Significance
Ebola virus (EBOV) infection is a severe, often fatal disease with poorly understood pathophysiology. Here, we performed [18F]-DPA-714 Positron Emission Tomography (PET) in a macaque model of EBOV infection to longitudinally track and quantify the translocator protein (TSPO), an immune cell marker. The imaging findings, along with immunohistology and other disease markers, showed increasing stem cell proliferation in the bone marrow, along with progressive monocytic and lymphocytic loss in the spleen and lungs. This study provides real-time noninvasive assessment of EBOV pathogenesis and disease progression, as well as the associated host responses, at the organ level. This approach can be similarly used to study other inflammatory and infectious diseases and to test the efficacy of newly developed therapeutics and vaccines.
Keywords: PET imaging, Ebola virus, DPA714, macaque
Abstract
Ebola virus (EBOV) disease is characterized by lymphopenia, breach in vascular integrity, cytokine storm, and multiorgan failure. The pathophysiology of organ involvement, however, is incompletely understood. Using [18F]-DPA-714 positron emission tomography (PET) imaging targeting the translocator protein (TSPO), an immune cell marker, we sought to characterize the progression of EBOV-associated organ-level pathophysiology in the EBOV Rhesus macaque model. Dynamic [18F]-DPA-714 PET/computed tomography imaging was performed longitudinally at baseline and at multiple time points after EBOV inoculation, and distribution volumes (Vt) were calculated as a measure of peripheral TSPO binding. Using a mixed-effect linear regression model, spleen and lung Vt decreased, while the bone marrow Vt increased over time after infection. No clear trend was found for liver Vt. Multiple plasma cytokines correlated negatively with lung/spleen Vt and positively with bone marrow Vt. Multiplex immunofluorescence staining in spleen and lung sections confirmed organ-level lymphoid and monocytic loss/apoptosis, thus validating the imaging results. Our findings are consistent with EBOV-induced progressive monocytic and lymphocytic depletion in the spleen, rather than immune activation, as well as depletion of alveolar macrophages in the lungs, with inefficient reactive neutrophilic activation. Increased bone marrow Vt, on the other hand, suggests hematopoietic activation in response to systemic immune cell depletion and leukocytosis and could have prognostic relevance. In vivo PET imaging provided better understanding of organ-level pathophysiology during EBOV infection. A similar approach can be used to delineate the pathophysiology of other systemic infections and to evaluate the effectiveness of newly developed treatment and vaccine strategies.
The Ebola virus (EBOV) is an extremely virulent pathogen with a very high mortality rate. The 2014 outbreak of EBOV in western Africa led to over 28,000 cases and 11,000 fatalities (1), with more recent resurgences reported in the Democratic Republic of the Congo and Guinea (2, 3).
EBOV has been shown to possess broad cell tropism and once inside the host, it infects multiple cell types and undergoes replication (4–7). It’s been shown that early infection of macrophages, dendritic cells, and monocytes is largely responsible for the systemic dissemination of the virus (8, 9). Clinically, following a brief incubation, EBOV infection causes acute fever, fatigue, dyspnea, vomiting, diarrhea, headaches, hypovolemia, and eventually multiorgan failure that generally leads to death about 7 to 14 d after the onset of symptoms (10–16). Despite differences in infection routes, the disease progression of EBOV disease in macaques has been found to closely mirror that of human disease and has been shown to be a good model to study the pathogenesis of EBOV (8).
At the molecular level, EBOV infection is characterized by a late-stage sustained inflammatory response in the form of a “cytokine storm” (9, 17–19). Previous studies using macaques have shown that the secretion of cytokines and chemokines, such as MIP-1α, MIP-1β, TNF, and interleukin (IL)-6 after initial infection leads to increased recruitment of neutrophils, which further aids in the propagation of an inflammatory response in a feedforward loop (20, 21). The continuing inflammatory response is believed to increase vascular permeability by weakening endothelial cell barriers with secondary disseminated vascular coagulation syndrome resembling a septic shock (7, 22).
The current body of literature documenting EBOV etiology is based on in vitro testing or postmortem specimen analysis from cross-sectional animal cohorts. However, in vivo molecular imaging with positron emission tomography (PET) (23) offers a unique opportunity to noninvasively observe disease progression in real time, at the organ level. In our study, we used [18F]-DPA-714 (DPA-714), a radioligand that binds the translocator protein 18 kDa (TSPO), an outer mitochondrial membrane receptor expressed in multiple cell types, especially macrophages. TSPO expression is up-regulated during immune cell activation and thus serves as a marker for not only measuring inflammation (immune cell activation) but also measuring immune cell loss (24–27).
In the present study, our goal was to use DPA-714 PET along with histopathology in a macaque model of Ebola infection, to better understand the pathophysiology of disease progression at the organ and cellular level. For this, we longitudinally imaged Rhesus macaques infected with EBOV at various time points between baseline and terminal endpoint (day 6/7). Whole-blood samples were collected before each imaging session to assess various disease biomarkers. DPA-714 volumes of distribution (Vt) values for different organs were measured over time. Postmortem spleen and lung tissue sections were stained for TSPO and other cell markers to better understand specific cell population involvement, in correlation with disease progression.
Materials and Methods
Flow Cytometry.
Whole-blood samples from eight healthy human subjects (six male and two females with an average age of 52.1 ± 16.4 y) and five healthy female macaques (4.4 ± 0.5 y) were obtained to determine the extent of TSPO expression in various immune cells of both species (Fig. 1 and SI Appendix, Fig. S2). See SI Appendix for additional experimental details and SI Appendix, Table S1 for antibodies used.
