Significance
Bacterial adaptation to the presence of an antibiotic often involves evolutionary trade-offs, such as increased susceptibility to other drugs (collateral sensitivity). Its exploitation to design improved therapeutic strategies is only feasible if collateral sensitivity is robust, reproducible, and emerges in resistant mutants; these issues are rarely addressed in available publications. We describe a robust collateral sensitivity phenotype that emerges in different antibiotic-resistance mutational backgrounds, due to different genetic events, and propose therapeutic strategies effective for treating infections caused by Pseudomonas aeruginosa antibiotic-resistant mutants. Since conserved collateral sensitivity phenotypes do not confer adaptation to the presence of antibiotics, our results are also relevant for understanding convergent evolution processes in which the force selecting the emerging phenotype remains unclear.
Keywords: collateral sensitivity, antibiotic resistance, convergent evolution, Pseudomonas aeruginosa, phenotypic convergence
Abstract
Collateral sensitivity is an evolutionary trade-off whereby acquisition of the adaptive phenotype of resistance to an antibiotic leads to the nonadaptive increased susceptibility to another. The feasibility of harnessing such a trade-off to design evolutionary-based approaches for treating bacterial infections has been studied using model strains. However, clinical application of collateral sensitivity requires its conservation among strains presenting different mutational backgrounds. Particularly relevant is studying collateral sensitivity robustness of already-antibiotic-resistant mutants when challenged with a new antimicrobial, a common situation in clinics that has hardly been addressed. We submitted a set of diverse Pseudomonas aeruginosa antibiotic-resistant mutants to short-term evolution in the presence of different antimicrobials. Ciprofloxacin selects different clinically relevant resistance mutations in the preexisting resistant mutants, which gave rise to the same, robust, collateral sensitivity to aztreonam and tobramycin. We then experimentally determined that alternation of ciprofloxacin with aztreonam is more efficient than ciprofloxacin–tobramycin alternation in driving the extinction of the analyzed antibiotic-resistant mutants. Also, we show that the combinations ciprofloxacin–aztreonam or ciprofloxacin–tobramycin are the most effective strategies for eliminating the tested P. aeruginosa antibiotic-resistant mutants. These findings support that the identification of conserved collateral sensitivity patterns may guide the design of evolution-based strategies to treat bacterial infections, including those due to antibiotic-resistant mutants. Besides, this is an example of phenotypic convergence in the absence of parallel evolution that, beyond the antibiotic-resistance field, could facilitate the understanding of evolution processes, where the selective forces giving rise to new, not clearly adaptive phenotypes remain unclear.
Besides its relevance for human health, antibiotic resistance (AR) is one of the few evolutionary processes that can be experimentally addressed. Consequently, the study of the evolution processes involved in the acquisition of resistance is relevant not just in the AR field, but also in that of evolution in general. Since AR is the result of bacterial evolution, evolution-based approaches aiming to find the Achilles’ heel associated with AR acquisition could be useful to tackle this relevant health problem (1). In this regard, evolutionary therapeutic strategies aiming to improve the efficacy of available antibiotics and to reduce the probability of selection of resistance are particularly interesting (2). One of the most promising trade-offs associated with the acquisition of resistance that could be exploited for implementing such novel therapeutic approaches is collateral sensitivity (CS), by which the acquisition of resistance to one drug renders an increased susceptibility to another (3).
Several studies, based on the alternation (4–7) or the combination of pairs of drugs (8–10), have tried to exploit CS (11, 12). However, for this exploitation to occur, a conserved CS phenotype must emerge when different strains become resistant to a particular drug. Unfortunately, although a few cases of robust CS have been described in model strains (5, 13, 14), this is not a common trait. Indeed, CS phenotypes are rarely conserved when different strains are compared; they differ not just among different isolates of the same species (15, 16), but also when replicated populations of the same strain and evolving in the presence of the same antibiotic are compared (17–20). While in the first case, epistasis and pleiotropy may shape the fitness effects associated with AR acquisition, restricting the type of mutations that can be selected in each genomic background (15, 21–23) and, therefore, the associated CS; in the second situation, genetic drift and population bottlenecks (24) might be responsible for the observed lack of conservation. This lack of conservation when different strains and replicated populations that present the same genetic background are analyzed would be a major drawback for implementing therapies based on the exploitation of CS networks since the emergence of this phenotype will be unpredictable if it is different for each isolate.
As mentioned above, most works in the field have focused on the evolution of model antibiotic-susceptible strains, despite the fact that antibiotic-resistant mutants are frequently encountered in clinics. When an isolate is resistant to one antibiotic, a different one is used for treating the infection that it produces—a common situation in clinics—with a possibility that resistance to this second drug could be selected. If this novel resistance is associated with a robust CS phenotype, this information may lead to prioritization of the use of a third antimicrobial, to which the newly selected resistant strain becomes hypersusceptible. Therefore, the identification of robust CS patterns, conserved in different antibiotic-resistant mutants, is a prerequisite for using this evolutionary trade-off for tackling AR. In the current work, we intend to fill this gap by analyzing the robustness of the CS phenotypes that may emerge in different Pseudomonas aeruginosa antibiotic-resistant mutants, previously identified in adaptive laboratory evolution (ALE) experiments of P. aeruginosa PA14 (13, 21), when they are submitted to short-term ALE experiments in the presence of antimicrobials belonging to different structural and functional categories.
The acquisition of robust CS patterns in different genomic backgrounds—understood as isolates with different genomes—and genetic mutational backgrounds—here understood as strains with the same genome, but presenting genetic changes that might impact AR evolution (21)—can be considered an example of convergent evolution, understood as the acquisition of the same phenotype by different organisms when confronted with similar selection pressures (25). Particularly relevant is the fact that any conserved phenotype emerging in different organisms, from bacteria to humans, as a result of convergent evolution is generally considered to be selected because it improves the adaptation of the evolved organism to the applied selective force (26, 27). Opposite to this situation, CS is not adaptive to the presence of antibiotics; it is a trade-off associated with the selection of the primary adaptive phenotype, which is resistance to the selective antimicrobial. One of the possible reasons behind convergent evolution is parallel evolution, defined as a situation in which the same genetic event responsible for the convergent phenotype (in our case, CS) is selected, irrespective of the genetic background (28). However, although one example of parallel evolution leading to robust CS has been recently reported (29), the genetic events behind CS robustness are not always conserved (30, 31).
P. aeruginosa is a relevant opportunistic pathogen with high prevalence at hospitals and the main cause of chronic infections in cystic fibrosis (CF) patients. It is included in the critical priority list of antibiotic-resistant pathogens (32), as well as in the group of ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter spp.) that comprises six of the organisms for which the development of novel antibiotics/therapeutic options is an urgent need (33). The identification of robust CS networks associated with the use of drugs commonly used to treat infections caused by P. aeruginosa, such as fluoroquinolones, may then be useful.
The fluoroquinolone ciprofloxacin is extensively used to treat a wide range of infections caused by P. aeruginosa (34, 35). The two principal mechanisms of acquisition of ciprofloxacin resistance in this bacterium include mutations in the ciprofloxacin target-encoding genes gyrAB (encoding DNA gyrase) and parCE (encoding DNA topoisomerase IV) (36–40) and the acquisition of mutations leading to overexpression of mexEF–oprN, mexAB–oprM, mexXY, and mexCD-oprJ (34, 38, 41–43), which encode efflux pumps able to extrude ciprofloxacin. Antibiotic-resistant P. aeruginosa clinical isolates often contain multiple ciprofloxacin-resistance mutations in gyrA, gyrB, mexA, mexB, nfxB, mexZ, or mexS (5, 43, 44). Importantly, ciprofloxacin-resistant clinical isolates of P. aeruginosa presenting mutations in gyrA, gyrB, mexB, and nfxB seem to display CS to aminoglycosides (5), which also form part of usual therapies against P. aeruginosa (45). Although the authors suggested that nfxB mutations might be responsible for the observed CS to aminoglycosides, an example of parallel evolution leading to phenotypic convergence, it remains to be firmly established to what extent different ciprofloxacin-resistance mutations, individually or jointly acquired in different mutational backgrounds, as preexisting antibiotic-resistant mutants, may be shaping the robustness of CS to aminoglycosides.
In a previous work, we determined that different antibiotic-resistant mutants of P. aeruginosa PA14 submitted to ALE in the presence of ceftazidime displayed a robust pattern of CS to the aminoglycoside tobramycin. This phenotype is associated with the deletion of large chromosomal regions—also found in isolates from CF patients (46)—containing mexXY, which encodes an intrinsic aminoglycosides-resistance efflux pump (29), hence being an example of parallel evolution. In this work, we go one step further and analyze the robustness of CS associated with short-term ALE assays performed by using preexisting antibiotic-resistant mutants in the presence of ciprofloxacin, tobramycin, or aztreonam, antibiotics widely used for treating P. aeruginosa infections. We found that the different preexisting resistant mutants that evolved under ciprofloxacin challenge present conserved, robust CS to tobramycin and aztreonam. However, unlike the situation in which ceftazidime was the selective force (29), the genetic events selected by ciprofloxacin were different depending on the AR mutational background. The conservation of an emerging CS phenotype in these different antibiotic-resistant mutants provides an example of phenotypic convergence in the absence of parallel evolution that can be exploited to tackle AR. Indeed, based on this information, we rationally designed and experimentally validated in vitro that evolution-based approaches can drive extinction of preexisting antibiotic-resistant mutants of P. aeruginosa, supporting the potential of exploiting CS convergence for tackling P. aeruginosa infections, including those due to antibiotic-resistant mutants.
