Significance
CHARGE syndrome is a complex developmental disorder caused by mutations in CHD7 (chromodomain helicase DNA-binding protein-7). We identified Caenorhabditis elegans chd-7 in a screen for suppressors of dauer formation, an alternative larval stage that develops under harsh environmental conditions. We found chd-7 regulates tumor growth factor-β (TGF-β) signaling pathways both for dauer diapause and for development of the cuticle, a specialized extracellular matrix. In frog embryos, Chd7 promotes Col2a1 expression, which is necessary and sufficient to prevent CHARGE features. These studies establish a conserved role for Chd7 from worms to vertebrates in regulating the TGF-β signaling pathway. Genetic dissection of chd-7’s role in C. elegans may help to define the molecular and cellular events that contribute to CHARGE syndrome.
Keywords: CHARGE syndrome, chd-7, dauer, TGF-β, Col2a1
Abstract
CHARGE syndrome is a complex developmental disorder caused by mutations in the chromodomain helicase DNA-binding protein-7 (CHD7) and characterized by retarded growth and malformations in the heart and nervous system. Despite the public health relevance of this disorder, relevant cellular pathways and targets of CHD7 that relate to disease pathology are still poorly understood. Here we report that chd-7, the nematode ortholog of Chd7, is required for dauer morphogenesis, lifespan determination, stress response, and body size determination. Consistent with our discoveries, we found chd-7 to be allelic to scd-3, a previously identified dauer suppressor from the DAF-7/ tumor growth factor-β (TGF-β) pathway. Epistatic analysis places CHD-7 at the level of the DAF-3/DAF-5 complex, but we found that CHD-7 also directly impacts the expression of multiple components of this pathway. Transcriptomic analysis revealed that chd-7 mutants fail to repress daf-9 for execution of the dauer program. In addition, CHD-7 regulates the DBL-1/BMP pathway components and shares roles in male tail development and cuticle synthesis. To explore a potential conserved function for chd-7 in vertebrates, we used Xenopus laevis embryos, an established model to study craniofacial development. Morpholino-mediated knockdown of Chd7 led to a reduction in col2a1 messenger RNA (mRNA) levels, a collagen whose expression depends on TGF-β signaling. Both embryonic lethality and craniofacial defects in Chd7-depleted tadpoles were partially rescued by overexpression of col2a1 mRNA. We suggest that Chd7 has conserved roles in regulation of the TGF-β signaling pathway and pathogenic Chd7 could lead to a defective extracellular matrix deposition.
When Caenorhabditis elegans encounter crowding, starvation, or high temperature during early development, worms can halt reproductive programs to enter an alternative larval stage, known as dauer. Dauers are long-lived, highly stress-resistant, and exhibit altered motility and metabolism (1–4). Upon return to normal growth conditions, the larvae exit dauer and develop into fertile adults. Study of dauer formation mutants has provided fundamental insights into pathways affecting longevity, neurodevelopment, metabolism, autophagy, and neurodegeneration (1, 4–7).
The DAF-2/insulin/IGF1 (IIS) signaling pathway controls the dauer entry decision by coupling external cues with neuroendocrine signaling (8). In favorable conditions, DAF-2 activity initiates a conserved kinase cascade, leading to phosphorylation and inhibition of the transcription factor DAF-16/FOXO. In harsh environments, a decrease in the activity of DAF-2 and downstream components of the pathway leads to activation of DAF-16 and causes animals to arrest as dauers (9, 10). In addition to the DAF-2 pathway, DAF-7/tumor growth factor-β (TGF-β) signaling also regulates dauer development (11). When worms sense suitable conditions for reproductive development, ASI neurosensory cells secrete the DAF-7 ligand, which binds to DAF-1/4 receptors, leading to activation and phosphorylation of the R-SMAD complex DAF-8/14, promoting reproductive programs and inhibiting the prodauer complex composed of the SMAD protein DAF-3 and repressor DAF-5. Conversely, absence of DAF-7 leads to activation of the DAF-3/DAF-5 complex to promote dauer entry (12, 13).
The DAF-7/TGF-β and DAF-2/IIS were initially described as parallel pathways to regulate dauer entry (14), but recent observations suggest a strong, positive feedback between these pathways for dauer entry and longevity (15–20). First, decreased signaling through the TGF-β pathway leads to differential expression of many DAF-16–regulated genes with functions in longevity and dauer entry, such as SOD-3 and insulin peptides. This cross-activation of target genes may be important to amplify weak signals from each sensory pathway in order to make an all-or-none decision to enter dauer (15–17). Second, the longevity of daf-2 mutants can be blocked or enhanced by daf-5 and daf-3, respectively, suggesting that transcriptional components of the TGF-β pathway can modulate IIS-dependent longevity genes. Third, for dauer development, daf-16 is epistatic to daf-7/8/14 daf-c mutants (17, 18). Finally, both signaling pathways converge on daf-9 and daf-12 to integrate outputs for diapause entry (19, 20). Indeed, daf-9 expression levels are critical for both entering and exiting diapause (4, 19).
In chromatin immunoprecipitation (ChIP)-chip studies, we identified chd-7 as target of DAF-12, a nuclear receptor whose loss causes defective execution of dauer morphogenesis programs (21). chd-7 is an ortholog of CHD7 (chromodomain-helicase-DNA binding 7), which is the primary locus associated with CHARGE syndrome, a rare and severe neurodevelopmental disorder that affects the neural tube and neural crest cell derivatives, leading to hypogonadism, heart defects, and craniofacial anomalies among other features (22). Inactivating mutations in CHD7 are the predominant cause of CHARGE, accounting for greater than 90% of the cases (23). CHD7 is also mutated in Kallmann syndrome, a milder neurodevelopmental disorder with features overlapping with CHARGE, including impaired olfaction and hypogonadism (24). Exome sequencing studies in patients with autism spectrum disorders identified recurrent disruptive mutations in the related gene CHD8 (25).
The CHD proteins comprise a highly conserved family of SNF2-related ATP-dependent chromatin remodelers that are involved in chromatin remodeling and transcriptional regulation (26). Despite the public health relevance of these cognitive disorders, the mechanism of disease pathology due to mutations in CHD7/8 is poorly understood. The development of fly, fish, and mouse models of CHARGE has enabled characterization of associated dysfunction in model organisms, but our understanding of the underlying pathology of CHARGE is still incomplete (27–32). In C. elegans, CHD-7 has functions in habituation learning, normal locomotion, body size, and fecundity (33, 34). It contains a conserved ATPase/SNF2 domain and two chromodomains for nucleosome interaction. Being the only worm homolog of the class III CHD family, it contains a signature BRK domain (Brahma and Kismet domain) (Fig. 1E).
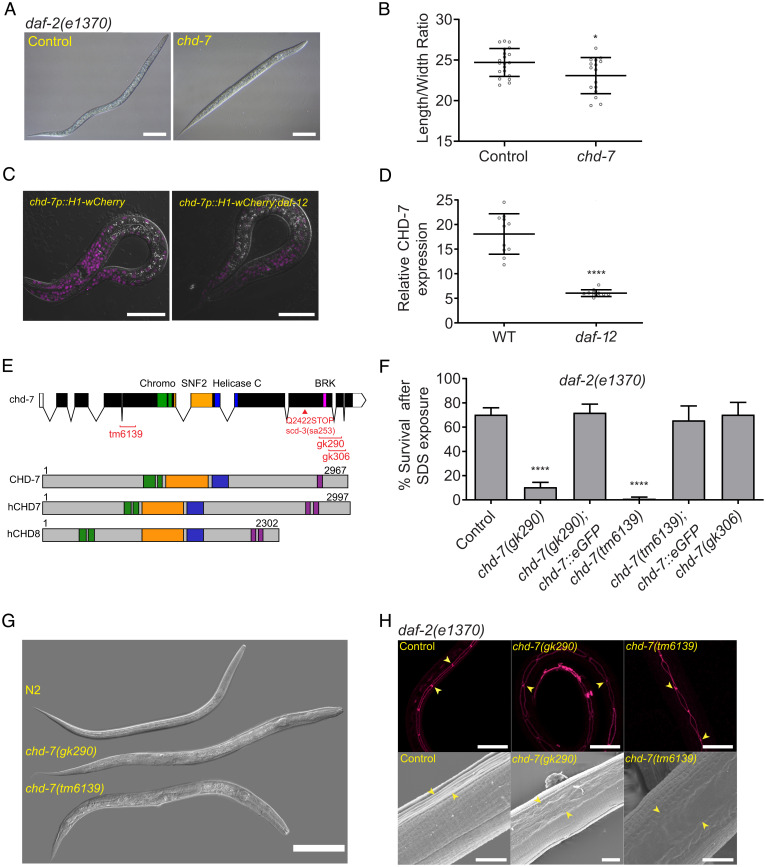
Fig. 1.
