.8,9 In CIV, rubella surveillance has not been established. The measles case-based surveillance system, established in 2005, provides an opportunity for confirmation of rubella cases by specific immunoglobulin M (IgM) antibody among persons with rash illness who are not positive for measles IgM. As part of this surveillance, samples collected from suspected measles cases are analyzed at the Pasteur Institute of Cote d’Ivoire (IPCI) laboratory in Abidjan and serologic data are shared with the Ministry of Health and WHO on a weekly basis.
The main objective of this study was to describe the epidemiology of rubella infection in CIV and characterize RV strains found in the country from 2012 to 2016.
2 |. MATERIALS AND METHODS
2.1 |. Study setting
CIV is located in West Africa and is bordered by five countries; Mali, Burkina Faso, Guinea, Liberia, and Ghana. The country has an area of 322 462 km2. Its population is estimated at 24 million inhabitants with 39.8% less than 15 years old. Recent studies on the Cote d’Ivoire’s population showed that life expectancy at birth was 50.4 years in 2012, and the total literacy rate of adult population was 43% 10
2.2 |. Specimen collection
Blood samples were collected from all measles suspected cases in the country as part of measles surveillance program using the guidelines from WHO/AFRO region. A suspected measles case was defined as any person in whom a clinician suspected measles or any person with fever and maculopapular rash and coryza or conjunctivitis or cough. Samples were sent to the National Reference Laboratory for Measles and Rubella at IPCI. For four districts of Abidjan (Abobo East, Abobo west, Yopougon East, and Yopougon West) oral fluid samples were systematically collected using Oracol devices (Malvern Medical, Worscester, UK), in addition to blood samples to be used as alternative samples for diagnosis of measles and rubella. Oral fluid, which is collected in a relatively noninvasive manner by rubbing an absorptive device between the gum and cheek, has the advantage that it can be used for both RV-specific antibody detection and RV RNA detection.11
2.3 |. Laboratory analysis
2.3.1 |. Serologic testing
Sera or oral fluid samples were first tested for the presence of measles specific IgM antibodies by enzyme-linked immunosorbent assay (ELISA) using the Enzygnost anti-measles virus IgM kit, (Siemens, Marburg, Germany) and microimmune measles IgM capture ELISA kit (Microimmune, Middx, UK), according to the manufacturer’s instructions. All samples that were determined to be measles IgM negative or indeterminate were tested for rubella IgM antibody using the Enzygnost anti-rubella virus IgM kit (Siemens) or Microimmune rubella IgM Capture Kits (Microimmune, Middx, UK). Patients with serum and/or oral fluid rubella IgM-positive samples were classified as laboratory confirmed rubella cases.
2.3.2 |. Molecular testing
RNA extraction
Viral RNA was extracted from serum or oral fluid samples with the automated QIAcube standard system (Qiagen, Hilden, Germany) using QiaAmp Viral RNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
2.4 |. Real-time RT-PCR amplification
The presence of RV RNA was detected by real-time reverse transcription polymerase chain reaction (RT-PCR) using the Super-Script III Platinum One-Step Quantitative RT-PCR System (Invitrogen, Carlsbad, CA) with rubella virus primers (RV11, RV12, and RV12-2) as previously described12 to amplify a 185 nucleotide region in the rubella virus structural protein E1 coding region. The thermal cycling was carried out with an ABI 7500 thermal cycler (Applied Biosystems, Foster City, CA) with 48°C for 30 minutes, 95°C for 5 minutes, and 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Only Rubella IgM-positive samples collected within 4 days of disease onset, were tested by real-time RT-PCR as recommended by Abernathy et al.11
2.5 |. Genotyping RT-PCR
All RNA samples that were positive by real-time RT-PCR were used as templates to amplify two over-lapping fragments whose sequences in combination contain the 739 nt region recommended by WHO for rubella genotyping. For oral fluids samples, genotyping reactions were performed using the Qiagen One-Step RT-PCR Kit (Qiagen, Germantown, MD) as previously described by Namuwulya et al.12 For serum samples, genotyping was performed using nested RT-PCR assays as previously described by Pukuta et al.13 PCR products were visualized using the FlashGel System from Lonza (Lonza Rockland, New York, NY).
