Significance
Subsistence shifts from hunting and gathering to agriculture over the last 12,000 y have impacted human culture, biology, and health. Although past human health cannot be assessed directly, adult stature variation and skeletal indicators of nonspecific stress can serve as proxies for health during growth and development. By integrating paleogenomic genotype and osteological stature data on a per-individual basis for 167 prehistoric Europeans, we observe relatively shorter than expected statures among early farmers after correcting for individual genetic contributions to stature. Poorer nutrition and/or increased disease burdens for early agriculturalists may partly underscore this result. Our integrated osteological–genetic model has exciting potential for studies of past human health and expansion into various other contexts.
Keywords: paleogenomics, stature variation, agriculture transition, health
Abstract
Human culture, biology, and health were shaped dramatically by the onset of agriculture ∼12,000 y B.P. This shift is hypothesized to have resulted in increased individual fitness and population growth as evidenced by archaeological and population genomic data alongside a decline in physiological health as inferred from skeletal remains. Here, we consider osteological and ancient DNA data from the same prehistoric individuals to study human stature variation as a proxy for health across a transition to agriculture. Specifically, we compared “predicted” genetic contributions to height from paleogenomic data and “achieved” adult osteological height estimated from long bone measurements for 167 individuals across Europe spanning the Upper Paleolithic to Iron Age (∼38,000 to 2,400 B.P.). We found that individuals from the Neolithic were shorter than expected (given their individual polygenic height scores) by an average of −3.82 cm relative to individuals from the Upper Paleolithic and Mesolithic (P = 0.040) and −2.21 cm shorter relative to post-Neolithic individuals (P = 0.068), with osteological vs. expected stature steadily increasing across the Copper (+1.95 cm relative to the Neolithic), Bronze (+2.70 cm), and Iron (+3.27 cm) Ages. These results were attenuated when we additionally accounted for genome-wide genetic ancestry variation: for example, with Neolithic individuals −2.82 cm shorter than expected on average relative to pre-Neolithic individuals (P = 0.120). We also incorporated observations of paleopathological indicators of nonspecific stress that can persist from childhood to adulthood in skeletal remains into our model. Overall, our work highlights the potential of integrating disparate datasets to explore proxies of health in prehistory.
The agricultural revolution—beginning ∼12,000 B.P. in the Fertile Crescent zone (1, 2) and then spreading (3–5) or occurring independently (6, 7) across much of the inhabited planet—precipitated profound changes to human subsistence, social systems, and health. Seemingly paradoxically, the agricultural transition may have presented conflicting biological benefits and costs for early farming communities (8, 9). Specifically, demographic reconstructions from archaeological and population genetic records suggest that the agricultural transition led to increased individual fitness and population growth (6, 10–12), likely due in part to new food production and storage capabilities. Yet, bioarchaeological analyses of human skeletal remains from this cultural period suggest simultaneous declines in individual physiological well-being and health, putatively from 1) nutritional deficiency and/or 2) increased pathogen loads as a function of greater human population densities, sedentary lifestyles, and proximity to livestock (9, 13–18).
To date, anthropologists have used two principal approaches to study health across the foraging-to-farming transition in diverse global regions (13, 19, 20). The first approach involves identifying paleopathological indicators of childhood stress that persist into adult skeletal remains. For example, porotic hyperostosis (porous lesions on the cranial vault) and cribra orbitalia (porosity on the orbital roof) reflect a history of bone marrow hypertrophy or hyperplasia resulting from one or more periods of infection, metabolic deficiencies, malnutrition, and/or chronic disease (21–26). Meanwhile, linear enamel hypoplasia (transverse areas of reduced enamel thickness on teeth) occurs in response to similar childhood physiological stressors (e.g., disease, metabolic deficiencies, malnutrition, weaning) that disrupt enamel formation in the developing permanent dentition (27–30). Broadly, these paleopathological indicators of childhood stress tend to be observed at higher rates among individuals from initial farming communities relative to earlier periods, potentially reflecting their overall “poorer” health (14, 31–36).
A second approach uses skeleton-based estimates of achieved adult stature as a proxy for health during childhood growth and development (37–39). Since stature is responsive to the influences of nutrition and disease burden alongside other factors, relatively short “height-for-age” (or “stunting”) has been used as an indicator of poorer health in both living and bioarchaeological contexts (39–43). When studying the past, individual stature can be estimated from long bone measurements and regression equations (44–47). Using these methods, multiple prior studies have reported a general profile of relatively reduced stature for individuals from early agricultural societies in Europe (15, 48–50), North America (51–53), the Levant (16, 32), and Asia (54, 55). For example, estimated average adult mean statures for early farmers are ∼10 cm shorter relative to those for preceding hunter-gatherers in both western Europe (females, −8 cm; males, −14 cm) (49, 50) and the eastern Mediterranean (females, −11 cm; males, −8 cm) (56). This pattern is not universal, as a few studies do not report such changes (57, 58); the variation could be informative with respect to identifying potential underlying factors (59).
However, in addition to environmental effects like childhood nutrition and disease, inherited genetic variation can have an outsized impact on terminal stature, with ∼80% of the considerable degree of height variation within many modern populations explainable by heritable genetic variation (60–63). Moreover, migration and gene flow likely accompanied many subsistence shifts in human prehistory. For example, there is now substantial paleogenomic evidence of extensive population turnover across prehistoric Europe (64–69). Therefore, from osteological studies alone, we are unable to quantify the extent to which temporal changes in height reflect variation in childhood health vs. changes/differences in the frequencies of alleles associated with height variation.
In this study, we have performed a combined analysis of ancient human paleogenomic and osteological data where both are available from the same n = 167 prehistoric European individuals representing cultural periods from the Upper Paleolithic (∼38,000 B.P.) to the Iron Age (∼2,400 B.P.). This approach allows us to explore whether “health,” as inferred from the per-individual difference between predicted genetic contributions to height and osteological estimates of achieved adult height, changed over the Neolithic cultural shift to agriculture in Europe. When craniodental elements were preserved and available for analysis (n = 98 of the 167 individuals), we also collected porotic hyperostosis, cribra orbitalia, and linear enamel hypoplasia paleopathological data in order to examine whether patterns of variation between osteological height and genetic contributions to height are explained in part by the presence/absence of these indicators of childhood or childhood-inclusive stress.
Results
We developed a database of n = 167 ancient European adult individuals (67 females, 100 males) with available genome-wide paleogenomic data (from either shotgun sequencing or DNA capture-based approaches and from both published and in-process studies) and stature estimates based on long bone measurements (either newly collected or published) (Fig. 1A and Dataset S1). The cultural and time periods represented in our dataset are Upper Paleolithic (38,000 to 12,000 B.P.; n = 8 individuals), Mesolithic (11,000 to 6,400 B.P.; n = 15), Neolithic (7,100 to 3,500 B.P.; n = 46), Copper Age (6,300 to 3,400 B.P.; n = 60), Bronze Age (4,500 to 2,500 B.P.; n = 31), and Iron Age (2,600 to 2,400 B.P.; n = 7).
Fig. 1.
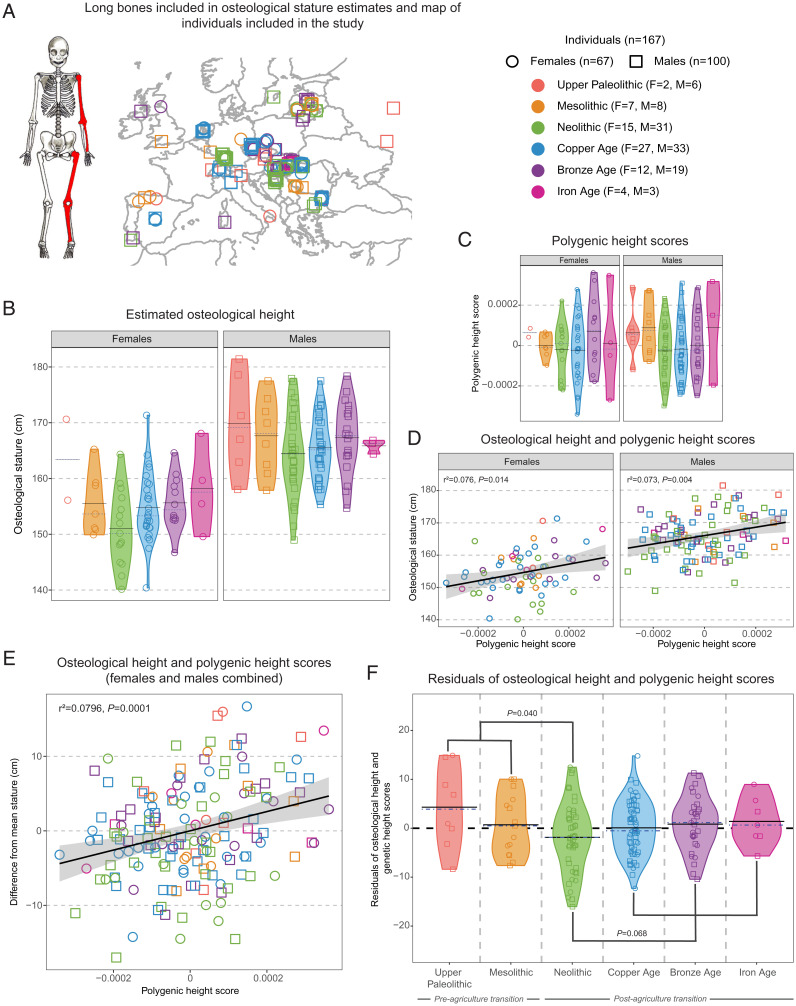
Osteological stature and ancient DNA–based polygenic height scores. (A) Map of the locations of the archaeological sites from which individuals included in the dataset were recovered. (B) Osteological height estimates generated using measurements of long bone lengths (highlighted in red on the illustration) and sex-specific regression equations (44). (C) Polygenic height scores generated using genome-wide association summary statistics for height-associated single-nucleotide polymorphisms and individual ancient DNA genotype data. (D and E) The relationship between polygenic height score and estimated osteological stature (centimeters) for females, for males, and for the full sample with height differences from mean stature calculated separately for females (mean = 154.64 ± 6.48 cm) and males (mean = 165.97 ± 6.60 cm), respectively (represented on the y axis in E). (F) Residuals of the relationship between polygenic height score and osteological height with sex as a covariate for all individuals by cultural period. Mean and median are represented by the black and blue dashed lines, respectively. Skeletal illustration in A image credit: Katharine Thompson (Stony Brook University, Stony Brook, NY).
