Significance
The connecting cilium (CC) of the photoreceptor provides the only route for the trafficking of the outer segment (OS) proteins. Failure of OS protein transport causes degenerative photoreceptor diseases, including retinitis pigmentosa. We demonstrate that Tmem138, a protein linked to ciliopathy, is localized to the photoreceptor CC. Germline deletion of Tmem138 abolished OS morphogenesis, followed by rapid photoreceptor degeneration. Tmem138 interacts with rhodopsin and two additional CC compartment proteins, Ahi1 and Tmem231, likely forming a membrane complex to facilitate trafficking of rhodopsin and other OS-bound proteins across the CC. The study thus implicates a new line of regulation on the delivery of OS proteins through interactions with CC membrane complex(es) and provides insights into photoreceptor ciliopathy diseases.
Keywords: Tmem138, photoreceptor, connecting cilium, outer segment, biogenesis
Abstract
Photoreceptor connecting cilium (CC) is structurally analogous to the transition zone (TZ) of primary cilia and gates the molecular trafficking between the inner and the outer segment (OS). Retinal dystrophies with underlying CC defects are manifested in a broad array of syndromic conditions known as ciliopathies as well as nonsyndromic retinal degenerations. Despite extensive studies, many questions remain in the mechanism of protein trafficking across the photoreceptor CC. Here, we genetically inactivated mouse Tmem138, a gene encoding a putative transmembrane protein localized to the ciliary TZ and linked to ciliopathies. Germline deletion of Tmem138 abolished OS morphogenesis, followed by rapid photoreceptor degeneration. Tmem138 was found localized to the photoreceptor CC and was required for localization of Ahi1 to the distal subdomain of the CC. Among the examined set of OS proteins, rhodopsin was mislocalized throughout the mutant cell body prior to OS morphogenesis. Ablation of Tmem138 in mature rods recapitulated the molecular changes in the germline mutants, causing failure of disc renewal and disintegration of the OS. Furthermore, Tmem138 interacts reciprocally with rhodopsin and a related protein Tmem231, and the ciliary localization of the latter was also altered in the mutant photoreceptors. Taken together, these results suggest a crucial role of Tmem138 in the functional organization of the CC, which is essential for rhodopsin localization and OS biogenesis.
Ciliopathies are pleiotropic genetic diseases affecting multiple organs and can be clinically classified as distinct entities, including Bardet–Biedl syndrome (BBS), Meckel–Gruber syndrome (MKS), Senior Loken syndrome, and Joubert syndrome (JBTS), among others. The retinal photoreceptors are affected in nearly all ciliopathies, as their light-sensing outer segments (OSs) are modified cilia and are thus highly vulnerable. About 10% of the OS is renewed daily, requiring active transport of large amounts of newly synthesized proteins and lipids across the cilium in support of OS renewal. Rhodopsin is the most abundant OS disc protein, mutations of which alone account for 10 to 20% of retinitis pigmentosa. Vectorial transport of rhodopsin from the biosynthetic inner segment (IS) to the OS is crucial for both photoreceptor function and homeostasis. Accordingly, ectopic rhodopsin localization is a major cause of photoreceptor death (1–4). Despite extensive studies, rhodopsin transport has only been partially understood. Trafficking of rhodopsin-carrying vesicles from trans-Golgi to the base of the CC is coordinated by GTPases, their effectors, and the intraflagellar transport (IFT) complexes (5–8). Recent evidence from live imaging in cultured cells and retinal explants suggests that IFT88 and Kif3a are involved in ciliary transport of rhodopsin (9), presumably along the axonemal microtubules. Although many mouse models with mutations in IFT subunits or associated motors exhibit mislocalization of rhodopsin (8, 10, 11), the causality between these transporting components and rhodopsin trafficking defects is inconclusive, often due to ciliary and/or OS structural defects of the mutant photoreceptors. Furthermore, aside from the proposed general protein-transporting machinery, the roles of ciliary membrane components in localizing rhodopsin to the OS remain largely unknown.
The photoreceptor OS is a modified sensory cilium that shares common structural features with those of other ciliated cells (5). At the base, it grows from the basal body (BB) derived from a mother centriole and further extends ciliary axonemes distally, which are microtubule arrays supporting OS membrane discs and providing routes for IFT. The most proximal zone of the axoneme is known as the connecting cilium (CC), structurally equivalent to the ciliary transition zone (TZ) of nonphotoreceptor cells. The CC and surrounding membranous structures function as the ciliary gate/diffusion barrier and play a crucial role in selecting cargo proteins allowed to enter the OSs (5, 12, 13). Genetic studies in Caenorhabditis elegans and vertebrate models have defined two functional groups, the NPHP (nephronophthisis) and MKS modules, in the TZ. The NPHP module includes NPHP-1, NPHP-4, and NPHP-5 (IQCB1) (14, 15), and the MKS module contains MKS-1, MKSR-1 (B9d1), MKSR-2 (B9d2), MKS-2 (TMEM216), MKS-3 (TMEM67/meckelin), MKS-6 (CC2D2A), TMEM231, JBTS-14 (TMEM237), TMEM107, TCTN-1, and AHI1/juberin (15–17). Both modules are assembled on the MKS5 (CEP290)/RPGRIP1L scaffold (17). Interestingly, two additional components, TMEM138 and CDKL-1, which are genetically dependent on MKS5/CEP290 but no other MKS or NPHP proteins, are considered as another potential distinct module (17, 18).
In the mammalian system, much has been learned about the RPGR/Cep290–centered interactome, including those of NPHP and BBS proteins (19–21). However, the MKS TZ module is relatively understudied in higher organisms, especially for a group of TZ-localized putative transmembrane proteins (TMEM17, TMEM67, TMEM107, TMEM138, TMEM216, TMEM218, TMEM231, and TMEM237) (17, 18, 22–26), the localization and ciliary function of which have been scarcely explored in either humans or mice. Thus, whether the data obtained from lower organisms and in vitro–cultured cells apply to mammals has yet to be verified.
In our attempts to investigate the potential roles of planar cell polarity (PCP) signaling in photoreceptor morphogenesis, we identified several genes encoding transmembrane proteins with altered expression in Prickle 1 (a core PCP component)–mutant retinas, including Tmem138 and Tmem216 known to underlie JBTS (27–29). Tmem138 and Tmem216 are arranged in a head-to-tail configuration on human chromosome 11 and mouse chromosome 19, sharing intergenic regulatory elements (27). Tmem138 is localized at the ciliary base and axoneme, while Tmem216 is primarily in the BBs in IMCD3 cells (27). Tmem138 was suggested to coordinate interdependent vesicular transport to the primary cilia in vitro, and its human mutations reduce the cilia length of the patient fibroblasts (27). To understand their functions in photoreceptor OS formation and maintenance, we ablated Tmem138 in mice either in the germline or in fully developed rod photoreceptors and report a complete arrest of OS formation in the mutants. Tmem138 interacts with rhodopsin and two CC-localized proteins, Ahi1 and Tmem231, which may together be a part of the CC membrane protein complex(es) crucial for delivering rhodopsin and/or other OS-bound proteins.
Results
Targeted Inactivation of Tmem138 Leads to Enlarged Brain Ventricles, Azoospermia, and Retinal Degeneration.
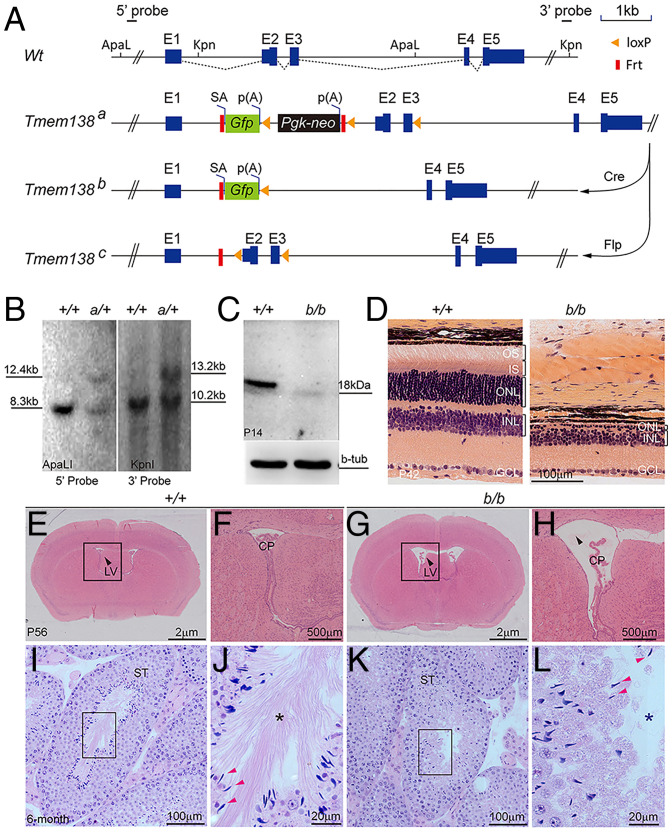
An early investigation on PCP in mouse retina identified several genes encoding transmembrane proteins and regulated by Prickle 1, a core PCP component (SI Appendix, Fig. S1A). One of them was Tmem138, mutations of which cause a form of ciliopathy known as JBTS spectrum disorder, which affects multiple organs, including the brain, testis, kidney, and retina (27, 28). To understand the mechanisms of retinal dystrophy caused by TMEM138 mutations, we genetically inactivated the Tmem138 gene using a knockout-first strategy in mice to create a gene-trap allele (Tmem138a; Fig. 1A and SI Appendix, Supplementary Materials and Methods). The gene-trap allele was confirmed by Southern blotting analysis of mouse genomic DNA (Fig. 1B). A germline Cre (Sox2-Cre; SI Appendix, Supplementary Materials and Methods) was crossed onto Tmem138a to generate a null allele (Tmem138b; SI Appendix, Fig. S1 B and C). Targeted RT-PCR was conducted to probe exonal transcripts from each allele (SI Appendix, Fig. S1 B, D, and E). Notably, the Tmem138a allele appeared to transcribe all exons indicated by the existence of the last exon E5 transcripts, whereas the Tmem138b allele was confirmed to be null, as it only expressed E1 and knock-in Gfp reporter (SI Appendix, Fig. S1 D and E). Western blotting further confirmed the loss of Tmem138 protein in Tmem138b/b mice (Fig. 1C). Tmem138b/b-mutant mice had a normal appearance and similar growth curves as wild-type mice (SI Appendix, Fig. S1F), and numbers of mice born for each genotype from Tmem138b/+ breeding approximately adhered to the Mendelian ratio (SI Appendix, Fig. S1G).