2.6 |. Sequencing
DNA products were purified using the Charge Switch-Pro PCR Cleanup Kit centrifugation protocol (Invitrogen). Sequences were determined bidirectionally using the Sanger dideoxy terminator sequencing method with a BigDye Terminator Version 3.1 Cycle Sequencing Kit (Applied Biosystems) and the 3500XL Genetic analyzer (ABI, Life Technology; Hitachi, Tokyo, Japan).
2.7 |. Sequences alignments and phylogenetic analysis
Sequence alignments and phylogenetic analysis were performed using MEGA version 6.011,14 Sequences were edited and assembled to obtain the 739 nt sequences required for rubella virus genotyping. Sequences from this study were aligned and analyzed together with 32 rubella virus reference strains recommended by WHO. RV sequences previously found in CIV in 2008,15 as well as sequences found in other African countries such as Uganda,9,12 Democratic Republic of the Congo (DRC),13 and Nigeria (exported to the United States),16 were also added to this analysis. The accuracy of the groupings was assessed using the bootstrap method with 1000 replicates. Phylogenetic trees were constructed using the neighbor-joining Kimura two-parameter distance method. Bootstrap values >80% were considered statistically significant for grouping. Sequence data were submitted to the rubella virus nucleotide sequence database (RubeNS) and to GenBank. Accession number is indicated in the phylogenetic tree.
2.8 |. Data management and epidemiological analysis
Data from serodiagnosis of rubella, between January 1, 2012 and December 31, 2016, were analyzed using Epi-info version 3.5 (US Center for Disease Control, Atlanta, Georgia, USA). The Epi-Info database included information on the type of sample, its date of collected and sociodemographic data for each patient such as age, sex, history of vaccination against rubella, date of onset of clinical signs, and dates of sampling. Descriptive analysis was performed for all the studied variables.
2.9 |. Ethical considerations
The CIV measles/Rubella surveillance is a national program approved by the Ministry of Health and supported by WHO/AFRO as part of the global goal to control and eliminate measles and rubella. Patient information and specimen collection respected the human subjects procedures stipulated in the WHO/AFRO measles/rubella surveillance protocol.
3 |. RESULTS
3.1 |. Epidemiology of rubella infection during 2012–2016
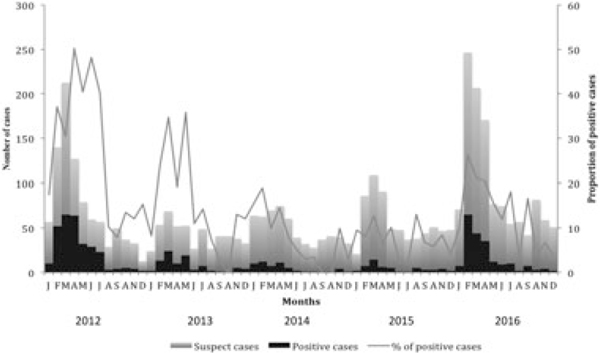
Between 2012 and 2016, 4121 sera and 126 oral fluids collected through the measles surveillance system were received from the 20 health regions in the country with 40% coming (1557 of 3823) from the capital Abidjan and its surroundings (Table 1 and Figure 2). Samples were first tested for the presence of measles IgM. Of the 4121 sera, 15 could not be tested for reasons such as inadequate sample volumes or lack of proper identification. Of 4106 sera tested, 3823 were measles IgM negative or indeterminate and were tested for the detection of rubella-specific IgM. Of these 3823 serum samples, 690 (18%) were rubella IgM-positive (Table 1) and were classified as rubella cases, 3006 were negative and 127 had indeterminate results. Of the 126 oral fluids samples, 108 were measles IgM negative; subsequent testing for rubella-specific IgM identified 14% (25 of 108) positive samples and these patients were classified as rubella cases. Of these 25 cases, 13 were rubella IgM-positive for both oral fluid and serum samples, 4 were rubella IgM-positive for oral fluid while their corresponding serum was negative. Conversely, eight sera were rubella IgM-positive while their corresponding oral fluid sample was negative. The percentage of rubella IgM-positive varied from 33% in 2012 to 8% in 2015 (Table 1). Yearly distribution showed that the highest number of rubella cases was found in 2012 (293 of 690) followed by 2016 (200 of 690) and 2013 (90 of 690) (Table 1). During the study period, rubella infections occurred year round but were more common from January through July with a peak from February to April (Figure 1).