Subsistence strategies in the Upper Paleolithic and Mesolithic focused on gathering, collecting, and hunting food. The Neolithic is marked by the emergence of plant cultivation and animal domestication (to varying degrees and tempos of integration), long-term settlements, larger populations, and increased social complexity—processes that then intensified and expanded in subsequent periods (70). The overlapping dates among the different cultural periods reflect both geographical variation in the timing of cultural change and the potential for co-occurrence of multiple cultural traditions within a single region.
Confirmation of an Average Osteological Stature Dip in the Neolithic.
We used an osteometric board to newly estimate the lengths of preserved long bones for n = 93 of the n = 167 total individuals (55.7%) in our database (Dataset S1). We also recorded published and unpublished (previously collected) long bone length estimates for an additional n = 54 individuals (32.3%). In these cases, we estimated osteological stature from the long bone length data (44). Finally, for n = 20 individuals, only precalculated terminal height estimates were available (12%).
We observed osteological stature variation among cultural periods (Fig. 1B). Reconstructions of osteological stature for females and males are lower during the initial shift to farming during the Neolithic compared with earlier and post-Neolithic periods. Specifically, individuals from the pre-Neolithic periods (female average stature = 157.28 ± 7.0 (SD) cm, males = 168.6 ± 7.6 cm) were ∼4 cm taller on average than those from the Neolithic (females = 151.0 ± 6.7 cm; males = 164.5 ± 7.4 cm; linear model including sex as a factor; P = 0.012). Then, Neolithic individuals were ∼2 to 5 cm shorter on average compared with all post-Neolithic individuals (females = 155.06 ± 5.7 cm; males = 166.16 ± 5.82 cm; P = 0.046), with average osteological stature steadily rebounding across the Copper Age (females = 154.8 ± 6.2 cm; males = 165.5 ± 5.5 cm), Bronze Age (females = 155.6 ± 4.8 cm; males = 167.3 ± 6.3 cm), and Iron Age (females = 158.2 ± 7.8 cm; males = 165.8 ± 1.3 cm) (SI Appendix, Tables S1 and S2).
The overall pattern from our data roughly parallels previously published reports. Specifically, 1) stature decreased slightly from the Upper Paleolithic to the Mesolithic (35, 36, 71), 2) marked stature reduction occurred during the initial agricultural transition in the Neolithic (14, 16, 34, 55) [although this is not universal (70, 72–74)], and 3) stature rebounded during subsequent post-Neolithic periods of agricultural intensification (14, 16).
Early Farmers Were Relatively Shorter than Expected Given Their Polygenic Height Scores.
We next considered the osteological height estimates in the context of ancient DNA-based polygenic height scores for the same individuals. Using an established approach for working with ancient DNA genotype data (65, 75, 76), we estimated a polygenic score for each prehistoric individual based on their available genome-wide genotypes in the context of results from a large-scale genome-wide association study (GWAS) of stature variation in modern Europeans (77) (data are from http://www.nealelab.is/uk-biobank/). For the results presented in the text and figures, we used a version of the dataset in which all variants possibly affected by deamination-based ancient DNA damage (78–80) were masked. We also performed all analyses with the unmasked dataset and obtained similar results (SI Appendix, Fig. S1 and Table S3).
While the polygenic scores that we estimated for the n = 167 individuals were somewhat variable across cultural periods (Fig. 1C and SI Appendix, Tables S4 and S5), as anticipated based on prior work (58), we were most interested in using these data to begin to account for genetic contributions to achieved adult (osteological) height on a per-individual basis. Polygenic height scores and osteological estimates of stature were positively correlated for females (n = 67 [Fig. 1D]; r2 = 0.076; P = 0.014), for males (n = 100 [Fig. 1D]; r2 = 0.073; P = 0.004), and for the combined dataset (Fig. 1E) (r2 = 0.0796; P = 0.0001). These results support the general biological plausibility of our integrative analysis of paleogenomic and osteological data.
Importantly and expectedly, there is still considerable interindividual variation in the relationship between polygenic height score and achieved adult stature, which could reflect any combination of incomplete genetic information; long bone measurement error; polygenic height score or stature estimate error; and the effects of childhood nutrition, disease, and other environmental variables on growth. Accordingly, we next analyzed the residuals from the combined sex osteological stature and ancient DNA-based polygenic score model (Fig. 1E) to test whether individuals tended to have taller or shorter adult stature relative to expectations given their individual polygenic scores across the different cultural periods. These residuals are expressed in plus or minus centimeters from the predicted stature per individual.
We present our results as differences between average residuals (reflecting the difference between osteological stature and genetic height score after accounting for sex) across cultural periods. As such, our below use of the term “expected” in relative statements in this context reflects a statistical property of the overall dataset rather than commentary on the broader anthropological hypotheses being tested.
When using our ancient DNA-based approach to partly account for the predicted contribution of genetic variation to adult stature, we observed that individuals from the Neolithic were indeed osteologically shorter than expected (i.e., based on their polygenic height scores and in the context of our overall sample) compared with individuals from other cultural periods (Fig. 1F and SI Appendix, Table S6). Specifically, pre-Neolithic individuals (average residual = +1.96 ± 7.06 cm) were +3.82 cm taller than expected on average relative to Neolithic individuals (average residual = −1.87 ± 7.08 cm; P = 0.040). Neolithic individuals were then −2.21 cm shorter than expected on average relative to post-Neolithic individuals (average residual = 0.341 ± 5.48 cm; P = 0.068). Neolithic individuals were the only group with a negative average residual.
We confirmed that these results cannot be explained by geographic variation (latitude and longitude) in our sample (SI Appendix, Fig. S2 and Table S7). We also obtained similar results when we separately analyzed females and males (SI Appendix, Fig. S3 and Table S6) and when we separately analyzed the lengths of individual long bones as opposed to the reconstructed stature estimates (for the femur and radius, in particular, although there are sample size limitations) (SI Appendix, Figs. S4 and S5 and Table S8).
In contrast, our results were partially muted when we included variables explicitly reflecting genetic ancestries in our model. Our primary approach (as above) already accounts for ancestry variation via individual-level calculations of polygenic scores. However, polygenic height scores explain only a proportion of total heritable variation (81–83). Therefore, we repeated our hypothesis tests after conducting a multidimensional scaling (MDS) analysis with the genome-wide genotype data for all n = 167 ancient individuals to then including the first four MDS components from this analysis as factors in an updated linear model, following Cox et al. (84). Similar to principal components analysis (PCA), MDS can be used to mitigate the effects of ancestry heterogeneity in a GWAS framework (85, 86). When including the MDS components in the model, pre-Neolithic individuals (average residual = +2.22 ± 6.98 cm) were +2.82 cm taller than expected on average relative to Neolithic individuals (average residual = −0.594 ± 6.9 cm; P = 0.12). Neolithic individuals were 0.38 cm shorter on average relative to post-Neolithic individuals (average residual = −0.21 ± 5.16 cm; P = 0.74) (SI Appendix, Fig. S6 and Table S9).
Paleopathological Indicators of Nonspecific Stress.
Adverse early life conditions may negatively impact adult stature. To begin to investigate whether individual-level early life effects on prehistoric stature could be identified, we incorporated observations of paleopathological indicators of nonspecific stress that can persist from childhood to adult skeletal remains into our analytical model. To do so, we characterized the presence/absence of one or more of cribra orbitalia (porosity on the orbital roof), porotic hyperostosis (porosity on the cranial vault), and linear enamel hypoplasia (reduced areas of enamel thickness) for n = 98 of the n = 167 (58.7%) individuals in our study (n = 82 newly characterized; n = 16 published/previously characterized) (SI Appendix, Table S10).