The DAF-12 regulated target chd-7 is required for proper dauer morphogenesis. (A) chd-7(RNAi) causes a partial dauer phenotype in daf-2(e1370). Representative DIC photomicrographs of normal and partial dauers from daf-2(e1370) exposed to Control (L4440) or chd-7 dsRNA, respectively. (Scale bars: 50 uM.) (B) Quantification of axial ratio of daf-2(e1370);control(RNAi) and daf-2(e1370);chd-7(RNAi) dauers. Three biological replicates were scored (n = 16 to 21 worms per replicate). Horizontal black lines represent mean with SD. Unpaired t test, *P < 0.05. (C) daf-12 regulates chd-7 expression. Representative images of chd-7 transcriptional reporter, chd-7p::H1-wCherry or chd-7p::H1-wCherry;daf-12(rh61rh411) worms at L2/L3 stage. (Scale bars: 20 uM.) (D) Relative expression of the transcriptional reporter (n > 10 per strain). Unpaired t test, ****P < 0.0001. (E) C. elegans chd-7 gene and protein. (Upper) chd-7 genomic region. UTR and exons shown as bars; introns by lines. In red, available chd-7 deletional alleles (data obtained from Caenorhadbitis Genome Center and National Bioresearch Project). (Lower) The predicted protein isoforms of C. elegans CHD-7, human CHD7, and human CHD8. Signature domains in CHD proteins: two N-terminal chromodomains for interaction with a variety of chromatin components (green), a SNF-2 like domain with ATPase activity (yellow), and a helicase domain (blue). The class III subfamily is defined by a BRK domain (purple). (F) chd-7(gk290);daf-2(e1370) and chd-7(tm6139);daf-2(e1370) develop as SDS-sensitive dauer larvae. n > 725 animals per strain tested. Bars and horizontal black lines represent mean percentage with SD. χ2 test with Bonferroni correction for multiple comparisons. ****P < 0.0001. Asterisks represent the comparison to daf-2(e1370). (G) chd-7(gk290) and chd-7(tm6139) prevent dauer development upon starvation of otherwise wild-type worms. Representative DIC photomicrographs of N2 dauers, arrested L3-like chd-7(gk290), and a small chd-7(tm6139) adult. (Scale bar: 100 uM.) (H) chd-7;daf-2 partial dauers fail to develop the dauer alae. (Upper) Representative photomicrographs of daf-2(e1370) dauers or chd-7;daf-2(e1370) partial dauers expressing the ajm-1::GFP reporter to delineate the seam cell borders (arrowheads mark a subset of junctions). (Scale bars: 20 uM.) (Lower) Scanning electron microscopy images of daf-2(e1370) dauers or chd-7;daf-2(e1370) partial dauers (arrowheads mark alae details). (Scale bars: 5uM.)
Here, we show that while CHD-7 can modulate multiple IIS-associated processes—including daf-2(e1370) dauer formation, longevity, and immunity—epistasis experiments place chd-7 in the DAF-7/TGF-β pathway that impinge on IIS signaling (15). In addition, CHD-7 affects daf-12, daf-14, daf-3, and daf-5 transcription levels, suggesting that it modulates the DAF-7/TGF-β pathway at multiple points. Whole-genome messenger RNA (mRNA) expression profiling of partial chd-7;daf-2 dauers show that chd-7 mutants fail to repress daf-9. We also found that CHD-7 binds and regulates dbl-1 and sma-2, key components of the BMP pathway, a conserved signaling pathway that regulates cuticle collagen expression from worms to mammals (35–39).
To explore a potential conserved function for chd-7 in vertebrates and study the relevance of our results for CHARGE etiology, we used Xenopus laevis. Disruption of Chd7 function in Xenopus embryos results in craniofacial defects that mimic CHD-dependent pathological phenotypes (40, 41). We demonstrate that Chd7 regulates expression of the collagen type-II α1 (col2a1), the main collagen protein of cartilage (42), whose expression depends on the TGF-β pathway (43, 44). Interestingly, craniofacial malformations and embryonic lethality due to chd7 knockdown can be rescued by col2a1 expression. These findings suggest a conserved function of Chd7/chd-7 in regulation of extracellular matrix (ECM) components and raise the intriguing possibility that defects in collagen expression may contribute to the craniofacial defects seen in Chd7/8-dependent syndromes.
Results
chd-7 Functions in Development of the Dauer Larva.
To identify novel regulators of dauer development, we previously used ChIP-chip to define DAF-12 target genes (21). Herein, we screened ∼300 targets, including chd-7, to determine which influenced dauer induction or morphogenesis. To identify such genes, we took advantage of the temperature-sensitive, constitutive dauer (daf-c) allele daf-2(e1371). Inactivation of chd-7, which contains seven DAF-12 binding sites in its promoter region (-4600, -4564, -2837, -2776, -1944, -1273, and -361) (21), led to development of defective, SDS-sensitive “partial” dauers in daf-2(e1371) animals grown at the nonpermissive temperature of 25 °C (9). To confirm these results, we assayed suppression of the more severe daf-2(e1370) mutation and discovered that chd-7(RNAi) produced a similar arrested partial dauer phenotype (45). As observed in Fig. 1 A and B, the axial ratio (length/width) of the partial dauers resulting from chd-7(RNAi) exhibited a significant reduction of these proportions and appeared to have defects in radial constriction of the dauer cuticle. By using a chd-7 transcriptional reporter (WBStrain00033709), we observed a substantial decrease in chd-7 expression in the daf-12(rh61rh411) background (Fig. 1 C and D). Thus, we infer that the binding of DAF-12 to the chd-7 promoter (21) up-regulates its expression.
To further validate our screen, we crossed daf-2(e1370) mutants with three chd-7 deletion alleles available from the Nematode Knockout Consortia (46). chd-7(tm6139) contains a 594-bp deletion that generates a frame shift and premature stop codon, eliminating all known protein domains (Fig. 1E). As shown in Fig. 1F, partial dauers were obtained when double mutants are grown at 25 °C, validating our interference RNA (RNAi) screen. Comparison of two C-terminal deletion alleles uncovered a critical role for the BRK domain in dauer formation. The chd-7(gk290) allele contains an 859-bp deletion that spans the BRK domain and introduces a frameshift that eliminates the last 356 aa. The chd-7(gk306) deletion is slightly more C-terminal, truncating the protein immediately after the BRK domain (Fig. 1E). When crossed into daf-2(e1370) worms, chd-7(gk306) developed normal dauer larvae, whereas chd-7(gk290) formed partial dauers (Fig. 1F). Importantly, a functional transgene expressing GFP-tagged CHD-7 protein (CHD-7::GFP) rescued the partial dauer phenotypes observed in chd-7(gk290);daf-2(e1370) and chd-7(tm6139);daf-2(e1370) mutants (Fig. 1F).
We then aimed to understand if loss of chd-7 also prevents normal dauer development in a daf-2(+) background. As shown in Fig. 1G, under starvation conditions that favor dauer arrest (47), N2 (wild-type) worms developed into dauers that were shorter than daf-2(e1370) dauers but, as expected, were resistant to SDS treatment (SI Appendix, Fig. S1). In contrast, chd-7 mutants showed complex phenotypes. Most of the chd-7(gk290) animals arrested as L3- and L4-like larvae (Fig. 1G and SI Appendix, Fig. S1). chd-7(tm6139) exhibited larval arrest and mortality at the L1 stage. Of the small fraction of animals that did progress, some developed into scrawny L3-like or L4-like larvae; some progressed to adulthood. These results are consistent with the requirement for chd-7 in the induction of dauer formation by starvation.
The dauer larva is characterized by a slim physique because of a reduction in the volume of ectodermal tissues, including the hypodermis, seam cells, and pharyngeal cells. In addition, the hypodermis produces the dauer cuticle, which confers protection against external damage and dehydration. During wild-type dauer formation, the seam cells adopt a stereotypical linear morphology and junctional association to produce the alae, bilateral ridges in the cuticle, that facilitate body motion. Dauer larvae also switch their metabolism to accumulate lipids to survive for longer periods (10). To further characterize the nature of the defects in the chd-7-induced partial dauers, we analyzed the seam using the adherens junction-associated protein marker AJM-1::GFP and interrogated the morphology of the cuticle using scanning electron microscopy. As shown in Fig. 1H, partial dauers exhibited defects in seam cell morphology and defective dauer alae formation. During the dauer transition, animals store fat in their intestinal and hypodermal cells, which is critical to survive during hibernation (48). We used the lipid-labeling dye Oil red O to examine fat storage. Unlike other partial dauer mutants (6), chd-7;daf-2 abnormal dauers did not exhibit fat-storage deficiencies (SI Appendix, Fig. S2).
During dauer, the global developmental arrest also impacts the germ cells, slowing their divisions and finally resulting in quiescence (49). We observed that the germline in chd-7;daf-2 mutant dauers was substantially larger than in control daf-2 dauers, arresting with a germline morphology that resembled the L3 larval stage (SI Appendix, Fig. S3). Therefore, we conclude that major morphological changes that occur during dauer formation of daf-2(e1370) fail to be executed in chd-7 mutants, including radial constriction of the body, formation of an SDS-resistant cuticle with dauer alae, and developmental arrest of the germline.
chd-7 Is Required for Longevity and Immunoresistance Induced by IIS Inactivation and Germline Removal.
In addition to dauer development, the IIS pathway also regulates longevity (1). Hence, we sought to investigate whether chd-7 also has roles in the determination of lifespan. First, we compared survival of wild-type (N2) worms with two chd-7 alleles and found that the dauer-defective allele chd-7(gk290) significantly shortened lifespan, whereas chd-7(gk306) had only a marginal effect on longevity (Fig. 2A). Surprisingly, the CHD-7::GFP rescue transgene also reduced N2 lifespan (Fig. 2B), suggesting that chd-7 copy number can influence longevity. We then analyzed how chd-7 affects longevity of IIS mutants. Remarkably, the dauer-defective allele chd-7(gk290), but not chd-7(gk306), shortened the lifespan extension of daf-2 mutants to an extent comparable with the null allele of daf-16, the key IIS downstream target (Fig. 2C) (1). Furthermore, daf-2 longevity was fully restored by CHD-7::GFP (Fig. 2D).