Fig. 1.
Targeted inactivation of Tmem138 leads to enlarged brain ventricles, azoospermia, and retinal degeneration. (A) Tmem138-targeting strategy. The Tmem138 gene has five exons, with protein coding starting at exon 2 (E2). The restriction enzyme sites ApaLI and Kpn1 were used for Southern blotting analysis in B with 5′ and 3′ probes, respectively; SA, splicing acceptor; p(A), polyadenylation signal; Gfp, green fluorescent protein; Pgk-neo, phosphoglycerate kinase promoter–driven neomycin gene; Flp, flippase; Frt, flippase recognition target. (B) Southern analysis of Tmem138 gene-trap allele (Tmem138a, a/+). The expected fragment sizes on Southern blot are 5′ probe (ApaLI), 8.30 kb (Wt: +/+) and 12.41 kb (a/+), and 3′ probe (KpnI), 10.26 kb (+/+) and 13.28 kb (a/+), respectively. (C) Western blotting of P14 retinal extracts using an anti-Tmem138 antibody; b-tub, β-tubulin. (D) The homozygous Tmem138b/b (b/b)-null mutant manifested severe retinal degeneration at P42; GCL, ganglion cell layer. (E and F) A coronal brain section from a wild-type mouse stained with hematoxylin and eosin at P56 (E). The arrow points to the lateral ventricle (LV). The boxed area is magnified in F; CP; choroid plexus. (G and H) Enlarged brain ventricles of the b/b mice (G). The boxed area is magnified in H. (I–L) Testis sections from 6-mo–old wild-type (I) and null mutant (K) mice, respectively. The boxed areas show the lumen of seminiferous tubes (ST) and are magnified in J and L, respectively. Red arrows point to sperm heads; asterisks indicate sperm tails, which are well developed in the +/+ mice but are absent from the b/b mice.
Histological examinations revealed rapid photoreceptor loss in the mutant retina with only one row of nuclei remaining at postnatal day 42 (P42; Fig. 1D). Mutant mice also exhibited hydrocephalus, as evidenced by enlarged brain ventricles (Fig. 1 E–H), and defective spermatozoa (Fig. 1 I–L). The homozygous gene-trap mice (Tmem138a/a) exhibited the same phenotypes as the Tmem138b/b-null mice (SI Appendix, Fig. S1 H and I). Western blotting also verified the absence of Tmem138 protein in the Tmem138a/a mutants (SI Appendix, Fig. S1J), probably due to truncation of Tmem138 mRNA translation from the stop codon in Gfp or neo. The described phenotypes were fully penetrant in the examined animals. Thus, the Tmem138 mutants recapitulated the disease manifestations of JBTS.
Early-Onset Rod Loss in Tmem138 Mutants.
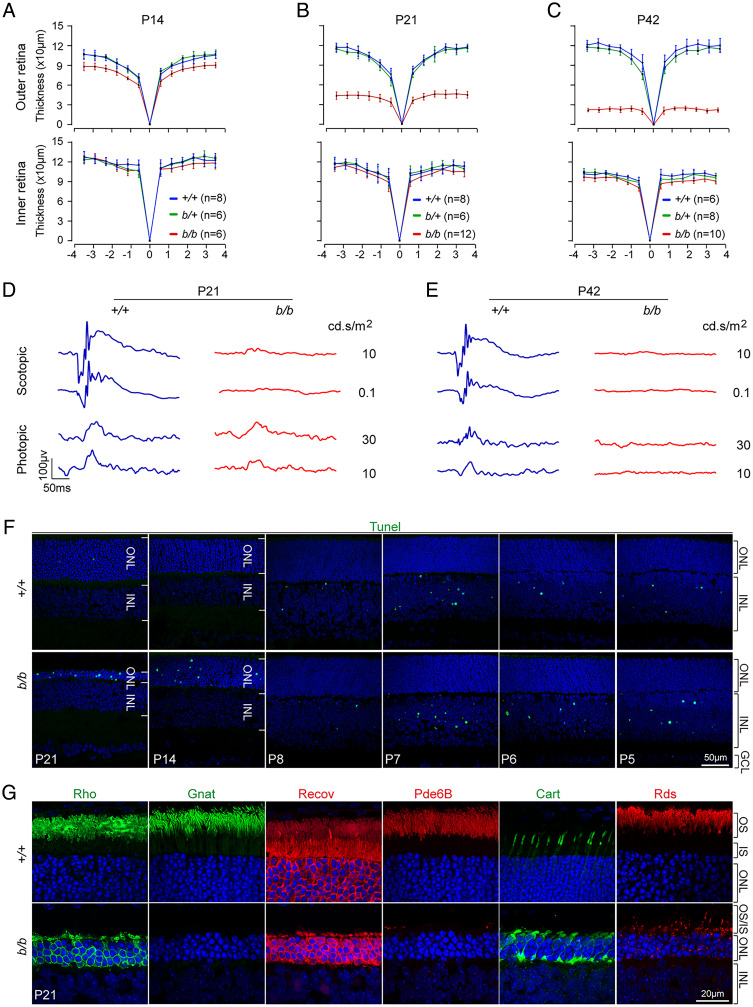
To determine the temporal windows of photoreceptor loss following Tmem138 disruption, we performed optical coherence tomography (OCT) in live mutant and control mice and followed the changing thickness in retinal layers longitudinally. Significant thinning of the mutant outer retina encompassing all segments of the photoreceptors was readily detectable as early as P14 (Fig. 2A and SI Appendix, Fig. S2 A and B). Rapid loss of the outer nuclear layer (ONL) occurred in the ensuing weeks, with the outer retinal thickness reduced to about 50% (∼50 μm) by P21 (Fig. 2B and SI Appendix, Fig. S2 C and D). Continued but slower thinning of the ONL was detected until its complete loss at P42 (Fig. 2C and SI Appendix, Fig. S2 E and F). A reduction of the inner nuclear layer (INL) was also observed at the examined ages but was much less severe than that of the outer retina (Fig. 1D and SI Appendix, Fig. S2 B, D, and F). Consistent with the thinning outer retina, electroretinography (ERG) measuring photoreceptor light responses revealed that the mutant rod response was abolished at P21, whereas the cone response was diminished but still measurable under higher light intensities (Fig. 2D and SI Appendix, Fig. S2G). By P42, both rod and cone responses were undetectable (Fig. 2E and SI Appendix, Fig. S2H).
Fig. 2.
Structural and functional retinal defects of Tmem138b/b mice. (A–C) OCT imaging analysis of retinal thickness at different ages. (D and E) Retinal ERG at P21 (D) and P42 (E). (F) Detection of cell death by TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) labeling at indicated ages. Note apoptosis at early ages from P5 to P8 in the INL is normal during retinal maturation. (G) Immunostaining with antibodies against rhodopsin (Rho), α-transducin (Gnat), recoverin (Recov), Pde6B, Cart, and peripherin/Rds.
To further corroborate the findings of OCT and ERG, we examined cell death at different ages. Extensive cell death was only detected in the mutant ONL after P14 by TUNEL labeling (Fig. 2F and SI Appendix, Fig. S2I), whereas early developmental cell death was comparable in control and the mutant INLs (from P5 to P8; Fig. 2F). A time series of examinations revealed that cell death started between P8 and P14 (Fig. 2F). Additionally, caspase-3, TUNEL, and phospho-histone 2A isoform X (for DNA damage) and LCA3A/B (for autophagy) were all elevated as early as P14 (SI Appendix, Fig. S2J). Muller glia response to retinal injury became apparent by P21, as indicated by up-regulation of glial fibrillary acidic protein (SI Appendix, Fig. S2K).
We next examined key phototransduction proteins by immunostaining in mutant and control retinas at P21 (Fig. 2G). Rhodopsin, α-transducin, recoverin, and phosphodiesterase 6B (Pde6B) were barely detected from the mutant rod OSs, with a considerable fraction of rhodopsin mislocalized in the cell bodies (Fig. 2G). The numbers of cones were comparable between the mutant and control retinas, although mislocalization of cone arrestin (Cart) was seen throughout the mutant cell bodies (Fig. 2G). OS disc protein peripherin 2 (Rds) essentially disappeared from the mutant OS, with punctate staining in the cell bodies (Fig. 2G). These data together demonstrated an early-onset photoreceptor degeneration upon disruption of Tmem138.
Localization of Tmem138 to the Proximal Compartment of the CC.