TABLE 1.
Distribution of rubella cases by year and by health region in Cote d'Ivoire, 2012–2016
| Variables | Category | Total cases tested for rubella IgM | Rubella IgM test results |
Percentage of rubella IgM-positive, %a | ||
|---|---|---|---|---|---|---|
| Positive | Negative | Indeterminate | ||||
| Year | 2012 | 895 | 293 | 554 | 48 | 33 |
| 2013 | 508 | 90 | 408 | 10 | 18 | |
| 2014 | 583 | 54 | 513 | 16 | 9 | |
| 2015 | 635 | 53 | 574 | 8 | 8 | |
| 2016 | 1202 | 200 | 957 | 45 | 17 | |
| Total | 3823 | 690 | 3006 | 127 | 18 | |
| Health regions | Abidjan 1-Grands Ponts | 324 | 69 | 243 | 12 | 21 |
| Abidjan 2 | 1233 | 196 | 1005 | 32 | 16 | |
| Agneby-Tiassa-Me | 101 | 27 | 68 | 6 | 27 | |
| Belier | 109 | 13 | 92 | 4 | 12 | |
| Bounkani-Gontougo | 109 | 33 | 74 | 2 | 30 | |
| Cavally-Guemon | 273 | 44 | 217 | 12 | 16 | |
| Gbeke | 144 | 29 | 111 | 4 | 20 | |
| Gbokle-Nawa-San Pedro | 139 | 22 | 111 | 6 | 16 | |
| Goh | 86 | 24 | 59 | 3 | 28 | |
| Hambol | 55 | 23 | 31 | 1 | 42 | |
| Haut Sassandra | 84 | 20 | 60 | 4 | 24 | |
| Indenie-Djuablin | 159 | 32 | 118 | 9 | 20 | |
| Kabadougou-Bafing-Folon | 108 | 9 | 97 | 2 | 8 | |
| Loh-djiboua | 95 | 16 | 74 | 5 | 17 | |
| Marahoue | 67 | 13 | 50 | 4 | 19 | |
| Nzi-iffou | 168 | 25 | 134 | 9 | 15 | |
| Poro-Tchologo-Bagoue | 187 | 31 | 152 | 4 | 17 | |
| Sud Comoe | 197 | 35 | 158 | 4 | 18 | |
| Tonkpi | 160 | 19 | 137 | 4 | 12 | |
| Worodougou-Bere | 25 | 10 | 15 | 0 | 40 | |
Abbreviation: IgM, immunoglobulin M.
Percentages of rubella-positive specimens may have been influenced by simultaneous measles activity, since specimens were obtained from measles surveillance.
