Significance
The steroid hormone progesterone is highly involved in different physiological–pathophysiological processes, including bone formation and cancer progression. Understanding the working mechanisms, especially identifying the receptors of progesterone hormones, is of great value. In the present study, we identified GPR126 as a membrane receptor for both progesterone and 17-hydroxyprogesterone and triggered its downstream G protein signaling. We further characterized the residues of GPR126 that interact with these two ligands and found that progesterone promoted the progression of a triple-negative breast cancer model through GPR126-dependent Gi-SRC signaling. Therefore, developing antagonists targeting GPR126-Gi may provide an alternative therapeutic option for patients with triple-negative breast cancer.
Keywords: progesterone, 17OHP, GPR126, Gi, TNBC
Abstract
GPR126 is a member of the adhesion G protein-coupled receptors (aGPCRs) that is essential for the normal development of diverse tissues, and its mutations are implicated in various pathological processes. Here, through screening 34 steroid hormones and their derivatives for cAMP production, we found that progesterone (P4) and 17-hydroxyprogesterone (17OHP) could specifically activate GPR126 and trigger its downstream Gi signaling by binding to the ligand pocket in the seven-transmembrane domain of the C-terminal fragment of GPR126. A detailed mutagenesis screening according to a computational simulated structure model indicated that K1001ECL2 and F1012ECL2 are key residues that specifically recognize 17OHP but not progesterone. Finally, functional analysis revealed that progesterone-triggered GPR126 activation promoted cell growth in vitro and tumorigenesis in vivo, which involved Gi-SRC pathways in a triple-negative breast cancer model. Collectively, our work identified a membrane receptor for progesterone/17OHP and delineated the mechanisms by which GPR126 participated in potential tumor progression in triple-negative breast cancer, which will enrich our understanding of the functions and working mechanisms of both the aGPCR member GPR126 and the steroid hormone progesterone.
GPR126, also called adhesion G protein-coupled receptor subfamily G, member 6 (ADGRG6), is a seven-transmembrane (7TM) receptor with important functions in tissue development and may be involved in cancer progression (1–14). Genome-wide association studies have revealed that GPR126 polymorphisms or mutations are closely associated with shortened height and a variety of diseases (1–3, 15–18). For example, a single nucleotide polymorphism (SNP, rs6570507) of GPR126 was reported to be associated with trunk length in European populations (1). GPR126 polymorphisms rs3817928, rs7776375, rs6937121, and rs11155242 were associated with normal pulmonary function (2, 15). The variants rs41289839, rs6570507, rs7774095, and rs7755109 of GPR126 were linked to adolescent idiopathic scoliosis (AIS), whereas a spontaneous mutation (p. Val769Glu [c.2306T > A]) of GPR126 was associated with arthrogryposis multiplex congenita in humans (3, 16–18). The important roles of GPR126 in tissue development were further supported by studies using animal models. In mice, genetically deleting Gpr126 led to embryonic lethality due to cardiovascular development failure, implying the importance of GPR126 for viability (4). The specific deletion of Gpr126 in osteoblasts resulted in delayed embryonic bone formation and development and consequently decreased body length (5). Targeted disruption of Gpr126 in chondrocytes resulted in apoptosis in the axial cartilage, followed by characteristics of AIS and pectus excavatum (6). Moreover, as in zebrafish, Gpr126 knockout mice exhibited insufficient axon myelination and multiple resulting ultrastructural abnormalities in the peripheral nervous system (7–9, 19). In addition to these pivotal roles of Gpr126 in the development of multiple tissues in mammals, emerging evidence reveals a close correlation between GPR126 and tumors (10–13, 20–22). GPR126 mutation and copy number variation are associated with tumor aggressiveness and reduced patient survival in bladder cancer and breast cancer (BC) (10–12, 21, 22). Additionally, GPR126 is reported to be overexpressed in colorectal cancer, and its knockdown significantly inhibits the cell growth of colorectal cancer cells (13). Despite these newly uncovered roles, the precise function and downstream signaling pathways of GPR126 in tumors are not fully established.
Structurally, GPR126 belongs to the adhesion G protein-coupled receptors (aGPCRs), most of whose members contain exceptionally long extracellular regions encompassing the GPCR autoproteolysis-inducing (GAIN) domain between the transmembrane region and the extracellular segment (23, 24). Similar to many other aGPCRs, GPR126 can be cleaved at a GPCR proteolytic site (GPS) in the GAIN domain to produce the N-terminal fragment (NTF) and C-terminal fragment (CTF) (14). In addition to the GAIN domain, the NTF of GPR126 contains a Complement, Uegf, Bmp1 (CUB) domain, a pentraxin (PTX) domain, a hormone binding domain, and 27 putative N-glycosylation sites. Even after cleavage, the NTF and CTF of aGPCRs are still associated with each other in many cellular conditions (23–26). Whereas the NTF of GPR126 is known to form docking sites for extracellular matrix proteins, the CTF of GPR126 forms a 7TM bundle, and its cellular part can engage with G proteins to transduce extracellular stimuli to intracellular signaling (27–29). Recently, laminin-211, type IV collagen, and the prion protein (PrPc) were identified as extracellular endogenous ligands for GPR126 and triggered cAMP signaling to induce biological effects in Schwann cells through Gs coupling (30–32). Biochemical characterization suggested that both laminin-211 and type IV collagen bind to GPR126 through the NTF. However, whether other endogenous ligands directly interact with the CTF of GPR126 and regulate its function is unclear.
Recently, we identified hydrocortisone, a steroid hormone or glucocorticoid, as the endogenous ligand of GPR97 (also called ADGRG3, another aGPCR) (33). As the residues contacting glucocorticoids in the ligand binding pocket of GPR97 are quite conserved among the aGPCR-G subfamily, we were interested in whether certain steroid hormones similar to hydrocortisone may act as ligands of GPR126 and trigger its downstream signaling. In the present study, through screening 34 steroid hormones or their derivatives for cAMP production, we found that progesterone and 17-hydroxyprogesterone (17OHP) could specifically activate GPR126 and trigger its downstream Gi signaling. Furthermore, we identified key interaction residues and pivotal conformational changes of GPR126 in response to these two steroid hormones by computer modeling, alanine scanning, and fluorescent arsenical hairpin bioluminescence resonance energy transfer (FlAsH-BRET) methods. Finally, we explored the biological consequences of GPR126 activation in response to progesterone in triple-negative breast cancer (TNBC) cells and found that progesterone-triggered GPR126 activation promoted cell growth in vitro and tumorigenesis in vivo at least partially through Gi-tyrosine-protein kinase SRC (SRC) pathways.
Results
Screening Steroid Hormone Ligands of GPR126.
Our recent studies found that GPR97 (also called ADGRG3) was able to recognize glucocorticoids, one type of steroid hormone, via its ligand pocket localized in its 7TM bundle and transduce signals via Go coupling (33). Importantly, the residues in contact with glucocorticoids in the ligand binding pocket of GPR97 are quite conserved among the aGPCR-G subfamily (33). In particular, among 13 residues of GPR97 that directly interact with cortisol, 10 residues were homologous with the other aGPCR-G subfamily member GPR126 (also called ADGRG6) (SI Appendix, Fig. S1). The glucocorticoids shared four ring steroid cores with other steroid hormones. We therefore suspected that certain steroid hormones may be able to bind to GPR126 and induce its downstream signaling.
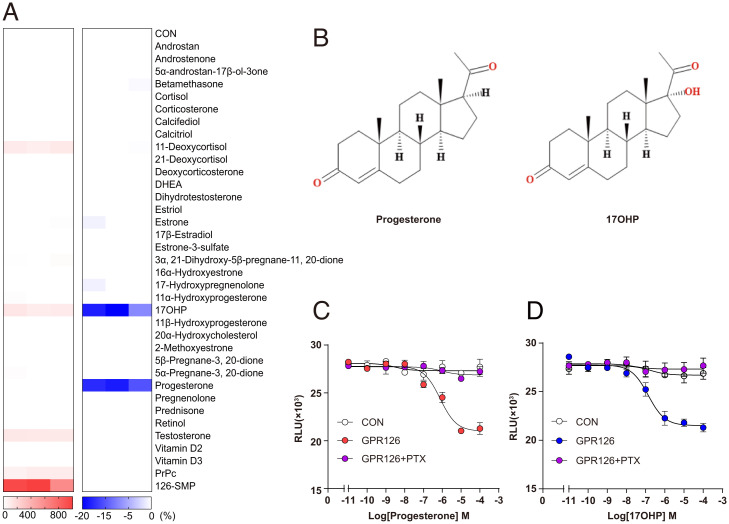
GPR126 was reported to primarily couple to Gs in response to agonist binding (30–32). We therefore screened 34 steroid hormones and their derivatives for Gs activation using GloSensor-cAMP assays. Despite that positive control GPR126-Stachel-mimicking peptide (126-SMP) was able to stimulate approximately ninefold of intracellular cAMP increase more than control vehicle in GPR126-overexpressing HEK293 cells, no steroid hormone showed an effect by increasing intracellular cAMP levels more than twofold. Notably, 17OHP, 11-deoxycortisol, and testosterone induced weak but detectable increase on intracellular cAMP accumulation. (Fig. 1A and SI Appendix, Fig. S2 A–F) (29). The EC50 of 17OHP, 11-deoxycortisol, and testosterone are 2.7 ± 0.5 μM, 1.4 ± 0.14 μM, and 0.8 ± 0.06 μM, respectively. The Emax of these three steroid hormones and a previously reported PrPc are less than one-sixth compared with 126-SMP, and thus are weak partial agonists of GPR126 for eliciting cAMP accumulation.
Fig. 1.