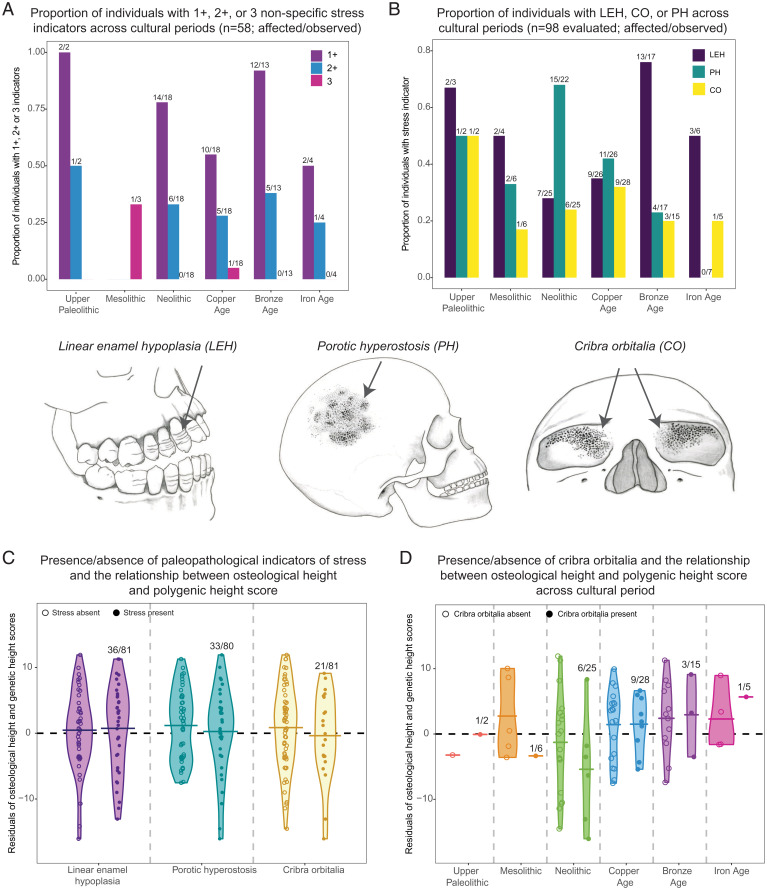
For 58 of these 98 individuals (59.2%), crania were sufficiently complete for assessment of the presence/absence of all three stress indicators (Fig. 2A). Of this subsample, one or more stress indicators were present for 41 (70.7%) of the individuals, two or more indicators were observed in 18 (31.0%) individuals, and all three paleopathological indicators were present in only 2 (3.4%) individuals. Thus, stresses on health were relatively common overall in prehistoric Europe.
Fig. 2.
Paleopathological indicators of stress. Paleopathological indicators of nonspecific stress evaluated in this study: linear enamel hypoplasia (LEH; bands of reduced enamel thickness on teeth), cribra orbitalia (CO; porosity on the orbits), and porotic hyperostosis (PO; porosity on the side of the skull). (A) The remains of 58 individuals were sufficiently complete to permit presence/absence assessment for all three paleopathologies. The proportions of individuals with one or more, two or more, and all three stress indicators are indicated across cultural periods. Numbers above the bars indicate sample sizes. (B) The presence/absence of at least one of the three paleopathological indicators of stress could be determined for 98 total individuals. Shown are the proportions of individuals (of those who could be assessed for that indicator) with LEH, CO, and PH across cultural periods. Numbers above the bars indicate sample sizes. (C) Residuals of the relationship between polygenetic height score and osteological height with sex as a covariate plotted separately for individuals with each paleopathological indicator of stress present vs. absent. Means are represented by the black lines. Numbers above the bars indicate sample sizes. (D) Residuals of the relationship between polygenetic height score and osteological height with sex as a covariate plotted separately for individuals with and without cribra orbitalia by cultural period. Means are represented by the black lines. Numbers above the bars indicate sample sizes. Skeletal illustrations image credit: Katharine Thompson (Stony Brook University, Stony Brook, NY).
A considerable 77.8% (14 of 18) of Neolithic individuals had one or more stress indicators (Fig. 2A). While the proportion of Copper Age individuals with one or more stress indicators (10 of 18; 55.6%) was lower compared with that for the Neolithic (Fisher’s exact test; P = 0.289), the Neolithic result is not unique, with one or more stress indicators also recorded for all but one individual in the Bronze Age sample (12 of 13; 92.3%).
Considering the larger dataset of n = 98 individuals with presence/absence data for at least one stress indicator to maximize sample sizes, we observed a distinct difference between the Neolithic and Bronze Age patterns (Fig. 2B). Specifically, porotic hyperostosis is common in the Neolithic sample (15 of 22; 68.2%), while linear enamel hypoplasia is relatively rare (7 of 25 individuals; 28.0%). The opposite is true in the Bronze Age, with 4 of 17 (23.5%) positive for porotic hyperostosis yet 13 of 17 (76.5%) with linear enamel hypoplasia.
We next tested whether the presence of paleopathological indicators of stress is predictive of individual-level deviations from the overall relationship between osteological stature and polygenic height score estimates. Based on the subset of individuals with presence/absence data for all three paleopathological indicators, the 41 individuals with one or more stress indicator were −0.917 cm shorter than expected on average compared with the 17 individuals without any stress indicators (SI Appendix, Fig. S7 and Table S11). With the moderate magnitude of this difference and the relatively small sample size available for this analysis, this result was not unlikely based on chance expectations (t test; P = 0.555). The 18 individuals with two or more stress indicators were −1.175 cm shorter than expected on average relative to individuals with no stress indicators (P = 0.561).
Using our larger dataset, we next separately analyzed the effect of each paleopathological stress indicator on the osteological stature–polygenic height score relationship (Fig. 2C). We found that the n = 21 individuals with cribra orbitalia were slightly but not significantly shorter (−1.23 cm) than expected on average compared with the n = 60 individuals without cribra orbitalia (P = 0.461; false discovery rate = 0.771). Linear enamel hypoplasia and porotic hyperostosis presence/absence were negligibly associated with osteological stature vs. polygenetic height score residual variation (Fig. 2D and SI Appendix, Table S12). These patterns did not change appreciably when excluding the few individuals with active cribra orbitalia or porotic hyperostosis lesions from their respective analyses (active lesions reflect adult stress while potentially masking evidence of lesions from childhood) (SI Appendix, Fig. S8 and Table S12).
Finally, we investigated the relationship between cribra orbitalia, porotic hyperostosis, and linear enamel hypoplasia presence/absence and osteological vs. polygenic height score residual in the context of cultural period (SI Appendix, Fig. S9 and Tables S13 and S14). Of note, the n = 6 Neolithic individuals with cribra orbitalia were −4.12 cm shorter than expected on average compared with the n = 19 individuals from the same cultural period without cribra orbitalia (Fig. 2D) (P = 0.329). This preliminary but suggestive effect was nearly absent in the Copper Age (+0.065 cm difference; P = 0.973), the only other period with sufficient presence/absence sample sizes for analysis.
Discussion
Bioarchaeologists have equated repeated observations of relatively shorter average adult statures in the Neolithic to a likely general health decline for individuals during this cultural period (13, 14, 16, 34, 55, 87). Combinations of reduced nutritional diversity, unpredictable food availability (e.g., crop failure, storage loss), and increased infectious disease burden may have negatively impacted childhood health and growth (18, 88, 89). Understandably, those prior studies did not account for the contribution of interindividual variation in the contribution of heritable genetic factors to adult stature. Yet, this consideration is especially important in light of updated understandings of considerable migration and gene flow processes associated with various farming transitions (90–93).
In our study, we sampled 167 prehistoric European individuals for whom both genome-wide ancient DNA data and intact long bones were available for analysis, making it possible to test whether Neolithic individuals were still osteologically shorter than expected when accounting (at least partly) for individual-level genetic contributions to height. Using this approach, we found that the average Neolithic farmer was indeed relatively shorter than expected compared with pre-Neolithic individuals (Fig. 1F). Average osteological vs. expected stature then increased over each post-Neolithic cultural period. This gradual recovery may reflect a history of continued (although variable) cultural and technological innovations that ameliorated and/or overpowered the initial nutritional and disease stressors faced by the earliest farmers (64, 66–69, 94).
Our framework is related to but differs from that of a previous study by Cox et al. (58), who compared population-level osteological height estimates [n = 1,159 total individuals; the osteological data were from Niskanen et al. (95)] and ancient DNA-based polygenic height scores (n = 1,071) across prehistoric Europe. These two estimates were computed separately (i.e., typically not for the same individuals), thereby facilitating the large sample sizes. In contrast, our approach expressly considers individual-level dynamics in the relationship between these two variables, which is sample size restrictive yet potentially insightful. Interestingly, Cox and colleagues observed that mean osteological stature estimates and polygenic height scores were both similar between the European Mesolithic and Neolithic (58, 95). This result is in contrast to our own osteological height estimate observations in this study and those of previous bioarchaeological studies (15, 48), which again may reflect potentially interesting interpopulation variability as part of the nuanced complexity underlying subsistence shifts (94).
Limitations.
One important potential limitation of our study, aside from the number of individuals, is uncertainty regarding the ultimate portability of polygenic scores over genetic, geographic, and temporal distances (81, 82, 96, 97). Our analyses were also based on incomplete ancient DNA genotype data (however, note that we did exclude potential error from deamination-based ancient DNA damage by masking all potentially affected sites in the primary versions of our analyses). Yet, the significant, positive relationship between polygenic height scores and estimated osteological statures across our overall sample (Fig. 1F) demonstrates the biological plausibility of our model. Moreover, our primary results were unchanged when we incorporated archaeological site latitude and longitude variables into our analyses.
However, even with complete genome-wide genotype data, polygenic height scores only capture a proportion of the heritable component of stature variation (83, 98–100). Furthermore, only a subset of variants is “available” for assessment in ancient genomic data. The predictive accuracy of polygenic scores is also higher when individuals are more closely related to the cohort used to develop the GWAS and generate the effect sizes of associated single nucleotide polymorphisms (SNPs), as increasing genetic distance from this cohort has been shown to negatively impact the predictive accuracy of polygenic scores (97, 100, 101). Therefore, for multiple reasons, our primary analytical approach might incompletely capture the stature-relevant effects of any genetic ancestry variation across our sample. That is, with respect to our hypotheses, we could only be partially accounting for any cultural period-confounded migration/gene flow among populations with different genetic height profiles. For example, gene flow as a result of the spreading of the Yamnaya/Corded Ware cultures (“steppe ancestries”) starting ∼4,000 to 5,000 y ago may have been associated with the introduction of relatively greater proportions of “tall” alleles into various regions of Europe (58, 64).