Results
Evolution of Resistance and CS Associated with Short-Term ALE in the Presence of Tobramycin, Aztreonam, or Ciprofloxacin.
In order to find out robust CS phenotypes, ALE experiments in the presence of antibiotics should be performed, using different antibiotics and in different genetic backgrounds. Indeed, it has been described that the loss-of-function of a single gene, not directly related with AR, modifies the evolutionary trajectories followed by P. aeruginosa PA14 in the presence of antibiotics and their patterns of CS (21), a feature that might compromise CS exploitation. Further, to have a situation closer to that found at clinics, strains to be tested should include antibiotic-resistant mutants, not only a model susceptible strain—an experimental approach that has been rarely addressed. Consequently, we analyzed the evolutionary conservation of CS associated with the short-term use of three different antibiotics—tobramycin, aztreonam, and ciprofloxacin—in antibiotic-resistant P. aeruginosa mutants presenting different AR mutational backgrounds. To such goal, we used a set of well-defined antibiotic-resistant mutants (29), derived from P. aeruginosa PA14 and containing single (nfxB, parR, orfN, and mexZ) and multiple (MDR6 and MDR12) mutations in genes encoding both, regulatory and nonregulatory proteins (Table 1). These different types of mutations were specifically chosen because, besides the fact that they are regularly found in clinical isolates, it has been previously described that AR mutations may lead to either robust or variable CS patterns in different genomic backgrounds, depending on whether they produce “target” or “regulatory” alterations, respectively (16).
Table 1.
Original genetic events of the parR87, orfN50, nfxB177, mexZ43, MDR6, and MDR12 mutational backgrounds
| Mutational background | Gene | Type of genetic modification | Amino acid change |
|---|---|---|---|
| parR87 | parR | SNP | Glu87Lys |
| orfN50 | orfN | Deletion | Val50fs |
| nfxB177 | nfxB | SNP | Phe177Ser |
| mexZ43 | mexZ | SNP | Val43Gly |
| MDR6 | mexC | SNP | Thr267Ala |
| pmrB | SNP | Leu87Gln | |
| frr | SNP | Ile98Ser | |
| phoQ | SNP | Val260Gly | |
| MDR12 | fusA | SNP | Tyr552Cys |
| fusA | SNP | Tyr683Cys | |
| orfN | Deletion | Val50fs | |
| pmrB | SNP | Met46Ile | |
| mexZ | SNP | Val43Gly | |
| gabP | SNP | Ser267Phe | |
| ptsP | SNP | Leu537Pro | |
| nuoC | SNP | Gln184* |
Four biological replicates of each single (nfxB177, parR87, mexZ43, and orfN50) and multiple antibiotic-resistant mutant (MDR6 and MDR12) strain and the wild-type PA14 strain were submitted to ALE in the presence of tobramycin, aztreonam, or ciprofloxacin, or in the absence of antibiotics (control populations) for 3 d (112 populations in total). As expected, a decrease in the susceptibility to each of the three drugs—tobramycin, aztreonam, and ciprofloxacin—was observed in all the populations evolved in their presence, respectively (Fig. 1 and SI Appendix, Tables S1–S3). Statistical analysis was performed by using both nonparametric tests on nontransformed data and parametric tests of log2(FC), where FC is fold change, as described in Materials and Methods. Nonparametric tests were used to assert significance in the differences between each combination of strain treatment with its respective control populations grown in the absence of any drug (SI Appendix, Table S4). Since statistically significant changes may be detected with a magnitude not associated with a relevant change in resistance, in the present study, we have settled for a threshold (above or below 2- or 0.5-fold, respectively) as the indicator of the potential biological significance. Differences surpassing this threshold were statistically validated by using log2(FC)-based parametric tests. In all cases, both types of statistical analyses (parametric and nonparametric) were consistent in detecting significant differences (P < 0.05).
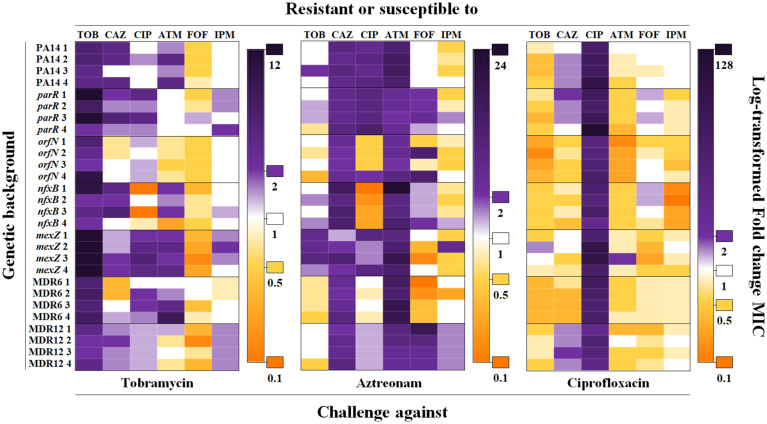
Fig. 1.
Diagram showing robustness of CS to tobramycin and aztreonam in PA14 and in different mutational backgrounds submitted to short-term ALE on ciprofloxacin. Cross-resistance and CS to antibiotics from different structural families were analyzed in PA14 and the mutational backgrounds parR87, orfN50, nfxB177, mexZ43, MDR6, and MDR12 (four replicate populations for each) submitted to ALE in the presence of tobramycin, aztreonam, or ciprofloxacin for 3 d. Intensity of the color is proportional to the log-transformed FC regarding the MIC of the respective parental strain. Since control populations evolved in the absence of antibiotics may present, on rare occasions, subtle changes (below or above 2- or 0.5-fold, respectively) in their susceptibility to antibiotics with respect to the MIC of parental strains, changes in MICs above or below 2- or 0.5-fold, respectively, were considered physiologically relevant to classify a population as “resistant” (purple) or “susceptible” (orange). MIC values (µg/mL) of populations evolved in the presence of tobramycin, aztreonam, or ciprofloxacin are included in SI Appendix, Tables S1–S3, respectively. MIC values (µg/mL) of control populations evolved in the absence of drugs are included in SI Appendix, Table S4. ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; FOF, fosfomycin; IPM, imipenem; TOB, tobramycin.
In the populations submitted to ALE in the presence of tobramycin, the tobramycin minimal inhibitory concentration (MIC) increased up to 4-fold in PA14, 16-fold in parR87, 8-fold in orfN50, 8-fold in nfxB177, 12-fold in mexZ43, 5.3-fold in MDR6, and 3-fold in MDR12 (P < 0.05 in all cases and using all approaches). Under aztreonam exposure, aztreonam MIC increased up to 8-fold in PA14; 2.7-fold in parR87; 4-fold in orfN50 and MDR12; 24-fold in nfxB177; 12-fold in mexZ43; and 12-fold in MDR6 (P < 0.005 in all cases). Finally, ALE challenge with ciprofloxacin increased the MIC to this drug up to 46.9-fold in PA14, 128-fold in parR87, 15.8-fold in orfN50, 16-fold in nfxB177, 48-fold in mexZ43, 24-fold in MDR6, and 10.5-fold in MDR12 (P < 0.005 in all cases). It is worth noting that the greatest increase of AR occurred in parR87 in the presence of tobramycin and ciprofloxacin and nfxB177 in the presence of aztreonam, indicating that some mutational backgrounds are more prone to evolve toward higher levels of resistance than others. The question remains whether these differences are due to the acquisition of different AR mutations or whether the same AR mutations have a different effect depending on the mutational background where they are acquired. Control populations that evolved in the absence of antibiotics generally did not present variations in their susceptibility to antibiotics with respect to the MIC of parental strains, and, in the few cases in which these changes occurred, they were minor, not statistically significant, changes (SI Appendix, Table S4). This feature supports that the changes in MICs (above or below 2- or 0.5-fold, respectively) observed in populations challenged with antibiotics were due to antibiotic selection, not to a nonspecific adaptation to the growth medium.