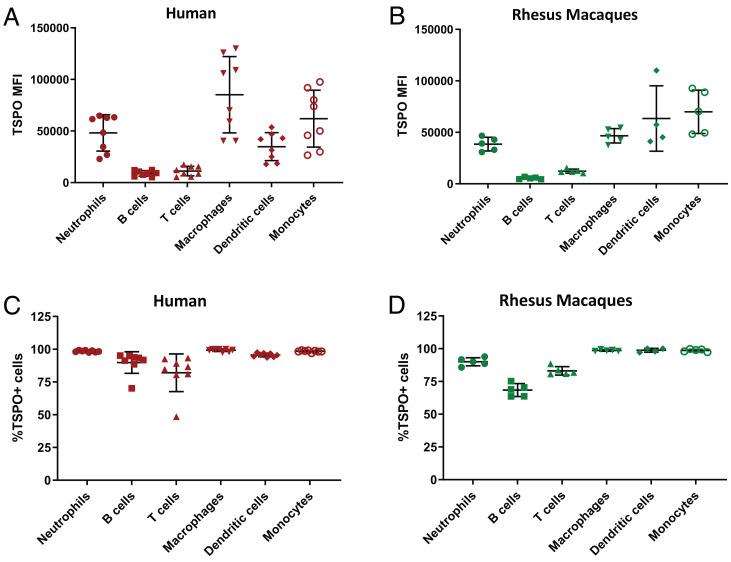
Fig. 1.
Distribution of TSPO expression in immune cell populations. Mean fluorescence intensity (MFI) indicating the levels of TSPO expression in various immune cell populations isolated from the whole blood of (A) healthy humans (n = 8) and (B) Rhesus macaques (n = 5), obtained by flow cytometry. (C and D) Percentage of cells staining positive for TSPO from each immune cell population is shown with red (human) (C) and green (macaque) (D) bars.
Animal Care and Use.
All animal experiments were performed in a biosafety level 4 (BSL-4) laboratory, approved by the National Institute of Allergy and Infectious Diseases Division of Clinical Research Institutional Animal Care and Use Committee, and performed in an American Association for Accreditation of Laboratory Animal Care International accredited facility in accordance with relevant NIH policies and the Animal Welfare Act and Regulations. Ten male rhesus macaques (weight: 5.4 ± 1.8 kg, age: 2.6 ± 1.3 y) were each inoculated with 1,000 plaque-forming units of Ebola virus/H.sapiens-tc/GIN/2014/Makona-C05 (EBOV/Mak) passage 3 virus (BEI resources) via intramuscular injection.
Infection and Sampling Timeline.
All animals had two baseline PET scans obtained between 6 and 12 d before EBOV inoculation (day 0). The macaques were split into three groups depending on the PET-imaging time points as follows: group 1, scanned on days 2 and 4 (n = 3); group 2, scanned on days 3 and 6 (n = 3); and group 3, scanned on days 1, 5, and 7 (n = 4). Blood collection for biomarker sampling was done after each scanning session. The animals were humanely killed after the final PET scan: group 1 on day 4, group 2 on day 6, while group 3 animals were humanely killed at the terminal time point on day 7 (SI Appendix, Fig. S1). The tissues collected postnecropsy were used for immunostaining or assessing viral titers.
PET/CT Imaging.
CT and PET scans were performed with a Gemini TF PET/CT scanner (Philips). Two baseline DPA-714 PET/CT scans were performed on each animal, and two to three scans were performed postinfection. One of the animals in group 3 died during the terminal stage (day 7) scan and so we have excluded the data for that time point from the analysis. The scan data from day 5 of another animal from group 3 was also excluded due to technical complications during the scan. For additional information, see SI Appendix.
Biomarker Testing.
For all animals, whole-blood samples were obtained at least twice prior to inoculation and subsequently on each postinfection scan day until being humanely killed. Complete blood counts with differential, as well as kidney and liver function panels, were performed. The total white blood cell (WBC) (×103/µL) and platelet counts (×103/µL) as well as percent lymphocytes (%lymphocytes), percent monocytes (%monocytes), and percent neutrophils (%neutrophils) were measured. Plasma viral loads were measured. Viral titers in different tissues were determined by plaque assay. For additional information, see SI Appendix.
PET/CT Image Analysis.
The PET image analysis was conducted using PMOD 3.8 software (PMOD Technologies). Forty-minute dynamic (42 frames) emission scan was merged with the single static scan acquired at 60 min to generate a 43-frame merged study. The merged PET images were then coregistered to the CT image (3-mm CT scans obtained through the chest and abdomen). Volumes of interest (VOIs) were drawn on the coregistered image for the liver, lungs, spleen, and bone marrow. The latter was measured using a VOI overlapping the L4 and L5 vertebral bodies in all animals, while avoiding the disk space, spinal canal, and paraspinal muscles. The vertebral bodies were used for this measurement instead of the iliac bones and femoral heads since the latter could not be included in the dynamic PET field-of-view, which encompassed the relevant organs: lungs, liver, spleen, and heart. For the lungs, 7 of 10 animals did not show any lung infiltrates/consolidations or atelectasis. In 3 of 10 animals, however, lung infiltrates and consolidations were seen. Care was made to avoid those regions when selecting the VOIs for the lungs. Additionally, a VOI was also drawn in the left ventricle to generate the whole-blood data for image-derived input function. Additionally, even though the axillary lymph nodes were included in the field-of-view, their Vt values could not be reliably measured due to increased signal in the surrounding fat planes, likely reflecting metabolically active brown fat, a phenomenon that has been already described with many TSPO ligands (28, 29).
The Vt values obtained from both preinoculation scans were averaged to get one baseline Vt value. Time activity curves were generated for tissues and whole-blood data. The kinetic tool of PMOD, PKIN was used to perform Logan analysis with the whole-blood data used as input function to generate Vt values for each organ. No correction for metabolites could be performed due to logistical limitations in the BSL-4 suite. Total spleen volumes of animals were manually segmented from the CT images using MIM Software (v6.9.4). Two scans with significant motion artifacts were excluded from the analyses.
Necropsy and Histopathology.
Tissue samples were obtained from humanely killed animals at various time points postinfection. Using a standard semiautomated rotary microtome and lighted water flotation bath (Leica Biosystems), tissue sections were cut to a thickness of 4 µm and mounted on positively charged uncoated glass slides (ThermoFisher), air-dried at room temperature, stained with H&E, and cover-slipped for microscopic evaluation by the pathologist. For additional information, see SI Appendix.
Immunostaining.
Multiplex fluorescence immunohistochemistry (MF-IHC) staining was performed on spleen tissue sections for the following targets: TSPO, CD3, CD20, CD68, CD31/PECAM-1, CD144/VE-cadherin, cleaved caspase-3/cleaved PARP1 [poly(ADP-ribose) polymerase] (CC3/PARP), and VP40 (SI Appendix, Table S2). Lung tissue sections were stained for the following targets: TSPO, CD68, CD66abce, and VP40 (SI Appendix, Table S3). The cell nuclei in the spleen and lung sections were counterstained using 1 μg/mL DAPI to facilitate cell counting. For additional information, see SI Appendix.