If this is a general phenomenon that extends to small effect and other loci not included in our individual-level polygenic height score calculations, then our cultural period–related inferences could be erroneous. Future improvements in the paleogenomics field related to low-coverage imputation and modeling the potential impacts of population turnovers and/or admixture events on polygenic scores will be important in driving forward genotype–phenotype trait reconstructions (86).
In the meantime, to help explore the potential effects of these processes on our results, we conducted a parallel set of analyses in which we included factors reflecting genome-wide genetic ancestries (from the first four components of a MDS analysis) in our model. When including both the polygenic height scores and MDS factors in the analysis, our downstream finding of shorter osteological statures than expected (i.e., based on polygenic height scores and MDS factors and in the context of the overall sample) in the Neolithic relative to other cultural periods was in fact attenuated. This approach does represent an overcorrection or more precisely, some level of double accounting of the genetic effect on height. That is, the MDS factors and the polygenic height scores are intertwined and cannot be deconvoluted readily due to the pervasive polygenicity of height (62, 102). Nonetheless, the signal attenuation result calls for cautious interpretations of the main results in our study while awaiting expanded future sample sizes.
To provide further insight into the relative magnitudes of these genetic components, we observed that 7.96% of the variation in the difference between individual and per-sex mean osteological stature is explained by individual-level polygenic height score alone (Fig. 1E), whereas 8.4% of the osteological variation is explained by MDS factors alone (i.e., without polygenic height score included) (SI Appendix, Table S15). Meanwhile, 14.2% of this variation is explained by a model including both individual polygenic height scores and the MDS components (SI Appendix, Table S16). While these comparisons are imperfect for the reasons noted above, our polygenic height score does appear to contain valuable biological information for our hypothesis-testing approach.
Additionally, we note that comparable r2 values for the relationship between polygenic height score and estimated stature between those for individual modern human populations and our temporally and culturally diverse ancient sample are not necessarily expected (SI Appendix, Fig. S10 and Table S17). That is, we have prior archaeological knowledge of major subsistence patterns and other cultural changes—with potentially large somatic effects on growth thereby reducing the strength of the relationship between polygenic height score and osteological adult stature—that occurred across our sample. How these cultural changes are associated with the (potentially highly variable) relationship between polygenic height scores and estimates of adult stature is the analytical essence of our study.
Toward the Integration of Paleopathological Data.
For a subset of the individuals in our study (n = 98), we were additionally able to consider the extent to which three paleopathological markers of nonspecific childhood and childhood-inclusive stress (linear enamel hypoplasia, cribra orbitalia, and porotic hyperostosis; each of which are maintained in the skeleton into adulthood) are associated with the relationship between osteological stature estimates and polygenic height scores. We observed a slight trend of relatively shorter than expected (given polygenic height score) adult statures among individuals with one or more childhood stress markers present (SI Appendix, Fig. S7).
However, larger sample sizes will be necessary to more fully explore interplays among specific paleopathological indicators, osteological vs. genetic height scores, and cultural periods. Still, our findings do at least suggest that factors underlying skeletal growth trajectories are separable, at least in part, from those leading to paleopathological indicators of stress. In particular, high rates of paleopathology are still observed in post-Neolithic cultural periods (for example, in the Bronze Age), even after absolute osteological stature and actual vs. expected stature averages have recovered.
Looking further and more generally forward to future studies aiming to integrate per-individual osteological and ancient DNA information, we note that genetic risk for certain diseases that can lead to paleopathology may vary among individuals within a population and on average between groups. For example, Berens et al. (103) reported higher genetic risk scores for dental/periodontal disease in the agriculturalist relative to the pastoral populations they studied.
Summary.
We united previously disparate osteological and paleogenomic datasets for 167 prehistoric European individuals on a per-individual basis. Our results represent an advance in the study of whether and how a major cultural transition in human evolution affected physiological health. In particular, we show that the average Neolithic individual may have been relatively short even when correcting for expected individual genetic contributions to adult stature. This result may reflect reduced nutrition and/or increased infectious disease burden. We also preliminarily developed a framework for further consideration of these results in the context of particular paleopathological indicators of childhood stress. Looking forward, our model can be expanded in various dimensions (for example, to different world regions or to more constrained spatial and temporal contexts) in order to further the study of emergent physiological trade-offs across periods of dramatic cultural or environmental change. Integrated osteological–genetic approaches will increasingly become important components of the tool kit for studying the dynamics of past human health.
Materials and Methods
The 167 individuals in our dataset have broad geographical, temporal, and cultural period ranges. Radiocarbon or archaeologically calibrated dates, latitude/longitude coordinates, genetic sex, and archaeological/cultural period were obtained from the original paleogenomic and archaeological publications of the paleogenomic data used for this study (Dataset S1).
Processing Ancient DNA Sequence Reads.
Published sequence data were downloaded from the Sequence Read Archive and the European Nucleotide Archive databases as indicated per the respective papers alongside sequence data from in-process studies (Dataset S1). FASTQ (65, 67, 104–107) and BAM (Binary Alignment Map) (66, 68, 69, 108–122) files from shotgun sequence (n = 26) and DNA capture-based datasets (n = 141) were aligned/realigned using the Burrows–Wheeler aligner aln (123) to the human reference genome (hg19, build 37) with seeding disabled. For unaligned shotgun sequence data (n = 7) (65, 104–107), leeHom (124) was used to trim adapters and merge reads. Reads from sequence libraries that were not treated to remove damage signatures typical of ancient DNA (“non-uracil–DNA–glycosylase”) were subject to rescaling using mapDamage 2.0 (125) after mapping (105, 121), which downscales quality values for likely ancient DNA misincorporations based on read position and damage pattern. For partial and full uracil–DNA–glycosylase–treated libraries, two base pairs (bp) at the 5′ and 3′ ends were trimmed (prior to mapping) using seqtk (source code from ref.126) so as to not confound downstream analyses with potential postmortem deamination at the terminal read ends (66, 68, 109, 127). SAMtools was used to sort mapped reads and filter for mapping quality 30 and minimum bp 30, with duplicates removed using SAMtools rmdup (128). Read groups were added using Picard Tools AddOrReplaceReadGroups function (129). Following the GATK (Genome Analysis Toolkit) workflow (130), realigning indels was performed using RealignerTargetCreator and IndelRealigner, followed by BaseRecalibrator to minimize sequence error introduced by potential mismatches to the reference (131).
Genotyping and Imputation.
We implemented GATKUnifiedGenotyper (130) followed by imputation to maximize the amount of genetic information for downstream analyses and since genotype likelihood scores can be generated for imputation. We opted to impute diploid genotypes and missing sites for the individuals in our dataset [using the 1000 Genomes Project Consortium (132) reference panel] to minimize potential reference bias that may otherwise occur when using an alternative approach of randomly sampling one allele at each site (133).
The 1000 Genomes phase 3 genetic variants reference panel was used for genotyping and imputation, as provided by BEAGLE (134, 135). After removing variants that are not SNPs, multiallelic SNPs, and X and Y chromosomes, 77,818,345 variants remained. UnifiedGenotyper (130) was used to obtain genotypes and likelihood scores with the following parameters: –genotyping_mode –alleles <1000 Genomes reference panel>, –GENOTYPE_GIVEN_ALLELES, –output_mode EMIT_ALL_SITES, –AllSitesPLs -R <hg19 reference FASTA> (131). Due to the potential for postmortem damage impacting C > T and G > A allele changes, the per-chromosome VCF (Variant Call Format) files of the called genotypes were filtered for potential postmortem damage (modifying source code from ref. 136 for multiindividual VCF files and removing deamination in both directions). Potential deamination signal C > T (T > C) and G > A (A > G) genotypes were replaced in ancient individuals heterozygous for these genotypes with “./.” similar to previous paleogenomic studies (65, 76).
Genotype likelihoods were then estimated using the per-chromosome VCF files, followed by imputation of missing SNPs based on the genotype probability score using the 1000 Genomes phase 3 haplotypes (137) and GRCh37 genomic maps (138). Parameters for estimating genotype likelihoods were gprobs = true, gl ≤ input genotypes from UnifiedGenotyper> ref ≤ Beagle imputation reference panel>, map ≤ hg19 recombination map>. Imputation parameters were gt ≤ GL_output_VCF>, gprobs = true, impute = true, ref ≤ Beagle imputation reference panel>, map ≤ GRCh37 recombination map> (131). This resulted in 30,761,499 markers imputed/genotyped across 167 individuals. Prior to downstream analyses, the imputed VCF was filtered for a minimum genotype probability of 0.99 to maximize confident genotype calls postimputation and filter out less confident calls. We repeated the above pipeline of genotyping calling and imputation without filtering for potential deamination signals, and the results were consistent with the deamination filtered data (SI Appendix, Fig. S1 and Table S3).
Assessing Accuracy of Imputation.
For the individuals in our study, the genomic/SNP capture sequence coverage mean for SNP positions analyzed was =1.595X, SD = 2.438, and median = 0.717, with a range of 0.002X to 22X. Prior to imputation, an average of 338,074 SNP sites per individual were covered by a minimum of one read with a median called genotype proportion =0.095 (i.e., median proportion of missing SNPs is =0.905) (SI Appendix, Table S18). Our imputation process generally increased the number of called/inferred variants available for study (mean = 29,497,786; SD = 246,336; median = 29,591,650), reduced the proportion of missing SNPs (mean = 0.041; SD = 0.008; median = 0.038), and increased the proportion of called/inferred genotypes (mean = 0.959; SD = 0.008; median = 0.962) (SI Appendix, Table S18). Specific to the genetic height scoring analysis, 1,658 SNPs were included, and of these markers, 155 were fully imputed variants (sites that were completely missing among all individuals).