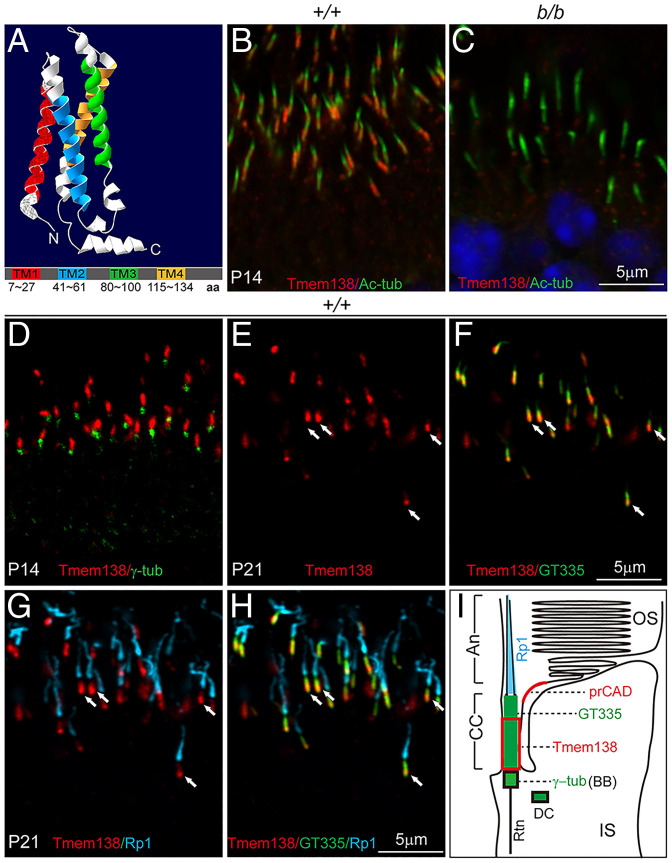
Tmem138 encodes a protein of 162–amino acid residues with an 18.4-kDa molecular weight and four transmembrane domains (Fig. 3A). Previous studies suggest that Tmem138 is a ciliary protein in mouse IMCD3 cells (27) and is localized in the TZ in C. elegans sensory neurons (17), but its localization in photoreceptors was unclear. Using the same antibody validated by Western blotting in Fig. 1C, we performed immunohistochemistry on retinal sections together with antibodies to acetylated α-tubulin (Ac-tub) and γ-tubulin labeling ciliary microtubules and BBs, respectively. Under light fixation (SI Appendix, Supplementary Materials and Methods), Tmem138 was found in the photoreceptor CC (Fig. 3 B and C and SI Appendix, Fig. S3 A–F) above the BB (Fig. 3D). Furthermore, Tmem138 was predominantly localized to the basal CC of the photoreceptors, as indicated by colocalization with the CC marker GT335 and the photoreceptor axonemal marker Rp1 (Fig. 3 E–H). A schematic illustration of Tmem138 localization in the photoreceptor is shown in Fig. 3I.
Fig. 3.
Tmem138 is localized to the CC. (A) Predicted Tmem138 three-dimensional structure and domains; TM, transmembrane; aa, amino acids. (B and C) P14 retinas. The CC staining of Tmem138 (red) (B) was abolished in the b/b mutant (C); Ac-tub, green. (D) Costaining of Tmem138 (red) with γ-tubulin (green) indicating the BB. (E–H) P21 retinas. Triple labeling of Tmem138 (red), GT335 (green), and Rp1 (turquoise). Arrows point to the Tmem138 domain overlapping with GT335 at the base of CC. (E) Tmem138 single channel. (F) Merged image of Tmem138 and GT335 (green) staining. (G) Merged image of Tmem138 and Rp1 (turquoise). (H) Merged image of the three channels of Tmem138, GT335, and Rp1. (I) A schematic drawing of a rod photoreceptor, with the red boxed area indicating the Tmem138 localization compartment relative to that of other ciliary markers; An, Axoneme; Rtn, rootlets. prCAD localization refers to SI Appendix, Fig. S4.
The extensive CC labeling of Tmem138 is consistent with a rod-dominant mouse retina. We then asked whether Tmem138 was also expressed in cones, of which the ERG function was less affected than rods (Fig. 2D). Limited by the lack of cone-specific ciliary markers and the small number of cones in the mouse retina, we performed immunohistochemistry on dissociated retinal cells with a mouse antibody against Ac-tub together with Tmem138 and Cart antibodies, both of which are of rabbit origin, to distinguish CC staining of Tmem138 from that of cone cell bodies stained by Cart. Examination of multiple cones indicated that the CC domain was free of Cart but not of Tmem138 staining (SI Appendix, Fig. S3 G–L). Additional analysis of retinal single-cell profiles from public databases (SI Appendix, Supplementary Materials and Methods) revealed relatively high-level cone expression of Tmem138 (SI Appendix, Fig. S3M). Thus, Tmem138 is localized to the CC of both rods and cones. The partial preservation of cone ERG function at early postnatal ages might reflect slower cone loss than rods, which is widely known among many mouse models of retinal degeneration.
Altered Molecular Domains of CC and Axonemal Compartments of the Mutant Photoreceptors.
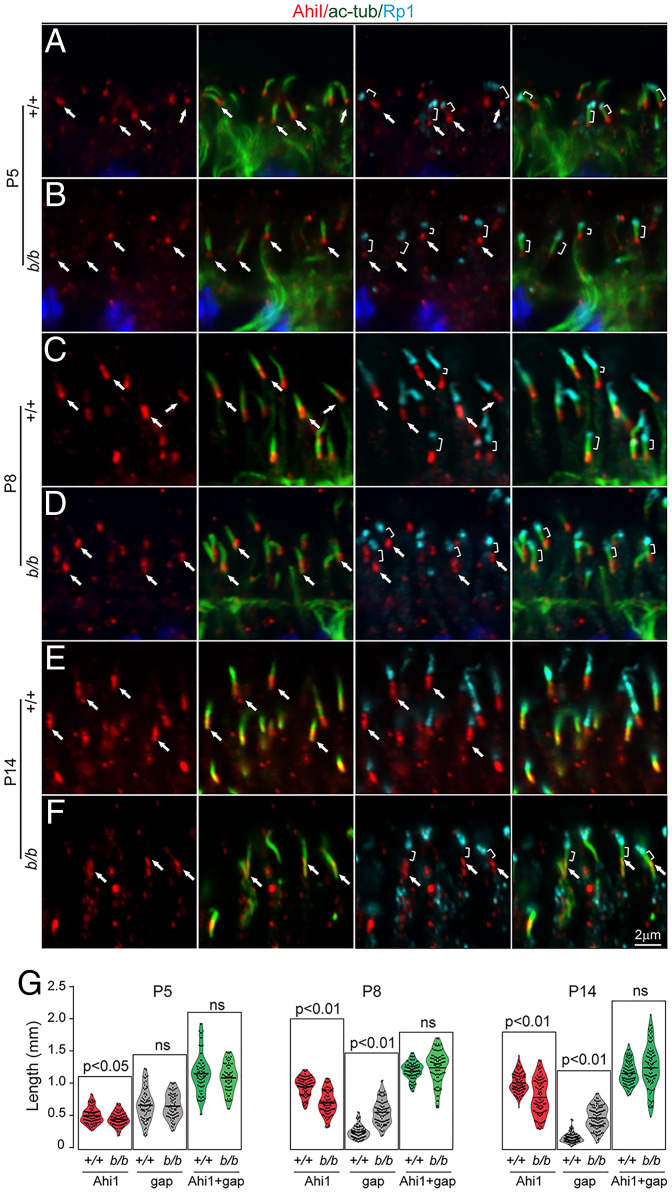
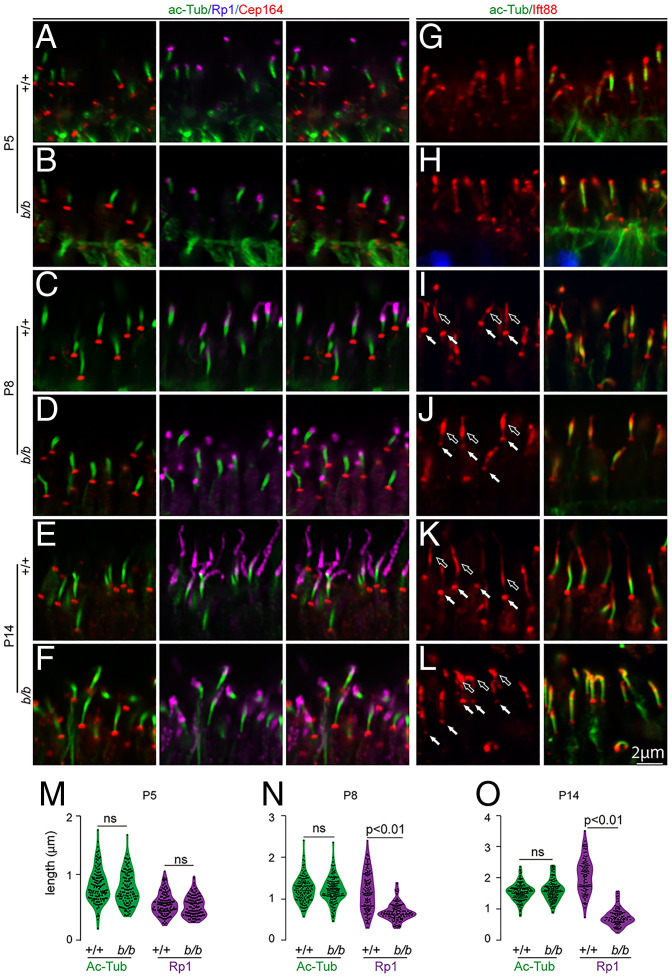
The CC localization of Tmem138 prompted us to investigate whether photoreceptor ciliary compartments are altered upon disruption of Tmem138. Triple labeling with Ahi1, a CC compartmental protein, Ac-tub, and Rp1 was performed on retinal sections of P5, P8, and P14 animals. At P5, basal CC staining of Ahi1 and axonemal staining of Rp1 were similar in wild-type and mutant retinas (Fig. 4 A and B). A gap in staining was present separating Ahi1 and Rp1 compartments at this age (Fig. 4 A and B). The Ahi1 domain extended distally, becoming contiguous with Rp1 in about two-thirds of wild-type cilia at P8 (Fig. 4C), whereas the gap between Ahi1 and Rp1 remained in most of the mutant cilia (Fig. 4D). Axonemal Rp1 extension was halted after P8 (Fig. 4 C–F), with further shortening of the Ahi1 domain in mutant photoreceptors at P14 (Fig. 4 E and F). Quantification of the Ahi1 domain lengths and the staining gaps between Ahi1 and Rp1 demonstrated significant differences between wild-type and mutant photoreceptors at P8 and P14 (P < 0.01), while the average length of the Ahi1 domain plus the gap between Rp1 and Ahi1 was statistically unaltered (Fig. 4G). Because the staining of photoreceptor-specific cadherin (prCAD), a nascent disc protein, was localized adjacent to the base of Rp1 in both the wild-type and the mutants (SI Appendix, Fig. S4), the mutant gap between Ahi1 and Rp1 (Fig. 4D) is likely a subdomain of the CC.
Fig. 4.