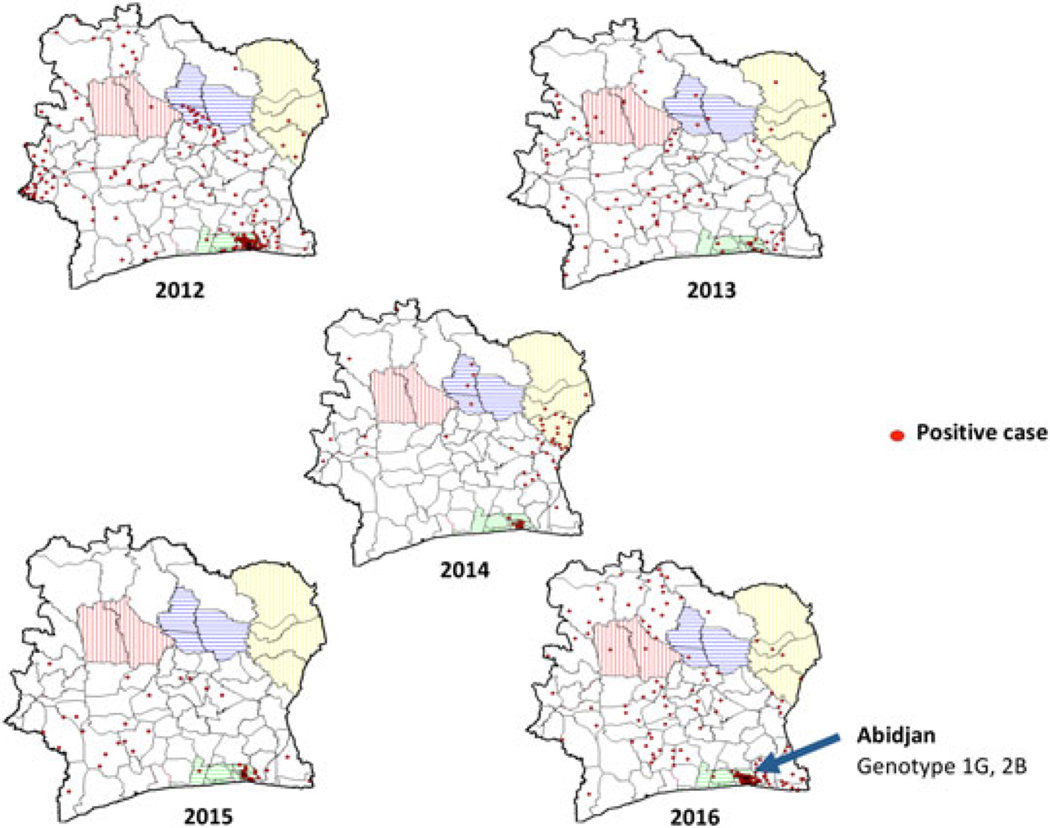
FIGURE 2.
Geographic distribution of rubella-positive cases and rubella genotypes in CIV (2012–2016). Health regions with highest percentage of rubella cases are indicated: Blue, Hambol; Red, Worodougou-Bere; Yellow, Bounkani-Gountougo. Genotypes were identified in the Abidjan (green) health region
FIGURE 1.
Distribution of rubella cases in Cote d’Ivoire by month (2012–2016). Suspected cases are indicated with gray color histogram, positives cases are in black and the proportion of positive cases is indicated with a grey line
3.2 |. Geographic distribution of rubella cases
Rubella cases were geographically distributed throughout the country with the highest-positive rate found in Hambol health region with 42%, followed by Worodougou-Bere with 40% and Bounkani-Gountougo with 30% (Table 1). The highest number of cases was found in Abidjan 2 health region, which accounted for 28% (196 of 690) of total cases followed by Abidjan 1 with 10% (69 of 690) and Cavally-Guemon (6%) (Table 1 and Figure 2). Each year there was a cluster of rubella cases in the city of Abidjan (Figure 2).
3.3 |. Sex and age distribution of rubella cases
Rubella cases were slightly higher in males with 53% (367 of 690) compared with 46% (316 of 690) in females (Table 2). The age distribution of rubella cases showed that the most affected age group was 5 to 9 years (39%), followed by 1 to 4 years (23%), and 10 to 14 years (21%) (Table 2). Among rubella-infected patients, 83% were between 1 to 14 years old. Only 2% of cases were observed in infants less than 1-year old, information on age was unknown for 8% of cases (Table 2). Seven percent (46 of 690) of rubella cases were in >15-year old of which 28 of 690 (4%) were female.
TABLE 2.