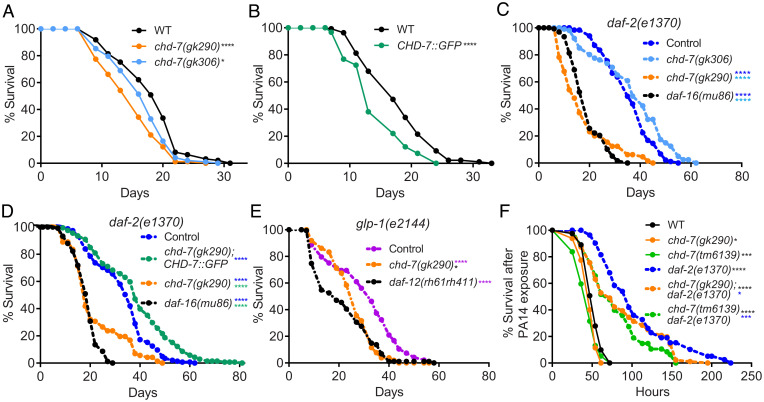
Fig. 2.
chd-7 affects longevity and response to pathogen. (A–E) chd-7 promotes longevity in wild-type, daf-2(e1370), and glp-1(e2144) mutants. Mean survival days on OP50-1; survival data analyzed using Kaplan−Meier test. For all experiments, the asterisk in color represents strain of reference for statistical analysis. Details of number of animals and additional data from replicates can be found in SI Appendix, Table S1. (A) WT (18.12), chd-7(gk290) (14.91), chd-7(gk306) (16.53). *P < 0.05 and ****P < 0.0001 compared to the wild-type, N2 strain. (B) WT (17.85), CHD-7::GFP (14.35). ****P < 0.0001 compared to the wild-type, N2 strain. (C) daf-2(e1370) (35.58), chd-7(gk306);daf-2(e1370) (37.18), chd-7(gk290);daf-2(e1370) (17.47), daf-16(mu86);daf-2(e1370) (18.85). ****P < 0.0001. (D) daf-2(e1370) (27.9), chd-7(gk290);daf-2(e1370);CHD-7::GFP (30.84), chd-7(gk290);daf-2(e1370) (19.13), daf-16(mu86);daf-2(e1370) (14.48). ****P < 0.0001. (E) glp-1(e2144) (30.4), chd-7(gk290);glp-1(e2144) (28.37), glp-1(e2144);daf-12(rh61rh411) (25.99). *P < 0.05 and ****P < 0.0001. (F) chd-7 mediates the response against the opportunistic bacteria P. aeruginosa. Mean lifespan in hours (m) ± SEM. n is the number of animals analyzed/total number in experiment. WT (mean = 52.11 ± 0.96, n = 94 of 162), chd-7(gk290) (mean = 47.45 ± 1.05, n = 54 of 130), chd-7(tm6139) (mean = 44.47 ± 1.57, n = 56 of 80), daf-2(e1370) (mean = 105.46 ± 5.61, n = 65 of 115), chd-7(gk290);daf-2(e1370) (mean = 86.74 ± 4.21, n = 103 of 120), and chd-7(tm6139);daf-2(e1370) (mean = 79.13 ± 4.9, n = 50 of 60). *P < 0.05; ***P < 0.001 and ****P < 0.0001. Survival data analyzed using Kaplan−Meier test.
To determine if the effects on lifespan were specific to the IIS pathway, we assayed whether chd-7 contributes to the longevity induced by germ-cell–less mutations, a longevity paradigm that operates in parallel to IIS (50). Temperature-sensitive glp-1(e2144) animals are sterile and long-lived at nonpermissive temperatures (51). This lifespan extension was dependent on chd-7, as chd-7(gk290);glp-1(e2144) double mutants had a mean lifespan significantly shorter than glp-1(e2144) single mutants (Fig. 2E). The impact was similar to that produced by absence of the nuclear receptor daf-12 (Fig. 2E), which is strictly necessary for longevity of germ-cell–less animals (51).
In addition to longevity, IIS reduction also enhances resistance against multiple stressors, including pathogen attack and starvation. We therefore tested if chd-7 inactivation impaired daf-2 immunoresistance. As shown in Fig. 2F, chd-7 mutants repressed the increased survival of daf-2 worms upon exposure to the human opportunistic pathogen, Pseudomonas aeruginosa strain PA14 (52). In contrast, chd-7 had only a modest effect on survival of wild-type worms to pathogen. Furthermore, the chd-7(gk290) and chd-7(tm6139) alleles reduced the survival of daf-2(e1370) L1 larvae subjected to starvation stress (SI Appendix, Fig. S4) (53). Taken together, these results suggest that chd-7 mediates the increased lifespan of at least two longevity paradigms (IIS mutants and germ-cellless animals), as well as the response to pathogens and starvation.
We then analyzed the ChIP-sequencing (ChIP-seq) datasets from CHD-7 and DAF-16 generated by ModEncode from YA/L4 larvae (Dataset S1). To our surprise, we found that both transcriptional regulators shared a significant number of genes (SI Appendix, Fig. S5). These data indicate that chd-7 might modulate longevity and pathogen resistance by regulation of the IIS pathway, possibly through direct regulation of target genes or through direct regulation of daf-16 (see below). Regulation of the IIS pathway could also be mediated by the TGF-β pathway (15), a hypothesis we examine below.
CHD-7 Functions in Multiple TGF-β Pathways.
We sought to understand whether chd-7 also modulates the TGF-β dauer pathway (14, 15). daf-7(1372) is dauer-constitutive at the restrictive temperature of 25 °C. As shown in Fig. 3A, chd-7;daf-7(e1372) double mutants bypassed the dauer arrest to become fertile adults at 25 °C. The downstream cofactor encoded by daf-5 also suppresses dauer-constitutive alleles in the TGF-β pathway at 25 °C, but is diminished in its ability to suppress at slightly higher temperatures (>25.8 °C) (13). Thus, we next asked whether chd-7 mutation also prevented the daf-7 dauer arrest at higher temperatures. As shown in Fig. 3A, chd-7 mutation could not bypass the daf-7–dependent arrest at higher temperatures (54). As expected for a putative transcriptional regulator, epistasis analysis placed CHD-7 downstream of the receptor DAF-1 and the R-Smad DAF-14 (Fig. 3B). Interestingly, at 26.5 °C, chd-7-defective dauers were typically surrounded by undetached cuticle (Fig. 3C), a phenotype previously associated with molting defects due to overexpression of daf-9 (55). Like chd-7;daf-2 dauers (Fig. 1F), chd-7;daf-7 dauers are sensitive to SDS exposure, indicating that there are cuticle defects in this daf-c genetic background (Fig. 3C).
Fig. 3.
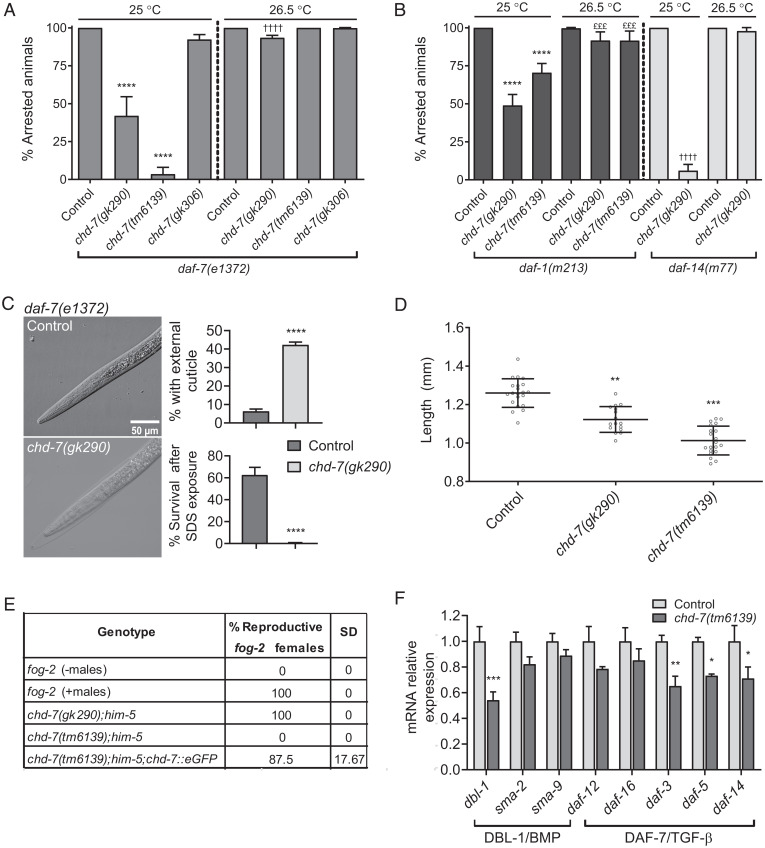
CHD-7 functions in the TGF-β signaling pathway. (A and B) Loss of chd-7 suppresses dauer arrest of TGF-β pathway mutants at 25 °C but not 26.5 °C. Seven L4s were plated individually and grown at the specified temperature for 1 wk when arrested and nonarrested progeny were scored. Bars and horizontal black lines represent mean percentage with SD. Statistical significance was calculated using χ2 test with Bonferroni correction for multiple comparisons. ****,††††P < 0.0001 and £££P < 0.001. (A) Quantification of dauer arrest in chd-7;daf-7(e1372) mutants. The asterisks represent comparison to daf-7(e1372) grown at 25 °C and the daggers represent comparison to daf-7(e1372) at 26.5 °C. (B) Dauer arrest in TGF-β pathway mutant backgrounds. The asterisks represent comparison to daf-1(m213) grown at 25 °C, and the daggers represent comparison to daf-14(m77) at 25 °C; the pound symbols represent comparison to daf-1(m213) at 26.5 °C. (C) High temperature chd-7(gk290);daf-7(e1372) dauers are surrounded by undetached cuticle. (Left) Representative DIC photomicrographs of dauers grown at 26.5 °C for 1 wk. (Upper Right) Quantification of the population of animals with undetached cuticle. Two biological replicates were scored (n > 398/replicate). (Lower Right) chd-7(gk290);daf-7(e1372) develop as SDS-sensitive dauer larvae at 26.5 °C. n > 667 animals per strain tested. Bars and horizontal black lines represent mean percentage with SD. Statistical analysis was calculated using two-tailed unpaired t test. ****P < 0.0001. (D) chd-7 regulates body size. Body length of day 1 adults at 20 °C (n > 16). One-way ANOVA, **P < 0.01, ***P < 0.001 compared to wild-type, N2 strain. (E) chd-7(tm6139) males do not mate. Eight males from each strain tested were plated with four fog-2 females on 10-cm plates. After 24 h, fog-2 females were transferred to new plates and within 48 h the proportion of fertile females were scored. The assay was repeated twice. (F) Relative mRNA levels of genes from the DBL-1/BMP and DAF-7/TGF-β pathways in chd-(tm6139) or N2 L4s determined by qRT-PCR. Error bars indicate SE from three biological repeats. cdc-42 was used as housekeeping gene. Statistical significance was calculated using t test for multiple comparisons. *P < 0.05, **P < 0.01, and ***P < 0.001.