Shortened Ahi1 domain in Tmem138b/b photoreceptor CC. Ahi1, Ac-tub, and Rp1 triple labeling for all images. Red, green, and turquoise channels represent Ahi1, Ac-tub, and Rp1, respectively. (A and B) P5 sections. Arrows point to Ahi1 staining, while brackets indicate gaps between Ahi1 and Rp1 staining. (C and D) P8 sections. (E and F) P14 sections. (G) Quantification of Ahi1 domain length, gap length between Ahi1 and Rp1, and Ahi1 domain length plus gap length. The total domain length of “Ahi1 + gap” is roughly equal between wild type and the mutants, suggesting that the gap of the mutant distal CC is a missing part of the Ahi1 domain; ns, no significance.
We next examined several other ciliary proteins, including Cep164, a protein in ciliary appendages (30), and IFT components Ift88 and Kif3a, for anterograde protein transport to the OS. The Cep164 staining appears identical in both the wild-type and mutant photoreceptors through the ages examined (Fig. 5 A–F). While appearing normal at P5 (Fig. 5 G and H), the basal cilia staining of Ift88 was attenuated in P8 and P14 mutant photoreceptors with increased intensity at the distal end (Fig. 5 I–L). Kif3a did not exhibit notable changes in the mutants even at the age of P14 (SI Appendix, Fig. S5 A–F). Quantification of the domain length of Ac-tubulin did not show significant differences in contrast to that of Rp1, which was consistently shortened at P8 and P14 (Figs. 4 and 5 M–O). Thus, the distal CC and axonemal compartments were severely perturbed upon disruption of Tmem138.
Fig. 5.
Altered IFT88 and Rp1 domains in the mutant photoreceptor cilia. (A–F) Triple staining of Ac-tub (spanning CC), Rp1(for axoneme), and Cep164 (for distal appendages). Colored protein names at the top correspond to the colors of the imaging channels. All three proteins showed similar expression and localization in both wild-type and mutant cilia at P5 (A and B). Shortened axonemes of the mutant cilia were observed at P8 and P14 (C–F). (G–L) Double staining of Ac-tub (for CC) and Ift88 (for CC and axonemes). Shortened axonemal Ift88 domain in the mutants was observed at P14 (E and F). (M–O) Quantification of CC (Ac-tub staining) and axonemal (Rp1 staining) lengths. A Student’s t test was performed to detect statistical power; ns, no significance (P > 0.01).
Photoreceptor Ciliogenesis Appears Normal in the Tmem138 Mutants at Ultrastructural Levels.
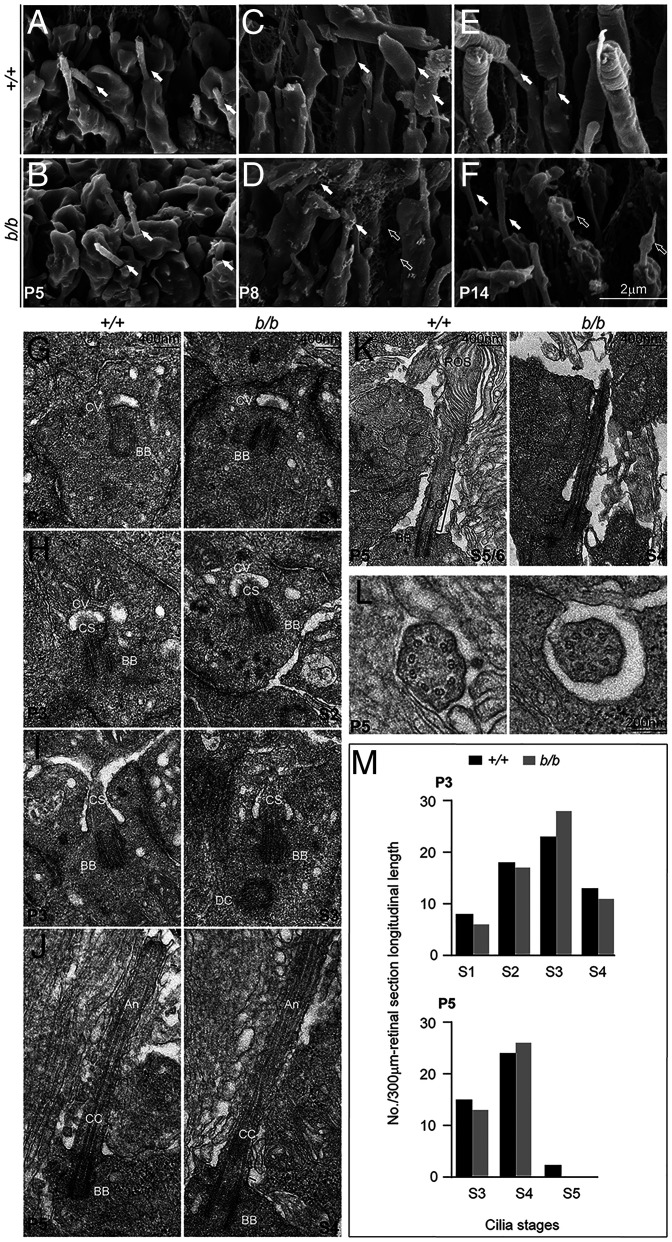
We next investigated whether ciliogenesis was affected by disruption of Tmem138 using scanning electron microscopy (SEM) and transmission electron microscopy (TEM). SEM imaging demonstrated similar ciliary morphology of both wild-type and mutant photoreceptor cells at P5 (Fig. 6 A and B), when photoreceptors were yet to develop OSs. At P8 and P14, wild-type photoreceptors began to grow OSs distally from the CC (Fig. 6 C and E), whereas no OSs were found in the mutant cells (Fig. 6 D and F). To obtain further details of photoreceptor ciliogenesis, we performed TEM to examine the early developmental ages of photoreceptors. Photoreceptor ciliogenesis can be divided into four stages (31): S1, ciliary vesicle docking on the distal end of the mother centriole (Fig. 6G); S2, ciliary shaft elongation within the ciliary vesicle (Fig. 6H); S3, membrane fusion of the ciliary vesicle with the apical surface of IS (Fig. 6I); and S4, elongation of the ciliary shaft to assemble axoneme (Fig. 6J). Multiple TEM sections collected from both wild-type and mutant retinas showed comparable ciliary features at each of the four stages (Fig. 6 G–J). Two additional stages S5 and S6, pertaining specifically to the photoreceptor OS morphogenesis between P5 and P7 (Fig. 6K), were not found in mutant photoreceptors. In contrast, the array of nine microtubule doublets of the mutant CC was similar to that of wild-type photoreceptors (Fig. 6L). Furthermore, the frequency in finding each stage of S1 to S4 ciliogenesis was comparable between wild-type and mutant photoreceptors (Fig. 6M).
Fig. 6.
Photoreceptor ciliogenesis revealed by SEM and TEM. (A and B) SEM images of wild-type (A) and mutant (B) photoreceptor cilia (arrows) at P5. (C and D) P8 photoreceptors with arrows pointing to the OS in C and membrane material in D. Open arrows point to the membrane vesicles embedded in the interphotoreceptor matrices. (E and F) P14 photoreceptors. Arrows point to the photoreceptor CC; open arrows point to the membranous materials on top of the mutant cilia. (G–J) P3 sections. (G) Stage I cilium (S1), ciliary vesicles docked on the distal end of the mother centrioles. (H) S2 cilium, ciliary shafts elongated within the ciliary vesicles. (I) S3 cilium, ciliary vesicle membrane fused with that of the IS at the apical surface. (J) S4 cilium, cilium grew to assemble axoneme. (K and L) P5 retinal sections. (K) S5/S6, rudimental OS emerged above the cilia in wild-type, but not mutant, photoreceptors; rOS, rudimentary OS. (L) Sections across the CC showed nine doublet microtubules in both wild-type and mutant photoreceptor cells. (M) Quantification of cilia numbers at each stage of P3 and P5. Cilia between RPE (retinal pigment epithelium) and the ONL at each stage of photoreceptor ciliogenesis were counted from three retinal sections/retina, each longitudinally spanning over a 100–μm-length in midperipheral retinal areas. The numbers of cilia from three sections (3 × 100 μm = 300 μm total span) for each stage were used for plotting the graphs. One retina for each genotype was counted. Brackets indicate CC; CV, ciliary vesicle; CS, ciliary shaft. Note that the axonemal, CC, and ciliary shaft are empirically defined according to their relative positions.
We further quantified cilia numbers by using Ac–tub-stained flat-mount retinas by fluorescence microscopy (SI Appendix, Fig. S5 G–I). No significant changes in cilia numbers were noted at P8 between the wild-type and mutant retinas (SI Appendix, Fig. S5 J–L and P). Reduction in cilia numbers of the mutants at central and midperiphery retinal areas at P14 (SI Appendix, Fig. S5 M–O and Q) likely reflects the previously observed photoreceptor death. Additionally, a widespread reduction in cilia density at P14 in comparison to that of P8 in both wild-type and mutant photoreceptors was likely a result of retinal growth and resulted expansion (SI Appendix, Fig. S5R). Thus, ciliogenesis at the early stages of the mutant photoreceptors appeared structurally normal, as revealed by SEM, TEM, and light microscopy.
Mislocalization of Rhodopsin Before the Onset of OS Growth.