Sex and age distribution of rubella cases in Cote d’Ivoire 2012–2016
| Variables | Category | Total cases tested for rubella IgM | Rubella IgM test results |
||
|---|---|---|---|---|---|
| Positive, % | Negative | Indeterminate | |||
| Sex | Male | 1995 | 367 (53) | 1565 | 63 |
| Female | 1802 | 316 (46) | 1423 | 63 | |
| Unknown | 26 | 7(1) | 18 | 1 | |
| Total | 3823 | 690 | 3006 | 127 | |
| Age group, year | < 1 | 360 | 17 (2) | 342 | 1 |
| 1–4 | 1385 | 158 (23) | 1202 | 25 | |
| 5–9 | 1028 | 272 (39) | 706 | 50 | |
| 10–14 | 483 | 144 (21) | 308 | 31 | |
| ≥ 15 | 335 | 46 (7) | 279 | 10 | |
| Unknown | 232 | 53 (8) | 169 | 10 | |
| Total | 3823 | 690 | 3006 | 127 | |
IgM, immunoglobulin M.
3.4 |. Rubella virus genotype phylogenetic analysis
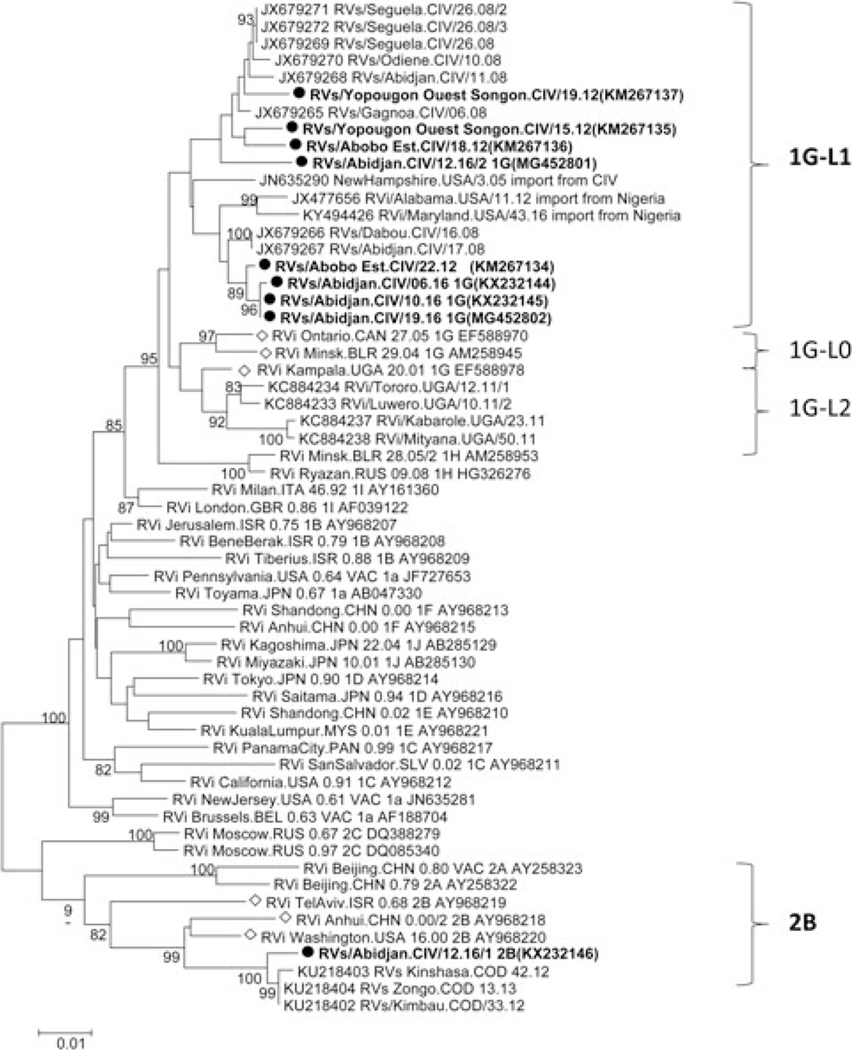
To identify the genotype of rubella strains, all 25 oral fluids and 27 serum samples that were positive for rubella-specific IgM were first analyzed by real-time RT-PCR to detect RNA from RV. Among these samples, 23 oral fluids were collected within 4 days of disease onset of which 11 were positive by real-time RT-PCR and 19 serum samples were collected within 3 days of disease onset of which two were positive by real-time PCR. The 739 nt segment of the E1 glycoprotein-coding region used for genotyping was successfully amplified and sequenced for seven oral fluids and two serum samples. PCR-positive samples were collected in 2012 and 2016; there were no PCR-positive samples available from 2013 to 2015. Phylogenetic analysis showed that eight of the rubella viruses from CIV belonged to lineage 1 of genotype 1G (1G-L1) and that one sample belonged to genotype 2B (Figure 3).16 Genotype 2B was identified from an oral fluid sample and its corresponding serum was also positive by real-time PCR. The eight genotype 1G sequences had an overall mean distance at the nucleotide level of 2.3% within the group, with a range of 0% to 3.5%. They varied from two WHO reference strains for 1G-L0 (Ontario.CAN 27.05 and Minsk.BLR 29.04) by 4% and from the third genotype 1G-reference strain for IG-L2 (Kampala.UGA/20.01) by 3.6%. The eight sequences differed from other 1G-L1 sequences by a mean of 2.4%. The genotype 2B sequence differed from the closest genotype 2B reference strain (RVi Washington. USA 16.00-AY968220) by 3.1%, but varied by only 0.5% compared with genotype 2B viruses found in the DRC in 2012 and 2013. The virus belongs to the L2 lineage of 2B viruses.16 All of the 2012 and 2016 CIV viruses sequences were obtained from samples collected in the Abidjan health region.
FIGURE 3.