Progesterone and 17OHP were identified as novel ligands of GPR126 through steroid hormones screening. (A) Heatmap representing the cAMP accumulation (Left) or inhibition ratio (Right) induced by steroid hormones (100 μM) in GPR126-overexpressing HEK293 cells using GloSensor assay. The red color scale indicates the steroid ligands-cAMP accumulation efficacy level (Emax value ratio) via GPR126 with Stachel-mimicking peptide (126-SMP) as positive control and the blue color indicates steroid ligands-cAMP inhibition efficacy level (Emax value ratio) with 5 μM Fsk-stimulated via GPR126. The values are generated according to the data shown in SI Appendix, Fig. S2 A–E and G–L. (B) Chemical structures of the identified GPR126 ligands: progesterone and 17OHP. (D and E) Representative dose–response curves that progesterone (C) and 17OHP (D) induced cAMP inhibition in control and GPR126-overexpressing HEK293 cells treated with DMSO or Gi inhibitor PTX (100 ng/mL) using a GloSensor assay.
In addition to Gs activity, previous reports indicated that GPR126 could stimulate Gi activity (7). We therefore stimulated HEK293 cells overexpressing GPR126 with Forskolin (Fsk) and determined whether the application of steroid hormones could dampen the cellular cAMP level by stimulating Gi activity. Notably, progesterone and 17OHP, but no other steroid hormones and the reported ligands (Stachel-mimicking peptide, collagen IV, PrPc) were able to inhibit Fsk-induced cAMP accumulation in HEK293 cells overexpressing GPR126 but not in HEK293 cells transfected with control vectors (Fig. 1 A and B and SI Appendix, Fig. S2 G–M). The EC50 values for full-length GPR126 activated by progesterone or 17OHP were 539.6 ± 3.1 or 95 ± 1.8 nM, respectively (SI Appendix, Fig. S2M). In addition, these signals were blocked by PTX, a selective Gi/o subtype inhibitor (Fig. 1 C and D and SI Appendix, Fig. S3 A and B). To investigate whether progesterone and 17OHP were able to activate GPR126 in native cells, we measured cAMP levels in GPR126 stable knocking down MDA-MB-231 cells or its control cells after administration of different concentrations of progesterone or 17OHP and Fsk (5 μM) for 10 min. The intracellular cAMP levels were detected using a cAMP enzyme linked immunosorbent assay (ELISA) kit. The results showed that Fsk induced cAMP up-regulation by approximately three times compared to cells treated with control vehicle. Both progesterone and 17OHP inhibited cAMP levels induced by Fsk in GPR126-expressing cells in a concentration-dependent manner. In the condition of GPR126 knocking down, progesterone and 17OHP showed no significant effects on cAMP levels, indicating that progesterone and 17OHP could inhibit cAMP levels via GPR126 (SI Appendix, Fig. S3 C and D).
Previous studies also suggested that several membrane-associated receptors, such as membrane bound progesterone receptors (also called progestin and adipoQ receptors, PAQRs), PGRMC1 (progesterone receptor membrane component 1), and GABA-A, have been involved in the rapid, cell surface-initiated progesterone actions (34). Among these membrane receptors, GABA-A is an ion channel and PGRMC1 is a single transmembrane protein, which have not been reported to couple to G proteins (35, 36). Despite distinct trans-7TM helical topology, the membrane progesterone receptors (PAQR5/6/7/8/9) have been reported to activate G protein signaling in response to progesterone stimulation (37–41). PAQR 5/7/8 were reported to couple to Gi, whereas PAQR 6/9 coupled to Gs (42). We therefore compared the potency and efficacy of progesterone in activation of GPR126 and PAQRs. The progesterone activated PAQR7 and PAQR8 ∼100-fold stronger than GPR126, but with significant lower efficacy once these membrane receptors have similar plasma membrane expression levels (SI Appendix, Fig. S3 E and F). Considering that progesterone concentration varied across a great range, especially during pregnancy and before a baby is newly born, these data indicated that GPR126 and PAQR 5/7/8 may mediate progesterone responses in different pathophysiological contexts.
Nascent aGPCRs in general underwent autocleavage at their GPS, which produced the NTF (α-subunit) and a CTF (β-subunit), harboring the 7TM bundle (23, 24, 26, 43, 44). The Stachel sequence that lies in the N terminus of aGPCRs, such as that in GPR126, is known to mediate the self-activation of these receptors and may interfere with steroid hormone binding. We therefore generated constructs of GPR126-β and GPR126-β-ΔGPS (whose Stachel sequence was deleted) and examined their activation in response to progesterone or 17OHP stimulation. Importantly, the EC50 for GPR126-β activated by progesterone or 17OHP was ∼10-fold lower than that of full-length GPR126 in cAMP inhibition assays, suggesting that the α-subunit of GPR126 may block steroid-induced GPR126 activation (SI Appendix, Fig. S4). Moreover, further deletion of the Stachel sequence by GPR126-β-ΔGPS increased the EC50 of 17OHP toward cAMP inhibition by approximately twofold, suggesting a positive regulatory role of the Stachel sequence for GPR126 activated by 17OHP (SI Appendix, Fig. S4). We next examined whether GPR126 could couple to Gq or induce β-arrestin-2 recruitment upon progesterone or 17OHP administration. The results showed no obvious Gq activity or β-arrestin-2 recruitment of GPR126 in response to progesterone or 17OHP stimulation, using angiotensin II and AT1R as controls (SI Appendix, Fig. S3 G and H) (45, 46). Collectively, these results indicated that progesterone selectively induces the Gi coupling of GPR126 but is not detectable with Gs, Gq, or β-arrestin-2, whereas 17OHP induces both Gs and Gi coupling of GPR126 but not Gq or β-arrestin-2 coupling.
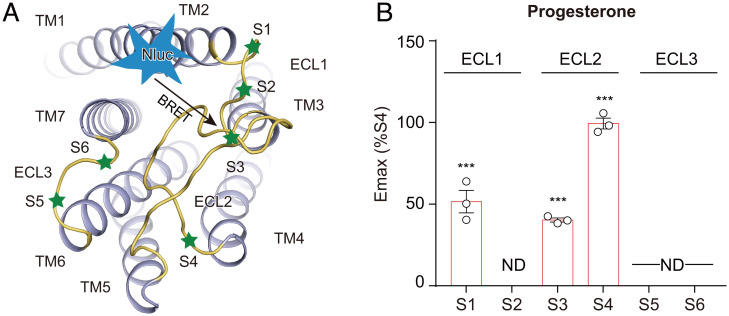
Conformational Changes within GPR126 Extracellular Domains as Ligand Binding.
Our recent study showed that the binding of steroid hormones to the orthosteric site was able to induce selective conformational changes in the extracellular domain of GPR97 (33). We therefore investigated the extracellular conformational changes of GPR126 by the FlAsH-BRET method (33, 47). Specific positions of each extracellular loop (ECL) were screened for FlAsH motif incorporation (Fig. 2A and SI Appendix, Fig. S5A). The Nluc and FlAsH fusions used in our assays didn’t significantly alter their responses to progesterone stimulation in the cAMP assay, suggesting their functional integrity (SI Appendix, Fig. S5 B–D). Importantly, the binding of both progesterone and 17OHP induced a significant increase in the BRET signal between the Nluc-tagged N terminus and FlAsH-labeled ECL2 in a concentration-dependent manner, suggesting the movement of ECL2 close to the N terminus in response to stimulation with these two agonists (Fig. 2B and SI Appendix, Fig. S5 E–G). The FlAsH-BRET experiments also revealed different conformational changes of GPR126 in response to these two steroid hormones. Whereas progesterone induced an increase in the BRET signal between the Nluc-N terminus and FlAsH-labeled S1 site of ECL1, 17OHP showed no significant difference. Instead, 17OHP promoted an increase in the BRET signal between the FlAsH-labeled S2 site of ECL1 and the S6 site of ECL3 (Fig. 2B and SI Appendix, Fig. S5 E–G).
Fig. 2.
Progesterone and 17OHP binding induced conformational changes within GPR126 extracellular domains. (A) Schematic representation of the FlAsH-BRET assay design. NanoLuc (Nluc) was inserted at the N terminus of GPR126-β, and the FlAsH motif (CCPGCC) was incorporated in the designated positions at the ECLs of the receptor. (B) The maximal response of GPR126 FlAsH-BRET sensor S4 (annotated as WT) and the sensor-based CCPGCC mutants upon progesterone stimulation. Data are derived from the dose–response curves in SI Appendix, Fig. S4E, normalized to the maximal response of sensor S4. Values are the mean ± SEM from three independent experiments performed in triplicate. Data information: (B) ***P < 0.001; ND, not detectable; FlAsH-BRET sensors stimulated with progesterone were compared with those stimulated with control vehicle.
Structural Model of the Interaction of Progesterone and 17OHP with GPR126.
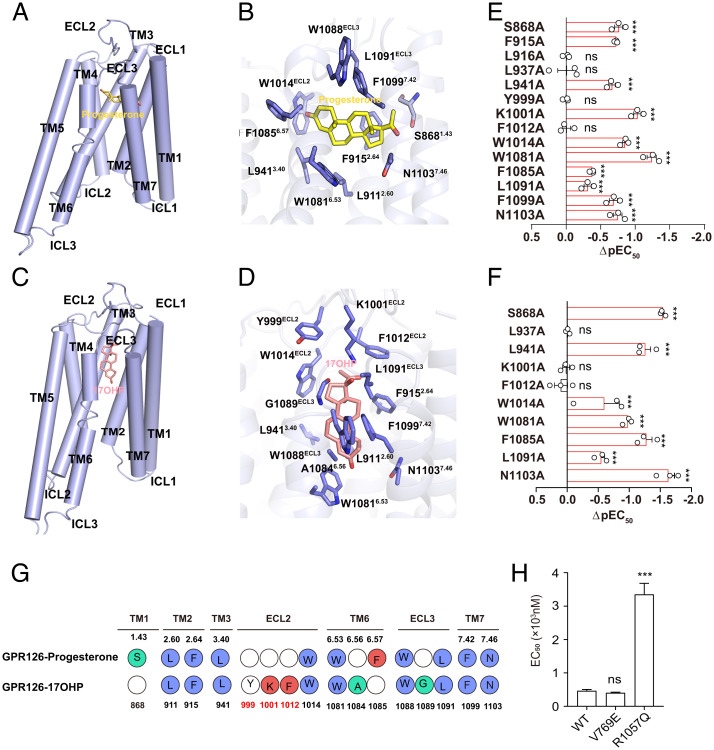
The binding mode of progesterone and 17OHP in the GPR126 ligand pocket was probed by computational simulation using the Lamarckian Genetic Algorithm and Auto Dock 4 (Fig. 3 A–D and SI Appendix, Fig. S6 A and B) (33, 47–49). GPR126 was modeled according to the Swiss model using the structure of GPR97 as a template (33, 50). The steroid core of progesterone sits flat and in a direction perpendicular to TM5, and at an angle of ∼60° from the central TM3, which was similar to the binding mode of cortisol bound with GPR97 in our recently solved complex structure (Fig. 3 A and B). Compared to progesterone, the modeled 17OHP assumed a conformation perpendicular to the plane of the plasma membrane, rotating by ∼90° compared with that of progesterone (Fig. 3 C and D).