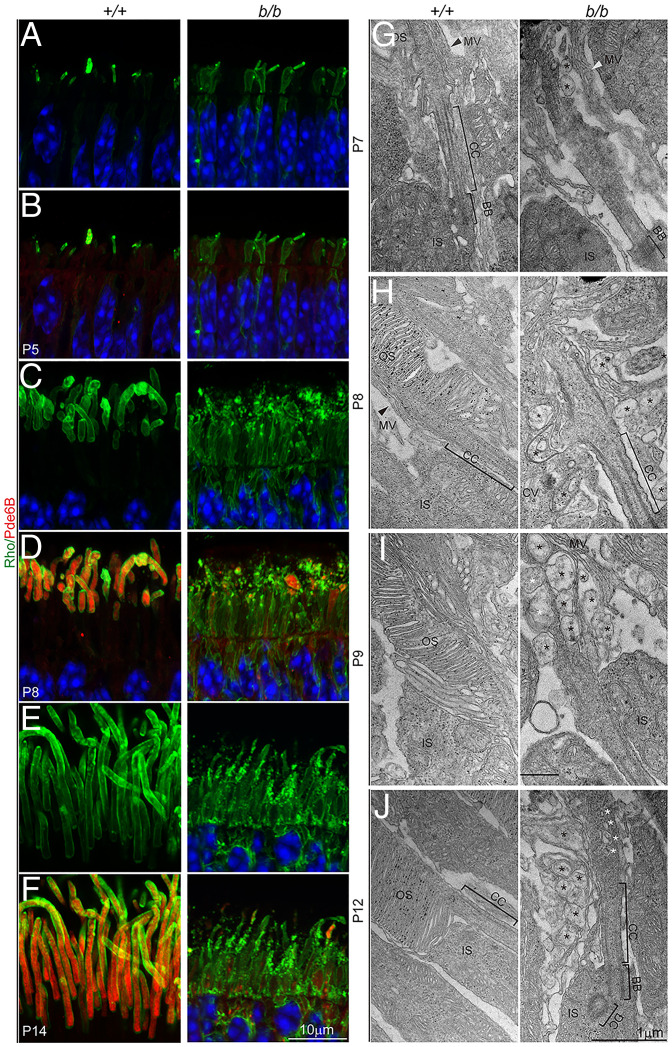
To obtain further insights into Tmem138 function in OS biogenesis, we first examined a series of photoreceptor developmental ages from P3 to P14 by monitoring the expression and localization of rhodopsin using light and electron microscopy. At P3, as the IS started to form, punctate rhodopsin staining appeared at apical domains of both wild-type and mutant photoreceptors (SI Appendix, Fig. S6 A and B). At P4, ciliary protrusions emerged with more intense rhodopsin labeling in the wild type (SI Appendix, Fig. S6 C and D). Starting at P5, rhodopsin localization was polarized to the ciliary protrusions in the wild-type photoreceptors in contrast to the mutant, which remained randomly distributed throughout cell bodies (Fig. 7 A and B). At P8 and P14, the emerging OS with compartmentalized rhodopsin and Pde6B were developed in control photoreceptors, whereas they appeared to be degenerating in the mutants with lower levels of rhodopsin and Pde6B (Fig. 7 C–F).
Fig. 7.
Failure of photoreceptor OS formation. (A–F) Photoreceptor OSs were labeled with antibodies to rhodopsin (Rho) and Pde6B. (A and B) Mislocalization of rhodopsin was detected as early as P5 in the mutant retina. Note that Pde6B was only weakly expressed at P5. (C–F) At P8 and P14, rhodopsin and Pde6B were strongly expressed and transported to the OS in the wild-type photoreceptors but were mislocalized throughout the mutant photoreceptor cell body. Note rhodopsin-positive puncta in the presumptive OS and IS regions and the absence of intact OS in the mutant retina. (G–J) TEM observations of photoreceptor OS development; MV, microvilli (arrowheads). Black asterisks indicate aberrant membrane-bound vacuoles; white asterisks indicate vesicles within the ciliary shaft. The black grains in H to J are from nonspecific staining due to sample processing.
On TEM sections, from P7 to P12, extracellular vesicles of varying sizes accumulated surrounding the mutant photoreceptors with occasional intraciliary vesicles (Fig. 7 G–J). Ciliary components, including daughter centrioles (DCs), BBs, and CC, appeared normal (Fig. 7J). On immunoelectron microscopy (immuno-EM) for rhodopsin, the rudimentary OSs of developing photoreceptors at P5 were densely labeled by immunogold particles (Fig. 8A). In contrast, disorganized membranous structures in place of OSs were observed, with less rhodopsin labeling in the mutant (Fig. 8A). Many immunogold particles were found at the IS plasma membrane (Fig. 8A), indicating ectopic localization of rhodopsin. At P8 and P14, the extracellular vesicle mutant photoreceptors were also heavily labeled with rhodopsin (Fig. 8 B and C). Interestingly, despite perturbations at the mutant CC as early as age P8, the distribution of rhodopsin gold particles along the ciliary membrane did not show significant differences from that of the wild type (SI Appendix, Fig. S6E). CC distribution of peripherin/Rds, another OS protein, was also similar in controls and mutants at these ages (SI Appendix, Fig. S6 F and G). Taken together, aberrant trafficking of rhodopsin and other OS-bound proteins might underlie failed OS morphogenesis of the mutant photoreceptors.
Fig. 8.
Mislocalized rhodopsin revealed by immuno-EM. (A) P5 retina sections. Immunogold-labeled rhodopsin was localized to the CC membrane and the rudimentary OS of the wild-type photoreceptors. It was mislocalized to the IS membrane of the mutant photoreceptors. Some rhodopsin labels were observed near the distal end of the CC, probably associated with the failed OS morphogenesis. Black arrowheads point to rhodopsin particles labeling the CC of the wild-type photoreceptors and the IS membrane of the mutants as well. Open arrows point to the disorganized mutant membrane. (B) P8 retina sections. Disc structures appeared in the wild-type OS packed with rhodopsin. Mutant photoreceptors did not develop OS but with extracellular vesicles, some of which were rhodopsin positive. (C) P14 sections. Wild-type OS was well developed, whereas mutant photoreceptors accumulated numerous rhodopsin-bearing extracellular vesicles.
Disc Proteins Appear on Ciliary Protrusions of Both Wild-Type and Mutant Photoreceptors at the Time of OS Genesis.
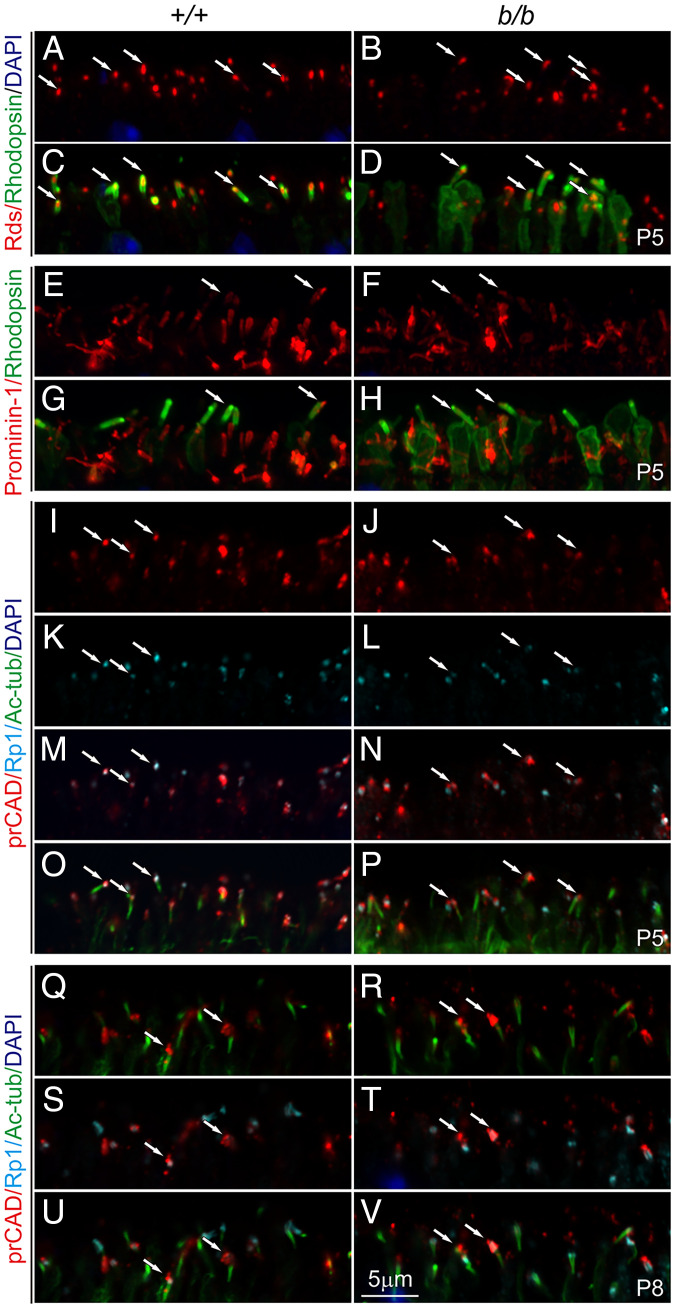
We next examined whether failure of OS formation is due to misrouting of crucial molecules for disc morphogenesis. Peripherin/Rds and prominin-1/CD133 are rim proteins, with the former localized to the photoreceptor disc rims and the latter to the base of OSs in rods during disc assembly (32–34). Ablation of either proteins leads to failure of OS biogenesis (32, 33). PrCAD localized to the nascent disc (35, 36) is also essential for disc integrity. Immunostaining showed that all three proteins were similarly localized to the tip of control and mutant ciliary protrusions at P5 (Fig. 9 A–P). At P8, localization of prCAD in the mutant remained comparable to the control, confined to a domain above the CC overlapping with Rp1 (Fig. 9 Q–V), whereas peripherin/Rds and prominin-1 were extensively mislocalized to the IS (SI Appendix, Fig. S7 A–D). Thus, all three disc proteins were transported to their prospective locations of the mutant photoreceptors at the initiation of OS genesis (P5; the IS mislocalization of peripherin/Rds or prominin-1 at P8 will be discussed later).
Fig. 9.
Localization of disc rim proteins in the early-age photoreceptors. (A–D) Rds (red; A and B) and rhodopsin (green; C and D) staining at P5. (E–H) Prominin-1 (red; E and F) and rhodopsin (green; G and H) staining at P5. (I–P) Triple labeling of prCAD (red), Rp1 (turquoise), and Ac-tub at P5. (Q–V) Triple labeling of prCAD (red), Rp1 (turquoise), and Ac-tub at P8. Arrows at P5 indicate the ciliary tips, whereas at P8, arrows indicate nascent disc positions.
Continued Requirement for Tmem138 in Photoreceptor Homeostasis.