A second TGF-β signaling pathway regulates body size and male tail development, mainly through the ligand DBL-1 and downstream Smads (56, 57). To test if chd-7 is required for these processes as well, we measured the length of chd-7 young adults and found them to be significantly shorter than wild-type animals (Fig. 3D). We also observed that males carrying the severe loss-of-function allele chd-7(tm6139) failed to mate with fog-2 mutant females due to defects in male tail development (see next paragraph) (18). This male infertility was rescued by the CHD-7::GFP transgene (Fig. 3E). Mating did occur with the weaker chd-7(gk290) allele.
More than 20 y ago, in a screen for suppressors of dauer formation within the TGF-β pathway, Inoue and Thomas (18) identified three complementation groups. We noticed that one of these, scd-3 (suppressor of constitutive dauer-3), was located between unc-11 and dpy-5 on chromosome I, in the same genetic region as chd-7. Features of scd-3(sa253) worms include low brood size, egg-laying defects (Egl), short body size (Dpy), and male abnormal defects (Mab), all of which are phenotypes shared with chd-7, described above. In addition, improper gonad migration is a common phenotype of scd-3 and chd-7(tm6139) mutant animals (SI Appendix, Fig. S6). To determine if chd-7 and scd-3 are allelic, we sequenced scd-3(sa253) and found a single G/A mutation in exon 8 of the chd-7 locus that introduces a premature STOP codon at position Q2422, eliminating the BRK domain of CHD-7 protein. This analysis confirmed that chd-7 is scd-3 (Fig. 1E).
Analysis of the male tail defect in scd-3 mutants showed missing, deformed, and fused rays, as well as defective hook and spicules (18). These phenotypes are more severe than the typical fused rays seen in animals carrying mutations in dbl-1, sma-2, sma-3, and sma-4, which encode the ligand and downstream signaling components of the Sma/Mab pathway, respectively (56–58). Thus, while chd-7 appears to impact many of the same developmental pathways as TGF-β mutants, the severity of its phenotypes may indicate that it has broader roles in developmental gene regulation.
chd-7 Regulates Expression of TGF-β Pathway Components.
Our phenotypic analysis suggested that CHD-7 may influence both the DAF-7/Dauer and Sma/Mab TGF-β pathways. One possibility is that CHD-7 might directly impact expression of the pathway members. By analyzing CHD-7’s CHIP-seq data from young adults generated by the ModEncode project, we noticed that CHD-7 associated with genes regions encoding TGF-β pathway components (Dataset S1). To determine whether chd-7 is required for proper expression of these genes in vivo, we performed qRT-PCR on wild-type and chd-7(tm6139) L4 larvae. As shown in Fig. 3F, chd-7(tm6139) worms presented reduced expression of dbl-1, daf-5, and daf-14 compared to N2. We also observed decreased expression of daf-3, although this was not identified in the ModEncode data, suggesting the effect of chd-7 may be indirect. When we analyzed gene expression relative to ama-1, a different housekeeping gene, we also observed a significant down-regulation of sma-2 and daf-12 (SI Appendix, Fig. S7). This latter result raises the intriguing possibility that chd-7 and daf-12 may be mutually transcribed in a regulatory feedback loop, which will be an area of future investigation. The impact of chd-7 on expression of multiple TGF-β pathway members, together with the cooccurrence of associated phenotypes, argues in favor of regulatory roles for chd-7 in both the DAF-7/dauer and the Sma/Mab pathways.
daf-9 and Cuticle Genes Are Misregulated in chd-7 Mutants.
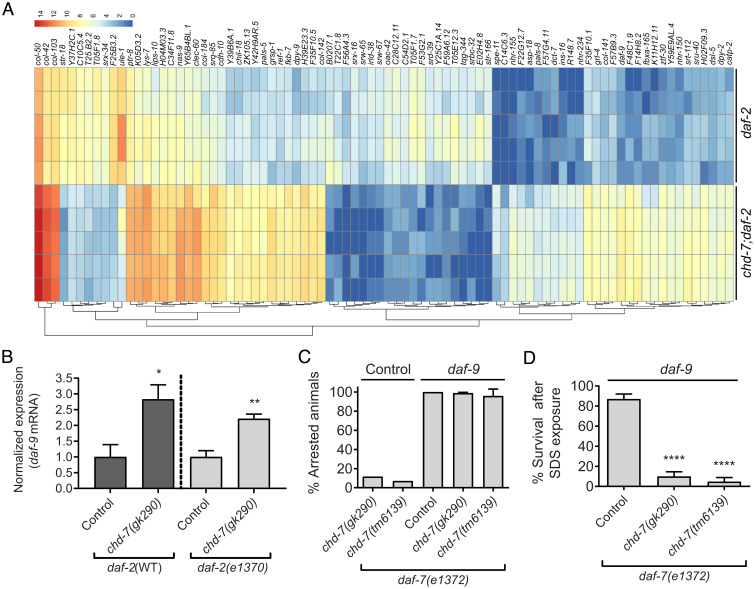
To gain further insight into the role of CHD-7 in dauer development, we performed RNA-sequencing (RNA-seq) analysis of daf-2(e1370) dauers and chd-7(gk290);daf-2(e1370) partial dauers. Differentially expressed gene (DEG) analysis revealed decreased expression of 28 genes and increased expression of 56 genes in the double mutants (Fig. 4A and SI Appendix, Table S2). Among the latter group, we found daf-9, encoding the cytochrome p450 that integrates inputs from TGF-β and insulin/IGF-II pathways to regulate DAF-12 activity during dauer development (59). We confirmed this increased expression of daf-9 by qRT-PCR of chd-7 natural dauers and chd-7(gk290);daf-2(e1370) partial dauers (Fig. 4B). We noted that the ModEncode CHD-7 ChIP-seq did not identify daf-9 as a potential target (Dataset S1). This apparent discrepancy may reflect differences in developmental timing since the ChIP-seq was performed on young adults or may suggest that daf-9 is an indirect target of CHD-7. Nonetheless, we hypothesized that since daf-9 expression levels are critical in the decision to either develop as fertile adults or enter diapause, the increased expression of daf-9 in the chd-7;daf-2 double mutants may be preventing full execution of the dauer program. Consistent with this hypothesis, we found that depletion of daf-9 in chd-7;daf-7 animals restored the dauer arrest phenotype at the nonpermissive temperature of 25 °C (Fig. 4C). However, these animals still appeared to be detergent-sensitive partial dauers (Fig. 4D), presumably because daf-9 itself is required for proper dauer morphogenesis (45). Thus, we posit that the inability of chd-7 mutants to fully repress daf-9 may be sufficient to activate DAF-12 to promote reproductive development. Consistent with a general role for chd-7 in regulation of daf-9, we also observed an undetached cuticle associated with the chd-7(gk290);daf-7(e1372) high-temperature dauers (Fig. 3B). Hypodermal daf-9 overexpression causes a similar undetached cuticle when it bypasses starvation-induced arrest of L3 or L4 larvae (55).
Fig. 4.
RNA-seq analysis of transcriptome changes in chd-7(gk290) mutant dauers. (A) Heat map of expression values for the 84 DEGs (cutoff of 0.05 on FDR). DEGs were determined using DESeq2 (v1.20.0). The color scale represents the normalized count values in log2 scale. Hierarchical clustering of the DEGs is represented by dendrograms at bottom. (B) Expression levels of daf-9 mRNA are increased in chd-7(gk290); daf-2(e1370) partial dauers and in L3-like arrested chd-7(gk290) animals upon starvation (Fig. 1G). Error bars indicate SE from three biological repeats. Two-tailed unpaired t test. *P < 0.05 and **P < 0.01. (C) daf-9 knockdown in chd-7;daf-7(e1372) animals rescues dauer arrest at 25 °C. Two to three young L4s were plated on to freshly seeded plates with either daf-9 or Control empty vector RNAi and allowed to lay eggs. After 72 h, the adults were removed and the proportion of progeny that arrested as dauers was calculated. Bars and horizontal black lines represent mean percentage with SD (n > 598 total animals per strain). (D) daf-9 RNAi rescues arrest in chd-7;daf-7(e1372) animals but leads to partial dauers. Arrested animals grown at 25 °C on daf-9 RNAi plates were treated with 1% SDS for 30 min and survival was scored. Bars and horizontal black lines represent mean percentage with SD (n > 159 total animals per strain). One-way ANOVA, ****P < 0.0001, compared to daf-7(e1372).
Further analysis of our transcriptomic data showed that 10% of the DEGs were collagens (col-103, col-50, dpy-2, col-184, col-141, col-142, col-42, and dpy-9), which are structural components of the cuticle. All of these collagens had increased expression in the chd-7-mutant dauer larvae (Fig. 4A). Fragments per kilobase of transcript per million mapped reads data from ModEncode libraries indicates that each of these collagens shows very low expression in dauers, but are expressed during various stages of reproductive development, suggesting that the cuticle signature of partial dauers is distinct from the normal dauer cuticle . Interestingly, col-141 and col-142 contain SMAD-binding elements and their expression is directly regulated by DBL-1 to determine body size (35). These results could suggest the intriguing possibility that CHD-7, through regulation of the DBL-1/BMP pathway, may contribute to dauer morphogenesis by ensuring the repression of reproductive-stage cuticles.