Statistical Analysis.
Mixed-effect linear regression models were used to evaluate the change in DPA-714 Vt values of different organs over the course of disease. The organ Vt values were used as the dependent variable while time was designated as the explanatory/independent variable, with corresponding baseline value as covariate. Because of the repeated-measures nature of the data, the optimal variance–covariance matrix structure for each response variable was identified based on the Bayesian Information Criterion, and was used in the final analysis.
Pearson partial correlations between DPA-714 Vt values in the organs and biological markers of disease progression were computed, accounting for repeated measures. Tests were performed on both nontransformed and log10-transformed data, depending on which followed the normal distribution more closely. The analysis provided both correlation coefficients and P values for each pair of variables. In order to account for multiple comparisons in this analysis, correlations with P ≥ 0.01 should be considered as reflecting no evidence of an association. All statistical analyses were performed using SAS v9.4 (SAS Institute).
Results
Distribution of TSPO Expression in the Macaque and Human Blood Cell Population.
On flow cytometry, the mean fluorescence intensity of TSPO staining in each of the human immune cell populations was calculated. All the immune cell types were shown to express TSPO, albeit to varying degrees. The highest TSPO expression was seen in macrophages followed by monocytes, dendritic cells, and neutrophils, while T and B cells had the lowest expression levels (Fig. 1A). Similar results were observed in the macaques where macrophages, monocytes, and dendritic cells seemed to have the highest expression levels of TSPO (Fig. 1B). In all cell types, over 70% of the cells in each isolated population expressed TSPO (Fig. 1C). Our flow results suggested that all the major immune cell populations and not just the monocytes and macrophages contribute to the total DPA-714 signal in the periphery. Therefore, changes in TSPO expression and DPA-714 signal by extension would likely reflect on the status of multiple immune cell populations over the course of EBOV infection.
Changes in Complete Blood Cell, Viral Loads, and Cytokines Over Time.
The complete blood counts showed a decrease in %lymphocyte, %monocyte, and total platelet counts while the total WBC counts and %neutrophil counts increased over time (SI Appendix, Fig. S3 A and B). The plasma viremia increased over time, as did the viral titers measured in various peripheral organs between day 4 and the terminal time points (SI Appendix, Fig. S3 C and D). Multiple plasma inflammatory cytokines dramatically increased starting day 4/5 postinoculation. There was a concurrent up-regulation of IL-10, an antiinflammatory cytokine that acts to dampen the immune-related inflammation (SI Appendix, Fig. S4).
Changes in Organ DPA-714 Binding during EBOV Infection.
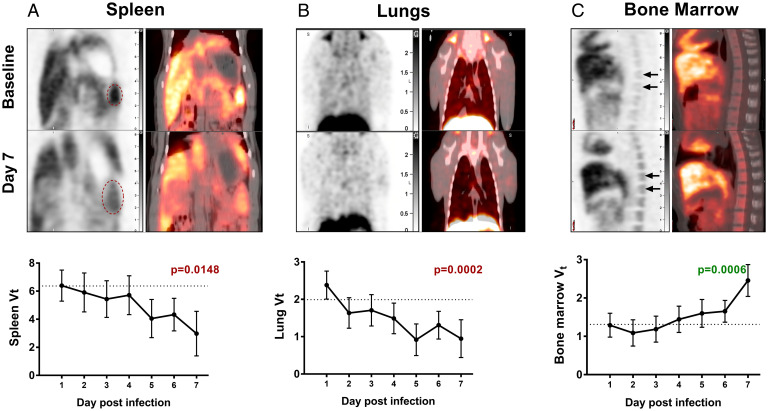
The liver Vt values showed slight drops at days 2 and 4 (SI Appendix, Table S4); however, there was no statistically significant overall change in Vt values over time. The Vt values of the spleen and lungs decreased as a function of time postinfection (P = 0.0148 and P = 0.0002, respectively) (Fig. 2 A and B), while the opposite trend was observed in the bone marrow (P = 0.0006) (Fig. 2C). Assessing individual time points, the lung Vt values were lower in each of days 2 through 7, relative to day 1 (P = 0.012, 0.0263, 0.0035, <0.0001, 0.0006, <0.0001, respectively), as did the spleen Vt values which decreased over time, with the largest reductions compared to day 1 occurring on days 5 (P = 0.0106), 6 (P = 0.0135), and 7 (P = 0.0014). The decrease in DPA-714 binding (Fig. 2A) occurred despite a general trend of splenomegaly seen in these animals (SI Appendix, Fig. S5). On the other hand, the bone marrow Vt values showed an increase on day 7 compared to day 1 (P < 0.0001).
Fig. 2.
DPA-714 binding in EBOV infection. The macaques underwent longitudinal DPA-714 PET imaging at baseline and at the indicated time points postinoculation with EBOV. The representative PET images (Upper) show the changes in DPA-714 binding at baseline and day 7 postinoculation in the (A) spleen, (B) lungs, and (C) bone marrow. The red ovals indicate the splenic region in A. The time course depicting the changes in TSPO binding (Vt) in the organs over time is shown below each image (least-square means adjusted for baseline with 95% confidence intervals). The dotted line represents the average baseline Vt. There is a progressive decrease in TSPO binding in both the spleen and the lungs over time, while there is increased binding seen in the bone marrow. Statistical analysis was performed using the mixed-effect linear regression model and the changes in Vt were found to be correlated with the duration of infection.
Biomarkers Correlate with DPA-714 Organ Vt.