Fig. 3.
Structural models of the interactions of progesterone and 17OHP within GPR126 ligand pocket. (A and C) Binding model of progesterone (A) and 17OHP (C) in GPR126 according to computational simulation. Note that progesterone was modeled in perpendicular to TM5 at an angle of ∼60° from the central TM3, and the modeled 17OHP assumed a conformation perpendicular to the plane of the plasma membrane. (B and D) Detailed interactions between progesterone and GPR126 (B) as well as 17OHP and GPR126 (D) ligand binding pocket residues according to computational simulation. (E and F) Alanine mutagenesis scanning of putative residues in GPR126 ligand binding pocket on progesterone induced cAMP inhibition using GloSensor assay (E) or progesterone-induced ECL2 conformational changes measured by FlAsH-BRET (F). (G) The residues in GPR126 that contact with progesterone or 17OHP. (H) EC50 values of GPR126 SNPs (V769E and R1057Q) in cAMP inhibition assay were analyzed upon progesterone stimulation. Data information: (E, F, and H) Values are the mean ± SEM of three independent experiments. **P < 0.01; ***P < 0.001; ns, no significant difference; GPR126-WT transfected cells were compared to GPR126 mutant transfected cells. All data were analyzed by one-way ANOVA with Tukey’s test. The ΔpEC50 values were obtained according to the data in SI Appendix, Figs. S7 B–E and H–J and S8 A–D and F–H.
In the modeled ligand binding pocket of GPR126, nine hydrophobic residues and two polar residues from TM1-TM3, TM6-TM7, and ECL2 contacted progesterone in the simulated model (Fig. 3 B and G and SI Appendix, Fig. S6C). The model of 17OHP shared nine common contacting residues of GPR126, compared with those of progesterone (Fig. 3 D and G and SI Appendix, Fig. S6D). Consistent with these two simulated models, mutations of eight common contacting residues of both steroid ligands, including F9152.64, L9413.40, W1014ECL2, W10816.53, L1091ECL3, F10997.42, and N11037.46 to Ala, but not several other randomly selected surrounding residues, such as L9162.65A or L9373.36A, significantly impaired GPR126 activities in response to progesterone or 17OHP stimulation in cAMP inhibition assays (Fig. 3 E and F and SI Appendix, Figs. S7 A–F and S8 A–E).
In particular, mutation of K1001ECL2 or F1012 ECL2 to Ala impaired only the extracellular conformational change of GPR126 in response to 17OHP, but showed no significant effects in response to progesterone stimulation. Conversely, mutation of F10856.57 impaired only the GPR126 response stimulated by progesterone but not by 17OHP (Fig. 3F and SI Appendix, Figs. S6F, S7 G and K, and S8 F–I). The basal BRET value of mutations of K1001ECL2, F1012 ECL2, or F10856.57 to Ala showed no significant changes (SI Appendix, Fig. S7L). These results indicated that these interactions, in particular the potential H-bond between K1001ECL2 and the 17-hydroxyl group of 17OHP, as indicated by the simulated model, are the governor of the differential recognition of steroid hormones by GPR126.
It’s worth noting that genome-wide association studies analysis has discovered dozens of SNPs in the GPR126 (ADGRG6) gene tightly associated with particular disorders or diseases, such as shortened height and AIS. Of the reported 22 SNPs in GPR126, rs17280293 (S123G), rs11155242 (K230Q), and rs2143390 (D373E) caused missense mutations (2, 51, 52). Importantly, one spontaneous mutation in GPR126 (V769E) has been reported to be highly associated with arthrogryposis multiplex congenita, whereas another mutation in GPR126 (R1057Q) was reported to be associated with aggressive periodontitis (AgP) in a Japanese population (18, 53). The S123G mutation is located in the CUB domain and K230Q mutation is located in the PTX domain, while the V769E mutation is located in the GAIN domain and R1057Q mutation is located in the intracellular loop (SI Appendix, Fig. S7M). Because only the positions of V769E and R1057Q mutations are located close to the 7TM domain, which senses the progesterone binding and couples to G proteins, we investigated the effects of these two mutations in response to progesterone stimulation in cAMP inhibition assays. Notably, the R1057Q mutant, but not the V769E, decreased the potency of progesterone (Fig. 3H and SI Appendix, Fig. S7 N and O).
GPR126 Expression Is Elevated in BC Tissues and Is Associated with a Poor Prognosis.
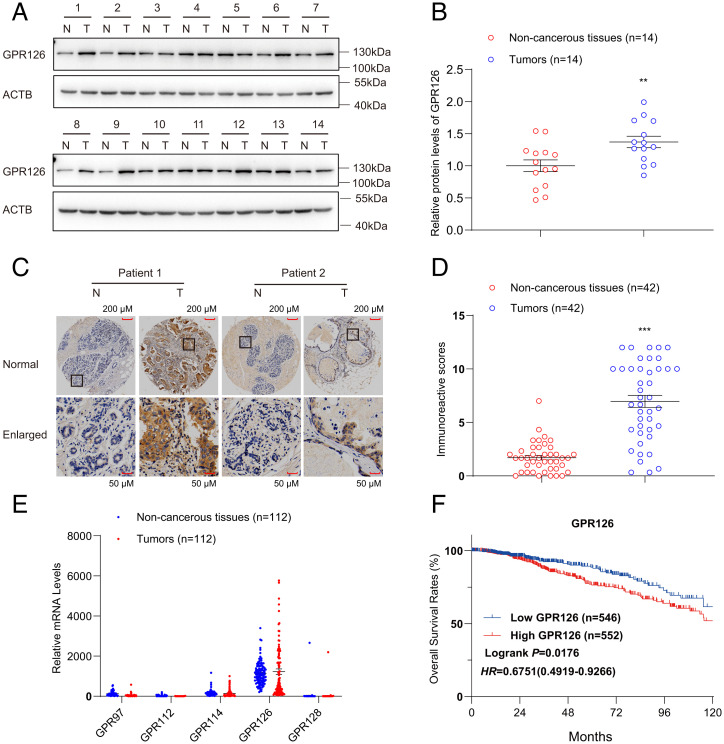
Progesterone is a key female hormone and plays essential roles in driving BC (54–57). We therefore investigated whether the activation of GPR126 by progesterone contributed to BC and how GPR126 correlates with BC progression. We examined the expression of GPR126 in 14 fresh BC tissues paired with adjacent noncancerous tissues (ANTs) by Western blot. Strikingly, 11 of 14 (78.6%) of the BC specimens had higher GPR126 protein levels than the ANT specimens (Fig. 4A). Quantification analysis showed that the mean GPR126 level was ∼1.40-fold higher in BC tissues than that in ANTs (Fig. 4B). Next, we performed immunohistochemistry (IHC) on another 42 BC tissue samples with paired ANT tissues to analyze GPR126 expression. Consistently, GPR126 expression was highly expressed in BC tissues (38 of 42: 90.48%, average IHC score 6.97) compared to ANT tissues (average IHC score 1.71) (P < 0.001) (Fig. 4 C and D). The aforementioned findings were further verified by analyzing the public clinical gene expression data of BC patients from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/) database (Fig. 4E). Importantly, we found that high GPR126 expression was associated with shorter overall survival by evaluating the prognostic value of GPR126 expression using the same database (Fig. 4F). Together, these results indicate the involvement of GPR126 in the occurrence and development of BC.
Fig. 4.
High GPR126 expression is observed in BC tissues and relates to a poor prognosis. (A and B) Western blot analysis of GPR126 in BC tissue (T) and adjacent nontumorous breast tissue (N). Fourteen representative paired samples were shown in A. The intensities of the immunoblot bands were quantified using ImageJ software and shown in B. The data are presented as the mean ± SEM from 14 paired patient samples and Student’s t test was used for comparisons between two groups. **P < 0.01. (C and D) IHC analysis of GPR126 in BC tissue (T) and adjacent nontumorous breast tissue (N). Two representative paired samples are shown in C. The statistic results showed that P < 0.001. The scale bar values were 0.216 for non-cancerous tissues and 0.566 for tumors (C). Immunoreactive score (IRS) of GPR126 in 42 paired BC samples were evaluated (D). The data are represented as the mean ± SEM and Student’s t test was used for comparisons between two groups. ***P < 0.001. (E) mRNA levels of GPR97, GPR112, GPR114, GPR126, and GPR128 in BC tissue (T) and adjacent nontumorous breast tissue (N) from 112 patients in TCGA database were calculated and analyzed. (F) Kaplan–Meier survival analysis of the relationship between GPR126 levels and the overall survival rates of breast cancer patients. The clinical data from 1,098 patients with breast cancer were analyzed and downloaded from TCGA database. High GPR126 expression was statistically associated with a shorter overall survival rate.
Progesterone Promotes BC Cell Growth In Vitro and Tumorigenesis In Vivo by Activating GPR126.