Chd7 Regulates col2a1 during Xenopus Embryogenesis.
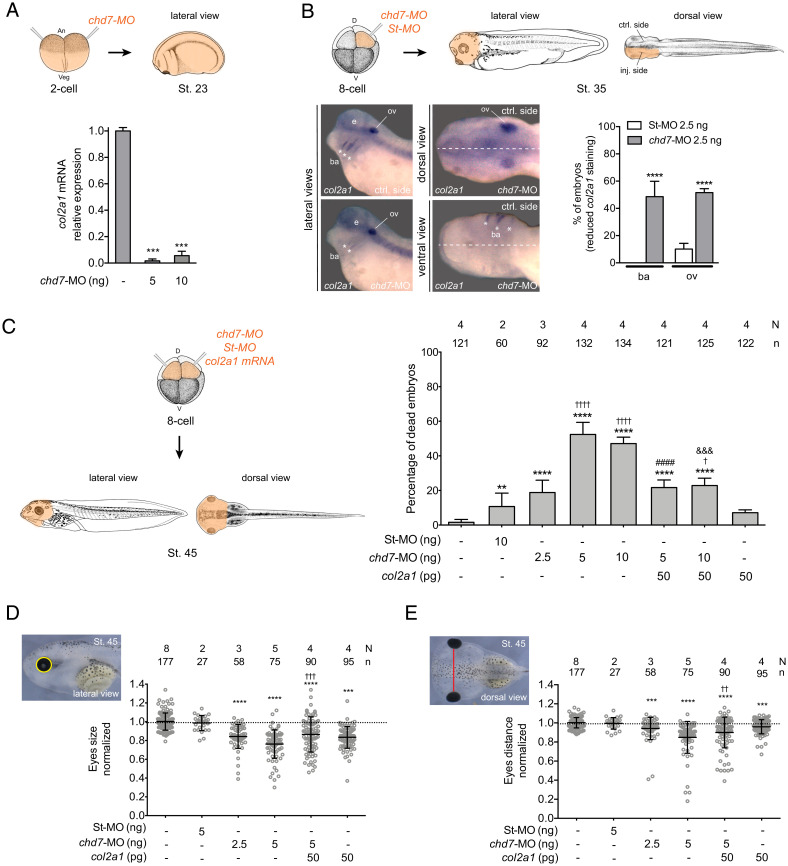
Type-II collagen is an ECM protein conserved in all multicellular animals, which forms fibrils (60) and has fundamental roles in development and tissue homeostasis (61). In vertebrates, the fibrillar type-II collagen is the major structural protein of cartilage and plays a prominent role in cranial development in multiple organisms (42, 62). Col2a1 is the major component of the cartilage matrix, having a structural function and being an important extracellular signaling molecule for regulation of chondrocyte proliferation, metabolism, and differentiation and its expression is regulated by TGF-β (43, 44, 63). The African frog X. laevis is a well-established model to study vertebrate facial disorders, which often arise from defects in neural crest development and migration (41). In Xenopus embryos, prior studies established that Chd7 regulates neural crest specification and migration and its depletion recapitulates craniofacial defects seen in CHARGE patients (40). To investigate whether a role for Chd7 in the regulation of collagen expression might be conserved in vertebrates, we used a previously validated morpholino to induce Xenopus Chd7 knockdown (chd7-MO) (40). In Xenopus, Col2a1 is essential for normal development of the skeleton and its expression is restricted to the cartilaginous skeleton of the tadpole and adult frog (64). In chd7-MO–injected embryos, qRT-PCR revealed a significant reduction of col2a1 mRNAs as compared to the uninjected, control animals (Fig. 5A).
Fig. 5.
Expression of col2a1 rescues Chd-7 knockdown in Xenopus embryos. (A) Expression levels of col2a1 mRNA are reduced in chd7-MO–injected embryos. Both blastomeres of two-cell–staged embryos were injected with 5 or 10 ng of chd7-MO and processed for RNA extraction at stage 23 (St. 23) (schematic above). Error bars indicate SE from two repeats of the PCR with different biological samples. One-tailed paired t test, ***P < 0.001. (B) col2a1 expression domain is altered in the branchial arches (ba) and otic vesicle (ov) of Chd7-depleted embryos. In situ hybridization of stage 35 embryos for col2a1. One D1 blastomere of eight-cell–staged embryos was injected with 2.5 ng of St-MO or chd7-MO (schematic above) (N = 2; 42 and n = 3; 71, respectively) (N = number of experiments; n = number of embryos). Lateral, dorsal, and ventral views, anterior to the left. e: eye. Lateral views: Upper and Lower are control (ctrl) and chd7-MO injected sides of the same representative injected embryo, respectively. Dorsal and ventral views: white dashed lines show the middle line of the embryo. Comparisons were done between the injected side and the contralateral control side of the same embryo (schematic above). The graph is a quantification of the results. Reduced col2a1 staining was observed in 18% of St-MO and 77% of chd7-MO–injected embryos that survived through stage 35. Data on graph is presented as means with SE. Fisher’s exact test (****P < 0.0001). Asterisks represent comparison to St-MO group. (C) Lethality in Chd7-depleted embryos is rescued by overexpression of col2a1. Graph showing the percentage of dead embryos by stage 45. Both D1 blastomeres of eight-cell–staged embryos were injected as indicated in the graph and scored for survival at stage 45 (schematically depicted above). Data on graph is presented as means with SE. Fisher’s exact test (†P < 0.05, **P < 0.01, &&&P < 0.001, ****,††††,####P < 0.0001). Asterisks represent comparison to uninjected group; daggers represent comparison to St-MO group; pound and ampersands represent comparison to chd7-MO 5 ng and 10 ng, respectively. (D and E) Craniofacial morphometric analysis of Xenopus tadpoles at stage 45. Embryos were injected as indicated in C. Each dot represents a single embryo. Means and SD are indicated. One-way ANOVA and Tukey’s multiple comparisons test. (D) Quantification of the eye size (Upper Left). ***,†††P < 0.001, ****P < 0.0001. (E) Quantification of the eye distances (Upper Left). ††P < 0.01, ***P < 0.001, ****P < 0.0001. Lateral views, anterior to the left. Asterisks represent comparison to uninjected group and daggers represent comparison to chd7-MO 5-ng injected group. N = number of experiments, n = number of embryos. The source of Xenopus stage illustrations in A–C is Xenbase (https://www.xenbase.org/entry/, RRID:SCR_003280). Xenopus illustrations ©: Natalya Zahn (90) and by Nieuwkoop and Faber (86). The 12.5x and 20X digital magnification zoom of Leica L2 stereoscope were used for images in B. The 12.5x digital magnification was used for images in D and E.
For targeted disruption of Chd7 function, we injected the dorsal-animal (D1) blastomeres of eight-cell–stage embryos fated to contribute to the dorsal anterior structures (Fig. 5C, schema). In situ hybridization of unilaterally injected embryos with chd7-MO (Fig. 5B, schema) showed alterations in col2a1 expression in the branchial arches and the otic vesicle (ear vesicle) on the injected side (64). In contrast, no defects were observed in standard control morpholino (St-MO)–injected embryos (Fig. 5B).
The injection of chd7-MO into both D1 blastomeres of eight-cell–stage embryos (Fig. 5C, schema) induced lethality: with doses between 5 ng and 10 ng, >50% lethality was seen (Fig. 5C). We next sought to investigate if the mortality associated with Chd7 loss was caused by downstream effects on collagen gene expression. Therefore, we conducted rescue experiments by coexpressing Xenopus col2a1 mRNA with the morpholino. As shown in Fig. 5C, coinjection of col2a1 mRNA substantially improved (∼50%) embryo survival relative to the injection of chd7-MO alone.
To further interrogate the ability of ectopic col2a1 to overcome the defects associated with chd-7 loss, we examined the extent of the craniofacial defects in chd7 loss-of-function. Initially, we analyzed the gross morphology of the surviving stage 45 tadpoles and observed a high incidence of craniofacial malformations (83%) in Chd7-depleted animals (SI Appendix, Fig. S8). These defects were significantly reduced upon col2a1 mRNA coinjection (43%) (SI Appendix, Fig. S8B). Next, we examined eye size and eye distance, since microphthalmia and midline defects are often associated with CHARGE syndrome (65) and are recapitulated in Xenopus embryos (40). Both eye size and distance between eyes were reduced in Chd7-depleted tadpoles and were partially rescued by col2a1 mRNA expression (Fig. 5 D and E). Therefore, expression of col2a1 ameliorated the phenotypes associated with pathogenic Chd7, suggesting that collagen is a conserved and important target of this protein.
Discussion
We initially identified chd-7 as a target of the nuclear receptor DAF-12 (21), a transcription factor regulating worm aging, development, and dauer formation (66). Here, we show that chd-7 expression is up-regulated by DAF-12 and has roles in dauer development, longevity, pathogen resistance, male fertility, and body size. Our mining of ChIP-seq data and qRT-PCR analyses found decreased expression of TGF-β components in both the DAF-7/dauer and the Sma/Mab branches, indicating that chd-7 is a regulator of the TGF-β pathways in C. elegans (Fig. 3F). While the preponderance of phenotypic and expression data supports a role for chd-7 in the TGF-β pathways, chd-7 might also function in TGF-β–independent mechanisms (67–69) that regulate these developmental processes.