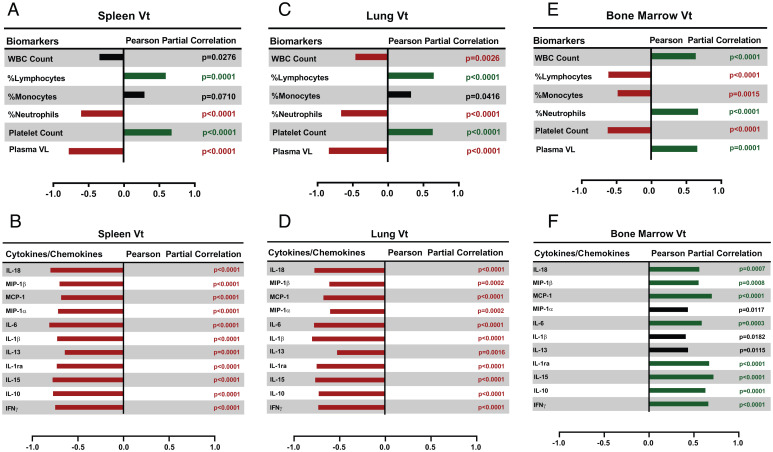
The spleen and lungs Vt were negatively correlated to %neutrophil counts and positively correlated to %lymphocyte counts (Fig. 3 A and C). Conversely, the bone marrow Vt showed positive correlations with WBC and %neutrophil counts and negative correlations with platelet, %lymphocyte, and %monocyte counts (all P ≤ 0.0015) (Fig. 3E). The plasma viremia was found to correlate negatively with spleen and lung Vt and positively with bone marrow Vt (all P ≤ 0.0001) (Fig. 3 A, C, and E). The spleen and lung Vt negatively correlated with interferon (IFN)-γ, IL-10, IL-15, IL-1Ra, IL-6, MCP-1, MIP-1α, IL-13, IL-1β, MIP-1β, and IL-18 (all P ≤ 0.0016) (Fig. 3 B and D). On the other hand, the bone marrow Vt showed positive correlations with a number of cytokines such as IFN-γ, IL-15, IL-1Ra, IL-6, MCP-1, MIP-1β, IL-18, and IL-10 (all P < 0.001) (Fig. 3F). The uptake of DPA-714 in the liver did not correlate with any of the cytokines or other biomarkers.
Fig. 3.
Biomarker correlations. Blood cell populations, plasma viral loads (VL), and cytokines measured at the imaging time points were correlated with (A and B) spleen, (C and D) lung, and (E and F) bone marrow DPA-714 Vt values. Positive and negative correlations with P < 0.01 are shown in dark green and red bars, respectively. The black bars indicate correlations that have a P > 0.01. Pearson partial correlations accounting for repeated measures were used to calculate the correlation coefficients and the P values. Plasma VL and cytokine values were log-transformed before performing the correlations in order to achieve a more normal distribution. Log-transformations were not needed for platelet counts, WBC counts, and differential.
Histopathology Results.
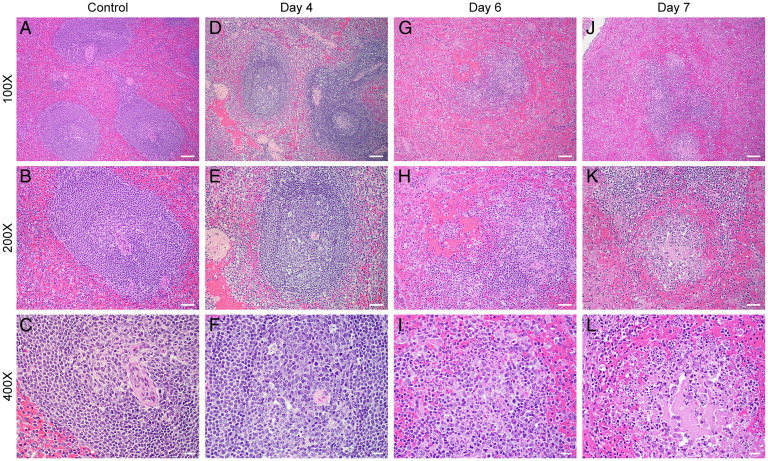
Histopathologic findings in the spleen appeared on day 4 necropsies and worsened by days 6 to 7. Those ranged from lymphoid depletion to necrosis of germinal centers (Fig. 4). In the liver, lesions ranged from mild hepatocellular degeneration with few intracytoplasmic viral inclusion bodies to hepatocellular necrosis by day 7. Mild tubular degeneration was noted in the kidneys by day 7, consistent with prerenal failure. In the lungs, focal areas of congestion and edema with few peribronchiolar inflammatory infiltrates were noted in 2 of the 10 subjects at day 7, which was believed to be due to intubation and aspiration and corresponded to infiltrates seen on the CT scans. The lungs otherwise showed no major abnormalities on H&E staining.
Fig. 4.
H&E spleen (mid disease and terminal). Histopathologic findings for the spleen are consistent with EBOV disease in the nonhuman primate (D–L). Negative controls (A–C, 100×, 200×, and 400×, respectively) show relatively normal splenic architecture. (D–F) 100×, 200×, and 400×, respectively, at day 4 postexposure show mild lymphoid depletion with occasional evidence of lymphocytolysis within germinal centers (white pulp). (G–I) 100×, 200×, and 400×, respectively, at day 6 postexposure show significant lymphoid depletion, with areas of lymphoid degeneration and necrosis within germinal centers, and fibrin with cellular debris in the red pulp. Similarly, in J–L 100×, 200×, and 400×, respectively, at day 7 postexposure, show lymphoid depletion, germinal center necrosis, with hemorrhage, fibrin, and cellular debris in the red pulp. (Scale bars: 100× bar is 100 μ, 200× is 50 μ, and 400× is 20 μ.)
Postmortem Immunostaining of Spleen and Lung Tissues.