In order to ascertain the evolutionary conservation of CS associated with the short-term use of tobramycin, aztreonam, or ciprofloxacin in the different mutational backgrounds, MICs to antibiotics from different structural families were determined for each of the final populations and their parental strains, making a total of 714 MICs (SI Appendix, Tables S1–S4). Short-term ALE in the presence of tobramycin or aztreonam resulted in cross-resistance to ceftazidime, ciprofloxacin, and aztreonam, at least in some of the analyzed mutational backgrounds (P < 0.05). The tested populations did not present a clear pattern of CS to other drugs, except for the case of fosfomycin (Fig. 1 and SI Appendix, Tables S1 and S2). In particular, CS to fosfomycin was observed in replicate populations from PA14 and four out of six mutational backgrounds (orfN50, nfxB177, mexZ43, and MDR12) submitted to short-term ALE in the presence of tobramycin (P < 0.05). The possibility of alternating or combining these two antibiotics has been proposed (13, 29, 31, 47, 48) and hence will not be discussed here. Interestingly, no cross-resistance to other drugs was observed in populations submitted to short-term ALE in the presence of ciprofloxacin, while CS to all the analyzed drugs was observed in replicate populations from at least three different mutational backgrounds. It is worth noting the robustness of CS toward two antibiotics (tobramycin and aztreonam), which was detected in replicate populations from PA14 and from six and five out of six mutational backgrounds analyzed, respectively (Fig. 1 and SI Appendix, Table S3). In particular, tobramycin MIC was reduced up to 2-fold in nfxB177, mexZ43, and MDR12; 2.6-fold in PA14; 3-fold in parR87 and MDR6; and 6-fold in orfN50 (P < 0.05 in all cases except in mexZ43). In the case of aztreonam, MIC was reduced up to 2-fold in PA14, nfxB177, and MDR6; 3-fold in parR87 and MDR12; and 6-fold in orfN50 (P < 0.05 in all cases except in MDR12). It is relevant to highlight that the observed CS was not reciprocal; while populations evolved in the presence of ciprofloxacin presented a robust CS to aztreonam and tobramycin, populations challenged with either aztreonam or tobramycin did not present an equivalent CS to ciprofloxacin. Further, while ciprofloxacin-resistant populations did not present cross-resistance to other antibiotics, cross-resistance was frequently observed in aztreonam- and tobramycin-resistant populations. This means that the order in which antibiotics are used might be fundamental for implementing evolution-based strategies to deal with infections and AR.
Evolutionary Strategies Based on Robustness of CS to Drive Extinction of Preexisting P. aeruginosa-Resistant Mutants.
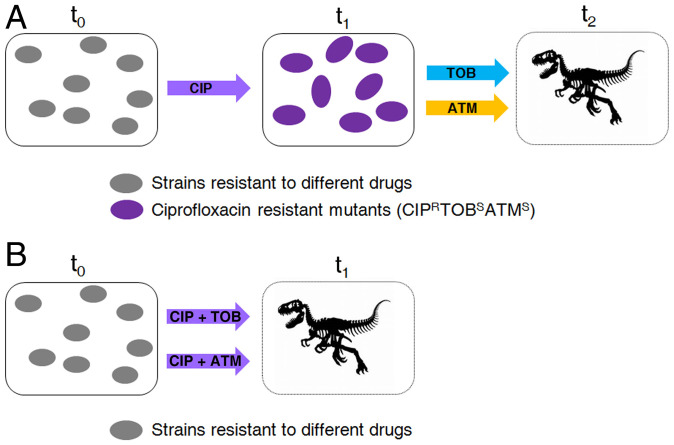
CS to aminoglycosides has been described in clinical strains of P. aeruginosa from CF patients treated with ciprofloxacin (5), and CS to tobramycin and aztreonam has been observed in PAO1 during ALE experiments in the presence of ciprofloxacin (5). Here, we have observed that ciprofloxacin rapidly selects mutants presenting CS to tobramycin and aztreonam in the P. aeruginosa PA14 genomic background. Since this strain presents genomic and physiological differences with PAO1 (49, 50), the conservation of the same CS pattern between them supports its robustness among strains presenting different genomic backgrounds. In addition, we found that the same robust CS phenotype was found when different mutants, already presenting AR mutations, were submitted to ciprofloxacin-selective pressure. Altogether, these findings support the robustness of the observed CS in different strains, including former antibiotic-resistant mutants. Therefore, we tested the possibility of alternating ciprofloxacin with these antibiotics. This strategy consisted of two stages: a first step on ciprofloxacin, which would drive evolution toward CS to tobramycin and aztreonam; and a second step on tobramycin or aztreonam, which would drive extinction of tobramycin–aztreonam-susceptible cells (Fig. 2A). Between treatments, cells were stocked in glycerol. This two-step design of the experiment allowed us to decouple CS (the aim of this work) from potential hysteresis situations (51) that might compromise the interpretation of the results.
Fig. 2.
General model illustrating evolution of antibiotic-resistant mutants of P. aeruginosa submitted to the alternation of ciprofloxacin with tobramycin or aztreonam or the combination of ciprofloxacin with tobramycin or aztreonam. (A) Evolution starts when different antibiotic-resistant mutants are treated with ciprofloxacin at time 0 (t0). Then, there is evolution toward ciprofloxacin resistance and CS to tobramycin and aztreonam (purple cells), rendering ciprofloxacin ineffective (t1). Subsequently, treatment is switched to tobramycin (TOB) or aztreonam (AZT) that may result in the elimination of cells susceptible to tobramycin and aztreonam (t2). (B) Evolution starts when different antibiotic-resistant mutants are treated with a ciprofloxacin–tobramycin or a ciprofloxacin–aztreonam combination at time 0 (t0). Since ciprofloxacin-resistance acquisition leads to CS to tobramycin and aztreonam, it may be expected that drug combinations result in a reduced rate of adaptation or the elimination of cells (t1).
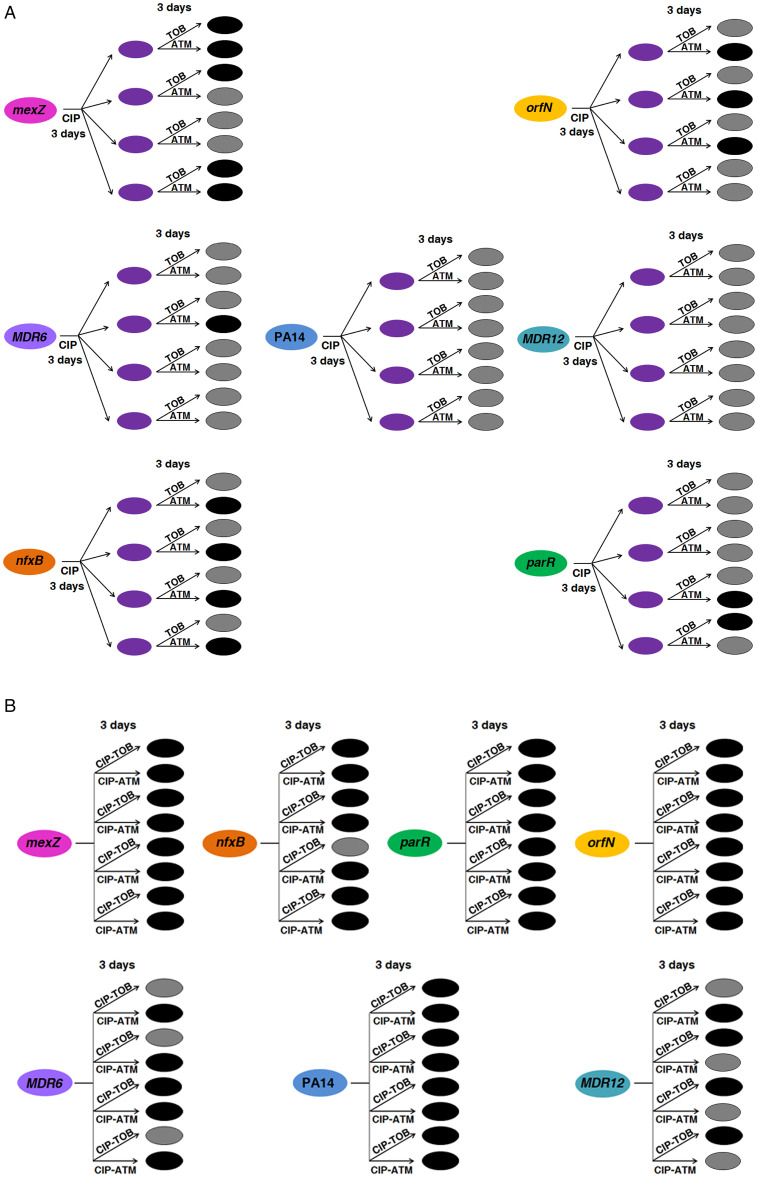
We started with the 28 ciprofloxacin-resistant populations (SI Appendix, Table S3) belonging to PA14 and to six different mutational backgrounds (parR87, orfN50, nfxB177, mexZ43, MDR6, and MDR12), four replicate populations of each, previously submitted to short-term ALE on ciprofloxacin, and 28 control populations (previously unchallenged with ciprofloxacin). As mentioned above, these ciprofloxacin-resistant populations became resistant to ciprofloxacin, being all MICs above the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoint (0.5 µg/mL) (SI Appendix, Table S3), and presented CS to aztreonam and tobramycin, being MICs below the EUCAST clinical breakpoint (16 and 2 µg/mL, respectively), with the exception of tobramycin ones in MDR12 (SI Appendix, Table S3). At this point, we focused on the switch from ciprofloxacin to tobramycin or aztreonam (Fig. 2A). Even though we had observed conservation of CS to tobramycin and aztreonam after evolution in the presence of ciprofloxacin within the analyzed set of mutational backgrounds (Fig. 1 and SI Appendix, Table S3), a critical point would be determining if the proposed strategies could be effective to eliminate cell viability in most of them. Hence, we switched the selective pressure from ciprofloxacin to tobramycin (28 ciprofloxacin-resistant populations and 28 control populations) or aztreonam (28 ciprofloxacin-resistant populations and 28 control populations) (Materials and Methods). As shown in Fig. 3A, 11 out of 28 ciprofloxacin-resistant populations submitted to short-term ALE in the presence of aztreonam became extinct, whereas this only occurred in 4 out of 28 ciprofloxacin-resistant populations submitted to short-term ALE on tobramycin. In both cases, the extinction differences were statistically significant (P < 0.05). These results suggest that exploiting the aztreonam CS associated with the use of ciprofloxacin by switching selective pressure from ciprofloxacin to aztreonam, although a feasible approach, could be ineffective in driving extinction of the model strain PA14 and of some mutational backgrounds, such as MDR12, at least at the concentrations tested.