Phylogenetic tree of rubella of rubella viruses found in Cote d’Ivoire during 2012–2016. Sequences obtained in this study are indicated in in bold with a black dot. Bootstraps values >80% are indicated. The WHO reference sequences for genotype 1G and 2B are marked with a diamond shape and GenBank accession numbers shown for reference sequences
4 |. DISCUSSION
This study reports the epidemiology of RV infections in CIV. The positivity rate of rubella infection in this study was 18%, which is slightly lower than the 24% prevalence of rubella previously reported in other African countries such as the DRC,13 Uganda,12 and in the African region as a whole.17 Epidemiologic data showed that an increase in the number of rubella cases was found in 2012 and again in 2016. The age distribution of rubella cases indicated that 4% of rubella cases occurred in women of reproductive age (>15 years old); therefore, the risk of CRS births in CIV is expected to be similar to the 5% previously reported in the African region.17,18 The seasonality of rubella cases is consistent with the pattern previously reported in the West African region with a sharp increase in rubella cases seen starting in January and peaks in March to April.17
Rubella virus RNA was amplified and sequence for nine samples, of which seven oral fluids collected within 4 days and two serum collected within 3 days of disease onset. This is in agreement with the findings described by Abernathy et al,11 and emphasizes that the timing of sample collection is crucial for the genotyping of rubella. Phylogenetic analyses indicate that genotypes 1G (eight viruses) and 2B (one virus) were found during the study time period. Genotype 1G can currently be divided into three lineages based on genetic diversity and geographic location16 Lineage 1G-L0 viruses are mainly from Europe, lineage 1G-L1 viruses are found in west Africa, and lineage 1G-L2 virus are found in east Africa. The 1G strains found in this study, clustered with other rubella viruses from West Africa, JN635290-NewHamshire.USA/3.05, and JX477656-RVi/Alabama. USA/11.12, and KY494426-RVi/Maryland.USA/43.16 (imported into the United States from CIV and Nigeria, respectively), as well as with other viruses found in CIV in 2008. Rubella genotype 1G strains were reported in other African countries including Uganda,12 Sudan,19 and Democratic Republic of the Congo (DRC).13 During this study, a genotype 2B virus was found for the first time in CIV. Genotype 2B viruses are known to have a wide geographical distribution.6,16 In sub-Saharan Africa, genotype 2B has been found in several countries, including Sudan in 200619 and in DRC 2012.13 The genotype 2B virus from CIV was closely related to the 2B viruses from the 2012 to 2013 DRC outbreak, varying by only 6 of 739 nucleotides.