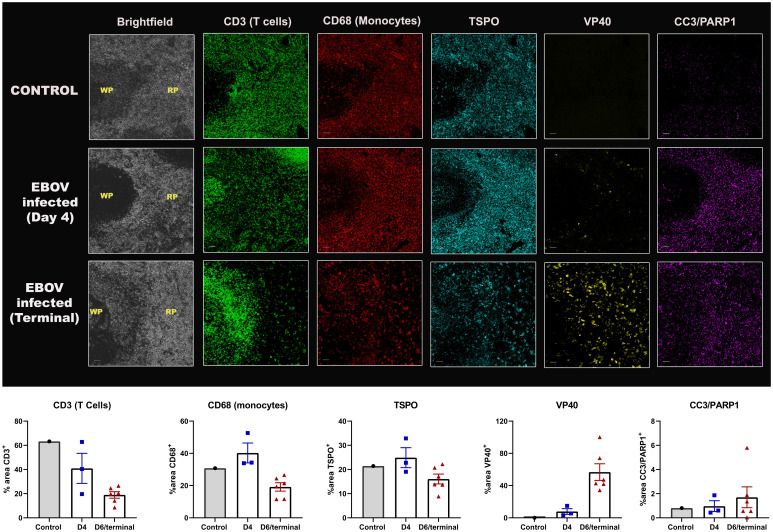
There was increased staining of VP40 (EBOV matrix protein) over time in the spleen of infected animals (Fig. 5). However, there was a progressive reduction in CD3 staining (T cell marker) starting from day 4 until the terminal endpoint, indicating widespread T cell death. On the other hand, B cells (CD20 staining) seemed to increase initially (day 3) before tapering off (terminal) (SI Appendix, Fig. S6). Such increase in CD20hi population of B cells has been previously reported in infected patients (30). There was an increase in apoptosis from day 4 to terminal point, as measured by CC3 and PARP1 staining. Concurrent decreases in CD68 expression at the terminal time point indicated that the macrophage population was also depleted in the later stages of the disease. The MF-IHC data suggest that a majority of the infected cells in the spleen were macrophages, since VP40 staining was mainly colocalized with CD68, and many of these cells also stained for CC3/PARP1. None of the surviving lymphocytes seemed to be infected with the virus, which has been reported before (20, 31, 32). In concordance with PET data, there was a decrease in TSPO staining at the terminal time point as well. This is further supported by decreased staining of CD68, a macrophage lineage marker, indicating that a loss of the macrophage population is what primarily led to a decline in the TSPO signal. The MF-IHC data suggest that as a result of EBOV infection, most immune cells in the spleen are depleted as a result of infection (monocytes), apoptotic cell death (monocytes and lymphocytes), and possibly peripheral migration, rather than an immune activation.
Fig. 5.
Postmortem immunostaining in the spleen. Representative MF-IHC images of a single spleen section from a control (Top), day 4-infected (Middle), and a terminally infected EBOV macaque (Bottom). The various stains are: CD3 (T cells), CD68 (Macrophages), TSPO, CC3/PARP1 (cell death markers), VP40 (EBOV marker). The regions shown here consist of both the red pulp (RP) and white pulp (WP) areas, as indicated in the brightfield images. (Scale bars, 50 μ.) The quantification data measuring percent area of positive staining for control (n = 1), day 4 (n = 3) and terminal animals (n = 6) is shown below each stain and was obtained by drawing multiple regions-of-interest across the entire splenic section using ImageJ.
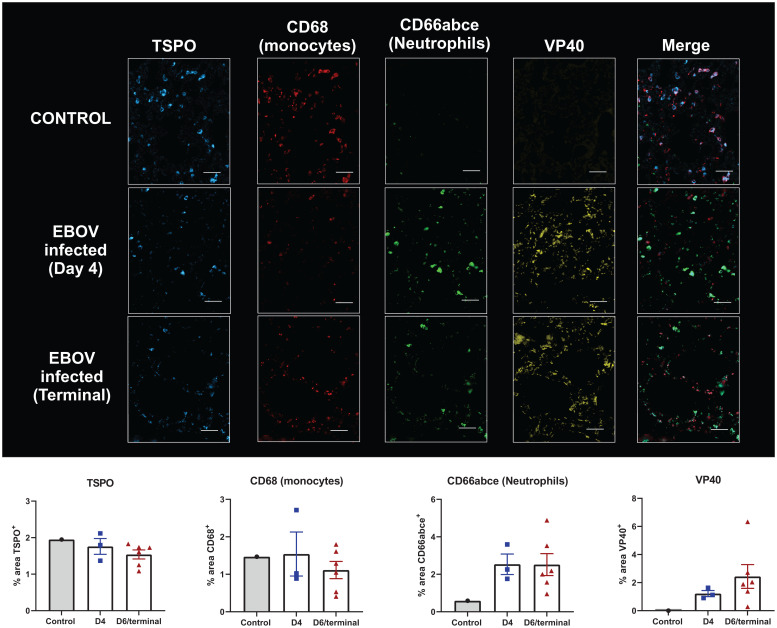
In the lungs, the CD68 expression increased in one and decreased in two animals at day 4 compared to the control, while a modest decrease of staining in most animals was observed at the terminal time point. A corresponding decrease in TSPO was observed at the terminal time point when compared to both day 4 and control macaques, although that was less impressive than the decreased CD68 staining. This could be due to concurrent increase in neutrophil infiltration in the lungs, which also express TSPO (compared to uninfected animals), thus partially offsetting the decreased monocyte counts (Fig. 6). As expected, the VP40 staining increased progressively throughout the infection (Fig. 6). Additionally, frequent costaining between neutrophils (CD66abce) and macrophages was observed, suggesting that either the neutrophils are coexpressing CD68, as they are known to do under inflammatory conditions (33), or that some macrophages may be getting phagocytosed by neutrophils, especially since the CD68 staining was sometimes seen within the cytoplasmic area of the neutrophils (SI Appendix, Fig. S7).
Fig. 6.
Postmortem immunostaining in the lungs. Representative MF-IHC images of a single lung section from an uninfected control (Top), an EBOV-infected macaque at day 4 (Middle), and a terminally infected EBOV macaque (Bottom). The various stains are: TSPO, CD68 (Macrophages), CD66abce (neutrophils), and VP40 (EBOV marker). (Scale bars, 50 μ.) The quantification data measuring percent area positive stain is shown for control (n = 1), day 4 (n = 3), and terminal (n = 6) below each stain and was obtained by drawing multiple regions-of-interest across the entire lung section using ImageJ.
Discussion
In this study, we used DPA-714 to characterize organ-level host immune response to EBOV infection over time. Since it was unclear as to which immune cell types express TSPO, flow cytometry was performed to analyze the degree of protein expression in human and macaque cells. Monocytes, macrophages, and dendritic cells expressed the highest amounts of TSPO. However, neutrophils, and to a lesser extent T cells and B cells, also expressed basal levels of TSPO (Fig. 1). Using DPA-714 PET imaging, we found a decrease in TSPO expression in both the spleen and lungs of animals over time after EBOV infection (Fig. 2 A and B). On the other hand, there was increase in binding in bone marrow TSPO expression of these animals (Fig. 2C). Taken together, these results indicate that at the spleen level, there is massive immune cell depletion rather than an active inflammatory process, while in the bone marrow there is a reactive hematopoietic activation.