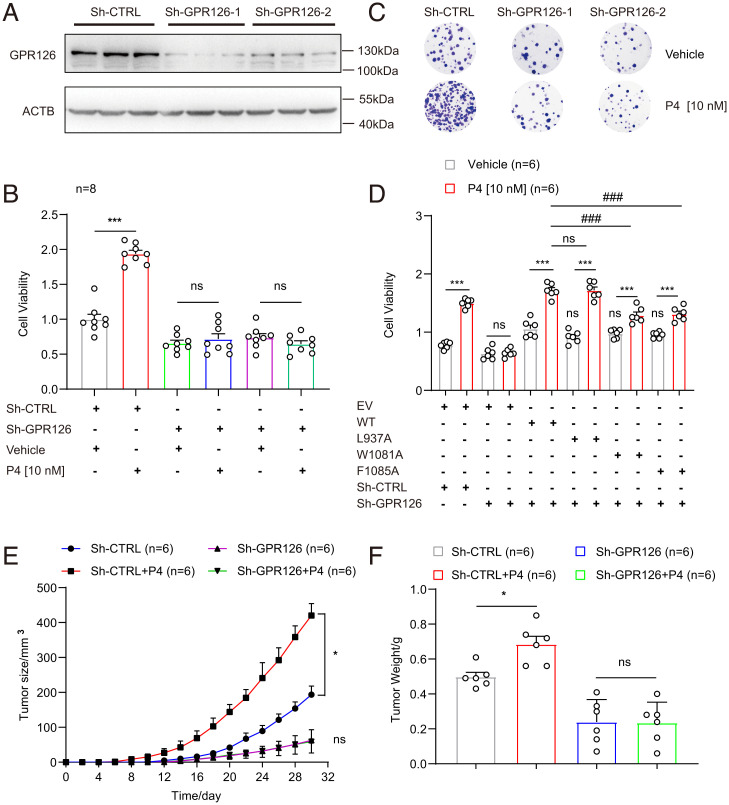
To elucidate the biological roles of progesterone-induced GPR126 activation in BC development, the TNBC MDA-MB-231 cell line, which expresses a relatively high level of GPR126 but lacks expression of nuclear progesterone receptor (PR), was exploited (58). Two independent GPR126 short-hairpin RNAs (shRNAs) (specific for GPR126, designated Sh-GPR126-1 and Sh-GPR126-2) and one control shRNA (nonspecific sequence, designated Sh-CTRL) were stably introduced into MDA-MB-231 cells by lentivirus infection and subjected to puromycin selection. GPR126 protein levels were reduced by ∼80% in both Sh-GPR126-1 and Sh-GPR126-2 cells compared with Sh-CTRL cells (Fig. 5A and SI Appendix, Fig. S9A). Then, these cells were treated with or without progesterone (P4, 10 nM) for the indicated time (SI Appendix, Fig. S9B). The MTT [3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide] assay showed that progesterone treatment led to an approximately twofold increase in the growth rate of Sh-CTRL cells, whereas such proliferation-promoting effects of progesterone were attenuated in GPR126-silenced cells (Fig. 5B). In accordance with this, colony-formation analysis revealed that progesterone treatment resulted in an increase in colony number and size in Sh-CTRL cells, whereas GPR126 knockdown largely abolished the increase in colony number and size in response to progesterone stimulation (Fig. 5C and SI Appendix, Fig. S9C). These results suggest that progesterone promoted the proliferation of BC cells by binding to GPR126 in vitro. At the same time, we detected the effects of the reported ligands (Stachel-mimicking peptide, collagen IV, PrPc) on the growth of MDA-MB-231 cells. Strikingly, different from progesterone, an MTT assay showed that the reported ligands had no significant effect on cell proliferation, indicating the different regulatory roles of GPR126 activation triggered by different ligands (SI Appendix, Fig. S9D).
Fig. 5.
GPR126 mediates the protumor activity of progesterone in BC cells both in vitro and in vivo. (A) Western blot analysis of GPR126 in MDA-MB-231 cells stably expressing sh-CTRL, sh-GPR126-1, or sh-GPR126-2. The results are representative of three independent experiments. (B–C) MDA-MB-231 cells stably expressing sh-CTRL, sh-GPR126-1 or sh-GPR126-2 were treated with progesterone (P4, 10 nM) for 72 h. Then cell proliferation was detected using either an MTT assay (B) or colony formation assay (C). Cell viability and colony numbers in P4-treated cells were compared to vehicle cells in B. Data are presented as the mean ± SEM from eight independent experiments and Student’s t test was used for comparisons between two groups. ***P < 0.001. (D) MDA-MB-231 cells with stably GPR126 silencing (Sh-GPR126) were respectively transfected with wild-type GPR126 (WT), L937A, W1081A, or F1085A mutants. Then the cells were treated with progesterone (P4, 10 nM) for 72 h. Cell proliferation was analyzed by MTT assay. The data are represented as the mean ± SEM from six independent experiments and Student’s t test was used for comparisons between two groups. Data in cells treated with progesterone (P4) were compared to data in cells treated with vehicle. ns, not significant, ***P < 0.001. Data in cells transfected with GPR126 mutants were compared to data in cells transfected with WT. ns, not significant, ###P < 0.001. (E and F) MDA-MB-231 cells (2 × 106 cells) that stably expressing sh-CTRL or sh-GPR126 were injected subcutaneously into the axilla of each nude mouse. Then, mice inoculated with sh-GPR126 or sh-CTRL cells were divided randomly into the following two groups that received either vehicle (DMSO) or progesterone (15 mg/kg) (n = 6) via intraperitoneal injection. Tumor sizes were measured every 2 d (E). After 32 d, the nude mice were killed. The dissected tumors were weighed (F). Tumor sizes and tumor weights in progesterone (P4) treated groups were compared to the vehicle groups. Data are presented as the mean ± SEM and all data were analyzed by one-way ANOVA with Tukey’s test. ns, not significant, *P < 0.05.
Our biochemical and computational simulation analysis indicated that the F10856.57 is the key residue to mediate progesterone but not 17OHP recognition, whereas the mutations of K1001A and F1012A disrupt cAMP inhibition induced by 17OHP but not progesterone. In addition, the L937A mutant showed no effect on GPR126 engagements by progesterone or 17OHP, whereas W1081A is a loss-of-function mutant in response to both progesterone and 17OHP stimulation (Fig. 3 A–G and SI Appendix, Fig. S6). We next investigated how these mutations affected the biological function of GPR126 in response to progesterone. MTT and colony formation assays showed that progesterone treatment significantly promoted the growth of GPR126-silenced MDA-MB-231 cells with ectopic overexpression of wild-type GPR126 or L937A mutant, but these promotions were significantly attenuated in cell overexpression with W1081A or F1085A mutants (Fig. 5D and SI Appendix, Fig. S9 F and G). Consistently, only exogenous overexpression of the W1081A mutant, but not L937A or F1085A mutants, inhibited 17OHP-induced cell growth in GPR126-silenced MDA-MB-231 cells (SI Appendix, Fig. S9 E, F, and H). Compared with wild-type GPR126, both K1001A and F1012A mutants alleviated cell growth promotion in response to 17OHP, but not progesterone (SI Appendix, Fig. S9 I and J–M). We also detected the effects of GPR126 SNPs on cell growth induced by progesterone. MTT assay showed that exogenous overexpression of R1057Q, but not V769E, inhibited progesterone-induced cell growth in GPR126-silenced MDA-MB-231 cells (SI Appendix, Fig. S10). Collectively, these data suggest that recognition of progesterone and 17OHP by specific residues in GPR126, or natural GPR126 missense mutation, contributed to its function in promotion of cell growth and colony formation.
To extend our in vitro observations to a more pathological context, we investigated whether progesterone-induced GPR126 activation regulated the tumorigenic capacity of MDA-MB-231 cells using an athymic nude mouse xenograft model. MDA-MB-231-Sh-GPR126-1 and Sh-CTRL cells were subcutaneously inoculated into the axillae of 12 BALE/c-nude mice, 6 for each group injected intraperitoneally with either physiological saline or progesterone (15 mg/kg, every 3 d; 32 d). The mouse weight and subcutaneous tumor size were measured every 2 d. To determine the plasma concentration of progesterone delivered through intraperitoneal injection, we measured plasma progesterone levels after administration. The results showed that the plasma concentration of progesterone initially increased rapidly, reached the maximum (151 nM) ∼1 h after injection, and declined thereafter until 8 h to maintain a steady level (4.547 nM) (SI Appendix, Fig. S9N). The effective plasma concentrations of progesterone after injection are in the range of the reported physiological levels during pregnancy from below 6.36 nM (2 ng/mL) to 477 nM (150 ng/mL) (59). Notably, although the rates of mouse weight gain were not significantly different between each group (SI Appendix, Fig. S7O), the tumors originating from MDA-MB-231-Sh-GPR126-1 cells grew more slowly than those originating from MDA-MB-231-Sh-CTRL cells at the implantation sites (Fig. 5E). Moreover, progesterone treatment significantly enhanced tumor growth in the Sh-CTRL group but had minimal effects on tumor growth in the Sh-GPR126-1 group (Fig. 5E). After 32 d, the mice were killed. We found that the volume and weight of xenografted tumors originating from MDA-MB-231-Sh-GPR126-1 cells were much smaller and lighter than those originating from MDA-MB-231-Sh-CTRL cells (Fig. 5F and SI Appendix, Fig. S7P). In addition, progesterone treatment significantly increased the size and weight of tumors in the Sh-CTRL group but had no significant effects on those in the Sh-GPR126-1 group, suggesting that progesterone promotes tumorigenesis through GPR126 in vivo (Fig. 5 E and F). Taken together, our results demonstrated that GPR126 activated by progesterone stimulated the proliferation of BC cells in vitro and tumor formation in vivo.
Gi-Dependent SRC Activation Contributes to Progesterone/GPR126-Mediated Cellular Function.