A remaining limitation impacts paleogenomic studies in general, whether diploid genotypes were inferred via imputation or a “pseudohaploid” approach related to the broadly low coverage of ancient genomic data, which could impact the predictive accuracy of polygenic scores. To consider the potential effects of this phenomenon on imputation and genotype accuracy in our dataset, we evaluated how sequence coverage may have impacted our imputation accuracy (i.e., whether imputation is outperforming under high vs. low sequence coverage conditions). Specifically, we compared our imputed genotype data in the full dataset of the high-coverage Loschbour individual (∼16×) (115) with down-sampled BAM files (using SAMtools -s parameter) (128) from 3× coverage to 0.3× for chromosome 1 using SnpSift (139). We obtained a concordance rate (in terms of total sites recovered) of ∼97 to 99%, suggesting that imputation accuracy is not dramatically lower in the low-coverage imputed genotype data (SI Appendix, Table S19). We also assessed the imputation accuracy of heterozygous sites by comparing the not imputed high-coverage genotype data for Loschbour with each down-sampled imputed BAM file from 0.3× to 3× coverage. At the lowest coverage of 0.3×, ∼85% of the heterozygous sites are still recovered with increase to ∼98% at 3× coverage (SI Appendix, Table S20).
Polygenic Height Scores.
Polygenic scores were computed by downloading a publicly available GWAS dataset from the UK Biobank (77), specifically genome-wide summary statistics available from the Neale Lab (140). The quality controls implemented for the publicly available UK Biobank dataset (e.g., minor allele frequency > 0.1%, Hardy–Weinberg equilibrium P value > 1e-10 in 337,199 individuals) contain 10.8 million analyzable SNPs (141). The variant identifications (“variants.tsv.bgz”) were merged with the “standing height GWAS” file (“50_irnt.gwas.imputed_v3.both_sexes.tsv.bgz”), where the beta values represent the effect size of the “ALT” (alternate) allele subsequently used for performing the polygenic scores.
For our data, polygenic height scores were estimated using PLINK 1.9 (142) with clumping of independent SNPs. Clumping was used to identify the SNP with the lowest P value in each linkage disequilibrium (LD) block (142). This approach retains SNPs with the strongest statistical evidence while reducing the correlation between the remaining SNPs (143). Although all common SNPs could be used in polygenic scoring, clumping to remove SNPs that have limited statistical association is also a practical approach (144). Clumping was performed at the genome-wide P value 5e-08 using PLINK 1.9 parameters “–clump-r2 0.1” and “–clump-kb 1000” with the 1000 Genomes “Europeans” reference population panel to retain the most correlated SNPs (“index SNPs”) from the UK Biobank height summary statistics, which were then used to calculate the polygenic height scores. Polygenic scores were calculated using “–geno 0” to exclude missing genetic markers and the “–score” flag, extracting the specific index SNPs (131).
To help assess whether the level of portability between modern reference (UK Biobank) and ancient (“genetically distant”) individuals in our study was in line with expectations, we created approximately equivalent SNP and sample size datasets for individuals who were excluded from the UK Biobank cohort (“non-GWAS” individuals; conducted using the UK Biobank Resource under application no. 31063 to R.M.). These excluded individuals have differing genetic relatedness from the UK Biobank cohort considered in the stature GWAS, the data that served as the basis for our per-individual polygenic height score estimates. This non-GWAS UK Biobank cohort is a very rough but conceptually similar approximation to the sample of ancient individuals analyzed in our study.
Specifically, we evaluated the predictive accuracy between phenotypic and predicted genetic height (r2) for two sample sets (n = 361,182 GWAS and n = 4,712 non-GWAS individuals), each stratified by sex and for both sexes combined. A dataset of 784,256 genotyped SNPs was subject to LD clumping with parameters similar to those in our ancient DNA analysis (using PLINK 1.9). Postclumping, 5,183 SNPs were retained at the genome-wide significance level. We also performed a down sampling (100 random draws) of the full set of GWAS and non-GWAS cohorts to 167 individuals.
For females and males combined, the non-GWAS individuals exhibit slightly lower variance in the relationship between phenotypic height and predicted genetic height (r2 = 0.031) relative to the GWAS cohort (r2 = 0.04) (SI Appendix, Fig. S10 and Table S17). For the down-sampled data (167 individuals), there is similarly a slightly lower variance in the relationship between stature differences and polygenic height scores among non-GWAS individuals (r2 = 0.037) relative to the GWAS cohort (r2 = 0.043). Relative to these results, the variance explained by polygenic height score in our ancient sample (r2 = 0.0796) is not exceedingly low.
Long Bone Measurement and Stature Reconstruction.
Both newly collected (n = 93 individuals) and previously collected/published (n = 54) osteological data were included in this study (Dataset S1). For n = 20 of the latter set of individuals, only precalculated terminal height estimates (based on unavailable long bone length measurements) were available (Dataset S1). Only adult individuals were included in our study. For newly collected data, this assessment was based on the complete fusion of all long bone epiphyses; we otherwise relied on classifications of “adult” in the published record (which were likely based on the same criterion).
Permissions to collect new long bone measurement data were coordinated with researchers (coauthors on this publication) affiliated with the museums and university departments housing the various individuals. An osteometric board was used to measure the maximum length measurements (to the nearest millimeter) of the femur, tibia, radius, and humerus following standard osteological methods (44, 144, 145). Intact long bones were selected, either the left or right side depending on availability and preservation; if both sides were available and fully preserved, then both were measured.
We used a regression-based approach to reconstruct osteological stature from Ruff et al. (44). These equations were developed using 501 individuals from across Europe ∼7,000 BC to AD 1900, broadly approximating the geographical and temporal span of the individuals in our dataset. Sex-specific regression equations for the femur, tibia, humerus, and radius were used (SI Appendix, Table S1), with SE estimates ranging from 1.66 to 2.73% (44). For the tibia, separate “north” and “south” equations are available (44). Given the potential for migration occurring across the temporal and cultural periods in our dataset, we computed estimates from both equations and averaged them for the tibia-derived stature estimate for all individuals in our study (with available tibia measurements) regardless of geographic origin. When measurements from multiple different bones from the same individual were available, stature estimates derived from each of the different bones were estimated separately and then averaged to obtain a single point estimate per individual.
Paleopathological Indicators of Nonspecific Stress.
For paleopathological evaluations, 82 individuals were newly characterized, and 16 were published/previously characterized (SI Appendix, Table S10). Crania with at least one permanent incisor were examined to record the presence or absence and severity of three skeletal indicators of nonspecific stress: porotic hyperostosis, cribra orbitalia, and linear enamel hypoplasia (21, 22, 27, 28). Cribra orbitalia was assessed according to Stuart-Macadam (21) on a five-stage scale of severity (n = 81 evaluated) and whether the lesions were healed or active (n = 80) (21). Porotic hyperostosis was evaluated on a three-stage scale (n = 80) and whether the lesions were healed or active (n = 79) (22), and linear enamel hypoplasia was assessed as present or absent (i.e., whether one or more linear bands of decreased enamel thickness were visible; n = 81) (27). The differences in the number of individuals assessed for healed or active lesions are because of the exclusion of a previously published individual (n = 1 Neolithic) (29) identified as having active lesions and n = 1 Mesolithic individual for whom the nature of the porotic hyperostosis lesions was unspecified (146).
Statistical Analyses.
Statistical analyses were conducted in RStudio (v1.2.5033). Our main linear model was generated using osteological height and genetic height scores with sex as a covariate. Data normality was assessed using “ggResidpanel” (v0.3.0) (147) (SI Appendix, Fig. S11). The residuals from this model were the basis for downstream analyses comparing patterns of stature variation across cultural periods as well as with the paleopathology data. Statistical analyses (t tests) on the residuals from various linear models (including those below) were performed, and the results are provided in SI Appendix, Tables S1–S21.
We performed two additional analytical iterations of our linear model to evaluate consistency of downstream results. First, we included latitude and longitude as additional factors in the linear model framework described above. Second, we included factors related to genetic ancestries variation into our linear model following the approach of Cox et al. (84) using MDS rather than PCA.
Following Cox et al. (84), we used an MDS-based rather than a PCA-based approach to account for population stratification. PCA uses genetic correlations among individuals to represent population structure (148, 149), while MDS converts population structure into a matrix of observed (pairwise) genetic distance among individuals (based on similarity/dissimilarity) (85, 142, 150). Essentially, MDS configures the coordinates of the data in a low-dimensional space in a way that reflects the pairwise relationships of the original data, which is appropriate for large genomic datasets, particularly when the data are nonlinear (i.e., missing information) (151). In this way, MDS (152) reduces the “noise” from the combined influences of genetic variation and phenotypic association on height by accounting for the potential risk of false positives due to residual population stratification from individual ancestries (85).
Specifically, we performed the MDS analysis in PLINK (v1.9) (142) using the full genome-wide SNP genotype data available for all of the individuals in our study. We generated the “plink.genome” file (plink –file <input_plink_files> –genome), which was used as input for the MDS analysis specifying four dimensions (plink –file <input_plink_files> –read-genome plink.genome –cluster –mds-plot 4). The first four components (C1, C2, C3, and C4) were each included in the linear model.
Since the selection of the number of MDS components is somewhat arbitrary (but following the methods of another study) (84), we also fit increasing numbers of components (from five to eight) and then performed all subsequent analyses accordingly. In each case, the signal of decreased but muted patterns of reduced stature among Neolithic individuals relative to other cultural periods is replicated (e.g., Neolithic average residual for MDS 8 = −0.589 ± 6.8 cm vs. −0.603 ± 6.9 cm for MDS 5) (SI Appendix, Fig. S12 and Table S21).