DPA-714 binding in the spleen decreased despite progressive enlargement of the organ as measured on CT (SI Appendix, Fig. S5) (34), likely due to congestion and necrosis (Fig. 4). The decreased spleen DPA-714 binding was validated with decreased TSPO staining on MF-IHC (Fig. 5). Additional staining indicated extensive T cell (CD3) and monocytic (CD68) depletion in the spleen. A majority of the monocytes costained with VP40, suggesting infection with the virus. There was also increased costaining of macrophages with cell death markers (CC3 and cleaved PARP1). However, there was no evidence of T cell infection in our animals (Fig. 5), although a recent study has shown that EBOV can induce abortive replication in T cells leading to lymphopenia (35). Peripheral cell death in the monocyte/macrophage population after direct infection with EBOV and bystander T cell death have been reported in previous studies (7, 31, 36–41). However, this has not been shown previously in vivo, dynamically, at the level of the organs such as the spleen and lungs. While the decrease in TSPO staining was not as precipitous as the drop of monocytes and T cells during the terminal stages, this could be attributed to the expression of TSPO in other cell types, such as B cells and neutrophils, in which the decline in TSPO expression is likely less impressive than that seen in monocytes. Interestingly, there was an increase in B cell staining (CD20+) in the spleen during the early stages of the infection, a phenomenon that has been reported previously in humanized mice (42) and humans (30) infected with EBOV (SI Appendix, Fig. S6). This could be attributed either to initial activation of B cells by secreted cytokines or the increasing exposure to viral antigens (43, 44) (SI Appendix, Fig. S6).
Similarly, in vivo, we found a progressive decrease in TSPO binding in the lungs of infected macaques, suggesting depletion of alveolar macrophages. It has been previously reported that EBOV infection leads to alveolar edema, hemorrhaging, and alveolar damage (45–47), which is believed to be a consequence of sustained inflammation (cytokine storm) and direct infection of endothelial cells and alveolar macrophages, supported by immunohistochemical evidence of viral presence in lung fibroblasts and alveolar macrophages (45, 46, 48). We validated our PET imaging results with MF-IHC data showing modest decrease in CD68 expression between day 4 and the terminal time points with concurrent reduction in TSPO staining, as well when compared to the control. The CD66abce (neutrophils) staining increased at day 4 and then stayed fairly constant on terminal day. These findings suggest a progressive loss of macrophages in the lungs during the course of the disease with associated decrease in TSPO expression (Fig. 6). On the other hand, concurrent neutrophilic infiltration into the lung tissues likely offset this decrease in TSPO staining since neutrophils also express TSPO, albeit to a lesser extent than monocytes/macrophages.
In the bone marrow, on the other hand, we found a progressive increase in DPA-714 binding, which correlated with time postinoculation (Fig. 2C). We hypothesize that this is due to hematopoietic activation as a means to offset the cellular loss in the organs and to control the ongoing infection. A couple studies have shown the bone marrow to be infected with EBOV (8, 45). In another study, however, McElroy et al. (49) analyzed peripheral blood mononuclear cells from patients infected with EBOV by mass cytometry and found larger cell populations of myeloid origin likely derived from the bone marrow due to increased hematopoiesis during the acute EBOV infection. More recently, Kotliar et al. (50) found evidence of immature neutrophils originating from the bone marrow in the EBOV-infected rhesus macaques using single-cell RNA and protein profiling. They also identified subsets of monocytes and macrophages that had a transcriptional profile similar to the monocyte precursors derived from the bone marrow, which further supports the prevalence of emergency myelopoiesis during the course of the disease. These observations suggest that even though there could be potential cellular loss due to viral infection in the bone marrow, the expansion of immature proliferating myeloid pool outpaces this loss, thus resulting in a progressive increase in DPA-714 binding over time. Indeed, it is well established that there is an emergency myelopoiesis in the bone marrow due to general infection-mediated inflammation (51, 52). This myeloid proliferation is mainly driven by growth factors, such as M-CSF, GM-CSF, and G-CSF, and our results also show modest increase in GM-CSF production in the plasma of the infected macaques over time. Additionally, we found several positive correlations of cytokines/chemokines with DPA-714 binding in the bone marrow of the infected animals (Fig. 3), indicating that the inflammatory milieu could contribute to hematopoietic activation during the disease course. This is an important observation since it depicts a systemic nature of the response to the infectious process, which could potentially be a prognostic indicator of outcome in patients.
One of the limitations of our study is that, while the macaque model faithfully recapitulates the acute phase of the disease course in humans, the accelerated pace and fatal nature of the infection prevents us from observing the long-term effects of the virus on the organs. Further studies thus need to be conducted to understand the interplay of cellular loss and hematopoietic activation in survivors. Another limitation is that the effect of lymphocyte, monocyte, and platelet depletion, as well as leukocytosis and neutrophilia in the vessels during the course of infection, could affect the radioactivity measures in vascular organs like the lungs and spleen. However, our analysis takes this into account since we used the left ventricle as the image-derived input function. We did not obtain an arterial input function in the animals due to logistical limitations associated with performing imaging studies in a BSL-4 suite, including the difficulty of obtaining arterial access and quickly acquiring blood samples while wearing positive pressure personnel suits. This would also be difficult to perform repeatedly in progressively sicker animals. We did not perform metabolite correction either, since we had no access to chromatographic analysis methods inside the BSL-4 space and transferring contaminated blood outside the space requires virus inactivation by chemical reagents (e.g., neutral-buffered formalin, glutaraldehyde, paraformaldehyde, or chaotropic agents) for at least 10 min, which would delay and affect the results of metabolite analysis afterward. We assume, however, that metabolism of the parent compound would not change significantly over time and is probably less important in longitudinal follow-up scans of the same animals. Due to the same logistical limitations mentioned above, and since the amount of blood drawn was limited to 3 to 5 mL per scanning session, we could not perform longitudinal flow cytometry assessment of TSPO expression in the blood of the infected animals. Another limitation is that we could only test for a limited number of immune cell markers by immunofluorescence staining, thereby not accounting for TSPO expression in nonimmune cells, especially in the lungs, considering prior work has shown that bronchial epithelial cells and to a lesser extent alveolar cells express TSPO (53). Longitudinal changes in TSPO expression in the latter cells types could have possibly affected our results; however, we believe this effect to be minimal since we did not identify appreciable changes to lung architecture during the course of the infection by H&E staining. One last limitation is that we could not perform immunostaining in the liver sections due to high levels of autofluorescence in those tissues, possibly because of the fact that the animals were not perfused during necropsy.