Increasing evidence has highlighted the importance of SRC in tumorigenesis and in Gi-coupled receptor signaling (60–66). We therefore sought to determine the involvement of SRC signaling in progesterone-stimulated cell proliferation through GPR126-Gi activation in BC cells. Phosphorylation of SRC at Y416 (p-SrcY416) acts as the known active form to trigger the cascade phosphorylation of a series of downstream effectors, including the phosphorylation of AKT at S473 (p-AKTS473) and phosphorylation of ERK at T202 and Y204 (p-ERKT202/Y204) (67, 68). Then the phosphorylation status of these proteins was detected in MDA-MB-231 cells treated with progesterone for different times. Strikingly, in MDA-MB-231 cells, progesterone promoted the phosphorylation of SRC at Y416 as well as p-AKTS473 and p-ERKT202/Y204 in a time-dependent manner and reached a maximum 5 min after progesterone treatment (SI Appendix, Fig. S10 A and B). Importantly, either GPR126 silencing or Gi inhibition abrogated increase of p-SRCY416, p-AKTS473, and p-ERKT202/Y204 induced by progesterone, indicating the essential role of the GPR126-Gi axis in SRC signaling activation in response to progesterone stimulation (SI Appendix, Fig. S11 C and E and SI Appendix, Fig. S11 D and F).
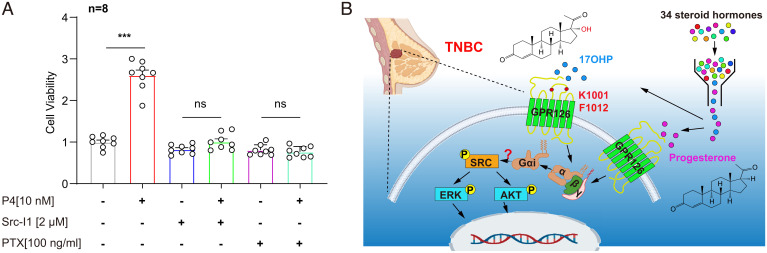
We next investigated GPR126 ligand pocket mutations on the p-SRCY416 level in response to progesterone or 17OHP stimulation. Progesterone treatment was able to promote the pY416 SRC phosphorylation in GPR126 wild-type or L937A mutant-overexpressed cells with GPR126 silencing, but failed to increase the p-SRCY416 level in GPR126-silenced MDA-MB-231 cells with ectopic overexpression of F1085A or W1081A mutants (SI Appendix, Fig. S12 A and B). Specifically, 17OHP treatment was able to promote the pY416 SRC phosphorylation in GPR126 wild-type, L937A, or F1085A mutants overexpressed cells with GPR126 silencing, but failed to increase the p-SRCY416 level in GPR126-silenced MDA-MB-231 cells with ectopic overexpression of W1081A mutant (SI Appendix, Fig. S12 C and D). These results support the correlation between the specific progesterone/17OHP recognition and the downstream SRC activation. Further MTT analysis revealed that both the SRC inhibitor (Src-I1, 2 μM) and Gi inhibitor (PTX, 100 ng/mL) significantly suppressed progesterone-induced cell growth (Fig. 6A). Collectively, these data suggested that progesterone binding to GPR126 promoted Gi coupling and downstream SRC activation, thus promoting cancer cell growth (Fig. 6B).
Fig. 6.
Progesterone/GPR126-mediated cellular function is dependent on Gi-triggered SRC activation. (A) Cell viability of MDA-MB-231 cells treated with progesterone (P4, 10 nM), Src-I1 (2 μM), and PTX (100 ng/mL) for 72 h alone or in combination was detected using MTT assay. Cell viability in P4 treated cells were compared to that in vehicle group cells, relatively. The data are presented as the mean ± SEM and Student’s t test was used for comparisons between two groups. ns, not significant, ***P < 0.001. (B) Schematic diagram of progesterone and 17OHP induced-GPR126 activation in promoting TNBC development via the Gi-SRC pathway. TNBC, cell membrane, G proteins and DNA cartoon elements were obtained from BioRender.
Discussion
GPR126 has been reported to play critical roles in the normal development of diverse tissues and is implicated in various pathological processes, such as scoliosis and carcinogenesis, through involvement in many different signaling networks (1–14). The identification of specific ligands for GPR126 in distinct physiological or pathophysiological processes is of great importance for elucidating its precise functions and corresponding mechanisms. Previous studies have identified at least three proteins or protein fragment—laminin-211, type IV collagen, and PrPc—as ligands for GPR126 (30–32). Laminin-211 and type IV collagen are major components of the extracellular matrix of Schwann cells and axonal neurons. Laminin-211 binds to a specific domain of GPR126, ranging across residue D446 to residue C807, whereas type IV collagen interacts with the CUB and PTX domains in GPR126-NTF. Similar to the GPRGKPG motif of type IV collagen, PrPc has a conserved sequence (KKRPKPG) in the amino-terminal flexible tail, which is necessary for the activation of GPR126 (30–32). All three protein ligands contribute to Schwann cell development and peripheral nervous system myelination by activating GPR126 signaling. Although it is known that both type IV collagen and laminin-211 require the extracellular domain of GPR126 for their efficient interactions, whether these three protein ligands of GPR126 directly interact with the 7TM domain of the CTF remains elusive.
In the present study, we found that progesterone and 17OHP are ligands of GPR126, both in vitro and particularly in a tumor progression model (Fig. 6B). Progesterone and 17OHP are canonical steroid hormones that play important roles in the development and functional maintenance of the female reproductive system. Our results suggest that progesterone and 17OHP bind to the ligand pocket in the 7TM domain of the CTF rather than the NTF of GPR126. Moreover, a detailed mutagenesis screening according to a computational simulated structure model indicates that K1001ECL2 and F1012 ECL2 are key residues that specifically recognize 17OHP but not progesterone, probably by interaction with the hydroxyl group. Recently, we identified that glucocorticoids, a group of steroid hormones, including hydrocortisone and cortisone, are able to activate GPR97 (33). Both GPR126 and GPR97 belong to the aGPCR subfamily (69, 70). Our work on GPR126, together with this previous study showing that GPR126 and GPR97 share the conserved ligand binding pocket, raises the possibility that multiple steroid hormones may function as ligands for different aGPCRs, especially members of the ADGRG subfamily.
Our data show that progesterone activated GPR126 with an EC50 of ∼539.6 nM, which is significantly differed from the reported EC50 of progesterone to activate its nuclear receptor (∼1 nM), suggesting these two receptors may respond to progesterone in different physiological contexts. It is known that concentrations of progesterone vary across a wide range during the menstrual period. In human, the concentration of progesterone in the premenstrual and follicular phase keeps below 0.636 nM (0.2 ng/mL), and then goes up to 63.6 nM (20 ng/mL) in the luteal phase (71). Notably, the concentrations of progesterone are significantly elevated during pregnancy. The highest concentration of progesterone could reach to ∼1.09 μM (342 ng/mL, covering its EC50 to activate GPR126) before a baby is newly born (71). Moreover, GPR126 expression profile analysis indicated the enrichment of Gpr126 in mice uterus (SI Appendix, Fig. S12E) and mammary gland (72). These data indicate that progesterone-GPR126 signaling may play important roles in modulation of pregnancy and mammary gland development, which deserve future investigation.
Compared with progesterone, 17OHP has been much less studied. Notably, patients with classic congenital adrenal hyperplasia had markedly higher plasma 17OHP concentration (mean plasma concentrations = 9,510 ng/dL, which corresponds to a concentration of 280 nM) compared to the normal people (mean plasma concentrations = 155 ng/dL, which corresponds to a concentration of 4.7 nM) (73). Moreover, recent reports have indicated that the hepatic levels of 17OHP were significantly increased in diabetic mice with a maximum concentration up to 1.8 µM and the accumulated 17OHP contributed to the pathogenesis of hyperglycemia and insulin resistance through activating glucocorticoid receptor (EC50 = 7.14 µM) (74). Given that our data show the EC50 values of 17OHP to activate full-length GPR126 and its β-subunit GPR126-β were 94 ± 0.63 nM and 10.0 ± 1.23 nM, respectively, the indicated concentrations of 17OHP in congenital adrenal hyperplasia patients and diabetic mice are sufficient to activate full-length GPR126, implying the possible involvement of 17OHP-mediated GPR126 activation in such pathological processes. Recent studies have also indicated the β-subunit of aGPCRs, such as GPR133, functions independently (75). Therefore, it doesn’t exclude the possibility that the 17OHP may regulate function of GPR126 β-subunit in normal physiological processes.
Although all previously reported agonists activate Gs signaling downstream of GPR126 and subsequently stimulate the production of cAMP (30–32), our studies demonstrate that GPR126 was able to stimulate Gi activity in response to progesterone or 17OHP stimulation. These results suggest that different ligands for the same aGPCR member, for example GPR126, may have distinct biased signaling properties that constitute the fine-tuning signaling network for these receptors with diverse functions. Recently, different biased properties of the G protein subtype or arrestins of GPCR ligands have been suggested to have great therapeutic potential, such as that for angiotensin type I receptors, opioid receptors, and adrenergic receptors (66, 76–90). Therefore, understanding the preferences of GPR126 may facilitate future therapeutic development targeting this particular receptor.
Given the critical roles of progesterone in regulating BC (54–56), we determined the involvement of GPR126 in BC progression and the functional consequence of progesterone-induced GPR126 activation. By examining the GPR126 expression levels in clinical samples and analyzing the public clinical gene-expression data of BC patients from the TCGA database, we found that GPR126 was highly expressed in the tumor tissue of BC patients. In addition, prognostic analysis revealed that high GPR126 expression correlates with unfavorable overall survival in BC patients, implicating GPR126 in the occurrence and progression of BC. Functionally, progesterone-triggered GPR126 activation significantly promoted the cell growth and tumorigenesis of TNBC cells both in vitro and in vivo. A preliminary mechanistic investigation demonstrated that Gi-dependent SRC activation downstream of the progesterone-GPR126 interaction contributes to these effects. TNBC is more aggressive and has a poorer prognosis than other types of BC because of the lack of expression of nuclear receptors for estrogen and progesterone, thus leading to refractoriness to hormone therapy (91). Our study showed that despite the absence of progesterone nuclear receptor, progesterone is still able to promote the development of BC by activating the membrane receptor GPR126 and its downstream Gi signaling. Therefore, developing antagonists targeting GPR126-Gi may provide an alternative therapeutic option for patients with TNBC, which requires further investigation.