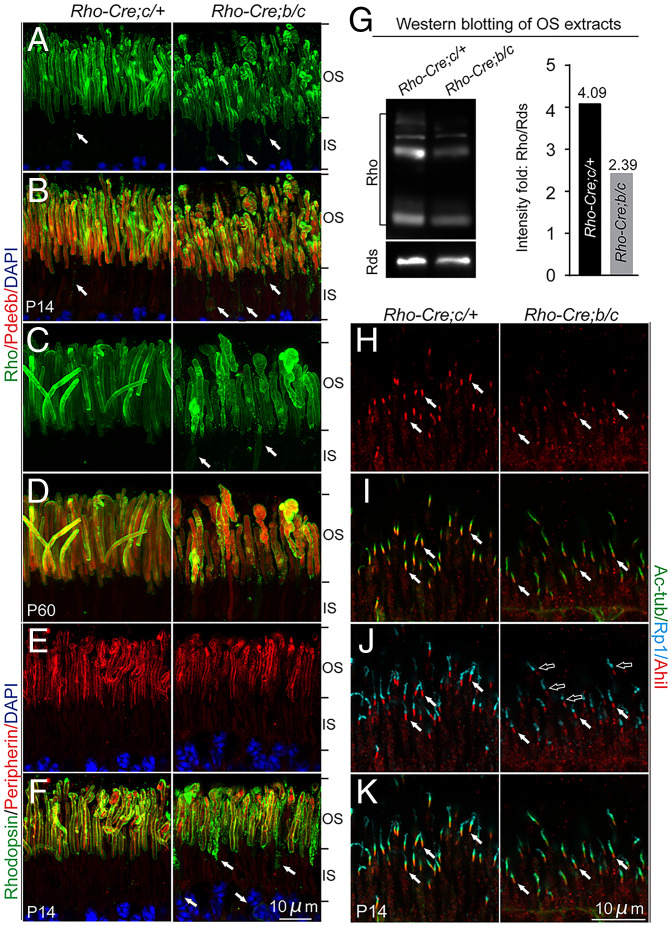
We next investigated whether Tmem138 was also crucial for OS maintenance once it had formed. A Rhodopsin-Cre line was used to delete floxed exons 2 and 3 of the Tmem138 gene (SI Appendix, Fig. S8 A and B). To increase excision efficiency, we produced Rhodopsin-Cre;Tmem138b/c compound mutants with one conditional allele (Tmem138c) on top of the germline null allele (Tmem138b). We first examined the timing of Cre expression and deletion efficacy of Tmem138 by performing immunohistochemistry using antibodies against Cre and Tmem138. Rhodopsin promoter–driven Cre expression was only detectable in a few cells at P8 with more detectable at P10 (SI Appendix, Fig. S8 C and D), and widespread expression was observed between P14 and P21 (SI Appendix, Fig. S8D). Accordingly, Tmem138 protein was nearly completely ablated after P14 (SI Appendix, Fig. S8D). Rhodopsin-stained OSs were disorganized in many mutant photoreceptors at P14, with increased rhodopsin puncta near the base of the OS (Fig. 10 A and B). Disorganized photoreceptors deteriorated further in 2-mo–old mutant retinas (Fig. 10 C and D). Interestingly, peripherin/Rds did not show notable changes in the mutant OS at P14 despite severe mislocalization of rhodopsin (Fig. 10 E and F). Quantification of rhodopsin and peripherin contents of the OS using Western blotting indicated a lower rhodopsin/Rds ratio in the mutants (Fig. 10G).
Fig. 10.
Photoreceptors of Tmem138b/c conditional knockout mice. (A and B) P14 photoreceptors. Disorganized OSs and mislocalization of rhodopsin to the IS were revealed by rhodopsin (green) and Pde6b (red) staining. (C and D) Two–mo-old photoreceptors. The mutant photoreceptors appeared swollen. Arrows in A to C point to mislocalized rhodopsin. (E and F) Colabeling of peripherin/Rds (red) and rhodopsin (green) at P14. (G) Western blotting of rhodopsin and Rds using P21 photoreceptor OS extracts (Materials and Methods). Right, quantification of the rhodopsin/Rds signal intensity fold. (H–K) P14 photoreceptors. Shortened and diminished Ahi1 (red) and Rp1 (turquoise) staining was observed. White arrows point to the Ahi1 staining. Open arrows point to the gap between Ahi1 and Rp1 in the mutants.
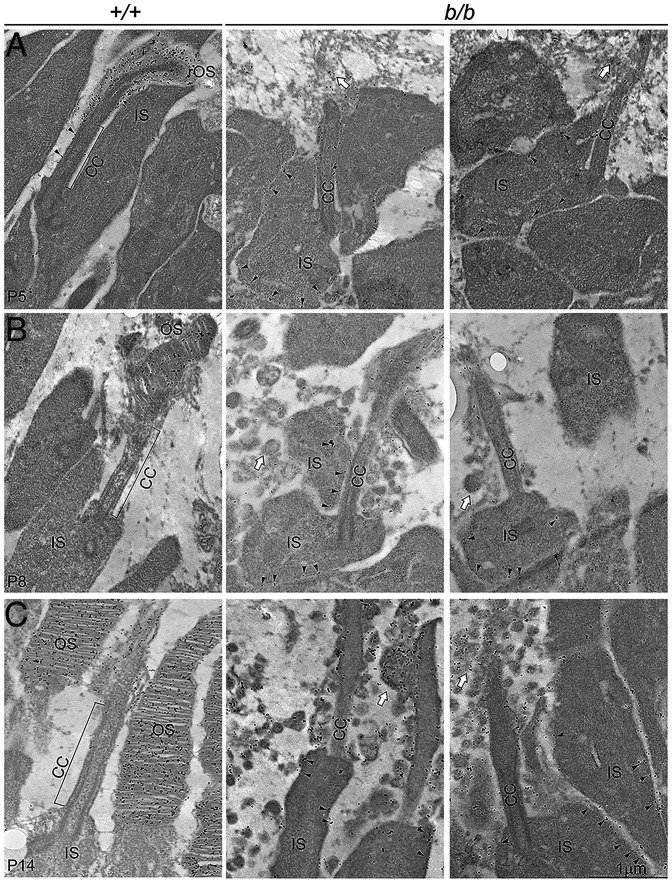
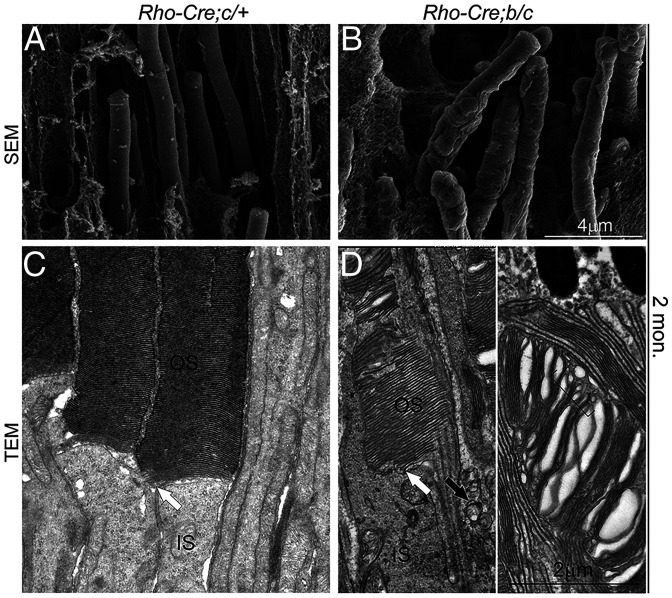
We next investigated whether ciliary compartments and protein-transporting machinery were altered as they were in the germline knockout. Similar to that of the germline knockout, diminished and shortened Ahi1 and Rp1 domains (Fig. 10 H–K) and apical axonemal accumulation of Ift88 (SI Appendix, Fig. S9 A, B, and E–H) were observed with unaltered Kif3a (SI Appendix, Fig. S9 C and D). At ultrastructural levels, 2-mo–old mutant photoreceptors exhibited bumpy outer surfaces in comparison to the smooth and uniform OSs in the wild type on SEM sections (Fig. 11 A and B). On TEM sections, rod OS discs showed variable morphological defects, including splitting and disorientated discs and intra- and extracellular vesicles (Fig. 11 C and D). These results suggest that Tmem138 functions not only in initial OS morphogenesis but also in OS renewal, likely through similar mechanisms as in the early phase of OS development.
Fig. 11.
SEM and TEM of the photoreceptors of Tmem138b/c conditional knockout mice. (A and B) SEM images of wild-type (A) and mutant (B) photoreceptor OSs. (C and D) TEM images of wild-type (C) and mutant (D) photoreceptor OSs. White arrows point to the base (nascent disc) of the OS. The black arrow in D indicates the mutant photoreceptor extracellular vesicles. The bracket indicates variable sizes of intracellular vesicles within the OS. Retinal sections were prepared from 2–mo-old mice.
Tmem138 Interacts with Rhodopsin, Ahi1, and Tmem231.