Our genetic epistasis analyses placed CHD-7 downstream of the TGF-β–like DAF-7, the type I receptor DAF-1, and the R-SMAD DAF-14 putting CHD-7 at the level of the Co-Smad DAF-3 and the Sno/Ski repressor DAF-5, which are also Daf-d (70). Supporting a role for CHD-7 at the DAF-3/DAF-5 step in the pathway, we observed that chd-7 completely suppressed dauer formation in a daf-7 background at 25 °C, and like daf-5 (13), failed to suppress dauer formation in TGF-β mutants at higher temperatures (Fig. 3 A and B). Interestingly, the daf-3 and daf-5 genes have opposite effects on daf-2–induced longevity: daf-3 enhances daf-2(e1370) longevity, while daf-5 mutations suppresse it (17). We observed that chd-7 suppresses daf-2 longevity (Fig. 2C), like daf-5(e1386). Since CHD-7 can associate with the daf-5 locus (ModEncode ChIP-seq data), and since both daf-3 and daf-5 expression are reduced when chd-7 function is compromised (Fig. 3F), one possibility is that CHD-7 simply functions to ensure proper expression of these critical TGF-β dauer regulators. However, CHD-7 may also directly interact with the DAF-3/DAF-5 complex to regulate downstream target genes. In mice, CHD7 was shown to physically interact with SMAD1 and form a transcriptional complex with SMAD4, the mammalian ortholog of DAF-3 (71). We therefore speculate that DAF-3, DAF-5, and CHD-7 may be in a ternary complex that regulates daf-9 expression for dauer entry. Interactome mapping of the TGF-β pathway previously identified SWSN-1, a SWI/SNF subunit component of the BAF complex, as a physical interactor of DAF-3 (72). CHD7 interacts with human and Xenopus PBAF (polybromo- and BRG1-associated factor-containing complex) to control neural crest genes expression (40). In worms, both swsn-1 and chd-7 fail to develop normal dauers in daf-2 and daf-7 mutants (73) (SI Appendix, Fig. S9). Thus, we envision that CHD-7 may work together with the BAF complex and DAF-3/DAF-5 to control gene expression of target genes critical for dauer formation.
DAF-9 activity is a critical determinant of the decision to enter diapause: reduced activity of TGF-β and IIS pathways leads to daf-9 repression and dauer entry. Conversely, daf-9 expression in the hypodermis is sufficient to inhibit diapause, driving reproductive programs in daf-7 mutants (16, 19). Ectopic daf-9 expression also drives reproductive programs in the weak daf-2(e1368) allele, but only partially suppresses daf-2(e1370) diapause, leading to arrest as L3 or early L4 larvae (19). Based on these observations, we speculate that daf-9 misexpression explains a subset of the chd-7 phenotypes observed herein, including the partial dauer phenotype, gonad migration defects, and vulval protrusions, all of which overlap with published daf-9 phenotypes (19, 20, 74). Interestingly, daf-9 is both upstream and downstream of daf-12 for dauer formation (19, 20). Our identification of chd-7 as both a downstream target of DAF-12 (21) and as an upstream component of the regulatory pathway that promotes DAF-12 expression suggests that these two genes may function in a regulatory feedback loop. Further dissection of the tissue- and temporal-specific requirements for these proteins is likely to further illuminate the interplay between these proteins. Notably, DAF-12 and CHD-7 both bind to sites in daf-5, daf-14, daf-16, daf-12, and daf-19 (21) (Dataset S1), suggesting that they could coregulate expression of target genes as a transcriptional complex. We speculate that the mutual regulation between chd-7 and daf-12 might be important to maintain the stoichiometry of this complex. In addition, the identification of shared binding sites in daf-16 provides a possible mechanism for the impact of chd-7 on the IIS pathway.
Both chd-7 mutation and chd-7 overexpression shortened the lifespan of otherwise wild-type worms, suggesting that CHD-7 protein levels must be tightly regulated to ensure proper development. Of note, Chd7 is the most commonly amplified gene in tumors among the CHD superfamily members, and its overexpression is associated with aggressive subtypes of breast cancer and poor prognosis (75). In glioblastoma cells, Chd7 overexpression leads to a 3×-fold down-regulation of BMPR1B, vertebrate’s ortholog of SMA-6 and DAF-1 (76). Thus, it’s possible that the TGF-β signaling pathway, which regulates longevity, is negatively impacted in worms overexpressing CHD-7. We note that autoregulatory roles for CHD-7 during adult life, as proposed for dauer, might also explain how overexpression could be pathogenic.
It was well-established that in multiple systems, TGF-β stimulates ECM deposition mainly by promoting expression of fibronectin, collagens, and other ECM components (37, 77, 78). In addition, TGF-β promotes the synthesis of inhibitors to enzymes that degrade the ECM, including the plasminogen activator inhibitor 1 (PAI-1) and the tissue inhibitor of matrix metalloproteinases (79, 80). Mutations in the TGF-β ligand dbl-1 and its downstream receptor and signaling components result in small body size due to transcriptional misregulation of cuticle collagen genes (35, 56, 57). Consistent with a role for CHD-7 in regulation of dbl-1 and sma-2 expression, chd-7(gk290) and chd-7(tm6139) are significantly shorter than wild-type worms (Fig. 3 D and F). In Xenopus, we show a role for Chd7 in regulating col2a1 expression, a type II collagen and the major component of cartilage (62). Interestingly, regulation of col2a1 by Chd7 is also observed in zebrafish (81). While additional studies are required, we speculate that Chd7 could regulate col2a1 in a complex with the transcription factor Sox10 (82, 83) or through the TGF-β signaling pathway (37, 39). Supporting the latter mechanism, it was demonstrated in chondrocytes that TGF-β regulates col2a1 expression (43, 44). Therefore, our results suggest that these evolutionary conserved helicases have roles in ECM deposition, supporting a model in which collagen misexpression by pathogenic Chd7 leads to craniofacial defects and embryonic lethality.
Comparison of the chd-7 alleles gk290 and gk306 showed a critical role for the BRK domain in dauer development and longevity (Figs. 1 F–H and 2 A, C, and E). In CHARGE patients, deletions or mutations within the BRK domains of CHD7 are sufficient to elicit all the features characteristic of the disease, underscoring the importance of this domain (84). The phenotypic differences between the worm alleles highlight the potential of the worm to delimit functional domains of CHD-7 that contribute to disease pathology. In mice, homozygous mutations in Chd7 lead to embryonic lethality at embryonic day 10.5, in part because Chd7 is necessary for early brain development (40). In worms, the presumptive null alleles chd-7(tm6139) and scd-3(sa253) were viable but showed reproductive defects, such as improper gonad proliferation and migration, reduced fecundity, male tail defects, and hermaphrodite vulval defects (18), indicating that C. elegans are more able to tolerate loss of CHD-7 than mice or humans. Thus, our studies establish C. elegans as an animal model to study the mechanisms underlying the developmental defects observed in pathogenic Chd7.
Materials and Methods
C. elegans Strains.
Strains utilized in this study are listed in SI Appendix, Table S3. Standard genetic crosses were used to make double or triple mutants. The presence of mutant alleles was confirmed 1) by the daf-c phenotypes in animals heterozygous for additional mutations and 2) by PCR and sequencing for all additional mutations. Details for RNAi screen for dauer suppresors can be found in SI Appendix.
RNAi Screen for Dauer Suppressors.
All RNAi clones were picked from the Ahringer bacterial feeding library. These E. coli clones were seeded on NGM plates supplemented with 1 mM of IPTG (Isopropyl β-d-1-thiogalactopyranoside) and 0.1 µg/mL ampicillin, and used for inducing RNAi by the feeding method.
GL228 [rrf-3(pk1426)] II;daf-2(e1371) III] eggs were placed in 24-well RNAi plates seeded with bacteria expressing the double-stranded RNA (dsRNA) of interest. Worms were maintained for 5 d at 15 °C until adulthood, then were transferred to an identical 24-well plate to lay eggs for 5 h. Adults were removed and the eggs were incubated at 25 °C for 4 d to allow formation of dauers. daf-16(RNAi) and the empty vector were used as controls. Proper dauer formation was assessed by observation in a dissecting microscope and by 1% SDS resistance. RNAi clones that caused abnormal dauer phenotypes were validated in daf-2(e1370) worms. Identity of the dsRNA was confirmed by sequencing (Macrogen).
Dauer Formation in Liquid Media.
We obtained dauers in liquid media following a protocol recently described (47). Details can be found in SI Appendix.
daf-9 Suppression Assays.
L4-stage animals were placed on 3-cm daf-9(RNAi) plates (see SI Appendix for preparation details). Two worms per plate were used for daf-7(e1372), while three worms were used for daf-7;chd-7(gk290) and daf-7;chd-7(tm6139). After 72 h, the adults were removed, and plates were replaced at 25 °C for 2 to 3 d. The total number of dauers, L4s, and adults were then assessed.
SDS Survival Assay.
Young adults were transferred to seeded plates and permitted to lay eggs for 5 d at 25 °C. The arrested progeny were then washed off plates with M9 (22 mM KH2PO4, 42 mM Na2HPO4, 85.5 mM NaCl, 1 mM MgSO4) into 15-mL glass conical tubes. Collected animals were washed two to three times with M9 and the excess liquid was aspirated off. Animals were then treated with 2 mL of 1% SDS for 30 min on a nutator at 25 °C. Following incubation, the samples were washed three times with M9 and any excess liquid was aspirated. Animals were aliquoted to five seeded plates with 50 to 70 worms per plate and allowed to recover at 16 °C. The recovered animals were then quantified, and the percentage recovered was calculated. This was repeated three times for each strain tested.
Microscopy and Fluorescence Imaging.
Different imaging modalities were used for fluorescent, DIC, and EM. Details for preparation and visualization can be found in SI Appendix.