In conclusion, despite multiple devastating EBOV epidemics, there is a dearth of understanding of disease progression and pathophysiology due to the resource-limited nature of endemic regions. Using DPA-714 PET imaging in the macaque model along with various disease biomarkers and IHC helped us better understand the systemic progression of EBOV infection and provided insight into organ-level pathophysiology. The results indicate that there is a sustained loss of TSPO expression in the lungs and spleen due to local depletion of monocytes, macrophages, and dendritic cells. On the other hand, hematopoietic activation seems to occur in the bone marrow, possibly as a compensatory mechanism. Therefore, having validated DPA-714 PET as a noninvasive method of following up changes in TSPO expression during EBOV infection, our technique can now be applied to evaluate the timing and efficacy of novel therapeutic interventions aimed at mitigating cellular loss or the aberrant inflammatory reaction during infection. This methodology can also be adapted to elucidate the host immune responses to other emerging pathogens with similar systemic debilitating effects, such as SARS-CoV2, and help assess the usefulness of various therapeutic and preventative strategies in animal models.
Supplementary Material
Acknowledgments
We thank the veterinary and comparative medicine staff of the Integrated Research Facility, National Institute of Allergy and Infectious Diseases (NIAID), for maintaining the nonhuman primates used in this study; the imaging team, including Dr. David Thomasson, Dr. Jurgen Seidel, and Philip Sayre, for meticulously planning and performing the scans; and the National Heart, Lung, and Blood Institute Flow core for their help with our flow cytometry experiments. Funding for this study was provided by the Intramural Research Program of the NIAID, NIH, Department of Health and Human Services, under Contract HHSN272201800013C, and by the Center for Infectious Disease Imaging, Clinical Center, NIH. It is also funded in part by federal funds from the National Cancer Institute, NIH, under Contract 75N910D00024, Task Order 75N91019F00130. This work was supported in part through Battelle Memorial Institute’s (BMI) former prime contract with the NIAID under Contract HHSN272200700016I and Laulima Government Solutions’s (LGS) current prime contract with NIAID under Contract HHSN272201800013C. K.R.H. performed this work as a former employee of BMI, and R.R. and L.M.H. performed this work as former employees of Charles River Laboratories, a subcontractor of BMI and current employees of LGS. J.H.L. performed this work as a former employee of MedRelief, a subcontractor of BMI, and as a current employee of Tunnell Government Services, a subcontractor of LGS. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mentioning of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110846119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or Supporting Information.
References
- 1.World Health Organization, Ebola situation report 2016. https://apps.who.int/ebola/current-situation/ebola-situation-report-30-march-2016.
- 2.Centers for Disease Control, 2021 Democratic Republic of the Congo, North Kivu Province. https://www.cdc.gov/vhf/ebola/outbreaks/drc/2021-february.html (Accessed 23 March 2022).
- 3.World Health Organization, Ebola—Guinea” (17 February 2021). https://www.who.int/emergencies/disease-outbreak-news/item/2021-DON312 (Accessed 23 March 2022).
- 4.Takada A., Filovirus tropism: Cellular molecules for viral entry. Front. Microbiol. 3, 34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowell S. F., et al. , Transmission of Ebola hemorrhagic fever: A study of risk factors in family members, Kikwit, Democratic Republic of the Congo, 1995. Commission de Lutte contre les Epidémies à Kikwit. J. Infect. Dis. 179 (suppl. 1), S87–S91 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Bausch D. G., et al. , Assessment of the risk of Ebola virus transmission from bodily fluids and fomites. J. Infect. Dis. 196 (suppl. 2), S142–S147 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Feldmann H., Geisbert T. W., Ebola haemorrhagic fever. Lancet 377, 849–862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geisbert T. W., et al. , Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: Evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 163, 2347–2370 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ploquin A., Zhou Y., Sullivan N. J., Ebola immunity: Gaining a winning position in lightning chess. J. Immunol. 201, 833–842 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Peters C. J., LeDuc J. W., An introduction to Ebola: The virus and the disease. J. Infect. Dis. 179 (suppl. 1), ix–xvi (1999). [DOI] [PubMed] [Google Scholar]
- 11.Wong K. K., et al. ; Monrovia Medical Unit, Supportive care of the first 2 Ebola virus disease patients at the Monrovia Medical Unit. Clin. Infect. Dis. 61, e47–e51 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Schieffelin J. S., et al. ; KGH Lassa Fever Program; Viral Hemorrhagic Fever Consortium; WHO Clinical Response Team, Clinical illness and outcomes in patients with Ebola in Sierra Leone. N. Engl. J. Med. 371, 2092–2100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin E., et al. , Clinical features of patients with Ebola virus disease in Sierra Leone. Clin. Infect. Dis. 61, 491–495 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Nanclares C., et al. , Ebola virus disease, Democratic Republic of the Congo, 2014. Emerg. Infect. Dis. 22, 1579–1586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ksiazek T. G., et al. , Clinical virology of Ebola hemorrhagic fever (EHF): Virus, virus antigen, and IgG and IgM antibody findings among EHF patients in Kikwit, Democratic Republic of the Congo, 1995. J. Infect. Dis. 179 (suppl. 1), S177–S187 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Hunt L., et al. , Clinical presentation, biochemical, and haematological parameters and their association with outcome in patients with Ebola virus disease: An observational cohort study. Lancet Infect. Dis. 15, 1292–1299 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Baize S., et al. , Inflammatory responses in Ebola virus-infected patients. Clin. Exp. Immunol. 128, 163–168 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruibal P., et al. , Unique human immune signature of Ebola virus disease in Guinea. Nature 533, 100–104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Younan P., Iampietro M., Bukreyev A., Disabling of lymphocyte immune response by Ebola virus. PLoS Pathog. 14, e1006932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebihara H., et al. , Host response dynamics following lethal infection of rhesus macaques with Zaire ebolavirus. J. Infect. Dis. 204 (suppl. 