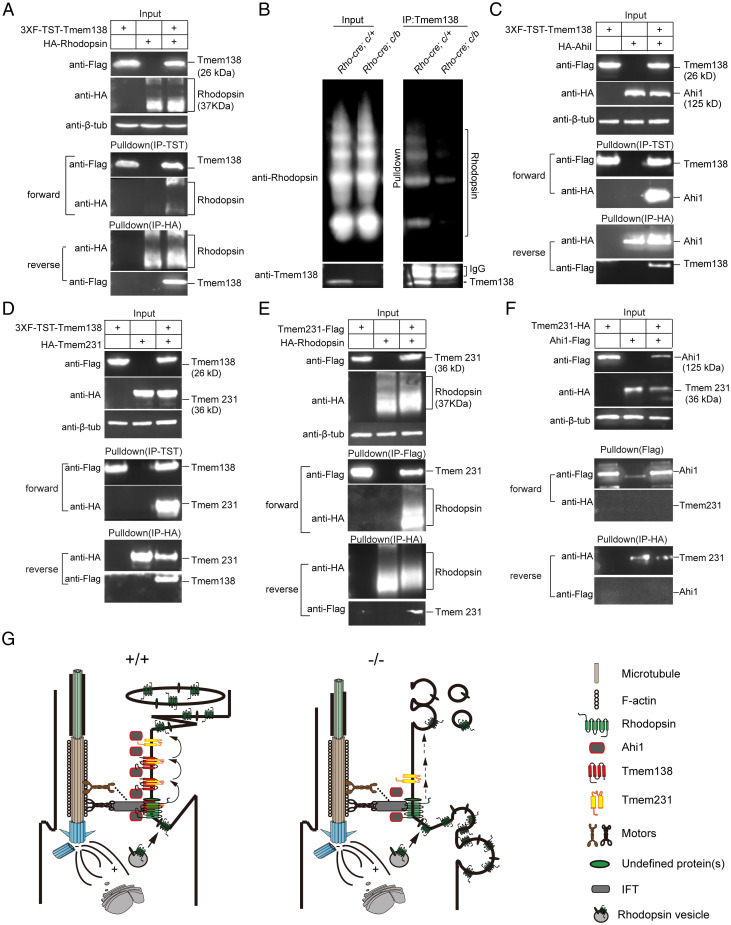
We next explored possible mechanisms by which Tmem138 could be involved in OS biogenesis. In light of rhodopsin mislocalization being the earliest event among other examined disc proteins in both germline and conditional mutants, we first tested whether Tmem138 directly interacts with rhodopsin by a protein pulldown assay. We coexpressed Flag–twin-strep-tag (Flag–TST)-tagged Tmem138 and hemagglutinin (HA)-tagged rhodopsin in HEK 293 cells and purified the protein extracts through columns loaded with strep-tactin resins and HA antibody–cross-linked Sepharose beads (SI Appendix, Supplementary Materials and Methods and Fig. S12 A and B). Tmem138 and rhodopsin reciprocally pulled down each other (Fig. 12A). Furthermore, rhodopsin coimmunoprecipitated with Tmem138 from the wild-type retinal extracts but little from the conditional mutants (Fig. 12B). The weak Tmem138 band (Fig. 12B) from the mutant retinal extracts is probably due to incomplete excision of the Tmem138 allele.
Fig. 12.
Tmem138 interacts with Ahi1, Tmem231, and rhodopsin. A hypothetical model for the CC membrane complex involved in rhodopsin trafficking. (A) Tmem138 interacts with rhodopsin revealed by protein pulldown assay in HEK293 cells in both forward and reverse directions. (B) Tmem138 coimmunoprecipitated with rhodopsin from P21 retinal extracts. The retinal extracts were prepared from rhodopsin–Cre-driven Tmem138b/c (conditional knockout) and Tmem138c/+ (control) retinas. (C) Tmem138 interacts with Ahi1. (D) Tmem138 interacts with Tmem231. (E) Tmem231 interacts with rhodopsin. (F) Tmem231 does not interact with Ahi1. (G) A hypothetical model. Tmem138 localized at the base of the CC membrane is crucial for molecular organization of the CC by formation of the Tmem138/Tmem231/Ahi1 complex. This complex transiently interacts with rhodopsin to facilitate IFT powered by different types of motors along actin and/or microtubule tracks. Disruption of Tmem138 results in a defective CC complex (mislocalization of Ahi1 and Tmem231), impeding rhodopsin (possibly some other OS proteins as well) entering and/or traveling across the CC. Eventually, rhodopsin is mislocalized, and the photoreceptors degenerate, releasing extracellular vesicles.
A literature search found two JBTS genes, Ahi1 and Arl13b, that, when mutated in mice, also lead to failed OS growth (3, 37, 38). Another TZ-localized JBTS/MKS protein Tmem231 has not been studied in photoreceptors, but mutations in mice caused microphthalmia during embryogenesis (22, 39). Using similar affinity approaches described above, we found that Ahi1 and Tmem231, but not Arl13b, interacted with Tmem138 (Fig. 12 C and D and SI Appendix, Fig. S10B). Moreover, Tmem231, but not Ahi1, interacted with rhodopsin (Fig. 12E and SI Appendix, Fig. S10A), and the two did not interact with each other (Fig. 12F). Furthermore, neither Tmem67/meckelin, which when mutated in mice blocked OS biogenesis (24), nor Rab8a, a GTPase important for rhodopsin transport from the Golgi to the base of the CC, interacted with Tmem138 (SI Appendix, Fig. S10 C and D).
Because Tmem231 interacted with both Tmem138 and rhodopsin, we asked whether its localization was also changed in the mutant photoreceptors. Tmem231 appeared to be localized to a broad area of photoreceptor cilia, including the ciliary base, CC, and proximal axoneme (SI Appendix, Fig. S11 A, B, E, F, I, J, M, N, Q, and R). Its staining was slightly weaker in the mutant CC than in the wild-type photoreceptors at the examined ages (SI Appendix, Fig. S11 B, D, F, H, N, and P). Together, these results suggest that Tmem138 may recruit a subset of JBTS proteins to regulate rhodopsin CC localization and probably trafficking across the cilium as well.
Discussion
Significant progress has been made during the past decades in understanding photoreceptor OS biogenesis and related protein transport. A great deal has been learned about these processes from studying rhodopsin trafficking and mistrafficking using various genetically engineered animal models (8). While frameworks emerge on the endomembrane rhodopsin trafficking within the IS (5, 7), the OS disc assembly and morphogenesis (40), and the segregation of plasma from disc membrane domain within the OS (8, 41), how rhodopsin travels through the photoreceptor CC remains largely unsolved. To obtain further mechanistic insights into this process and related disease mechanisms, we conducted the current study focusing on Tmem138, a putative transmembrane protein implicated in JBTS (27–29) with associated retinal dystrophy. Inactivation of Tmem138 in mice led to ciliopathy-like phenotypes, consistent with previous reports of Tmem138 as a ciliary protein in worm and cultured mammalian cells and the related ciliary defects in zebrafish Tmem138 morphants (17, 18, 27). Among a subset of JBTS-related defects of the Tmem138-mutant mice, the complete failure of photoreceptors to develop OS and, consequently, early-onset retinal degeneration is prominent.
Failure of OS biogenesis could result from disruption of ciliogenesis, protein trafficking, or OS disc assembly. To explore through which process Tmem138 may contribute to OS morphogenesis, we first examined its protein localization in photoreceptors. Although Tmem138 has been reported to localize at the base and axoneme of the cilia in cultured mouse IMCD3 cells (27) and in the TZ (equivalent to the photoreceptor CC) of C. elegans sensory neurons (17, 18), its localization in photoreceptors had not been examined previously. We show that Tmem138 is localized to the proximal CC of the photoreceptor. Accordingly, an altered ciliary compartment was observed upon disruption of Tmem138. However, disruption of Tmem138 does not appear to affect photoreceptor ciliogenesis, since the structural features of ciliogenesis (31), the number of cilia, and the length of the CC all appear normal before P8. Furthermore, substantial changes were not found in localization of Ift88 or Kif3a, two crucial components of the anterograde transporting machinery of the photoreceptors required for CC and axoneme formation (42). The actin-based ciliary transport system is proposed for rhodopsin trafficking across the cilium in several studies (43, 44), and actin assembly is also crucial for initiation of nascent discs, disc expansion, and OS membrane compartmentalization (41, 44–46). However, photoreceptors of Tmem138 mutants do not resemble those with a compromised actin system in general (45), and the CC accumulation of rhodopsin in Myosin VIIa mutants (43) is not observed in Tmem138 mutants before photoreceptor degeneration. Thus, ciliogenesis and protein-transporting machinery may not be the primary targets of Tmem138.
OS disc morphogenesis requires a set of membrane proteins to assemble rims and central discs (8, 40). Several known crucial proteins for disc assembly include Rds, prominin-1, and prCAD (32–34). Disruption of these genes shows a range of OS defects (47); in particular, deletion of Rds abolishes OS genesis, leading to release of photoreceptor extracellular vesicles (33), which is also seen in theTmem138 mutants. All three proteins are normally placed to the nascent disc domain at P5, and the delivery of nascent disc protein prCAD is largely normal even at P8. Additionally, rhodopsin, prominin-1, and Rds have different localization compartments in the IS (Fig. 9 and SI Appendix, Fig. S7), implying their different secretory pathways, consistent with previous studies on Rds and rhodopsin trafficking (48, 49). The observed mislocalization of peripherin and prominin-1 in P14 mutant photoreceptors might be secondary to the failed OS disc formation or due to mistrafficking caused by disruption of Tmem138 at late ages. Nonetheless, their mislocalization may further exacerbate extracellular vesicle formation and photoreceptor degeneration.
The earliest and consistently observed molecular event in the Tmem138 mutants is the severe mislocalization of rhodopsin, which coincides with the appearance of ciliary protrusions and OS outgrowth thereafter. Rhodopsin-null mutants do not elaborate rod OSs (50, 51), similar to Tmem138. Transgenic mice overexpressing rhodopsin wild-type and mutated alleles exhibit varied degrees of OS degeneration depending on expression dosage, with some forming extracellular vesicles (2, 4). The partial overlap in phenotypes between rhodopsin and Tmem138 mutants suggests, on one hand, a regulatory role of Tmem138 in rhodopsin localization or trafficking, and, on the other hand, that Tmem138 may also regulate delivery of other OS proteins responsible for its distinct phenotypes. The regulation of rhodopsin localization by Tmem138 is further supported by its direct interaction with rhodopsin using protein pulldown assays in cultured cell and retinal extracts.
A literature search for other ciliary proteins that might interact with Tmem138 based on phenotypical similarities of genetic mutants gave rise to a list of candidates: Arl13b, Ahi1/juberin, Tmem231, and Tmem67/meckelin. Arl13b and Ahi1 are JBTS-associated genes, mutants of which do not form OSs, like Tmem138 (3, 37, 38). Tmem231 genetically interacts with multiple MKS proteins (22) and is associated with OFD6 (Orofaciodigital syndrome type 6) syndrome, and so does Tmem138 (17). Tmem231-mutant mice also develop microphthalmia during early embryogenesis (22, 52), while Tmem67/Meckelin mutants have severe photoreceptor degeneration and lack OSs (24). Among these candidates, Ahi1 and Tmem231 were identified by in vitro protein pulldown assay to interact with Tmem138. Tmem231 is found localized to the ciliary base, CC, and proximal region of the photoreceptor axoneme. Its CC localization is also altered in Tmem138 mutants, like Ahi1. Furthermore, Tmem138, Tmem231, and rhodopsin interact reciprocally. Altogether, these data suggest that Tmem138 is crucial for the molecular organization of a complex in the CC that interacts with intraciliary and ciliary membrane proteins, which might regulate rhodopsin delivery and OS biogenesis.