Supplementary Material
Acknowledgments
We thank the many researchers and skeletal collections curators for their positive responses to us and their interest in this study, even when materials or data were unavailable for inclusion. Components of this work were supported by Wenner-Gren Foundation Grant 222377 (to S.M.), NIH Grant R01-GM115656 (to G.H.P.), and the Harry J. and Elissa M. Sichi Early Career Professorship in Anthropology (to G.H.P.). P.V. was supported by Ministry of Culture of the Czech Republic Grant DKRVO/2019-2023/7.I.d, National Museum, 00023272. J Kolář was supported by Long-Term Development Project RV67985939 of the Institute of Botany of the Czech Academy of Sciences and Czech Science Foundation Grant 19-20970Y. B.G. was supported by Marie Skłodowska-Curie Grant H2020-MSCA-IF-2015 -703373. T.S., K.K., and T.H. were supported by a grant of Hungarian Research, Development and Innovation Office Projects FK128013 and K124326. M.N. was supported by Croatian Science Fund Grant HRZZ IP-2016-06-1450. G.H.P. thanks the DFG Center for Advanced Studies “Words, Bones, Genes, Tools” at the University of Tübingen for support. Computations for this research were performed on the Pennsylvania State University’s Institute for Computational and Data Sciences’ supercomputing cluster. This content is solely the responsibility of the authors and does not necessarily represent the views of the Institute for Computational and Data Sciences.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106743119/-/DCSupplemental.
Data Availability
Imputed genotypes for the 167 ancient individuals included in the dataset are available at Dryad (https://doi.org/10.5061/dryad.b5mkkwhfp). Although no paleogenomic data were newly generated directly for this study, for n = 28 individuals the analyzed ancient DNA data are from primary manuscripts in preparation (data were generated using laboratory methods as in ref. 68 and processed using publicly available software at GitHub [https://github.com/DReichLab/ADNA-Tools]); while a formal description of the data from these 28 individuals from a population genetic point of view will come in future work, these data have been made available as aligned sequence files (.bam files) through the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/view/PRJEB51250). All scripts related to genotype calling, imputation, polygenic scoring, and statistical analyses used throughout the study are available at GitHub (https://github.com/smmarciniak/aDNA_osteo_height) (131). All other data (osteological measurements, final stature estimates, and other skeletal individual-level information; e.g., identification, sex, radiocarbon dates, archaeological/cultural period, geographical coordinates, publication sources, and accession numbers for the ancient DNA data) are in the manuscript, SI Appendix, or Dataset S1. Previously published ancient DNA sequence data were used from the following studies: refs. 65–69 and 104–122.
References
- 1.Zeder M. A., The origins of agriculture in the Near East. Curr. Anthropol. 52, 221 (2011). [Google Scholar]
- 2.Bar-Yosef O., Meadow R. H., “The origins of agriculture in the Near East” in Last Hunters, First Farmers: New Perspectives on the Prehistoric Transition to Agriculture, Price T. D., Gebauer A.-B., Eds. (School of American Research Press, 1995), pp. 39–94. [Google Scholar]
- 3.Pinhasi R., von Cramon-Taubadel N., Craniometric data supports demic diffusion model for the spread of agriculture into Europe. PLoS One 4, e6747 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogucki P., The spread of early farming in Europe. Am. Sci. 84, 242–253 (1996). [Google Scholar]
- 5.Fort J., Demic and cultural diffusion propagated the Neolithic transition across different regions of Europe. J. R. Soc. Interface 12, 20150166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gignoux C. R., Henn B. M., Mountain J. L., Rapid, global demographic expansions after the origins of agriculture. Proc. Natl. Acad. Sci. U.S.A. 108, 6044–6049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith B. D., The origins of agriculture in the Americas. Evol. Anthropol. 3, 174–184 (2005). [Google Scholar]
- 8.Page A. E., et al. , Reproductive trade-offs in extant hunter-gatherers suggest adaptive mechanism for the Neolithic expansion. Proc. Natl. Acad. Sci. U.S.A. 113, 4694–4699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert P. M., Health vs. fitness: Competing themes in the origins and spread of agriculture? Curr. Anthropol. 50, 603–608 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Armelagos G. J., Goodman A. H., Jacobs K. H., The origins of agriculture: Population growth during a period of declining health. Popul. Environ. 13, 9–22 (1991). [Google Scholar]
- 11.Zimmermann A., Hilpert J., Wendt K. P., Estimations of population density for selected periods between the Neolithic and AD 1800. Hum. Biol. 81, 357–380 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Bocquet-Appel J.-P., The Neolithic demographic transition, population pressure and cultural change. Comp. Civilizations Rev. 58, 36–49 (2008). [Google Scholar]
- 13.Cohen M. N., Armelagos G. J., Paleopathology at the Origins of Agriculture, Cohen M. N., Armelagos G. J., Eds. (University Press of Florida, 1984). [Google Scholar]
- 14.Macintosh A. A., Pinhasi R., Stock J. T., Early life conditions and physiological stress following the transition to farming in Central/Southeast Europe: Skeletal growth impairment and 6000 years of gradual recovery. PLoS One 11, e0148468 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennike P., Alexandersen V., “Population plasticity in southern Scandinavia: From oysters and fish to gruel and meat” in Ancient Health: Skeletal Indicators of Agricultural and Economic Intensification, Cohen M. N., Crane-Kramer G., Eds. (University Press of Florida, 2007), pp. 130–148. [Google Scholar]
- 16.Smith P., Horwitz L., “Ancestors and inheritors: A bioanthropological perspective on the transition to agropastoralism in the Southern Levant” in Ancient Health: Skeletal Indicators of Agricultural and Economic Intensification, Cohen M. N., Crane-Kramer G., Eds. (University Press of Florida, 2007), pp. 207–222. [Google Scholar]
- 17.Larsen C. S., The agricultural revolution as environmental catastrophe: Implications for health and lifestyle in the Holocene. Quat. Int. 150, 12–20 (2006). [Google Scholar]
- 18.Larsen C. S., Biological changes in human populations with agriculture. Annu. Rev. Anthropol. 24, 185–213 (1995). [Google Scholar]
- 19.Steckel R. H., Rose J. C., Eds., The Backbone of History: Health and Nutrition in the Western Hemisphere (Cambridge University Press, 2002). [Google Scholar]
- 20.Pinhasi R., Stock J. T., Human Bioarchaeology of the Transition to Agriculture, Pinhasi R., Stock J., Eds. (John Wiley & Sons, 2011). [Google Scholar]
- 21.Stuart-Macadam P., “Anemia in Roman Britain: Poundbury Camp” in Health in Past Societies. Biocultural Interpretations of Human Skeletal Remains in Archaeological Contexts, Bush H., Zvelebil M., Eds. (BAR International Series, Tempus Reparatum, Oxford, United Kingdom, 1991), vol. 567, pp. 101–113. [Google Scholar]
- 22.Stuart-Macadam P., Porotic hyperostosis: Representative of a childhood condition. Am. J. Phys. Anthropol. 66, 391–398 (1985). [DOI] [PubMed] [Google Scholar]
- 23.Rivera F., Mirazón Lahr M., New evidence suggesting a dissociated etiology for cribra orbitalia and porotic hyperostosis. Am. J. Phys. Anthropol. 164, 76–96 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Walker P. L., Bathurst R. R., Richman R., Gjerdrum T., Andrushko V. A., The causes of porotic hyperostosis and cribra orbitalia: A reappraisal of the iron-deficiency-anemia hypothesis. Am. J. Phys. Anthropol. 139, 109–125 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Wapler U., Crubézy E., Schultz M., Is cribra orbitalia synonymous with anemia? Analysis and interpretation of cranial pathology in Sudan. Am. J. Phys. Anthropol. 123, 333–339 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Brickley M. B., Cribra orbitalia and porotic hyperostosis: A biological approach to diagnosis. Am. J. Phys. Anthropol. 167, 896–902 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Goodman A. H., Armelagos G. J., Rose J. C., Enamel hypoplasias as indicators of stress in three prehistoric populations from Illinois. Hum. Biol. 52, 515–528 (1980). [PubMed] [Google Scholar]
- 28.Goodman A. H., Rose J. C., “Dental enamel hypoplasias as indicators of nutritional status” in Advances in Dental Anthropology, Kelley M., Larsen C. S., Eds. (Wiley-Liss Inc., 1991), pp. 279–293. [Google Scholar]
- 29.Ash A., et al. , Regional differences in health, diet and weaning patterns amongst the first Neolithic farmers of central Europe. Sci. Rep. 6, 29458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holder S., Miliauskienė Ž., Jankauskas R., Dupras T., An integrative approach to studying plasticity in growth disruption and outcomes: A bioarchaeological case study of Napoleonic soldiers. Am. J. Hum. Biol. 33, e23457 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Dabbs G. R., Health status among prehistoric Eskimos from Point Hope, Alaska. Am. J. Phys. Anthropol. 146, 94–103 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Eshed V., Gopher A., Pinhasi R., Hershkovitz I., Paleopathology and the origin of agriculture in the Levant. Am. J. Phys. Anthropol. 143, 121–133 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Starling A. P., Stock J. T., Dental indicators of health and stress in early Egyptian and Nubian agriculturalists: A difficult transition and gradual recovery. Am. J. Phys. Anthropol. 134, 520–528 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Zakrzewski S. R., Variation in ancient Egyptian stature and body proportions. Am. J. Phys. Anthropol. 121, 219–229 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Formicola V., Giannecchini M., Evolutionary trends of stature in upper Paleolithic and Mesolithic Europe. J. Hum. Evol. 36, 319–333 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Holt B. M., Formicola V., Hunters of the Ice Age: The biology of Upper Paleolithic people. Am. J. Phys. Anthropol. 137 (suppl. 47), 70–99 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Hoppa R. D., Saunders S. R., Human Growth in the Past: Studies from Bones and Teeth, Hoppa R. D., Saunders S. R., Eds. (Cambridge University Press, 1999). [Google Scholar]