3), S991–S999 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hensley L. E., Young H. A., Jahrling P. B., Geisbert T. W., Proinflammatory response during Ebola virus infection of primate models: Possible involvement of the tumor necrosis factor receptor superfamily. Immunol. Lett. 80, 169–179 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Baseler L., Chertow D. S., Johnson K. M., Feldmann H., Morens D. M., The pathogenesis of Ebola virus disease. Annu. Rev. Pathol. 12, 387–418 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Gordon O., Ruiz-Bedoya C. A., Ordonez A. A., Tucker E. W., Jain S. K., Molecular imaging: A novel tool to visualize pathogenesis of infections in situ. MBio 10, e00317-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vicidomini C., et al. , In vivo imaging and characterization of [(18)F]DPA-714, a potential new TSPO ligand, in mouse brain and peripheral tissues using small-animal PET. Nucl. Med. Biol. 42, 309–316 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Arlicot N., et al. , Initial evaluation in healthy humans of [18F]DPA-714, a potential PET biomarker for neuroinflammation. Nucl. Med. Biol. 39, 570–578 (2012). [DOI] [PubMed] [Google Scholar]
- 26.James M. L., et al. , DPA-714, a new translocator protein-specific ligand: Synthesis, radiofluorination, and pharmacologic characterization. J. Nucl. Med. 49, 814–822 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Largeau B., Dupont A. C., Guilloteau D., Santiago-Ribeiro M. J., Arlicot N., TSPO PET imaging: From microglial activation to peripheral sterile inflammatory diseases? Contrast Media Mol. Imaging 2017, 6592139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran C., et al. , PET imaging of human brown adipose tissue with the TSPO tracer [11C]PBR28. Mol. Imaging Biol. 20, 188–193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh C., et al. , Brown adipose tissue imaging using the TSPO tracer [18F]fluoromethyl-PBR28-d2: A comparison with [18F]FDG. Nucl. Med. Biol. 90–91, 98–103 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Ellebedy A. H., et al. , Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol. 17, 1226–1234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iampietro M., et al. , Ebola virus glycoprotein directly triggers T lymphocyte death despite of the lack of infection. PLoS Pathog. 13, e1006397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geisbert T. W., et al. , Apoptosis induced in vitro and in vivo during infection by Ebola and Marburg viruses. Lab. Invest. 80, 171–186 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Chistiakov D. A., Killingsworth M. C., Myasoedova V. A., Orekhov A. N., Bobryshev Y. V., CD68/macrosialin: Not just a histochemical marker. Lab. Invest. 97, 4–13 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Smither S. J., et al. , Experimental respiratory infection of marmosets (Callithrix jacchus) with Ebola virus Kikwit. J. Infect. Dis. 212 (suppl. 2), S336–S345 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Younan P., et al. , Ebola virus-mediated T-lymphocyte depletion is the result of an abortive infection. PLoS Pathog. 15, e1008068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baize S., Leroy E. M., Mavoungou E., Fisher-Hoch S. P., Apoptosis in fatal Ebola infection. Does the virus toll the bell for immune system? Apoptosis 5, 5–7 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Reed D. S., Hensley L. E., Geisbert J. B., Jahrling P. B., Geisbert T. W., Depletion of peripheral blood T lymphocytes and NK cells during the course of Ebola hemorrhagic fever in cynomolgus macaques. Viral Immunol. 17, 390–400 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Gupta M., Spiropoulou C., Rollin P. E., Ebola virus infection of human PBMCs causes massive death of macrophages, CD4 and CD8 T cell sub-populations in vitro. Virology 364, 45–54 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Wauquier N., Becquart P., Padilla C., Baize S., Leroy E. M., Human fatal Zaire Ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl. Trop. Dis. 4, e837 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olejnik J., et al. , Ebola virus does not block apoptotic signaling pathways. J. Virol. 87, 5384–5396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falasca L., et al. , Molecular mechanisms of Ebola virus pathogenesis: Focus on cell death. Cell Death Differ. 22, 1250–1259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Escudero-Pérez B., Volchkova V. A., Dolnik O., Lawrence P., Volchkov V. E., Shed GP of Ebola virus triggers immune activation and increased vascular permeability. PLoS Pathog. 10, e1004509 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moens L., Tangye S. G., Cytokine-mediated regulation of plasma cell generation: IL-21 takes center stage. Front. Immunol. 5, 65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lam J. H., Smith F. L., Baumgarth N., B cell activation and response regulation during viral infections. Viral Immunol. 33, 294–306 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martines R. B., Ng D. L., Greer P. W., Rollin P. E., Zaki S. R., Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J. Pathol. 235, 153–174 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Osterholm M. T., et al. , Transmission of Ebola viruses: What we know and what we do not know. MBio 6, e00137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lalle E., et al. , Pulmonary involvement during the Ebola virus disease. Viruses 11, E780 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf T., et al. , Severe Ebola virus disease with vascular leakage and multiorgan failure: Treatment of a patient in intensive care. Lancet 385, 1428–1435 (2015). [DOI] [PubMed] [Google Scholar]
- 49.McElroy A. K., et al. , Immunologic timeline of Ebola virus disease and recovery in humans. JCI Insight 5, 137260 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kotliar D., et al. , Single-cell profiling of Ebola virus disease in vivo reveals viral and host dynamics. Cell 183, 1383–1401.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takizawa H., Boettcher S., Manz M. G., Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood 119, 2991–3002 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Furusawa J., et al. , Promotion of expansion and differentiation of hematopoietic stem cells by interleukin-27 into myeloid progenitors to control infection in emergency myelopoiesis. PLoS Pathog. 12, e1005507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hatori A., et al. , PET imaging of lung inflammation with [18F]FEDAC, a radioligand for translocator protein (18 kDa). PLoS One 7, e45065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or Supporting Information.