GPR126 is expressed in a wide range of tissues and functions in a variety of physiological and pathological processes, such as the development of bone, ear, and heart, myelination of the peripheral nervous system, and neuromuscular diseases (2, 5, 7, 9, 51, 92, 93). In addition to the contributions of progesterone-GPR126 signaling to BC, further elucidation is needed to examine whether these widely distributed steroid hormones regulate other pathophysiological processes through GPR126 and whether the mechanism of GPR126 signaling activation by progesterone deciphered here has broader applications.
In summary, our present study found that the steroid hormones progesterone and 17OHP can activate the Gi signaling of GPR126 by binding to its 7TM domain, and the interaction between progesterone and GPR126 has important roles in a tumor progression model (Fig. 6B). The identification of a membrane receptor for progesterone other than classic nuclear receptors and the definition of the mechanisms of GPR126 involved in potential tumor progression in TNBC will enrich our understanding of the functions and working mechanisms of both the aGPCR member GPR126 and progesterone.
Materials and Methods
Cell Lines.
HEK293 cells were obtained from the Cell Resource Center of Shanghai Institute for Biological Sciences (Chinese Academy of Science) and grown in Dulbecco's Modified Eagle Medium (DMEM) culture with 10% FBS (Gibco). MDA-MB-231 cells were originally obtained from the American Type Culture Collection (ATCC) and grown in monolayer culture in Roswell Park Memorial Institute (RPMI) 1640 with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin. Both kinds of cells were maintained at 37 °C in a humidified atmosphere consisting of 5% CO2 and 95% air.
Chemicals, Reagents, and Peptides.
Cortisol, corticosterone, calcifediol, calcitriol, 11-deoxycortisol, deoxycorticosterone, DHEA (dehydroepiandrosterone), estriol, 2-methoxyestrone, pregnenolone, progesterone, vitamin D3, 16α-hydroxyestrone, 11β-hydroxyprogesterone, vitamin D2, prednisone, betamethasone were purchased from Med Chem Express. Androstan, 20α-hydroxycholesterol, dihydrotestosterone, 11α-hydroxyprogesterone, 5α-androstan-17β-ol-3one, 21-deoxycortisol, 17β-Estradiol, 17-hydroxypregnenolone, 17OHP, 3α,21-dihydroxy-5β-pregnane-11,20-dione, retinol were from Sigma-Aldrich. 5β-pregnane-3,20-dione, estrone-3-sulfate were from Santa Cruz. Testosterone, estrone, androstenone, 5α-pregnane-3,20-dione were from Aladdin.
Monoclonal ANTI-FLAG M2 antibody (Cat #F1804) and anti-Rabbit IgG-HRP secondary antibody (Cat #A6154) were from Sigma Aldrich. Goat anti-mouse secondary antibody (Cat #A-21235) was from Thermo Fisher Scientific. Rabbit polyclonal anti-GPR126 antibody (Cat #TA315653) and β-actin mouse monoclonal antibody (Cat #TA811000) were from ORIGENE. GPR126 rabbit polyclonal antibody (Cat #17774-1-AP), Phospho-ERK1/2 (Thr202/Tyr204) Rabbit Polyclonal antibody (Cat #28733-1-AP), phospho-AKT(Ser473) mouse monoclonal antibody (Cat #66444-1-lg) and AKT mouse monoclonal antibody (Cat #60203-2-lg) were from Proteintech. Phospho-Src family (Tyr416) antibody (Cat #2101) was from Cell Signaling Technology. ERK2 antibody (sc-1647) was from Santa Cruz Biotechnology.
The high purity (above 95%) 126-SMP [VHF(4-Me)GVLMDLPRSASQI] were chemically synthesized at Chinapeptides (Shanghai, China, http://www.chinapeptides.com). All the steroid chemicals or peptides were dissolved in dimethyl sulfoxide (DMSO) to make stock solutions at final concentrations of 100 mM, which was further diluted to working concentrations with phosphate buffer solution (PBS).
Constructs.
The human GPR126 gene (ADGRG6) was cloned into the pcDNA3.1 vector with an N-terminal Flag epitope tag (DYKDDDDK). The CTF of GPR126 (GPR126-β) and GPR126-β-ΔGPS (whose Stachel sequence was deleted) were subcloned into pcDNA3.1 vector with an N-terminal Flag tag. The GPR126 mutations S868A, F915A, L916A, L937A, L941A, Y999A, K1001A, F1012A, W1014A, W1081A, F1085A, L1091A, F1099A, N1103A, S123G, K230Q, D373E, V769E, and R1057Q were generated using the Quikchange mutagenesis kit (Stratagene). The human PAQR5, PAQR6, PAQR7, PAQR8, and PAQR9 genes were cloned into the pcDNA3.1 vector with an N-terminal Flag epitope tag (DYKDDDDK). For detection of conformational changes of extracellular region using FlAsH-BRET assay, NanoLuc (Nluc) and a four-amino acid linker (Gly-Ser-Ser-Gly) were fused to the N-terminal of GPR126. FlAsH sequence (Cys-Cys-Pro-Gly-Cys-Cys) was inserted into the ECL1, ECL2 or ECL3 region of Nluc-GPR126, as indicated in SI Appendix, Fig. S5A. All primers used above are listed in SI Appendix, Tables S1 and S2. The GloSensor plasmid was obtained from Promega. All constructs were generated using Quikchange site-directed mutagenesis kit (Stratagene) and verified by DNA sequencing.
Measurement of Receptor Expression at Cell Surface by ELISA.
To evaluate the expression level of wild-type GPR126 and its mutants, HEK293 cells were transfected with wild-type and mutant GPR126 or vehicle (pcDNA3.1) using PEI reagent in six-well plates, according to the manufacturer’s instructions (33, 47). After incubation at 37 °C for 16 h, cells were then seeded into 24-well plates at a density of 105 cells per well for another 16-h incubation at the same temperature. Next, cells were fixed with 4% (wt/vol) paraformaldehyde for 15 min and blocked with 5% (wt/vol) BSA for 1 h at room temperature. Cells were then incubated with the monoclonal anti-FLAG primary antibody (Cat #F1804, Sigma Aldrich) overnight at 4 °C and followed by incubation of a secondary goat anti-mouse antibody (Cat #A-21235, Thermo Fisher) conjugated to horseradish peroxide for 1 h at room temperature. After washing, TMB (3, 3′, 5, 5′-tetramethylbenzidine) solution was added into each well for color reaction. An equal volume of 0.25 M HCl was used for stopping the reactions. The optical density at 450 nm was measured using the TECAN (Infinite M200 Pro NanoQuant) luminescence counter for calculating and analyzing the relative expression levels of corresponding plasmids.
cAMP Accumulation and Inhibition Assay.
To detect the cAMP accumulation of GPR126 and the inhibitory effects on Fsk-induced cAMP accumulation of different GPR126 constructs or mutants in response to different ligands, GloSensor cAMP assay (Promega) was performed according to previous publications (33). HEK293 cells were transfected with wild-type or mutant GPR126 or vehicle (pcDNA3.1) and GloSensor plasmid using PEI reagent in six-well plates according to the manufacturer’s instructions. After incubation at 37 °C for 24 h, cells were then seeded into 96-well plates at a density of 4 × 104 cells per well for another 24-h incubation at the same temperature. The cells were preincubated with serum-free medium (Gibco) containing 5% (vol/vol) dilution of the GloSensor cAMP reagent stock solution (Progema) for 2 h. After that, Fsk (5 μM, using in cAMP inhibition assay) and different concentrations of ligands were added into each well. Then, the luminescence intensity was examined immediately using an EnVision multilabel microplate detector (Perkin-Elmer). Data were analyzed using the sigmoidal dose–response function in GraphPad Prism 7.0.
FlAsH-BRET Assay.
To monitor the extracellular conformational changes of GPR126 deduced by progesterone and 17OHP binding, a FlAsH-BRET assay was performed as previously described (33, 45, 94). In brief, HEK293 cells were seeded in six-well plate and transfected with GPR126 FlAsH-BRET sensors (S1 to S6, as indicated in SI Appendix, Fig. S5A) or relative mutants based on sensor S4 and incubated for 48 h at 37 °C in 5% CO2. Then the cells were labeled with 2.5 µM FlAsH EDT2 (Thermo Fisher Scientific) in accordance with the manufacturer’s specification. After that, the cells expressing the labeled GPR126 FlAsH-BRET sensors were seeded into a 96-well plate at a density of 5 × 104 cells per well. The indicated concentrations of progesterone and 17OHP were added into each well together with luciferase substrate coelenterazine-h (5 μM). The BRET signal was calculated as the ratio of FlAsH to Nluc emission. The change in BRET signal due to ligand addition was recorded as ΔBRET.
Molecular Dynamics Simulation of Progesterone-GPR126 Complex and 17OHP-GPR126 Complex.
The initial binding pose of progesterone or 17OHP with GPR126 was determined by Auto Dock4 (48). The size of docking box was set to 60 × 60 × 60 Å, the number of GA runs was set to 200 with the 0.375-Å grid spacing, and the center of mass of progesterone or 17OHP was set to the docking center. We analyzed the molecular docking results of progesterone or 17OHP with GPR126, the conformation with the lowest binding energy was selected for the next molecular dynamics (MD) simulation. All the MD simulation input files of progesterone or 17OHP with GPR126 complex were generated by the CHARMM-GUI website (95). We embedded the progesterone-GPR126 complex or 17OHP-GPR126 complex into the double-layer POPC membrane, adding with 0.15 M NaCl to balance the system charge, and incorporation the ions into the system using the Monte Carlo method. The TIP3P model was used for hexagonal type water box. The steepest descent method was used to minimize the progesterone or 17OHP with GPR126 complex simulation system with a series of restraints for the GPR126, 17OHP, or progesterone and lipid atoms to ensure that the energy of the entire system converges to 500 kJ/(mol⋅nm). The simulation system was heated from 0 K to 310 K in the NVT ensemble (number of particles, volume and temperature are conserved) for 1,000 ps. Following this, the NPT ensemble simulations were run at 1 atm for 1,000 ps with 10.0 kcal mol−1 Å−2 harmonic restraints. The 200-ns MD simulation was carried out by Gromacs2019.5 (96) with a Charmm36m all-atom force field (97). In the MD simulation, the LINCS algorithm was used to constrain all the bonds involving hydrogen. The cutoff value for nonbonding interactions was 12 Å. A time step of 2 fs was used and the trajectories were saved every 10 ps.