Fig. 3.
Diagram showing the efficacy of the alternation of ciprofloxacin with tobramycin or aztreonam and the combination of ciprofloxacin with tobramycin or aztreonam for driving extinction of P. aeruginosa PA14 and different antibiotic-resistant mutants. (A) Short-term evolution of PA14 and six mutational backgrounds (nfxB177, parR87, mexZ43, orfN50, MDR6, or MDR12), four replicate populations of each parental strain, was performed during 6 d: 3 d in the presence of ciprofloxacin (CIP) or the absence of antibiotic (control populations), leading to ciprofloxacin-resistant populations (purple cells), and 3 d in the presence of tobramycin (TOB) or aztreonam (AZT). CS to tobramycin and aztreonam was observed in 21 and 19 out of 28 populations after a first step on ciprofloxacin (SI Appendix, Table S3). Populations that were extinct at the end of the experiment are represented in black, while surviving populations are colored in gray. Most of the populations (24 out of 28) submitted to short-term ALE in the presence of tobramycin grew after 3 d. However, short-term ALE in the presence of aztreonam led to extinction of 11 out of 28 populations. This evolutionary strategy was efficient in driving extinction of ciprofloxacin-resistant mutants belonging to nfxB177, parR87, mexZ43, orfN50, and MDR6, but ineffective in driving extinction of PA14 and MDR12. (B) Short-term evolution of PA14 and six mutational backgrounds (nfxB177, parR87, mexZ43, orfN50, MDR6, or MDR12), four replicate populations of each parental strain, was performed during 3 d in the presence of the ciprofloxacin–tobramycin (CIP-TOB) or the ciprofloxacin–aztreonam (CIP-ATM) combination. Growth of the 84 control populations was confirmed in the three drugs independently used at the concentrations present in the drugs combinations. A total of 25 out of 28 populations submitted to short-term ALE in the presence of the ciprofloxacin–aztreonam combination were extinct, while it occurred in 23 out of 28 populations submitted to short-term ALE in the presence of the ciprofloxacin–tobramycin combination. These results indicate that CS may not only improve treatment when drugs are applied sequentially, but it may also serve to optimize combinatory therapy.
It has been suggested that CS may not only improve treatment when drugs are applied sequentially, but it may also serve to optimize combinatory therapy, as it has been described for the combinations ciprofloxacin–aminoglycosides and ciprofloxacin–β-lactams (8). Thus, we decided to analyze the antibiotic-combination efficacy of the ciprofloxacin–tobramycin and ciprofloxacin–aztreonam pairs of drugs in our set of preexisting antibiotic-resistant mutants. We submitted the populations belonging to PA14 and six different mutational backgrounds (parR87, orfN50, nfxB177, mexZ43, MDR6, and MDR12), four replicate populations of each, to the drug combinations ciprofloxacin–tobramycin (28 populations) or ciprofloxacin–aztreonam (28 populations) at the concentrations used for previous ALE assays in the presence of ciprofloxacin, aztreonam, or tobramycin (Fig. 1 and SI Appendix, Tables S1–S3) (Materials and Methods). As shown in Fig. 3B, 23 and 25 out of 28 populations submitted to short-term ALE in the presence of either ciprofloxacin–tobramycin or ciprofloxacin–aztreonam became extinct. In both cases, the extinction differences were statistically significant (P < 0.05). To further analyze if the high efficiency of these combinatory therapies could be influenced by a synergistic antibiotic interaction between the antibiotics used, we performed 14 checkerboard analyses (Materials and Methods) for PA14 and the six mutational backgrounds here analyzed and the two pairs of drugs tested. We did not observe synergy (neither antagonism) between ciprofloxacin and tobramycin or between ciprofloxacin and aztreonam in any of the genetic backgrounds analyzed (fraction inhibitory concentration [FIC] index ≥ 0.5 and ≤ 4 in all cases) (SI Appendix, Table S5). This observation, in addition to the above-mentioned results (Fig. 3B), reinforces that CS to tobramycin and aztreonam associated with the use of ciprofloxacin optimizes the efficiency of the ciprofloxacin–tobramycin and ciprofloxacin–aztreonam pairs of drugs. These results suggest that exploiting the CS associated with the use of ciprofloxacin is a possibility that should be considered, particularly in the case of preexisting antibiotic-resistant mutants, a feature in agreement with previous data indicating that quinolone-resistant subpopulations isolated from CF patients may be eradicated using aminoglycoside and β-lactam drugs (5).
Genetic Variations Associated with both Ciprofloxacin Resistance and CS in P. aeruginosa PA14 and in Different Antibiotic-Resistant Mutants.
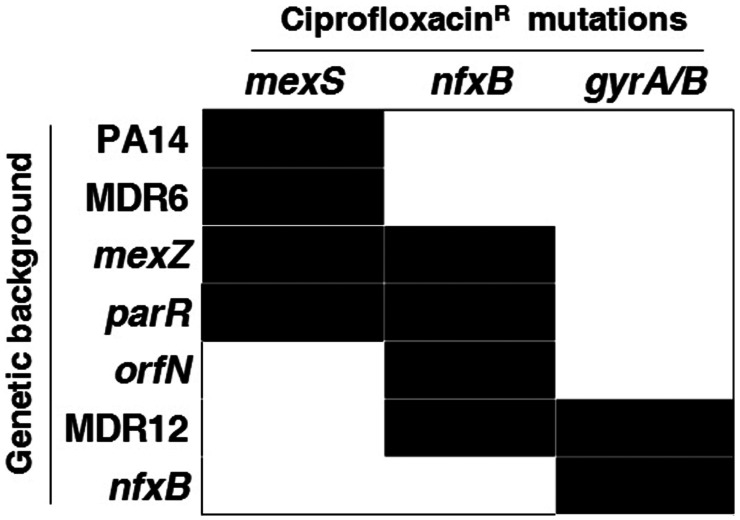
To gain insights into the genetic causes of the acquisition of ciprofloxacin resistance and the robust CS pattern observed in the populations submitted to short-term ALE in the presence of ciprofloxacin, the 28 populations independently evolved during 3 d and their parental strains (PA14, parR87, orfN50, nfxB177, mexZ43, MDR6, and MDR12) were subjected to whole-genome sequencing. Control populations evolved in the absence of drugs were not whole-genome-sequenced since they did not present any relevant change in the MICs of the tested antibiotics. The genome of the 28 ciprofloxacin-resistant populations was compared to the ones of their respective parental strains in order to determine newly genetic events acquired during evolution. A total of 50 genetic events were identified (Table 2): 8 in PA14, 8 in parR87, 5 in orfN50, 4 in nfxB177, 11 in mexZ43, 9 in MDR6, and 5 in MDR12. All detected gene variants were located just within five different genes, and their acquisition was dependent on the genetic background (Table 2): mexS variants were acquired in PA14, parR87, mexZ43, and MDR6; nfxB variants were acquired in parR87, orfN50, mexZ43, and MDR12; gyrAB variants were acquired in nfxB177 and MDR12; and an orfN variant was acquired in mexZ43.
Table 2.