Lifespan Assays.
All lifespan experiments were conducted by transferring 1-d-old adults from 15 °C to 20 °C for the remainder of the lifespan assay. NGM plates were seeded with E. coli OP50-1. ∼150 L4 hermaphrodites were transferred to five plates per experiment. Every 48 h, animals were scored as alive, dead, or censored (animals that exploded, died from bagging or dried out at the edges of the plates). Animals were considered dead when they did not respond to a soft touch to the head with a pick. To prevent the progeny from interfering with the assay, adults were transferred to fresh plates every 48 h until egg production ceased. For glp-1(e2144) assays, eggs were kept at 20 °C for 4 h and then transferred at 25.5 °C for 72 h to induce sterility and switched to 20 °C for the remainder of the experiment. Lifespan data were analyzed using the Kaplan–Meier method. Statistics were calculated using the Mantel–Cox nonparametric log-rank method using OASIS2 (85).
Library Preparation and RNA-Seq.
daf-2(e1370) and chd-7(gk290);daf-2(e1370) synchronized eggs were kept at 25 °C for 10 d and resulting dauers were collected and frozen. Total RNA was extracted with TRIzol (Invitrogen) following the kit’s protocol. The cDNA library was prepared with NEBNext Ultra II RNA library prep kit for Illumina (New England Biolabs), and the sequencing carried out using Illumina’s HiSeq-2500 sequencer with single-end mode and read length of 50 bp. Five replicates for daf-2(e1370) vs. chd-7(gk290);daf-2(e1370) were sequenced. For data assessment, a quality control with FastQC software (v0.11.5) was used. First, the raw reads that aligned against the E. coli genome (K12 genome) were removed. The remaining sequences were aligned against the reference genome of C. elegans WS260 using STAR (v2.5.4a). The number of mapped reads to genes was counted using Htseq (v0.9.1). Finally, the DEGs were determined using DESeq2 (v1.20.0) with a cutoff of 0.05 on false discovery rate (FDR). R v3.5.0 (2018-04-23) and Bioconductor v3.7 with BiocInstaller v1.30.0 were used. Principal component analysis was performed using plotPCA function in DESEQ2 package. For visualization in two dimensions, we used the top two principal component axes, PC1 and PC2 (SI Appendix, Fig. S10). Heatmaps were generated using pheatmap package (v1.0.12) with hierarchical clustering on the rows with the default options.
X. laevis Embryo Manipulation and Microinjections.
Xenopus embryos were obtained by natural mating. Adult frogs’ reproductive behavior was induced by injection of human chorionic gonadotropin hormone. Embryos were collected, de-jellied in 3% cysteine (pH 8.0), maintained in 0.1× Marc’s Modified Ringer’s (MMR) solution, and staged according to Nieuwkoop and Faber (86). The embryos were placed in 3% ficoll prepared in 1× MMR for microinjection. Chd7 morpholino (chd7-MO: 5′-AACTCATCATGCCAGGGTCTGCCAT-3′) specificity has been previously characterized (40). Chd7-MO and St-MO were provided by Gene Tools. The cDNA of X. laevis col2a1 was amplified by PCR from pCMV-Sport 6-col2a1 (Dharmacon) with primers M13F and M13R. The PCR fragment was digested with EcoRV and NotI and cloned into pCS2+ previously digested with StuI and NotI. Capped mRNAs for col2a1 were transcribed in vitro with SP6 using the mMessage mMachine kit (Ambion) following linearization with NotI. Morpholinos and col2a1 mRNA were injected into both D1 blastomeres of eight-cell staged embryos (81, 87) for lethality and morphometrics analysis. Chd7-MO was injected into one D1 blastomeres of eight-cell staged embryos for analysis of col2a1 expression. For the evaluation of col2a1 staining in the branchial arches and the otic vesicle, the injected side was compared against the contralateral control side of the same embryo. Whole-mount in situ hybridization was carried out as previously described (88) and specific details can be found in SI Appendix.
Ethics Statement.
X. laevis experiments were carried out in strict accordance with Guide for the Care and Use of Laboratory Animals of the NIH (89) and the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines. The animal care protocol was approved by the Comisión Institucional para el Cuidado y Uso de Animales de Laboratorio of the School of Applied and Natural Sciences, University of Buenos Aires, Argentina (Protocol #64).
Supplementary Material
Acknowledgments
The authors thank Bruno Moretti, Hernan Grecco, Mario Rossi, and Julie Kocherzat for experimental support. D.M.J., A.S.C., and L.F.G. were supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) Doctoral Fellowship Program. The D.H. laboratory was supported by the Agencia Nacional de Promoción Científica y Tecnológica of Argentina (PICT-2016-0269) and CONICET (PIP 1122015 0100731 CO). The M.C.C. laboratory was supported by the Agencia Nacional de Promoción Científica y Tecnológica of Argentina (PICT-2013-0381). Funding for this work was also provided by grants from the CHARGE Syndrome Foundation (to J.L.Y. and D.H.), The Company of Biologists (D.M.J.), National Institute of General Medical Sciences R01GM104007 (to J.L.Y.), and National Institute on Aging R01AG051659 (to A.G.). This work was supported in part by the Intramural Research Program of the NIH and the National Institute of Diabetes and Digestive and Kidney Diseases (S.Y.). Some strains used in this study were obtained from the Caenorhadbitis Genome Center housed at the University of Minnesota and supported by a grant from the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109508119/-/DCSupplemental.
Data Availability
All study data are included in the main text and supporting information. RNA-seq data has been uploaded to National Center for Biotechnology Information Gene Expression Omnibus, ID GSE199192.
References
- 1.Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R., A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Larsen P. L., Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 90, 8905–8909 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaglia M. M., Kenyon C., Stimulation of movement in a quiescent, hibernation-like form of Caenorhabditis elegans by dopamine signaling. J. Neurosci. 29, 7302–7314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J., Kim S. K., Global analysis of dauer gene expression in Caenorhabditis elegans. Development 130, 1621–1634 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Cohen E., Bieschke J., Perciavalle R. M., Kelly J. W., Dillin A., Opposing activities protect against age-onset proteotoxicity. Science 313, 1604–1610 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Meléndez A., et al. , Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Christensen R., de la Torre-Ubieta L., Bonni A., Colón-Ramos D. A., A conserved PTEN/FOXO pathway regulates neuronal morphology during C. elegans development. Development 138, 5257–5267 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolkow C. A., Kimura K. D., Lee M.-S., Ruvkun G., Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290, 147–150 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Cassada R. C., Russell R. L., The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46, 326–342 (1975). [DOI] [PubMed] [Google Scholar]
- 10.P. J. Hu, Dauer, “The C. elegans research community” in WormBook (August 8, 2007). 10.1895/wormbook.1.144.1. Accessed 29 March 2022. [DOI] [Google Scholar]
- 11.Ren P., et al. , Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274, 1389–1391 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Park D., Estevez A., Riddle D. L., Antagonistic Smad transcription factors control the dauer/non-dauer switch in C. elegans. Development 137, 477–485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Graca L. S., et al. , DAF-5 is a Ski oncoprotein homolog that functions in a neuronal TGF beta pathway to regulate C. elegans dauer development. Development 131, 435–446 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Thomas J. H., Birnby D. A., Vowels J. J., Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134, 1105–1117 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaw W. M., Luo S., Landis J., Ashraf J., Murphy C. T., The C. elegans TGF-β Dauer pathway regulates longevity via insulin signaling. Curr. Biol. 17, 1635–1645 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T., Zimmerman K. K., Patterson G. I., Regulation of signaling genes by TGFbeta during entry into dauer diapause in C. elegans. BMC Dev. Biol. 4, 11 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narasimhan S. D., et al. , PDP-1 links the TGF-β and IIS pathways to regulate longevity, development, and metabolism. PLoS Genet. 7, e1001377 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue T., Thomas J. H., Suppressors of transforming growth factor-beta pathway mutants in the Caenorhabditis elegans dauer formation pathway. Genetics 156, 1035–1046 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerisch B., Antebi A., Hormonal signals produced by DAF-9/cytochrome P450 regulate C. elegans dauer diapause in response to environmental cues. Development 131, 1765–1776 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Mak H. Y., Ruvkun G., Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development 131, 1777–1786 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Hochbaum D., et al. , DAF-12 regulates a connected network of genes to ensure robust developmental decisions. PLoS Genet. 7, e1002179 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vissers L. E. L. M., et al. , Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 36, 955–957 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Balasubramanian R., et al. , Functionally compromised CHD7 alleles in patients with isolated GnRH deficiency. Proc. Natl. Acad. Sci. U.S.A. 111, 17953–17958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H.-G., et al. , Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am. J. Hum. Genet. 83, 511–519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernier R., et al. , Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marfella C. G. A., Imbalzano A. N., The Chd family of chromatin remodelers. Mutat. Res. 618, 30–40 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patten S. A., et al. , Role of Chd7 in zebrafish: A model for CHARGE syndrome. PLoS One 7, e31650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosman E. A., et al. , Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 14, 3463–3476 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Tian C., et al. , Otitis media in a new mouse model for CHARGE syndrome with a deletion in the Chd7 gene. PLoS One 7, e34944 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daubresse G., et al. , The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 126, 1175–1187 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Melicharek D. J., Ramirez L. C., Singh S., Thompson R., Marenda D. R., Kismet/CHD7 regulates axon morphology, memory and locomotion in a Drosophila model of CHARGE syndrome. Hum. Mol. Genet. 19, 4253–4264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asad Z., et al. , Rescue of neural crest-derived phenotypes in a zebrafish CHARGE model by Sox10 downregulation. Hum. Mol. Genet. 25, 3539–3554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDiarmid T. A., et al. , Systematic phenomics analysis of autism-associated genes reveals parallel networks underlying reversible impairments in habituation. Proc. Natl. Acad. Sci. U.S.A. 117, 656–667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong W.-R., et al. , Autism-associated missense genetic variants impact locomotion and neurodevelopment in Caenorhabditis elegans. Hum. Mol. Genet. 28, 2271–2281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madaan U., et al. , BMP signaling determines body size via transcriptional regulation of collagen genes in Caenorhabditis elegans. Genetics 210, 1355–1367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madaan U., et al. , Feedback regulation of BMP signaling by Caenorhabditis elegans cuticle collagens. Mol. Biol. Cell 31, 825–832 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ignotz R. A., Massagué J., Transforming growth factor-beta stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J. Biol. Chem. 261, 4337–4345 (1986). [PubMed] [Google Scholar]