IHC and Scoring.
Samples of breast tumor and adjacent tissues were obtained from 14 patients undergoing surgical excision of tumors at Shandong Provincial Hospital (Jinan, China). The use of pathological specimens and the review of all pertinent patient records were approved by the Shandong Provincial Hospital Ethical Review Board. Informed consent was obtained from the patients. Frozen tissues were subjected to Western blot analysis. Breast tissue microarray was purchased from Wuhan Servicebio Technology, which contains 42 cases of breast adenocarcinoma with paired paraneoplastic tissues. The clinic pathological features of the samples were available on the company’s website. IHC was performed following standard protocol which was described in detail in SI Appendix files.
Establishment of GPR126 Stable Knockdown Cell Lines.
Two shRNA oligos targeting human GPR126 including 5′-GCCTCACAGTTAGATGCAA-3′(Sh1) and 5′-GCTCAACTCTAATGAATAT-3′ (Sh2) were synthesized and fused into pLKO.1 vector (Addgene) as instructions and a nonspecific sequence (5′- TTCTCCGAACGTGTCACGT-3′) was used as negative control. Lentivirus expressing GPR126 shRNAs or control shRNA was prepared as described previously (98). MDA-MB-231 cells were infected with the indicated lentiviral supernatant along with polybrene (4 μg/mL; Millipore/Chemicon), followed by selection with puromycin (2 μg/mL; Solarbio). Finally, the monoclonal cell lines stably expressing GPR126 shRNAs or control shRNAs were obtained. The expression levels of GPR126 in GPR126 knockdown cells (designated as sh-GPR126-1, and sh-GPR126-2) and their controls (designated as sh-CTRL) were confirmed by Western blot analysis.
Western Blot Analysis.
For protein phosphorylation detection, MDA-MB-231 cells were starved in serum free medium for 8 h and then treated with progesterone (10 nM) for 5 min. Then cells were harvested and lysed using lysis buffer (50 mM Tris, pH 8.0; 150 mM NaCl; 1 mM NaF; 1% Nonidet P-40; 2 mM EDTA; 10% glycerol; 0.25% sodium deoxycholate; 1 mM Na3VO4; 0.3 μM aprotinin; 130 μM bestiatin; 1 μM leupeptin; 1 μM pepstatin; 5 mM iodoacetate; 10 mM pyrophosphate; 1 mM PMSF) on ice for 30 min. Then, 30 µg whole-cell protein lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS/PAGE) and transferred to polyvinylidene fluoride (PVDF) membrane by electroblotting. The membrane was immunoblotted with the indicated primary and secondary antibodies. Protein bands from the Western blot were quantified using ImageJ software (NIH). For GPR126 expression detection, breast tumor tissues and adjacent nontumorous tissues were grinded in the above-mentioned lysis buffer and lysed on ice for 1 h. Then the samples were further analyzed using Western blot analysis.
cAMP Level Detection by ELISA Kit.
cAMP levels in MDA-MB-231 cells were detected using the cAMP Parameter Assay Kit (R&D Systems) according to the manufacturer’s instruction. Detailed protocol was described in SI Appendix files.
Cell Proliferation Assay.
Cell proliferation were detected using MTT assay which was described in detail in SI Appendix files.
Colony Formation Assays.
Colony formation assays were held follwed by protocol described in SI Appendix files.
Nude Mice Xenograft Model.
Nude mice xenograft experiments were carried on followed by protocol described in SI Appendix files.
Statistical Analysis.
All data are representative of at least three independent experiments. All statistical analyses were performed using GraphPad Prism 7.0. The data are presented as the mean ± SEM. Student’s t test or one-way ANOVA with Tukey’s test was used for comparisons among groups. Survival data were analyzed using Kaplan–Meier statistical method. P < 0.05 was considered statistically significant. In the graphed data, *P < 0.05, **P < 0.01, and ***P < 0.001, and #P < 0.05, ##P < 0.01, and ###P < 0.001.
Supplementary Material
Acknowledgments
The scientific calculations in this paper have been done on the HPC Cloud Platform of Shandong University. This work was supported by National Key R&D Program of China Grant 2019YFA0904200 (to P.X. and J.-P.S.); National Science Fund for Excellent Young Scholars Grant 81822008 (to X.Y.); National Science Fund for Distinguished Young Scholars Grant 81773704 (to J.-P.S.); National Natural Science Foundation of China Grants 92057121 (to X.Y.), 31870781 (to P.Z.), and 31971195 (to P.X.); Shandong Provincial Natural Science Fund for Excellent Young Scholars ZR2021YQ18 (to P.X.); Key Research and Development Program of Shandong Province 2021CXGC011105 (to J.-P.S.); Major Basic Research Project of Shandong Natural Science Foundation ZR2020ZD39 (to J.-P.S.); Key Research Project of the Natural Science Foundation of Beijing, China Z200019 (to J.-P.S.); Innovative Research Team in University Grant IRT_17R68 (to X.Y.); and Shandong Provincial Natural Science Foundation ZR2020QH057 (to W.A.)
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117004119/-/DCSupplemental.
Data Availability
All study data are included in the main text and SI Appendix.
References
- 1.Kou I., et al. , Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nat. Genet. 45, 676–679 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Hancock D. B., et al. , Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 42, 45–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu E., et al. , A genetic variant in GPR126 causing a decreased inclusion of exon 6 is associated with cartilage development in adolescent idiopathic scoliosis population. BioMed Res. Int. 2019, 4678969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waller-Evans H., et al. , The orphan adhesion-GPCR GPR126 is required for embryonic development in the mouse. PLoS One 5, e14047 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun P., et al. , Regulation of body length and bone mass by Gpr126/Adgrg6. Sci. Adv. 6, eaaz0368 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karner C. M., Long F., Solnica-Krezel L., Monk K. R., Gray R. S., Gpr126/Adgrg6 deletion in cartilage models idiopathic scoliosis and pectus excavatum in mice. Hum. Mol. Genet. 24, 4365–4373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mogha A., et al. , Gpr126 functions in Schwann cells to control differentiation and myelination via G-protein activation. J. Neurosci. 33, 17976–17985 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monk K. R., et al. , A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science 325, 1402–1405 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monk K. R., Oshima K., Jörs S., Heller S., Talbot W. S., Gpr126 is essential for peripheral nerve development and myelination in mammals. Development 138, 2673–2680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing X., et al. , Regulatory region mutations of TERT, PLEKHS1 and GPR126 genes as urinary biomarkers in upper tract urothelial carcinomas. J. Cancer 12, 3853–3861 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garinet S., et al. , High prevalence of a hotspot of noncoding somatic mutations in intron 6 of GPR126 in bladder cancer. Mol. Cancer Res. 17, 469–475 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Wu S., et al. , Whole-genome sequencing identifies ADGRG6 enhancer mutations and FRS2 duplications as angiogenesis-related drivers in bladder cancer. Nat. Commun. 10, 720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui H., et al. , GPR126 regulates colorectal cancer cell proliferation by mediating HDAC2 and GLI2 expression. Cancer Sci. 112, 1798–1810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patra C., Monk K. R., Engel F. B., The multiple signaling modalities of adhesion G protein-coupled receptor GPR126 in development. Receptors Clin. Investig. 1, 79 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall R. J., et al. , Functional genomics of GPR126 in airway smooth muscle and bronchial epithelial cells. FASEB J. 35, e21300 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Kou I., et al. ; Japan Scoliosis Clinical Research Group (JSCRG); Texas Scottish Rite Hospital for Children Clinical Group (TSRHCCG), A multi-ethnic meta-analysis confirms the association of rs6570507 with adolescent idiopathic scoliosis. Sci. Rep. 8, 11575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J. F., et al. , Association of GPR126 gene polymorphism with adolescent idiopathic scoliosis in Chinese populations. Genomics 105, 101–107 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Ravenscroft G., et al. , Mutations of GPR126 are responsible for severe arthrogryposis multiplex congenita. Am. J. Hum. Genet. 96, 955–961 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogha A., et al. , Gpr126/Adgrg6 has Schwann cell autonomous and nonautonomous functions in peripheral nerve injury and repair. J. Neurosci. 36, 12351–12367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musa G., et al. , Gpr126 (Adgrg6) is expressed in cell types known to be exposed to mechanical stimuli. Ann. N. Y. Acad. Sci. 1456, 96–108 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Nik-Zainal S., et al. , Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fatima A., Tariq F., Malik M. F. A., Qasim M., Haq F., Copy number profiling of MammaPrint™ genes reveals association with the prognosis of breast cancer patients. J. Breast Cancer 20, 246–253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassilana F., Nash M., Ludwig M. G., Adhesion G protein-coupled receptors: Opportunities for drug discovery. Nat. Rev. Drug Discov. 18, 869–884 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Liebscher I., Schöneberg T., Tethered agonism: A common activation mechanism of adhesion GPCRs. Handb. Exp. Pharmacol. 234, 111–125 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Paavola K. J., Stephenson J. R., Ritter S. L., Alter S. P., Hall R. A., The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J. Biol. Chem. 286, 28914–28921 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Q. X., et al. , Constitutive Gαi coupling activity of very large G protein-coupled receptor 1 (VLGR1) and its regulation by PDZD7 protein. J. Biol. Chem. 289, 24215–24225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriguchi T., et al. , DREG, a developmentally regulated G protein-coupled receptor containing two conserved proteolytic cleavage sites. Genes Cells 9, 549–560 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Stehlik C., Kroismayr R., Dorfleutner A., Binder B. R., Lipp J., VIGR—A novel inducible adhesion family G-protein coupled receptor in endothelial cells. FEBS Lett. 569, 149–155 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Liebscher I., et al. , A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 9, 2018–2026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen S. C., et al. , The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron 85, 755–769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paavola K. J., Sidik H., Zuchero J. B., Eckart M., Talbot W. S., Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci. Signal. 7, ra76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Küffer A., et al. , The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature 536, 464–468 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ping Y. Q., et al. , Structures of the glucocorticoid-bound adhesion receptor GPR97-Go complex. Nature 589, 620–626 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Garg D., Ng S. S. M., Baig K. M., Driggers P., Segars J., Progesterone-mediated non-classical signaling. Trends Endocrinol. Metab. 