Newly acquired genetic events after ciprofloxacin ALE by replicates of PA14 and of parR87, orfN50, nfxB177, mexZ43, MDR6, and MDR12 mutational backgrounds
| Mutational background | Gene | Position | Genetic change | Nucleotide location* | Amino acid change | Replicate | Coverage,%† |
|---|---|---|---|---|---|---|---|
| PA14 | mexS | 2820352 | SNP | 379T > G | Tyr127Asp | 1, 2, 3, 4 | 43, 28, 23, 50 |
| mexS | 2820074 | SNP | 101G > C | Arg34Pro | 1, 2, 3 | 20, 10, 14 | |
| mexS | 2820118 | SNP | 145C > T | Gln49* | 3 | 17 | |
| parR87 | mexS | 2820695 | SNP | 722T > A | Val241Glu | 1, 2 | 17 |
| mexS | 2820327 | Deletion | 355delA | Thr119fs | 2, 3, 4 | 86, 10, 23 | |
| mexS | 2820695 | SNP | 722T > G | Val241Gly | 2 | 23 | |
| nfxB | 5428144 | SNP | 115A > C | Thr39Pro | 1, 3 | 39, 18 | |
| nfxB | 5428448 | Deletion | 421delG | Ala141fs | 4 | 80 | |
| orfN50 | nfxB | 5428228 | SNP | 199C > T | Gln67* | 1, 2, 4 | 16, 37, 87 |
| nfxB | 5428148 | SNP | 119T > G | Leu40Arg | 2 | 49 | |
| nfxB | 5428091 | SNP | 62A > C | His21Pro | 3 | 100 | |
| nfxB177 | gyrB | 5671 | SNP | 1397C > T | Ser466Phe | 1, 2 | 99, 95 |
| gyrA | 2015001 | SNP | 248C > T | Thr83Ile | 3 | 84 | |
| gyrA | 2015289 | SNP | 536C > T | Ala179Val | 4 | 93 | |
| mexZ43 | mexS | 2820463 | SNP | 490C > T | Gln164* | 1 | 36 |
| mexS | 2820619 | SNP | 646A > C | Asn216His | 2, 3 | 10, 31 | |
| mexS | 2820782 | SNP | 809T > G | Leu270Arg | 2, 4 | 10, 23 | |
| mexS | 2820380 | SNP | 407G > T | Gly136Val | 2, 3, 4 | 10, 11, 21 | |
| mexS | 2820020 | SNP | 47T > C | Leu16Pro | 4 | 15 | |
| orfN | 2040286 | Insertion | 148insG | Val50fs | 2 | 79 | |
| nfxB | 5428091 | SNP | 62A > C | His21Pro | 2 | 26 | |
| MDR6 | mexS | 2820797 | SNP | 824G > C | Gly275Ala | 1, 2, 3, 4 | 38, 28, 11, 36 |
| mexS | 2820239 | SNP | 266T > C | Phe89Ser | 1, 2, 4 | 30, 29, 28 | |
| mexS | 2820529 | SNP | 556C > T | Leu186Phe | 3 | 22 | |
| mexS | 2820758 | SNP | 785A > T | His262Leu | 3 | 20 | |
| MDR12 | gyrA | 2015013 | SNP | 260A > G | Asp87Gly | 1, 2, 3, 4 | 18, 100, 100, 100 |
| nfxB | 5428091 | SNP | 62A > C | His21Pro | 1 | 31 |
Del, deletion; fs, frameshift; ins, insertion.
*Nucleotide location of the mutations referred to the specific gene in which they are located and their associated amino acid changes.
†The coverage indicates the percentage of reads of each mutant allele in the total number of reads, corresponding to the same region of the genome, within each population at the end of the ALE assay.
Despite orfN mutations having been described in ALE assays in the presence of ciprofloxacin (52), only a single replicate of mexZ43 presented a mutation in this gene. Instead, the most prevalent mutations were found in gyrAB, encoding the quinolones’ target (36–40), in mexS, encoding a regulator of the expression of mexEF–oprN, and in nfxB, encoding a regulator of the expression of mexCD–oprJ, which may lead to overexpression of these efflux pumps (34, 38, 41–43) (Table 2 and Fig. 4). Importantly, P. aeruginosa isolates from chronically infected CF patients treated with ciprofloxacin frequently present mutations in these genes (5, 43, 44). In particular, we identified 15 different variants of mexS, 5 different variants of nfxB, 3 different gyrA variants, and a single gyrB variant. Notably, variant NfxBHis21Pro was acquired in three out of the four mutational backgrounds presenting mutations in this gene, suggesting an important phenotypic impact, and variants NfxBThr39Pro, GyrAThr83Ile, GyrAAsp87Gly, and GyrBSer466Phe have already been described as clinically relevant (34, 36, 40, 43), further supporting our experimental approach.
Fig. 4.
Diagram showing genetic causes of ciprofloxacin-resistance acquisition and mutational background dependence. Classical mutations regularly found in clinical isolates from patients treated with ciprofloxacin, within gyrAB, nfxB, and mexS, were acquired (black boxes) during ALE in the presence of ciprofloxacin for 3 d in 28 populations belonging to PA14 and 6 different mutational backgrounds (parR87, orfN50, nfxB177, mexZ43, MDR6, and MDR12). We identified 15, 5, 3, and 1 different variants of mexS, nfxB, gyrA, and gyrB, respectively. As observed, early steps of ciprofloxacin-resistance evolution are dependent on the mutational background (Table 2).
Variants in which an isoleucine replaces the threonine at position 83 of GyrA are among the most frequent alterations associated with ciprofloxacin resistance in P. aeruginosa occurring in clinical and in vitro-selected resistant mutants (34, 36, 38–40, 53–55). The second most frequent variant in GyrA is the replacement of aspartate at position 87 with asparagine, glycine, or tyrosine residues (34, 36, 38–40, 43, 55). It is important to notice that, while new mutations were acquired in nfxB, a gene encoding a regulator of mexCD–oprJ expression, in parR87, orfN50, mexZ43, and MDR12, target mutations in gyrAB were the only ones acquired in nfxB177, and, with the exception of MDR12, no mutations were found in genes encoding the quinolone targets in the other mutational backgrounds (Table 2 and Fig. 4). It is remarkable noting that MDR12 originally presented a mutation in mexZ, which encodes the negative regulator of the expression of the mexXY efflux pump encoding genes (56). Altogether, these results suggest that mutations in regulators of the expression of efflux pumps may precede target mutations in the acquisition of quinolones’ resistance, something that has already been reported in clinical strains of patients treated with ciprofloxacin, in which mutations in nfxB were generally acquired early in the lineage evolution and were less persistent than mutations in gyrAB (57). According to what is found in clinical strains presenting mutations in both target-encoding genes and genes encoding negative regulators of efflux pumps (40, 58), the ciprofloxacin resistance level of the ciprofloxacin-resistant nfxB177 populations is higher than the ones of ciprofloxacin-resistant populations selected from other mutational backgrounds (SI Appendix, Table S3).
In this work, we describe a conserved CS to tobramycin in the populations submitted to short-term ALE on ciprofloxacin (Fig. 1). This phenotypic convergence toward CS to aminoglycosides has been described in ciprofloxacin-resistant clinical isolates of P. aeruginosa presenting mutations in gyrA, gyrB, mexB, and nfxB (5). In agreement with the findings of this work, our results indicate that mutations in nfxB are clearly associated with CS to tobramycin. This is easily deduced from the ciprofloxacin-resistant orfN50 populations, which only acquired mutations in nfxB and whose tobramycin MIC was reduced up to sixfold, with respect to the parental strain orfN50. To further analyze these genotype–phenotype relationships, individual clones, with each one presenting single mutations in either nfxB, mexS, gyrA, or gyrB, were isolated from the ciprofloxacin-resistant populations as described in Materials and Methods, and their susceptibility to antibiotics was determined. In particular, a NfxBHis21Pro ciprofloxacin-resistant mutant was isolated from an orfN50 population (replicate 3). Confirming our hypothesis, the mutant presents a reduction of tobramycin MIC, from 3 to 0.75 µg/mL. Moreover, a ciprofloxacin-resistant mutant containing the single-mutation NfxBAla141fs was isolated from a parR87 population (replicate 4), presenting a reduction of tobramycin MIC, from 1.5 to 0.5 µg/mL These results indicate that nfxB mutations cause CS to tobramycin (at least the ones selected in orfN50 and parR87). Further, mutations in mexS have been associated with an increased susceptibility to aminoglycosides and β-lactams (59). Taking into consideration those observations, mutations in mexS are likely responsible for the CS to tobramycin observed in PA14 and MDR6. This conclusion is further supported by the fact that the ciprofloxacin-resistant populations from these mutational backgrounds only acquired mutations in mexS and showed a 2.6-fold and 3-fold reduction of tobramycin MIC, respectively. To further analyze this possibility, a ciprofloxacin-resistant mutant containing the single-mutation MexSThr119fs was isolated from the parR87 replicate population 4. The mutant presents a reduction of tobramycin MIC, from 1.5 to 0.75, confirming the role of the mexS mutation in tobramycin CS. In the case of ciprofloxacin-resistant populations derived from nfxB177, which only acquired mutations in gyrAB, their tobramycin MICs were reduced in twofold, with respect to the parental strain, indicating that mutations in gyrAB may as well be responsible for tobramycin CS in this mutational background. Two different ciprofloxacin-resistant mutants containing the single-mutations GyrAThr83Ile or GyrBSer466Phe were isolated from two different nfxB177 populations (replicate 3 and 1, respectively), presenting a reduction of tobramycin MIC from 1 to 0.5. This denotes that phenotypic convergence toward CS to aminoglycosides in clinical isolates from patients treated with ciprofloxacin may be associated with the selection of mutations in different genes that lead to an increased efflux of antibiotics or to alterations in drug targets, although the strength of SC to aminoglycosides associated with nfxB mutations is considerably higher than that of mutations in the other genes. Besides tobramycin CS, we observed that CS to aztreonam is also associated with the acquisition of resistance to ciprofloxacin in the different mutational backgrounds tested. In this regard, the contribution of mexS, nfxB, gyrA, or gyrB mutations in CS to aztreonam is observed in the mutants MexSThr119fs, NfxBAla141fs, NfxBHis21Pro, GyrAThr83Ile, and GyrBSer466Phe isolated from ciprofloxacin-resistant populations derived from parR87, orfN50, and nfxB177. The NfxBHis21Pro ciprofloxacin-resistant mutant isolated from the orfN50 population presented a reduction of aztreonam MIC from 6 to 1.5 µg/mL, and the NfxBAla141fs ciprofloxacin-resistant mutant isolated from the parR87 population presented a reduction of aztreonam MIC from 3 to 0.5 µg/mL, indicating that nfxB mutations cause CS to aztreonam (at least the ones of orfN50 and parR87). The ciprofloxacin-resistant mutant isolated from the parR87 population containing MexSThr119fs as a single mutation presented a reduction of aztreonam MIC from 3 to 1 µg/mL, indicating that in this mutational background—at least—this mutation leads to CS to this antibiotic. Finally, the GyrAThr83Ile and GyrBSer466Phe ciprofloxacin-resistant mutants isolated from two different nfxB177 populations presented a reduction of aztreonam MIC from 2 to 1 µg/mL, indicating that gyrA and gyrB mutations cause CS to aztreonam (at least the ones selected in nfxB177), but that the strength of this phenotype is considerably lower than the ones associated with mexS and nfxB mutations.