The main limitation of this study is that samples were collected as part of measles case-based surveillance; and therefore, the number of rubella cases detected may have been influenced by the number of measles cases occurring simultaneously. As consequence, the number of rubella cases in this study may not represent the real number of rubella infections in CIV. Only a few genotypes could be identified compared with the number of rubella-positive cases. This is due to the fact that the virus load in rubella samples is very low compared with other viruses such as measles virus or poliovirus. As a result, it is very difficult to amplify rubella virus by PCR. All rubella virus genotypes were found in Abidjan region and therefore may not reflect the full genotype diversity in the country. The reference laboratory where samples were analyzed is located in Abidjan. Samples from other regions may have suffered from inappropriate storage/transportation condition and/or freeze/thaw cycles that may have affected the viral RNA integrity and contribute to negative RT PCR results. Some serum samples were IgM-positive while their corresponding oral fluids were negative and vice versa. This may be due to the timing of sample collection. A study by Abernathy et al,11 suggested that for sample collected early during the course of infection, within 4 days after rash onset, the detection rate of IgM antibody in oral fluid sample may be lower than the rate of IgM antibody detection in serum. In regard to samples with serum IgM-positive and oral fluid IgM negative, it is not clear what this is due to; perhaps it is due to limited transcription errors in specimen identification. The second limitation is that there are gaps in the surveillance for vaccine preventable diseases in CIV, especially in molecular epidemiology for both the country and the entire West African region, and this has limited our ability to identify the source of the rubella viruses described here. However, based on previous data, it is likely that viruses of genotype 1G-L1 are endemic in CIV and have been circulating there for at least a decade. The single detection of a 2B virus, which was not previously found in this country, suggests that this might have been an importation. Continued collection of samples for genotyping from suspected rubella cases will be necessary to confirm whether this was a sporadic case of 2B or whether the new genotype is circulating in CIV.
This study investigated the epidemiology of rubella infection and contributed to the establishment of baseline data of RV genotypes in CIV. These data are of critical importance as they will contribute to WHO control and elimination plans for rubella and CRS surveillance in Africa and globally. It will be important to continue to monitor changes in rubella virus genotypes in CIV as this will help to measure the impact of rubella vaccine once introduced the in country. Strengthening laboratory and epidemiologic surveillance of rubella in CIV will be vital for rubella control and elimination in CIV.
ACKNOWLEDGMENTS
We are grateful to personnel of hospital and Public Health Department for sending specimens of suspected measles cases. We are grateful to WHO Cote d’Ivoire office for logistical help in the country. We would like to thank Raydel Anderson, CDC Atlanta and the Pasteur Institute of Cote d’Ivoire personnel; Ibrahima Koné, Brou Martial, Tiemoko Gislain, Kra Eric, Yavo Albert, Kouamé Hilaire for technical assistance. We are grateful to Sie Kabran from WHO office in Abidjan for his help on data management. We want to thank Paul Rota, CDC Atlanta, for guidance leading to the completion of this study. Funding for this study was provided by the Ministry of Health of Cote D’Ivoire, the World Health Organization and the US Centers for Diseases Control and Prevention.
Funding information
U.S. Centers for Diseases Control and Prevention; Ministry of Health of Cote D’Ivoire; World Health Organization AFRO (WHO-AFRO)
Footnotes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Rubella WHO. 2017. http://www.who.int/en/news-room/fact-sheets/detail/rubella. Accessed 2017.