28, 656–668 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Ben-Ari Y., Gaiarsa J. L., Tyzio R., Khazipov R., GABA: A pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol. Rev. 87, 1215–1284 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Rohe H. J., Ahmed I. S., Twist K. E., Craven R. J., PGRMC1 (progesterone receptor membrane component 1): A targetable protein with multiple functions in steroid signaling, P450 activation and drug binding. Pharmacol. Ther. 121, 14–19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagiwara A., Ogiwara K., Takahashi T., Expression of membrane progestin receptors (mPRs) in granulosa cells of medaka preovulatory follicles. Zool. Sci. 33, 98–105 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Thomas P., Rapid steroid hormone actions initiated at the cell surface and the receptors that mediate them with an emphasis on recent progress in fish models. Gen. Comp. Endocrinol. 175, 367–383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Y., Rice C. D., Pang Y., Pace M., Thomas P., Cloning, expression, and characterization of a membrane progestin receptor and evidence it is an intermediary in meiotic maturation of fish oocytes. Proc. Natl. Acad. Sci. U.S.A. 100, 2231–2236 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y., Bond J., Thomas P., Identification, classification, and partial characterization of genes in humans and other vertebrates homologous to a fish membrane progestin receptor. Proc. Natl. Acad. Sci. U.S.A. 100, 2237–2242 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas P., et al. , Steroid and G protein binding characteristics of the seatrout and human progestin membrane receptor alpha subtypes and their evolutionary origins. Endocrinology 148, 705–718 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Pang Y., Dong J., Thomas P., Characterization, neurosteroid binding and brain distribution of human membrane progesterone receptors δ and epsilon (mPRδ and mPRepsilon) and mPRδ involvement in neurosteroid inhibition of apoptosis. Endocrinology 154, 283–295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun J. P., et al. , The very large G protein coupled receptor (Vlgr1) in hair cells. J. Mol. Neurosci. 50, 204–214 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Zhang D. L., et al. , Gq activity- and β-arrestin-1 scaffolding-mediated ADGRG2/CFTR coupling are required for male fertility. eLife 7, e33432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li T., et al. , Homocysteine directly interacts and activates the angiotensin II type I receptor to aggravate vascular injury. Nat. Commun. 9, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu Y., et al. , Cartilage oligomeric matrix protein is an endogenous β-arrestin-2-selective allosteric modulator of AT1 receptor counteracting vascular injury. Cell Res. 31, 773–790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang F., et al. , Structural basis of GPBAR activation and bile acid recognition. Nature 587, 499–504 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Morris G. M., et al. , AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao P., et al. , Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes. Cell 184, 943–956.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biasini M., et al. , SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soler Artigas M., et al. ; UK BiLEVE, Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat. Commun. 6, 8658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsson H., et al. , Targeted high-throughput sequencing of candidate genes for chronic obstructive pulmonary disease. BMC Pulm. Med. 16, 146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitagaki J., et al. , A putative association of a single nucleotide polymorphism in GPR126 with aggressive periodontitis in a Japanese population. PLoS One 11, e0160765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diep C. H., Daniel A. R., Mauro L. J., Knutson T. P., Lange C. A., Progesterone action in breast, uterine, and ovarian cancers. J. Mol. Endocrinol. 54, R31–R53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J. J., Kurita T., Bulun S. E., Progesterone action in endometrial cancer, endometriosis, uterine fibroids, and breast cancer. Endocr. Rev. 34, 130–162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shamseddin M., et al. , Contraceptive progestins with androgenic properties stimulate breast epithelial cell proliferation. EMBO Mol. Med. 13, e14314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conneely O. M., Mulac-Jericevic B., Lydon J. P., Progesterone-dependent regulation of female reproductive activity by two distinct progesterone receptor isoforms. Steroids 68, 771–778 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Rust S., et al. , Combining phenotypic and proteomic approaches to identify membrane targets in a ‘triple negative’ breast cancer cell type. Mol. Cancer 12, 11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.López-García C., et al. , Molecular and morphological changes in placenta and embryo development associated with the inhibition of polyamine synthesis during midpregnancy in mice. Endocrinology 149, 5012–5023 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Martellucci S., et al. , Src family kinases as therapeutic targets in advanced solid tumors: What we have learned so far. Cancers (Basel) 12, 1448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berndt S., Liebscher I., New structural perspectives in G protein-coupled receptor-mediated src family kinase activation. Int. J. Mol. Sci. 22, 6489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith H. W., et al. , An ErbB2/c-Src axis links bioenergetics with PRC2 translation to drive epigenetic reprogramming and mammary tumorigenesis. Nat. Commun. 10, 2901 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pagano E., et al. , Activation of the GPR35 pathway drives angiogenesis in the tumour microenvironment. Gut 71, 509–520 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lyu C., et al. , Targeting Gi/o protein-coupled receptor signaling blocks HER2-induced breast cancer development and enhances HER2-targeted therapy. JCI Insight 6, e150532 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma Y. C., Huang J., Ali S., Lowry W., Huang X. Y., Src tyrosine kinase is a novel direct effector of G proteins. Cell 102, 635–646 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Yang F., et al. , Allosteric mechanisms underlie GPCR signaling to SH3-domain proteins through arrestin. Nat. Chem. Biol. 14, 876–886 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Lavoie H., Gagnon J., Therrien M., ERK signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 21, 607–632 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Hua H., et al. , Targeting Akt in cancer for precision therapy. J. Hematol. Oncol. 14, 128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamann J., et al. , International union of basic and clinical pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 67, 338–367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langenhan T., Aust G., Hamann J., Sticky signaling—Adhesion class G protein-coupled receptors take the stage. Sci. Signal. 6, re3 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Brisken C., Scabia V., 90 years of progesterone: Progesterone receptor signaling in the normal breast and its implications for cancer. J. Mol. Endocrinol. 65, T81–T94 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Tabula Muris C., et al. ; Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators, Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang H. J., et al. , Metabolic disorders in newly diagnosed young adult female patients with simple virilizing 21-hydroxylase deficiency. Endocrine 38, 260–265 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Lu Y., et al. , Obesity-induced excess of 17-hydroxyprogesterone promotes hyperglycemia through activation of glucocorticoid receptor. J. Clin. Invest. 130, 3791–3804 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frenster J. D., et al. , Functional impact of intramolecular cleavage and dissociation of adhesion G protein-coupled receptor GPR133 (ADGRD1) on canonical signaling. J. Biol. Chem. 296, 100798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith J. S., Lefkowitz R. J., Rajagopal S., Biased signalling: From simple switches to allosteric microprocessors. Nat. Rev. Drug Discov. 17, 243–260 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wacker D., Stevens R. C., Roth B. L., How ligands illuminate GPCR molecular pharmacology. Cell 170, 414–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu C. H., et al. , Arrestin-biased AT1R agonism induces acute catecholamine secretion through TRPC3 coupling. Nat. Commun. 8, 14335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang F., et al. , Phospho-selective mechanisms of arrestin conformations and functions revealed by unnatural amino acid incorporation and (19)F-NMR. Nat. Commun. 6, 8202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Q., et al. , DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1H-NMR probe. Nat. Commun. 11, 4857 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Z., et al. , Phosphorylation of G protein-coupled receptors: From the barcode hypothesis to the flute model. Mol. Pharmacol. 92, 201–210 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Wootten D., Christopoulos A., Marti-Solano M., Babu M. M., Sexton P. M., Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 19, 638–653 (2018). [DOI] [PubMed] [Google Scholar]
- 83.Wingler L. M., et al. , Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR. Science 367, 888–892 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Suomivuori C. M., et al. , Molecular mechanism of biased signaling in a prototypical G protein-coupled receptor. Science 367, 881–887 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rakesh K., et al. , beta-Arrestin-biased agonism of the angiotensin receptor induced by mechanical stress. Sci. Signal. 3, ra46 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim K. S., et al. , β-Arrestin-biased AT1R stimulation promotes cell survival during acute cardiac injury. Am. J. Physiol. Heart Circ. Physiol. 303, H1001–H1010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Violin J. D., Lefkowitz R. J., Beta-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol. Sci. 28, 416–422 (2007). [DOI] [PubMed] [Google Scholar]
- 88.Violin J. D., Crombie A. L., Soergel D. G., Lark M. W., Biased ligands at G-protein-coupled receptors: Promise and progress. Trends Pharmacol. Sci. 35, 308–316 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Wingler L. M., Lefkowitz R. J., Conformational basis of G protein-coupled receptor signaling versatility. Trends Cell Biol. 30, 736–747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wisler J. W., Rockman H. A., Lefkowitz R. J., Biased G protein-coupled receptor signaling: changing the paradigm of drug discovery. Circulation 137, 2315–2317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bianchini G., Balko J. M., Mayer I. A., Sanders M. E., Gianni L., Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 13, 674–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Geng F. S., et al. , Semicircular canal morphogenesis in the zebrafish inner ear requires the function of gpr126 (lauscher), an adhesion class G protein-coupled receptor gene. Development 140, 4362–4374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Todd E. J., et al. , Next generation sequencing in a large cohort of patients presenting with neuromuscular disease before or at birth. Orphanet J. Rare Dis. 10, 148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee M. H., et al. , The conformational signature of β-arrestin2 predicts its trafficking and signalling functions. Nature 531, 665–668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee J., et al. , CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Van Der Spoel D., et al. , GROMACS: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005). [DOI] [PubMed] [Google Scholar]
- 97.Huang J., et al. , CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y., et al. , CUL4A overexpression enhances lung tumor growth and sensitizes lung cancer cells to erlotinib via transcriptional regulation of EGFR. Mol. Cancer 13, 252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.