It is worth emphasizing that the mutations that have been selected during our ALE experiments in the presence of ciprofloxacin are the most frequently found in patients infected by P. aeruginosa and treated with ciprofloxacin (34, 36, 38–40, 53–55, 57) and that mutations in nfxB, the ones that present a higher CS to tobramycin and aztreonam, are generally early acquired during ciprofloxacin treatment (57), supporting the potential impact of the results reported in the present work, regarding robustness of CS in clinical settings.
Discussion
Phenotypic convergence is a common process in evolution that may be the consequence of parallel evolution: The same genetic events are selected when organisms confront the same selective force (60). In this regard, we have recently described that parallel evolution underlies the acquisition of a robust pattern of CS to certain antibiotics, associated with the short-term ALE in the presence of ceftazidime in different mutational backgrounds of P. aeruginosa (29). However, besides being the result of parallel evolution, phenotypic convergence can also emerge, even when different genetic events, giving up to the same phenotype, are selected during evolution (61). In this case, it is usually argued that the emergent, conserved phenotype is selected because it is adaptive to the selective pressure, despite different genetic solutions being taken to deal with the challenge. However, it has been discussed that, on occasion, the observed convergent phenotypes do not provide direct adaptation to the selective force (62), and CS, a phenotype that is maladaptive to the presence of antibiotics, is an example of this situation. We have identified a robust, convergent CS pattern in different antibiotic-resistant mutants, associated with the use of ciprofloxacin, which is not caused by parallel evolution. In particular, we observed the selection of mutations in different genes driven by the same selective force (ciprofloxacin) in different mutational backgrounds, leading to a functionally equivalent change in susceptibility to second drugs (tobramycin and aztreonam). The current study describes a robust pattern of CS associated with the use of a specific drug that is not caused by parallel evolution in different genetic backgrounds, including preexisting antibiotic-resistant mutants.
Parallel evolution leading to the emergence of a conserved maladaptive phenotype was proposed by Haldane (63), in order to explain the high prevalence of some inherited diseases. The hypothesis, which was later experimentally validated (64), is that the selective force behind the unexpectedly high prevalence of those inherited diseases was infection; the selected mutations protect from infection, and the inherited disease was just the associated, maladaptive, trade-off. However, when parallel evolution is not the cause of convergent evolution, phenotypic convergence is usually explained as a mechanism of adaptation to the selective force (26, 27); a maladaptive, convergent phenotype is not expected to be selected. Differing from this situation, CS is maladaptive to antibiotic challenge; it is an evolutionary trade-off, whose emergence is hardly inferred, even when the selective force underlying evolution is known. Our results hence provide an example of phenotypic convergence (CS) unlinked to parallel evolution and not providing direct adaptation to the antibiotic-selective force. Besides the relevance concerning AR, our findings have consequences for understanding processes of phenotypic convergence without a clear link between selection and adaptation (62).
As stated above, the exploitation of the information concerning CS requires this phenotype to be conserved among different strains; otherwise, the phenotype will be unpredictable and with no use in clinics. Despite efforts in the field, few cases of robust CS have been reported, and, with the exception of one recent study from our laboratory (29), none of them has been performed using well-defined antibiotic-resistant mutants as a starting point. We believe that this aspect is relevant because infections by antibiotic-resistant organisms are common, and they need to be treated with another different drug, which can lead to the selection of additional resistance phenotypes. Knowing if the novel resistance concurs with a robust CS phenotype, shared in different resistant mutants, is needed to implement evolution-based strategies to fight multidrug resistance. Herein, we found a robust CS phenotype to aztreonam and tobramycin, associated with the acquisition of ciprofloxacin resistance by different P. aeruginosa mutants, formerly resistant to antibiotics, which constitutes an important step forward in this endeavor.
The exploitation of in vitro-generated knowledge regarding AR mutations requires that these mutations are also present in clinical isolates. Notably, mutations in the same genes reported in the present article to be involved in ciprofloxacin resistance, such as mutations in target-encoding genes gyrAB or in efflux pumps’ regulators encoding genes mexS and nfxB, have been ascertained in P. aeruginosa clinical isolates from patients treated with ciprofloxacin (5, 43, 44). Further, variants of clinical relevance, such as NfxBThr39Pro, GyrAThr83Ile, GyrAAsp87Gly, or GyrBSer466Phe (34, 36, 40, 43), were acquired in the populations submitted to short-term ALE in the presence of ciprofloxacin during this study, indicating that our results are not only close to reality, in terms of clinical relevance, but that these clinically relevant mutations might be associated with CS to tobramycin and aztreonam in clinical isolates, a feature that has not been addressed yet. This led us to consider the alternation of ciprofloxacin with tobramycin or aztreonam and the use of a ciprofloxacin–tobramycin or a ciprofloxacin–aztreonam combination. We observed that 25 out of 28 populations (belonging to PA14 and 6 different preexisting antibiotic-resistant mutants) challenged with the ciprofloxacin–aztreonam combination and 23 out of 28 populations challenged with the ciprofloxacin–tobramycin combination were extinct after 3 d of short-term ALE. This is in agreement with a previous work performed with the PA14 model strain of P. aeruginosa that pointed out the efficacy of the combination of drug pairs containing ciprofloxacin and aminoglycosides or β-lactams (8). In addition, we observed that 11 out of 28 populations were also extinct after the alternation of ciprofloxacin with aztreonam. Therefore, our results support that it might be possible to exploit CS associated with ciprofloxacin-resistance acquisition to design evolution-based approaches to tackle P. aeruginosa infections containing preexisting antibiotic-resistant mutants. In this regard, it is worth mentioning that blind—not evolutionary-based—clinical trials of antibiotic combinations for fighting P. aeruginosa infections are regularly performed (65), while rational evolutionary-based information, as the results presented here, supporting these trials are generally absent. However, we are aware that testing the proposed treatments in different infection models, as well as confirming the robustness of CS in clinical strains of P. aeruginosa, presenting different genomic backgrounds, not just different mutational backgrounds, would also be needed for the translation of the results into clinical practice. Besides, the fact that the acquisition of quinolone resistance, when due to overexpression of MDR efflux pumps, can be associated with cross-resistance to other drugs (66) and that, on occasion, growing in biofilms may hide the CS phenotype (67) should also be taken into consideration.
We conclude that robust CS patterns may result not just from parallel evolution, as we recently described (29), but also upon the selection of different genetic mechanisms, as we describe here. We therefore propose that the search for robust CS patterns in different genetic backgrounds (including antibiotic-resistant isolates) of a species could pave the way for the design of new evolutionary strategies to promote the extinction of bacteria causing infections. The finding of these robust CS networks is particularly relevant in the case of already-resistant mutants challenged with another antibiotic, a situation rarely explored until now, which is explored here and that is rather common and highly relevant in clinical settings.
Materials and Methods
Growth Conditions and Antibiotic-Susceptibility Assays.
Bacteria were grown in glass tubes in Luria–Bertani (LB) broth at 37 °C with shaking at 250 rpm. MICs of ceftazidime, aztreonam, imipenem, tobramycin, ciprofloxacin, and fosfomycin were determined at 37 °C in Mueller Hinton (MH) agar using E-test strips (MIC Test Strip, Liofilchem).
Short-Term ALE Experiments in the Presence of Ciprofloxacin, Tobramycin, or Aztreonam.
Four single mutants, two multiple mutants, and PA14 were subjected to short-term ALE—four replicates of each—in the presence of ciprofloxacin, tobramycin, or aztreonam or the absence of antibiotic (control populations), resulting in a total of 112 independent bacterial populations. Cultures were grown at 37 °C and 250 rpm for 3 d in independent glass tubes to avoid cross-contamination. Every day, the cultures were diluted (1/100), adding 10 µL of bacteria in 1 mL of fresh LB containing the concentration of antibiotic (close to MIC) that hinders the growth of P. aeruginosa PA14 and each mutational background under these culture conditions (ciprofloxacin: 0.1 µg/mL for MDR6; 0.2 µg/mL for MDR12; 0.3 µg/mL for PA14, orfN50, nfxB177, and mexZ43; and 0.4 µg/mL for parR87; tobramycin: 0.75 µg/mL for PA14 and nfxB177; 1.5 µg/mL for MDR6 and mexZ43; 2 µg/mL for orfN50; 2.5 µg/mL for parR87; and 12 µg/mL for MDR12; aztreonam: 1 µg/mL for MDR12; 4 µg/mL for PA14, nfxB177, and MDR6; 6 µg/mL for parR87; 7 µg/mL for orfN50; and 8 µg/mL for mexZ43) or without antibiotics (control populations). During the 3 d, the concentration of ciprofloxacin, tobramycin, or aztreonam was maintained. Every replicate population was preserved at −80 °C at the end of the experimental evolution. In addition, the MIC of the antibiotic used for selection and of antibiotics from other structural families was determined at 37 °C in MH agar using E-test strips.