- 38.Neves W., Wesolowski V., “Economy, nutrition, and disease in prehistoric coastal Brazil: A case study from the state of Santa Catarina” in The Backbone of History: Health and Nutrition in the Western Hemisphere, Steckel R., Rose J. C., Eds. (Cambridge University Press, 2002), pp. 376–400. [Google Scholar]
- 39.Vercellotti G., et al. , Exploring the multidimensionality of stature variation in the past through comparisons of archaeological and living populations. Am. J. Phys. Anthropol. 155, 229–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eveleth P. B., Tanner J. M., Worldwide Variation in Human Growth (Cambridge University Press, ed. 2, 1991). [Google Scholar]
- 41.Eveleth P. B., “Population differences in growth: Environmental and genetic factors” in Human Growth, Falkner F., Tanner J. M., Eds. (Springer, 1979), pp. 373–394. [Google Scholar]
- 42.Tanner J. M., “Introduction: Growth in height as a mirror of the standard of living.” in Stature, Living Standards, and Economic Development: Essays in Anthropometric History, Komlos J., Ed. (Chicago Press, Chicago, IL, 1994), pp. 1–6. [Google Scholar]
- 43.Steckel R. H., Stature and the standard of living. J. Econ. Lit. 33, 1903–1940 (1995). [Google Scholar]
- 44.Ruff C. B., et al. , Stature and body mass estimation from skeletal remains in the European Holocene. Am. J. Phys. Anthropol. 148, 601–617 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Raxter M. H., Ruff C. B., Auerbach B. M., Technical note: Revised fully stature estimation technique. Am. J. Phys. Anthropol. 133, 817–818 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Vercellotti G., Agnew A. M., Justus H. M., Sciulli P. W., Stature estimation in an early medieval (XI-XII c.) Polish population: Testing the accuracy of regression equations in a bioarcheological sample. Am. J. Phys. Anthropol. 140, 135–142 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Formicola V., Franceschi M., Regression equations for estimating stature from long bones of early holocene European samples. Am. J. Phys. Anthropol. 100, 83–88 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Sladek V., et al. , “Central European human postcranial variation” in Skeletal Variation and Adaptation in Europeans: Upper Paleolithic to the Twentieth Century, Ruff C. B., Ed. (Wiley-Blackwell, 2018), pp. 315–354. [Google Scholar]
- 49.Hermanussen M., Stature of early Europeans. Hormones (Athens) 2, 175–178 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Piontek J., Vančata V., Transition to agriculture in Europe: Evolutionary trends in body size and body shape. Ecol. Asp. Past Hum. Settlements Eur. Bienn. Books EAA 2, 61–92 (2002). [Google Scholar]
- 51.Goodman A. H., Lallo J. W., Armelagos G. J., Rose J. C., “Health change at Dickson Mounds, Illinois (A.D. 950–1300).” in Paleopathology at the Origins of Agriculture, Cohen M. N., Armelagos G. J., Eds. (Academic Press, 1984), pp. 271–306. [Google Scholar]
- 52.Walker P. L., Thornton R., “Health, nutrition, and demographic change in native California” in The Backbone of History: Health and Nutrition in the Western Hemisphere, Steckel R. H., Rose J. C., Eds. (Cambridge University Press, 2002), pp. 506–523. [Google Scholar]
- 53.Lambert P. M., Health in prehistoric populations of the Santa Barbara Channel Islands. Am. Antiq. 58, 509–522 (1993). [Google Scholar]
- 54.Temple D. H., Patterns of systemic stress during the agricultural transition in prehistoric Japan. Am. J. Phys. Anthropol. 142, 112–124 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Pechenkina E. A., Benfer R. A. Jr., Zhijun W., Diet and health changes at the end of the Chinese neolithic: The Yangshao/Longshan transition in Shaanxi province. Am. J. Phys. Anthropol. 117, 15–36 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Angel J. L., “Health as a crucial factor in the changes from hunting to developed farming in the eastern Mediterranean” in Paleopathology at the Origins of Agriculture, Armelagos G. J., Cohen M. N., Eds. (Academic Press, 1984), pp. 51–74. [Google Scholar]
- 57.Meiklejohn C., Key P., “Socioeconomic change and patterns of pathology and variation in the Mesolithic and Neolithic of Western Europe: Some suggestions” in Paleopathology at the Origins of Agriculture, Cohen M. N., Armelagos G. J., Eds. (University Press of Florida, 1984), pp. 75–100. [Google Scholar]
- 58.Cox S. L., Ruff C. B., Maier R. M., Mathieson I., Genetic contributions to variation in human stature in prehistoric Europe. Proc. Natl. Acad. Sci. U.S.A. 116, 21484–21492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meiklejohn C., Zvelebil M., “Health status of European populations at the agricultural transition and the implications for the adoption of farming” in Health in Past Populations, Bush H., Zvelebil M., Eds. (Oxford, 1991), pp. 129–144. [Google Scholar]
- 60.Lango Allen H., et al. , Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silventoinen K., Determinants of variation in adult body height. J. Biosoc. Sci. 35, 263–285 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Yang J., et al. , Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stulp G., Barrett L., Evolutionary perspectives on human height variation. Biol. Rev. Camb. Philos. Soc. 91, 206–234 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Mathieson I., et al. , Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martiniano R., et al. , The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods. PLoS Genet. 13, e1006852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olalde I., et al. , The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 555, 190–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brace S., et al. , Ancient genomes indicate population replacement in Early Neolithic Britain. Nat. Ecol. Evol. 3, 765–771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathieson I., et al. , The genomic history of southeastern Europe. Nature 555, 197–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haak W., et al. , Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larsen C. S., et al. , Bioarchaeology of Neolithic Çatalhöyük reveals fundamental transitions in health, mobility, and lifestyle in early farmers. Proc. Natl. Acad. Sci. U.S.A. 116, 12615–12623 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holt B. M., Mobility in Upper Paleolithic and Mesolithic Europe: Evidence from the lower limb. Am. J. Phys. Anthropol. 122, 200–215 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Larsen C. S., Hutchinson D. L., Stojanowski C. M., “Health and lifestyle in Georgia and Florida: Agricultural origins and intensification in regional perspective” in Ancient Health: Skeletal Indicators of Economic and Political Intensification, Cohen M. N., Crane-Kramer G. M. M., Eds. (University Press of Florida, 2007), pp. 20–34. [Google Scholar]
- 73.Cardoso H. F. V., Gomes J. E. A., Trends in adult stature of peoples who inhabited the modern Portuguese territory from the Mesolithic to the late 20th century. Int. J. Osteoarchaeol. 19, 711–725 (2009). [Google Scholar]
- 74.Roberts C. A., Cox M., “The impact of economic intensification and social complexity on human health in Britain from 6000 BP (Neolithic) and the introduction of farming to the mid-nineteenth century AD” in Ancient Health: Skeletal Indicators of Agricultural and Economic Intensification, Cohen M. N., Crane-Kramer G. M. M., Eds. (University Press of Florida, 2007), pp. 149–163. [Google Scholar]
- 75.Hui R., D’Atanasio E., Cassidy L. M., Scheib C. L., Kivisild T., Evaluating genotype imputation pipeline for ultra-low coverage ancient genomes. Sci. Rep. 10, 18542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gelabert P., Olalde I., de-Dios T., Civit S., Lalueza-Fox C., Malaria was a weak selective force in ancient Europeans. Sci. Rep. 7, 1377 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bycroft C., et al. , The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Briggs A. W., et al. , Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 38, e87–e87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Briggs A. W., et al. , Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U.S.A. 104, 14616–14621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dabney J., Meyer M., Pääbo S., Ancient DNA damage. Cold Spring Harb. Perspect. Biol. 5, a012567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mostafavi H., et al. , Variable prediction accuracy of polygenic scores within an ancestry group. eLife 9, e48376 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duncan L., et al. , Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 10, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zaidi A. A., Mathieson I., Demographic history mediates the effect of stratification on polygenic scores. eLife 9, e61548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cox S. L., et al. , Predicting skeletal stature using ancient DNA. Am. J. Biol. Anthropol. 177, 162–174 (2022). [Google Scholar]
- 85.Wang D., et al. , Comparison of methods for correcting population stratification in a genome-wide association study of rheumatoid arthritis: Principal-component analysis vs. multidimensional scaling. BMC Proc. 3 (suppl. 7), S109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Irving-Pease E. K., Muktupavela R., Dannemann M., Racimo F., Quantitative human paleogenetics: What can ancient DNA tell us about complex trait evolution? Front. Genet. 12, 703541 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mummert A., Esche E., Robinson J., Armelagos G. J., Stature and robusticity during the agricultural transition: Evidence from the bioarchaeological record. Econ. Hum. Biol. 9, 284–301 (2011). [DOI] [PubMed] [Google Scholar]
- 88.Larsen C. S., “Foraging to farming transition: Global health impacts, trends, and variation” in Encyclopedia of Global Archaeology, Smith C., Ed. (Springer, New York, NY, 2014), pp. 2818–2824. [Google Scholar]
- 89.Milner G. R., Early agriculture’s toll on human health. Proc. Natl. Acad. Sci. U.S.A. 116, 13721–13723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barker G., Richards M. B., Foraging–farming transitions in Island Southeast Asia. J. Archaeol. Method Theory 20, 256–280 (2013). [Google Scholar]
- 91.Lander F., Russell T., The archaeological evidence for the appearance of pastoralism and farming in southern Africa. PLoS One 13, e0198941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fuller D. Q., Kingwell-Banham E. J., Lucas L., Murphy C., Stevens C., Comparing pathways to agriculture. Archaeol. Int. 18, 61–66 (2015). [Google Scholar]