- 2.Yazigi A, De Pecoulas AE, Vauloup-Fellous C, Grangeot-Keros L, Ayoubi JM, Picone O. Fetal and neonatal abnormalities due to congenital rubella syndrome: a review of literature. J Matern Fetal Neonatal Med. 2017;30(3):274–278. [DOI] [PubMed] [Google Scholar]
- 3.Reef SE, Redd SB, Abernathy E, Zimmerman L, Icenogle JP. The epidemiological profile of rubella and congenital rubella syndrome in the United States, 1998–2004: the evidence for absence of endemic transmission. Clin Infect Dis. 2006;43(Suppl 3):S126–S132. [DOI] [PubMed] [Google Scholar]
- 4.WHO. Standardization of the nomenclature for genetic characteristics of wild-type rubella viruses. Wkly Epidemiol Rec. 2005;80 (14):126–132. [PubMed] [Google Scholar]
- 5.WHO. Rubella virus nomenclature update: 2013. Wkly Epidemiol Rec. 2013;88(32):337–343. [PubMed] [Google Scholar]
- 6.Abernathy ES, Hübschen JM, Muller CP, et al. Status of globalvirologic surveillance for rubella viruses. J Infect Dis. 2011;204(Suppl 1):S524–S532. [DOI] [PubMed] [Google Scholar]
- 7.Ouattara SA, Brettes JP, Kodjo R, et al. Seroepidemiology of rubellain the Ivory Coast. Geographic distribution. Bull Soc Pathol Exot Filiales. 1987;80(4):655–664. [PubMed] [Google Scholar]
- 8.Vrinat M, Dutertre J, Helies H, Ropero P. A serological survey of rubella among pregnant women in Abidjan (author’s transl). Med Trop (Mars). 1978;38(1):53–57. [PubMed] [Google Scholar]
- 9.Faye-Kette YH, Sylla-Koko DJ, Akoua-Koffi GC, et al. Seroprevalence of rubella in 461 pregnant women in Abidjan (Cote d’Ivoire). Bull Soc Pathol Exot. 1993;86(3):185–187. [PubMed] [Google Scholar]
- 10.Countrymeters. Côte d’Ivoire Population. 2017; http://countrymeters.info/en/Cote_d‘Ivoire#population_2017. Accessed 16/08/2017.
- 11.Abernathy E, Cabezas C, Sun H, et al. Confirmation of rubella within 4 days of rash onset: comparison of rubella virus RNA detection in oral fluid with immunoglobulin M detection in serum or oral fluid. J Clin Microbiol. 2009;47(1):182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Namuwulya P, Abernathy E, Bukenya H, et al. Phylogenetic analysisof rubella viruses identified in Uganda, 2003–2012. J Med Virol. 2014;86(12):2107–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pukuta E, Waku-Kouomou D, Abernathy E, et al. Genotypes of rubella virus and the epidemiology of rubella infections in the Democratic Republic of the Congo, 2004–2013. J Med Virol. 2016;88(10):1677–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Q, Abernathy ES, Sun H, et al. Genotyping of rubella virusRNA in sera and dried blood spots collected during routine surveillance and in archival sera. J Virol Methods. 2013;187(2): 284–287. [DOI] [PubMed] [Google Scholar]
- 16.Rivailler P, Abernathy E, Icenogle J. Genetic diversity of currently circulating rubella viruses: a need to define more precise viral groups. J Gen Virol. 2017;98(3):396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodson JL, Masresha B, Dosseh A, et al. Rubella epidemiology in Africain the prevaccine era, 2002–2009. J Infect Dis. 2011;204(Suppl 1): S215–S225. [DOI] [PubMed] [Google Scholar]
- 18.Alleman MM, Wannemuehler KA, Hao L, et al. Estimating the burdenof rubella virus infection and congenital rubella syndrome through a rubella immunity assessment among pregnant women in the Democratic Republic of the Congo: Potential impact on vaccination policy. Vaccine. 2016;34(51):6502–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adam O, El hussein A, El eragi A, Jin L. Primary investigation of 31 infants with suspected congenital rubella syndrome in Sudan. Clin Microbiol Infect. 2010;16(6):678–682. [DOI] [PubMed] [Google Scholar]