Alternation of Ciprofloxacin with Tobramycin or Aztreonam Using Short-Term ALE Experiments.
Short-term ALE experiments in the presence of tobramycin or aztreonam were performed for 3 d at 37 °C and 250 rpm using the 28 populations previously challenged with ciprofloxacin for 3 d and belonging to PA14 and six different mutational backgrounds (parR87, orfN50, nfxB177, mexZ43, MDR6, and MDR12). The 28 ciprofloxacin-resistant populations and the 28 control populations (not challenged with ciprofloxacin) were grown from glycerol stocks, and every day, during 3 d, the cultures were diluted (1/100) in fresh LB containing tobramycin or aztreonam at the concentration that hinders—but allows—the growth of each P. aeruginosa mutational background under these culture conditions, which are described in Short-Term ALE Experiments in the Presence of Ciprofloxacin, Tobramycin, or Aztreonam. Extinction of the populations was determined by measuring the absorbance OD600nm of 100 μL of bacterial cultures the last day of ALE in a 96-well microtiter plate (NUNC) in a Tecan Infinite 200 plate reader and by plating out final cultures on LB to look for viable cells.
Combination of Ciprofloxacin with Tobramycin or Aztreonam Using Short-Term ALE Experiments.
Four replicate populations from PA14 and six different mutational backgrounds (parR87, orfN50, nfxB177, mexZ43, MDR6, and MDR12) were grown from glycerol stocks. Every day, during 3 d, the cultures were diluted (1/100) in fresh LB medium containing a combination of ciprofloxacin–tobramycin (28 populations), ciprofloxacin–aztreonam (28 populations), or each single drug (84 control populations). Each antibiotic was added at the concentration that hinders—but allows—the growth of each P. aeruginosa mutational background under these culture conditions, which are described in Short-Term ALE Experiments in the Presence of Ciprofloxacin, Tobramycin, or Aztreonam. Extinction of the populations was determined by measuring the absorbance OD600nm of 100 μL of bacterial cultures the last day of ALE in a 96-well microtiter plate (NUNC) in a Tecan Infinite 200 plate reader and by plating out final cultures on LB to look for viable cells.
Whole-Genome Sequencing and Analysis of Genetic Changes.
The genomic DNA of each ciprofloxacin-resistant population and parental strain was extracted with the Gnome DNA kit (MP Biomedicals). The assay of DNA quality, libraries’ construction, and whole-genome sequencing was performed by Macrogen. Pair-end libraries (2 × 150) were constructed with Truseq DNA PCR-free and sequenced by using an Illumina NovaSeq6000 system. Coverage was greater than 300x for all samples. Genome sequence, gene coordinates, and annotations were obtained from the nucleotide database GenBank. The quality of Illumina short reads was verified by using FASTQC (68). RNA-STAR was used to align reads against P. aeruginosa genome UCBPP-PA14 (NC_008463.1) (69). Optical and PCR duplicates were detected by using the MarkDuplicates (Picard) function of The Genome Analysis Toolkit (70). SAMtools was used to index alignment files in BAM format (71). Single-nucleotide polymorphism (SNPs) and small insertions and deletions (INDELs) were detected by using freebayes (72). The impact of SNPs and INDELs was evaluated by using SnpEff (73), and annotated results were saved in the VCF format. Genetic variants were detected by using SNPer viewer (74) and the IGV browser (75).
Isolation of Ciprofloxacin-Resistant Mutants.
Clones presenting either of the single mutations NfxBAla141fs, NfxBHis21Pro, MexSThr119fs, GyrAThr83Ile, or GyrBSer466Phe were isolated from the ciprofloxacin-resistant populations parR87.4, orfN50.3, parR87.4, nfxB177.3, or nfxB177.1, respectively. The presence or absence of mutations in the quinolone-resistance-determining regions of gyrA and gyrB in mexS or in nfxB were searched by PCR in each individual clone by using the oligonucleotides described in SI Appendix, Table S5 and Sanger sequencing. Four pairs of primers, which amplify 378 bp of gyrA, 511 bp of gyrB, 627 bp of nfxB, and 1,076 bp of mexS, were used (SI Appendix, Table S6). After PCR, the amplicons were purified by using the QIAquick PCR purification kit (QIAGEN) and Sanger sequenced at Macrogen.
Synergy Assessed by Checkerboard Analysis.
Standard checkerboard broth microdilution assays were performed in PA14 and in six different mutational backgrounds (parR87, orfN50, nfxB177, mexZ43, MDR6, and MDR12) using 10 serially diluted concentrations of ciprofloxacin, seven of aztreonam or tobramycin, and a no-drug control, a total of 14 96-U-well plates. First, 90 μL of MH medium with ciprofloxacin (0.025 to 12.8 μg/mL), aztreonam (1 to 64 μg/mL), or tobramycin (0.125 to 8 μg/mL) was added to each well of two different 96-U-well plates for the analysis of each mutational background, with the exception of MDR12, for which tobramycin concentrations ranged between 4 and 256 μg/mL Ten microliters of cells were inoculated into each well to a final OD600 of 0.01. Bacteria were grown at 37 °C for 48 h without sacking. The FIC of ciprofloxacin, aztreonam, and tobramycin was calculated as the MIC of the combination of ciprofloxacin with each of the other drugs divided by the MIC of each of the drugs alone. The FIC index was calculated as the addition of the FICs of both drugs. An FIC index value of <0.5 was considered to indicate synergy, and an FIC index of >4 was considered to indicate antagonism (76).
Statistical Analysis.
Bidirectional nonparametric tests on the raw data and parametric bidirectional and unidirectional tests on log2-transformed data were performed.
Data were first subjected to Sapiro–Wilk and Levene’s tests to assert normality and homocedasticity. Subsequently, each combination of strain and treatment was compared against its respective control. Nonparametric Kruskal–Wallis, Dwass–Steel–Critchlow–Fligner, and Mann–Whitney U tests were applied to the untransformed data to assert significance of the difference. Additionally, FC was calculated, and the significance of the FCs observed was asserted by using ANOVA and bidirectional t tests on log2-transformed data (log2 FC). The significance of the variation in the number of extinct populations in alternated and combined evolution experiments was estimated by using a likelihood-ratio test. All statistical analyses were performed by using the R package (https://www.R-project.org/).