- 93.Cameron M. E., The Riet River sites: Positioning regional diversity in the introduction of domesticated livestock to southern Africa. J. Archaeol. Sci. Rep. 23, 72–79 (2019). [Google Scholar]
- 94.Rosenstock E., et al. , Human stature in the Near East and Europe ca. 10,000–1000 BC: Its spatiotemporal development in a Bayesian errors-in-variables model. Archaeol. Anthropol. Sci. 11, 5657–5690 (2019). [Google Scholar]
- 95.Niskanen M., Ruff C. B., Holt B., Sladek V., Berner M., “Temporal and geographic variation in body size and shape of Europeans from the Late Pleistocene to recent times” in Skeletal Variation and Adaptation in Europeans: Upper Paleolithic to the Twentieth Century, Ruff C. B., Ed. (John Wiley & Sons, 2018), pp. 49–90. [Google Scholar]
- 96.De La Vega F. M., Bustamante C. D., Polygenic risk scores: A biased prediction? Genome Med. 10, 100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martin A. R., et al. , Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sohail M., et al. , Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. eLife 8, e39702 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berg J. J., et al. , Reduced signal for polygenic adaptation of height in UK Biobank. eLife 8, e39725 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bitarello B. D., Mathieson I., Polygenic scores for height in admixed populations. G3 (Bethesda) 10, 4027–4036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Refoyo-Martínez A., et al. , How robust are cross-population signatures of polygenic adaptation in humans? Peer Community J. 1, e22 (2021). [Google Scholar]
- 102.Yair S., Coop G., Population differentiation of polygenic score predictions under stabilizing selection. bioRxiv [Preprint] (2022). 10.1101/2021.09.10.459833 (Accessed 10 February 2022). [DOI] [PMC free article] [PubMed]
- 103.Berens A. J., Cooper T. L., Lachance J., The genomic health of ancient hominins. Hum. Biol. 89, 7–19 (2017). [DOI] [PubMed] [Google Scholar]
- 104.González-Fortes G., et al. , Paleogenomic evidence for multi-generational mixing between Neolithic farmers and Mesolithic hunter-gatherers in the Lower Danube Basin. Curr. Biol. 27, 1801–1810.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cassidy L. M., et al. , Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc. Natl. Acad. Sci. U.S.A. 113, 368–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jones E. R., et al. , The Neolithic transition in the Baltic was not driven by admixture with early European farmers. Curr. Biol. 27, 576–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Skoglund P., et al. , Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science 344, 747–750 (2014). [DOI] [PubMed] [Google Scholar]
- 108.Lipson M., et al. , Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature 551, 368–372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mittnik A., et al. , The genetic prehistory of the Baltic Sea region. Nat. Commun. 9, 442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Allentoft M. E., et al. , Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- 111.Keller A., et al. , New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat. Commun. 3, 698 (2012). [DOI] [PubMed] [Google Scholar]
- 112.Seguin-Orlando A., et al. , Paleogenomics. Genomic structure in Europeans dating back at least 36,200 years. Science 346, 1113–1118 (2014). [DOI] [PubMed] [Google Scholar]
- 113.Sikora M., et al. , Ancient genomes show social and reproductive behavior of early Upper Paleolithic foragers. Science 358, 659–662 (2017). [DOI] [PubMed] [Google Scholar]
- 114.Fu Q., et al. , The genetic history of Ice Age Europe. Nature 534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lazaridis I., et al. , Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Olalde I., et al. , Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507, 225–228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sikora M., et al. , The population history of northeastern Siberia since the Pleistocene. Nature 570, 182–188 (2019). [DOI] [PubMed] [Google Scholar]
- 118.Marcus J. H., et al. , Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia. Nat. Commun. 11, 939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Furtwängler A., et al. , Ancient genomes reveal social and genetic structure of Late Neolithic Switzerland. Nat. Commun. 11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rivollat M., et al. , Ancient genome-wide DNA from France highlights the complexity of interactions between Mesolithic hunter-gatherers and Neolithic farmers. Sci. Adv. 6, eaaz5344 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jones E. R., et al. , Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 6, 10.1038/ncomms9912 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Freilich S., et al. , Reconstructing genetic histories and social organisation in Neolithic and Bronze Age Croatia. Sci. Rep. 11, 16729 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Renaud G., Stenzel U., Kelso J., leeHom: Adaptor trimming and merging for Illumina sequencing reads. Nucleic Acids Res. 42, e141–e141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jónsson H., Ginolhac A., Schubert M., Johnson P. L. F., Orlando L., mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.H. Li, Seqtk. https://github.com/lh3/seqtk. Accessed 8 May 2018. [Google Scholar]
- 127.Gamba C., et al. , Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 5, 5257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Broad Institute, Picard. http://broadinstitute.github.io/picard. Accessed 14 May 2018. [Google Scholar]
- 130.McKenna A., et al. , The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.S. Marciniak, aDNA_osteo_height. GitHub. https://github.com/smmarciniak/aDNA_osteo_height. Deposited 31 March 2021. [Google Scholar]
- 132.Auton A., et al. ; 1000 Genomes Project Consortium, A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Günther T., Nettelblad C., The presence and impact of reference bias on population genomic studies of prehistoric human populations. PLoS Genet. 15, e1008302 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Browning S. R., Browning B. L., Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.1000 Genomes Project, Phase 3 dataset. https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/. Accessed 6 June 2018. [Google Scholar]
- 136.R. Hui, pmd_filter.py. https://github.com/ryhui/imputation-pipeline/blob/master/pmd_filter.py. Accessed 1 November 2020. [Google Scholar]
- 137.1000 Genomes Project, Phase 3 reference panel (version 5a). http://bochet.gcc.biostat.washington.edu/beagle/1000_Genomes_phase3_v5a/. Accessed 6 June 2018. [Google Scholar]
- 138.HapMap, genomic maps. http://bochet.gcc.biostat.washington.edu/beagle/genetic_maps/. Accessed 6 June 2018. [Google Scholar]
- 139.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Neale Lab, GWAS of the UK Biobank, round 2. http://www.nealelab.is/uk-biobank. Accessed 8 May 2019. [Google Scholar]
- 141.D. Howrigan, Details and considerations of the UK Biobank GWAS. Neale Lab Blog, 20 September 2017. http://www.nealelab.is/blog/2017/9/11/details-and-considerations-of-the-uk-biobank-gwas. Accessed 5 July 2018. [Google Scholar]
- 142.Purcell S., et al. , PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Marees A. T., et al. , A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 27, e1608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Buikstra J. E., Ubelaker D. H., Standards for Data Collection from Human Skeletal Remains (Arkansas Archeological Survey Research Series (Arkansas Archeological Survey, 1994).
- 145.Ruff C., Skeletal Variation and Adaptation in Europeans: Upper Paleolithic to the Twentieth Century (Wiley-Blackwell, 2018). [Google Scholar]
- 146.Serrulla Rech F., Sanin Matias M., Forensic anthropological report of Elba. Cadernos do Laboratorio Xeolóxico de Laxe. Revista de Xeoloxía Galega e do Hercínico Peninsular 39, 35–72 (2017). [Google Scholar]
- 147.Goode K., Rey K., ggResidpanel: Panels and interactive versions of diagnostic plots using 'ggplot2.' R Package Version 0.3.0. https://cran.r-project.org/web/packages/ggResidpanel/index.html. Accessed 10 December 2020.
- 148.Patterson N., Price A. L., Reich D., Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Price A. L., et al. , Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 150.Li Q., Yu K., Improved correction for population stratification in genome-wide association studies by identifying hidden population structures. Genet. Epidemiol. 32, 215–226 (2008). [DOI] [PubMed] [Google Scholar]
- 151.Tzeng J., Lu H. H.-S., Li W.-H., Multidimensional scaling for large genomic data sets. BMC Bioinformatics 9, 179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Borg I., Groenen P. J. F., Modern Multidimensional Scaling: Theory and Applications (Springer, ed. 2, 2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Imputed genotypes for the 167 ancient individuals included in the dataset are available at Dryad (https://doi.org/10.5061/dryad.b5mkkwhfp). Although no paleogenomic data were newly generated directly for this study, for n = 28 individuals the analyzed ancient DNA data are from primary manuscripts in preparation (data were generated using laboratory methods as in ref. 68 and processed using publicly available software at GitHub [https://github.com/DReichLab/ADNA-Tools]); while a formal description of the data from these 28 individuals from a population genetic point of view will come in future work, these data have been made available as aligned sequence files (.bam files) through the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/view/PRJEB51250). All scripts related to genotype calling, imputation, polygenic scoring, and statistical analyses used throughout the study are available at GitHub (https://github.com/smmarciniak/aDNA_osteo_height) (131). All other data (osteological measurements, final stature estimates, and other skeletal individual-level information; e.g., identification, sex, radiocarbon dates, archaeological/cultural period, geographical coordinates, publication sources, and accession numbers for the ancient DNA data) are in the manuscript, SI Appendix, or Dataset S1. Previously published ancient DNA sequence data were used from the following studies: refs. 65–69 and 104–122.