- 38.Streuli C. H., Schmidhauser C., Kobrin M., Bissell M. J., Derynck R., Extracellular matrix regulates expression of the TGF-beta 1 gene. J. Cell Biol. 120, 253–260 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts A. B., McCune B. K., Sporn M. B., TGF-β: Regulation of extracellular matrix. Kidney Int. 41, 557–559 (1992). [DOI] [PubMed] [Google Scholar]
- 40.Bajpai R., et al. , CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463, 958–962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubey A., Saint-Jeannet J.-P., Modeling human craniofacial disorders in Xenopus. Curr. Pathobiol. Rep. 5, 79–92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seufert D. W., Hanken J., Klymkowsky M. W., Type II collagen distribution during cranial development in Xenopus laevis. Anat. Embryol. (Berl.) 189, 81–89 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Tekari A., Luginbuehl R., Hofstetter W., Egli R. J., Transforming growth factor beta signaling is essential for the autonomous formation of cartilage-like tissue by expanded chondrocytes. PLoS One 10, e0120857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chadjichristos C., et al. , Down-regulation of human type II collagen gene expression by transforming growth factor-beta 1 (TGF-beta 1) in articular chondrocytes involves SP3/SP1 ratio. J. Biol. Chem. 277, 43903–43917 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Albert P. S., Riddle D. L., Mutants of Caenorhabditis elegans that form dauer-like larvae. Dev. Biol. 126, 270–293 (1988). [DOI] [PubMed] [Google Scholar]
- 46.Haag E. S., Dial-a-mutant: Web-based knockout collections for model organisms. Biol. Cell 99, 343–347 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Hibshman J. D., Webster A. K., Baugh L. R., Liquid-culture protocols for synchronous starvation, growth, dauer formation, and dietary restriction of Caenorhabditis elegans. STAR Protoc 2, 100276 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narbonne P., Roy R., Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 457, 210–214 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Kadekar P., Roy R., AMPK regulates germline stem cell quiescence and integrity through an endogenous small RNA pathway. PLoS Biol. 17, e3000309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berman J. R., Kenyon C., Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 124, 1055–1068 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Hsin H., Kenyon C., Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399, 362–366 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Garsin D. A., et al. , Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300, 1921 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Lee I., Hendrix A., Kim J., Yoshimoto J., You Y.-J., Metabolic rate regulates L1 longevity in C. elegans. PLoS One 7, e44720 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ailion M., Thomas J. H., Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics 156, 1047–1067 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schindler A. J., Baugh L. R., Sherwood D. R., Identification of late larval stage developmental checkpoints in Caenorhabditis elegans regulated by insulin/IGF and steroid hormone signaling pathways. PLoS Genet. 10, e1004426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Savage C., et al. , Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc. Natl. Acad. Sci. U.S.A. 93, 790–794 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki Y., et al. , A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126, 241–250 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Morita K., Chow K. L., Ueno N., Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-beta family. Development 126, 1337–1347 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Gerisch B., Weitzel C., Kober-Eisermann C., Rottiers V., Antebi A., A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell 1, 841–851 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Boot-Handford R. P., Tuckwell D. S., Fibrillar collagen: The key to vertebrate evolution? A tale of molecular incest. BioEssays 25, 142–151 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Wilcox W. R., Connective tissue and its heritable disorders: Molecular, genetic, and medical aspects. Am. J. Hum. Genet. 72, 503–504 (2003). [Google Scholar]
- 62.Wood A., Ashhurst D. E., Corbett A., Thorogood P., The transient expression of type II collagen at tissue interfaces during mammalian craniofacial development. Development 111, 955–968 (1991). [DOI] [PubMed] [Google Scholar]
- 63.Baugé C., Cauvard O., Leclercq S., Galéra P., Boumédiene K., Modulation of transforming growth factor beta signalling pathway genes by transforming growth factor beta in human osteoarthritic chondrocytes: Involvement of Sp1 in both early and late response cells to transforming growth factor beta. Arthritis Res. Ther. 13, R23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kerney R., Hall B. K., Hanken J., Regulatory elements of Xenopus col2a1 drive cartilaginous gene expression in transgenic frogs. Int. J. Dev. Biol. 54, 141–150 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Siebert J. R., Graham J. M. Jr., MacDonald C., Pathologic features of the CHARGE association: Support for involvement of the neural crest. Teratology 31, 331–336 (1985). [DOI] [PubMed] [Google Scholar]
- 66.Antebi A., Yeh W. H., Tait D., Hedgecock E. M., Riddle D. L., daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 14, 1512–1527 (2000). [PMC free article] [PubMed] [Google Scholar]
- 67.So S., Miyahara K., Ohshima Y., Control of body size in C. elegans dependent on food and insulin/IGF-1 signal. Genes Cells 16, 639–651 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Del Rio-Albrechtsen T., Kiontke K., Chiou S.-Y., Fitch D. H. A., Novel gain-of-function alleles demonstrate a role for the heterochronic gene lin-41 in C. elegans male tail tip morphogenesis. Dev. Biol. 297, 74–86 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Fujiwara M., Sengupta P., McIntire S. L., Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron 36, 1091–1102 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Murakami M., Koga M., Ohshima Y., DAF-7/TGF-β expression required for the normal larval development in C. elegans is controlled by a presumed guanylyl cyclase DAF-11. Mech. Dev. 109, 27–35 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Liu Y., et al. , CHD7 interacts with BMP R-SMADs to epigenetically regulate cardiogenesis in mice. Hum. Mol. Genet. 23, 2145–2156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tewari M., et al. , Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGF-beta signaling network. Mol. Cell, 13, 469–482. [DOI] [PubMed] [Google Scholar]
- 73.Riedel C. G., et al. , DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 15, 491–501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jia K., Albert P. S., Riddle D. L., DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development 129, 221–231 (2002). [DOI] [PubMed] [Google Scholar]
- 75.Chu X., et al. , Genotranscriptomic meta-analysis of the CHD family chromatin remodelers in human cancers - initial evidence of an oncogenic role for CHD7. Mol. Oncol. 11, 1348–1360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Machado R. A. C., et al. , CHD7 promotes glioblastoma cell motility and invasiveness through transcriptional modulation of an invasion signature. Sci. Rep. 9, 3952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sporn M. B., et al. , Polypeptide transforming growth factors isolated from bovine sources and used for wound healing in vivo. Science 219, 1329–1331 (1983). [DOI] [PubMed] [Google Scholar]
- 78.Roberts A. B., et al. , Transforming growth factor type beta: Rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc. Natl. Acad. Sci. U.S.A. 83, 4167–4171 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laiho M., Saksela O., Andreasen P. A., Keski-Oja J., Enhanced production and extracellular deposition of the endothelial-type plasminogen activator inhibitor in cultured human lung fibroblasts by transforming growth factor-beta. J. Cell Biol. 103, 2403–2410 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Overall C. M., Wrana J. L., Sodek J., Independent regulation of collagenase, 72-kDa progelatinase, and metalloendoproteinase inhibitor expression in human fibroblasts by transforming growth factor-β. J. Biol. Chem. 264, 1860–1869 (1989). [PubMed] [Google Scholar]
- 81.Breuer M., Rummler M., Zaouter C., Willie B. M., Patten S. A., Abnormal craniofacial and spinal bone development with col2a1a depletion in a zebrafish model of CHARGE syndrome. bioRxiv [Preprint] (2020). https://www.biorxiv.org/content/10.1101/2020.07.10.197533v3 (Accessed 29 March 2022).
- 82.Suzuki T., Sakai D., Osumi N., Wada H., Wakamatsu Y., Sox genes regulate type 2 collagen expression in avian neural crest cells. Dev. Growth Differ. 48, 477–486 (2006). [DOI] [PubMed] [Google Scholar]
- 83.He D., et al. , Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nat. Neurosci. 19, 678–689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hale C. L., Niederriter A. N., Green G. E., Martin D. M., Atypical phenotypes associated with pathogenic CHD7 variants and a proposal for broadening CHARGE syndrome clinical diagnostic criteria. Am. J. Med. Genet. A. 170A, 344–354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han S. K., et al. , OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7, 56147–56152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nieuwkoop P. D., Faber J., Normal Table of Xenopus laevis (Daudin) (Garland Publishing, 1994). [Google Scholar]
- 87.Huang S., Johnson K. E., Wang H. Z., Blastomeres show differential fate changes in 8-cell Xenopus laevis embryos that are rotated 90 degrees before first cleavage. Dev. Growth Differ. 40, 189–198 (1998). [DOI] [PubMed] [Google Scholar]
- 88.Gawantka V., et al. , Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech. Dev. 77, 95–141 (1998). [DOI] [PubMed] [Google Scholar]
- 89.National Research Council, Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 90.Zahn N., Levin M., Adams D. S., The Zahn drawings: New illustrations of Xenopus embryo and tadpole stages for studies of craniofacial development. Development 144, 2708–2713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and supporting information. RNA-seq data has been uploaded to National Center for Biotechnology Information Gene Expression Omnibus, ID GSE199192.