Supplementary Material
Acknowledgments
We thank our colleagues and friends Fernando Baquero, Jesús Blázquez, and Fernando Rojo for carefully reading the manuscript and providing useful comments for its improvement. This work was supported by Instituto de Salud Carlos III Grant RD16/0016/0011—cofinanced by the European Development Regional Fund “A Way to Achieve Europe”; by Grant S2017/BMD-3691 InGEMICS-CM, funded by Comunidad de Madrid (Spain) and European Structural and Investment Funds; by MCIN/AEI/10.13039/501100011033 (PID2020-113521RB-I00); and by the Spanish Ministry of Economy and Competitivity (BIO2017-83128-R). P.L. is recipient of an Formación de Profesorado Universitario fellowship from the Spanish Ministry of Economy and Competitivity. We thank Juan C. Oliveros, from Bioinformatics for Genomics and Proteomics Service of Centro Nacional de Biotecnología, for his support in the whole-genome sequencing analysis.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.O. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109370119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Andersson D. I., et al. , Antibiotic resistance: Turning evolutionary principles into clinical reality. FEMS Microbiol. Rev. 44, 171–188 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Laxminarayan R., Antibiotic effectiveness: Balancing conservation against innovation. Science 345, 1299–1301 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Szybalski W., Bryson V., Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J. Bacteriol. 64, 489–499 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imamovic L., Sommer M. O., Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 5, 204ra132 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Imamovic L., et al. , Drug-driven phenotypic convergence supports rational treatment strategies of chronic infections. Cell 172, 121–134.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S., Lieberman T. D., Kishony R., Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc. Natl. Acad. Sci. U.S.A. 111, 14494–14499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbosa C., Römhild R., Rosenstiel P., Schulenburg H., Evolutionary stability of collateral sensitivity to antibiotics in the model pathogen Pseudomonas aeruginosa. eLife 8, e51481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbosa C., Beardmore R., Schulenburg H., Jansen G., Antibiotic combination efficacy (ACE) networks for a Pseudomonas aeruginosa model. PLoS Biol. 16, e2004356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munck C., Gumpert H. K., Wallin A. I., Wang H. H., Sommer M. O., Prediction of resistance development against drug combinations by collateral responses to component drugs. Sci. Transl. Med. 6, 262ra156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jahn L. J., et al. , Compatibility of evolutionary responses to constituent antibiotics drive resistance evolution to drug pairs. Mol. Biol. Evol. 38, 2057–2069 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baym M., Stone L. K., Kishony R., Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351, aad3292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pál C., Papp B., Lázár V., Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 23, 401–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanz-García F., Hernando-Amado S., Martínez J. L., Mutational evolution of Pseudomonas aeruginosa resistance to ribosome-targeting antibiotics. Front. Genet. 9, 451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanz-García F., Hernando-Amado S., Martínez J. L., Mutation-driven evolution of Pseudomonas aeruginosa in the presence of either ceftazidime or ceftazidime-avibactam. Antimicrob. Agents Chemother. 62, e01379-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apjok G., et al. , Limited evolutionary conservation of the phenotypic effects of antibiotic resistance mutations. Mol. Biol. Evol. 36, 1601–1611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knopp M., Andersson D. I., Predictable phenotypes of antibiotic resistance mutations. MBio 9, ••• (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podnecky N. L., et al. , Conserved collateral antibiotic susceptibility networks in diverse clinical strains of Escherichia coli. Nat. Commun. 9, 3673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbosa C., et al. , Alternative evolutionary paths to bacterial antibiotic resistance cause distinct collateral effects. Mol. Biol. Evol. 34, 2229–2244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lázár V., et al. , Bacterial evolution of antibiotic hypersensitivity. Mol. Syst. Biol. 9, 700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lázár V., et al. , Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat. Commun. 5, 4352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernando-Amado S., Sanz-García F., Martínez J. L., Antibiotic resistance evolution is contingent on the quorum-sensing response in Pseudomonas aeruginosa. Mol. Biol. Evol. 36, 2238–2251 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Vogwill T., Kojadinovic M., MacLean R. C., Epistasis between antibiotic resistance mutations and genetic background shape the fitness effect of resistance across species of Pseudomonas. Proc. Biol. Sci. 283, 20160151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schenk M. F., de Visser J. A., Predicting the evolution of antibiotic resistance. BMC Biol. 11, 14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes D., Andersson D. I., Evolutionary trajectories to antibiotic resistance. Annu. Rev. Microbiol. 71, 579–596 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Sackton T. B., Clark N., Convergent evolution in the genomics era: New insights and directions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi E., et al. , Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. 19, 331–342 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Witt K. E., Huerta-Sánchez E., Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood T. E., Burke J. M., Rieseberg L. H., Parallel genotypic adaptation: When evolution repeats itself. Genetica 123, 157–170 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernando-Amado S., Sanz-García F., Martínez J. L., Rapid and robust evolution of collateral sensitivity in Pseudomonas aeruginosa antibiotic-resistant mutants. Sci. Adv. 6, eaba5493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roemhild R., Linkevicius M., Andersson D. I., Molecular mechanisms of collateral sensitivity to the antibiotic nitrofurantoin. PLoS Biol. 18, e3000612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laborda P., Martínez J. L., Hernando-Amado S., Convergent phenotypic evolution towards fosfomycin collateral sensitivity of Pseudomonas aeruginosa antibiotic-resistant mutants. Microb. Biotechnol. 15, 613–629 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tacconelli E., et al. ; WHO Pathogens Priority List Working Group, Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Boucher H. W., et al. , Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Rehman A., Patrick W. M., Lamont I. L., Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: New approaches to an old problem. J. Med. Microbiol. 68, 1–10 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Høiby N., Recent advances in the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. BMC Med. 9, 32 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J. K., Lee Y. S., Park Y. K., Kim B. S., Alterations in the GyrA and GyrB subunits of topoisomerase II and the ParC and ParE subunits of topoisomerase IV in ciprofloxacin-resistant clinical isolates of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 25, 290–295 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Feng X., et al. , Mutations in gyrB play an important role in ciprofloxacin-resistant Pseudomonas aeruginosa. Infect. Drug Resist. 12, 261–272 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgins P. G., Fluit A. C., Milatovic D., Verhoef J., Schmitz F. J., Mutations in GyrA, ParC, MexR and NfxB in clinical isolates of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 21, 409–413 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Pasca M. R., et al. , Evaluation of fluoroquinolone resistance mechanisms in Pseudomonas aeruginosa multidrug resistance clinical isolates. Microb. Drug Resist. 18, 23–32 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Bruchmann S., Dötsch A., Nouri B., Chaberny I. F., Häussler S., Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob. Agents Chemother. 57, 1361–1368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Köhler T., et al. , Characterization of MexE–MexF–OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23, 345–354 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Masuda N., et al. , Substrate specificities of MexAB–OprM, MexCD–OprJ, and MexXY–oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44, 3322–3327 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu C., et al. , Mechanisms for development of ciprofloxacin resistance in a clinical isolate of Pseudomonas aeruginosa. Front. Microbiol. 11, 598291 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marvig R. L., Sommer L. M., Molin S., Johansen H. K., Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 47, 57–64 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Cheer S. M., Waugh J., Noble S., Inhaled tobramycin (TOBI): A review of its use in the management of Pseudomonas aeruginosa infections in patients with cystic fibrosis. Drugs 63, 2501–2520 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Mayer-Hamblett N., et al. , Pseudomonas aeruginosa in vitro phenotypes distinguish cystic fibrosis infection stages and outcomes. Am. J. Respir. Crit. Care Med. 190, 289–297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Díez-Aguilar M., et al. , Use of Calgary and microfluidic BioFlux systems to test the activity of fosfomycin and tobramycin alone and in combination against cystic fibrosis Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 62, e01650-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCaughey G., et al. , Fosfomycin and tobramycin in combination downregulate nitrate reductase genes narG and narH, resulting in increased activity against Pseudomonas aeruginosa under anaerobic conditions. Antimicrob. Agents Chemother. 57, 5406–5414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee D. G., et al. , Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7, R90 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carilla-Latorre S., et al. , Dictyostelium transcriptional responses to Pseudomonas aeruginosa: Common and specific effects from PAO1 and PA14 strains. BMC Microbiol. 8, 109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roemhild R., et al. , Cellular hysteresis as a principle to maximize the efficacy of antibiotic therapy. Proc. Natl. Acad. Sci. U.S.A. 115, 9767–9772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong A., Rodrigue N., Kassen R., Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Genet. 8, e1002928 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wydmuch Z., et al. , GyrA mutations in ciprofloxacin-resistant clinical isolates of Pseudomonas aeruginosa in a Silesian Hospital in Poland. Pol. J. Microbiol. 54, 201–206 (2005). [PubMed] [Google Scholar]
- 54.Yang X., Xing B., Liang C., Ye Z., Zhang Y., Prevalence and fluoroquinolone resistance of Pseudomonas aeruginosa in a hospital of South China. Int. J. Clin. Exp. Med. 8, 1386–1390 (2015). [PMC free article] [PubMed] [Google Scholar]
- 55.Mouneimné H., Robert J., Jarlier V., Cambau E., Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43, 62–66 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuo Y., Eda S., Gotoh N., Yoshihara E., Nakae T., MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ–mexX intergenic DNA. FEMS Microbiol. Lett. 238, 23–28 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Bartell J. A., et al. , Evolutionary highways to persistent bacterial infection. Nat. Commun. 10, 629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dunham S. A., McPherson C. J., Miller A. A., The relative contribution of efflux and target gene mutations to fluoroquinolone resistance in recent clinical isolates of Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 29, 279–288 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Sobel M. L., Neshat S., Poole K., Mutations in PA2491 (mexS) promote MexT-dependent mexEF–oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 187, 1246–1253 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stern D. L., The genetic causes of convergent evolution. Nat. Rev. Genet. 14, 751–764 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Manceau M., Domingues V. S., Linnen C. R., Rosenblum E. B., Hoekstra H. E., Convergence in pigmentation at multiple levels: Mutations, genes and function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 2439–2450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Losos J. B., Convergence, adaptation, and constraint. Evolution 65, 1827–1840 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Haldane J. B. S., Disease and evolution. Ricera Sci Suppl A 19, 68–76 (1949). [Google Scholar]
- 64.Navas A., et al. , Experimental validation of Haldane’s hypothesis on the role of infection as an evolutionary force for Metazoans. Proc. Natl. Acad. Sci. U.S.A. 104, 13728–13731 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elphick H. E., Scott A., Single versus combination intravenous anti-pseudomonal antibiotic therapy for people with cystic fibrosis. Cochrane Database Syst. Rev. 12, CD002007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laborda P., Sanz-García F., Hernando-Amado S., Martínez J. L., Pseudomonas aeruginosa: An antibiotic resilient pathogen with environmental origin. Curr. Opin. Microbiol. 64, 125–132 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Mulet X., et al. , Antagonistic interactions of Pseudomonas aeruginosa antibiotic resistance mechanisms in planktonic but not biofilm growth. Antimicrob. Agents Chemother. 55, 4560–4568 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wingett S. W., Andrews S., FastQ Screen: A tool for multi-genome mapping and quality control. F1000 Res. 7, 1338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McKenna A., et al. , The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garrison E., Marth G., Haplotype-based variant detection from short-read sequencing. arXiv [Preprint] (2012). https://arxiv.org/abs/1207.3907 (Accessed 20 October 2021).
- 73.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.BioinfoGP, SNPer, version 0.8. https://bioinfogp.cnb.csic.es/tools/snper/. Accessed 13 October 2021. [Google Scholar]
- 75.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Odds F. C., Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 52, 1 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.