Based on our results and current knowledge about OS biogenesis and rhodopsin trafficking (8), we propose a model highlighting rhodopsin carried by IFT trafficking across the CC, coordinated by transient interaction with the CC membrane complex–Tmem138/Tmem231/Ahi1 (Fig. 12G). Disruption of this CC complex leads to rhodopsin IS mislocalization and defective OS membranes, releasing extracellular vesicles. This model is drawn compatible with the prevailing “evagination model” for OS biogenesis (53), in which rhodopsin moves within the CC membrane, while it does not refute the “endosome” model (54–57), in which intraciliary traffic of rhodopsin vesicles is suggested. In the latter case, rhodopsin might still interact with the CC membrane complex to facilitate IFT.
An unresolved issue of this model is how rhodopsin interacts with the CC membrane complex(es) (or some other OS proteins) while moving across the CC. One scenario could be that rhodopsin powered by IFT motors moves stepwise through the CC membrane. The distance for each moving step matches that of the spatial arrangement of CC membrane complex(es) (e.g., Tmem138/Tmem231/Ahi1), which serve as “guideposts” and “monkey bars” for rhodopsin movement. Transient interaction of rhodopsin with these monkey bars would assist its vectorial delivery. Interestingly, by superresolution microscopy, a recent study showed a periodic arrangement of ciliary membrane complex(es) of the MKS module containing Tmem107, another JBTS and TZ protein (25). There are many other important questions to be addressed in our model, including defining rhodopsin-interacting domains with the CC membrane proteins and crystal structures of the rhodopsin–CC membrane complex(es) eventually. Further studies are needed to test and modify this model in the future and to address the mechanisms of the Tmem138–CC membrane complex in OS protein transport.
Materials and Methods
Details on animal husbandry, generation of Tmem138 mutant alleles, genotyping and detection of Tmem138 allelic transcripts and proteins, retinal cell dissociation, histology, immunohistochemistry, TUNEL labeling, performing OCT and ERG, TEM, SEM, immuno-EM, detection of protein interaction, quantification and statistics, prediction of Tmem138 tertiary structure, analysis of single-cell RNA-sequencing data, constructs, and antibodies are all provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Tiansen Li from the National Eye Institute for providing valuable suggestions in the preparation of the manuscript. We thank Tiansen Li and Rong Ju for critical reading of the manuscript and helpful comments. This work was supported by grants from the National Natural Science Foundation of China (NSFC; 31571077; Beijing, China), the Guangzhou City Sciences and Technologies Innovation Project (201707020009; Guangzhou, Guangdong Province, China), “100 People Plan” from Sun Yat-sen University (8300-18821104; Guangzhou, Guangdong Province, China), the State Key Laboratory of Ophthalmology at Zhongshan Ophthalmic Center (303060202400339; Guangzhou, Guangdong Province, China) to C.L., the State Key Laboratory of Ophthalmology at Zhongshan Ophthalmic Center (303060202400373; Guangzhou, Guangdong Province, China) to Y.W. and C.L., the National NSFC (21976216, Beijing, China) to Y.W., and the National NSFC (32000553, Beijing, China) and China Postdoctoral Science Foundation (2020M683033) to D.G.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. C.-H.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109934119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Grimm C., et al. , Protection of Rpe65-deficient mice identifies rhodopsin as a mediator of light-induced retinal degeneration. Nat. Genet. 25, 63–66 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Sung C. H., Makino C., Baylor D., Nathans J., A rhodopsin gene mutation responsible for autosomal dominant retinitis pigmentosa results in a protein that is defective in localization to the photoreceptor outer segment. J. Neurosci. 14, 5818–5833 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louie C. M., et al. , AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat. Genet. 42, 175–180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li T., Snyder W. K., Olsson J. E., Dryja T. P., Transgenic mice carrying the dominant rhodopsin mutation P347S: Evidence for defective vectorial transport of rhodopsin to the outer segments. Proc. Natl. Acad. Sci. U.S.A. 93, 14176–14181 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J., Deretic D., Molecular complexes that direct rhodopsin transport to primary cilia. Prog. Retin. Eye Res. 38, 1–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keady B. T., Le Y. Z., Pazour G. J., IFT20 is required for opsin trafficking and photoreceptor outer segment development. Mol. Biol. Cell 22, 921–930 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearring J. N., Salinas R. Y., Baker S. A., Arshavsky V. Y., Protein sorting, targeting and trafficking in photoreceptor cells. Prog. Retin. Eye Res. 36, 24–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemet I., Ropelewski P., Imanishi Y., Rhodopsin trafficking and mistrafficking: Signals, molecular components, and mechanisms. Prog. Mol. Biol. Transl. Sci. 132, 39–71 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Trivedi D., Colin E., Louie C. M., Williams D. S., Live-cell imaging evidence for the ciliary transport of rod photoreceptor opsin by heterotrimeric kinesin-2. J. Neurosci. 32, 10587–10593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marszalek J. R., et al. , Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell 102, 175–187 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Pazour G. J., et al. , The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 157, 103–113 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leroux M. R., Taking vesicular transport to the cilium. Cell 129, 1041–1043 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Nachury M. V., Seeley E. S., Jin H., Trafficking to the ciliary membrane: How to get across the periciliary diffusion barrier? Annu. Rev. Cell Dev. Biol. 26, 59–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkelbauer M. E., Schafer J. C., Haycraft C. J., Swoboda P., Yoder B. K., The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J. Cell Sci. 118, 5575–5587 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Barker A. R., Renzaglia K. S., Fry K., Dawe H. R., Bioinformatic analysis of ciliary transition zone proteins reveals insights into the evolution of ciliopathy networks. BMC Genomics 15, 531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams C. L., et al. , MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192, 1023–1041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C., et al. , MKS5 and CEP290 dependent assembly pathway of the ciliary transition zone. PLoS Biol. 14, e1002416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen V. L., et al. , Formation of the transition zone by Mks5/Rpgrip1L establishes a ciliary zone of exclusion (CIZE) that compartmentalises ciliary signalling proteins and controls PIP2 ciliary abundance. EMBO J. 34, 2537–2556 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rachel R. A., Li T., Swaroop A., Photoreceptor sensory cilia and ciliopathies: Focus on CEP290, RPGR and their interacting proteins. Cilia 1, 22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baehr W., et al. , Insights into photoreceptor ciliogenesis revealed by animal models. Prog. Retin. Eye Res. 71, 26–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rachel R. A., et al. , CEP290 alleles in mice disrupt tissue-specific cilia biogenesis and recapitulate features of syndromic ciliopathies. Hum. Mol. Genet. 24, 3775–3791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberson E. C., et al. , TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J. Cell Biol. 209, 129–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L., et al. , TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am. J. Hum. Genet. 89, 713–730 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collin G. B., et al. , Meckelin is necessary for photoreceptor intraciliary transport and outer segment morphogenesis. Invest. Ophthalmol. Vis. Sci. 53, 967–974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambacher N. J., et al. , TMEM107 recruits ciliopathy proteins to subdomains of the ciliary transition zone and causes Joubert syndrome. Nat. Cell Biol. 18, 122–131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Gonzalo F. R., et al. , A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43, 776–784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J. H., et al. , Evolutionarily assembled cis-regulatory module at a human ciliopathy locus. Science 335, 966–969 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki T., et al. , Molecular genetic analysis of 30 families with Joubert syndrome. Clin. Genet. 90, 526–535 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Tuz K., et al. , The Joubert syndrome-associated missense mutation (V443D) in the Abelson-helper integration site 1 (AHI1) protein alters its localization and protein-protein interactions. J. Biol. Chem. 288, 13676–13694 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graser S., et al. , Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179, 321–330 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sedmak T., Wolfrum U., Intraflagellar transport proteins in ciliogenesis of photoreceptor cells. Biol. Cell 103, 449–466 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Zacchigna S., et al. , Loss of the cholesterol-binding protein prominin-1/CD133 causes disk dysmorphogenesis and photoreceptor degeneration. J. Neurosci. 29, 2297–2308 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salinas R. Y., et al. , Photoreceptor discs form through peripherin-dependent suppression of ciliary ectosome release. J. Cell Biol. 216, 1489–1499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Z., Anderson D. W., Papermaster D. S., Prominin-1 localizes to the open rims of outer segment lamellae in Xenopus laevis rod and cone photoreceptors. Invest. Ophthalmol. Vis. Sci. 53, 361–373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rattner A., et al. , A photoreceptor-specific cadherin is essential for the structural integrity of the outer segment and for photoreceptor survival. Neuron 32, 775–786 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Burgoyne T., et al. , Rod disc renewal occurs by evagination of the ciliary plasma membrane that makes cadherin-based contacts with the inner segment. Proc. Natl. Acad. Sci. U.S.A. 112, 15922–15927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dilan T. L., et al. , ARL13B, a Joubert syndrome-associated protein, is critical for retinogenesis and elaboration of mouse photoreceptor outer segments. J. Neurosci. 39, 1347–1364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanke-Gogokhia C., et al. , The guanine nucleotide exchange factor Arf-like protein 13b is essential for assembly of the mouse photoreceptor transition zone and outer segment. J. Biol. Chem. 292, 21442–21456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srour M., et al. ; FORGE Canada Consortium, Mutations in TMEM231 cause Joubert syndrome in French Canadians. J. Med. Genet. 49, 636–641 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Goldberg A. F., Moritz O. L., Williams D. S., Molecular basis for photoreceptor outer segment architecture. Prog. Retin. Eye Res. 55, 52–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemet I., Tian G., Imanishi Y., Submembrane assembly and renewal of rod photoreceptor cGMP-gated channel: Insight into the actin-dependent process of outer segment morphogenesis. J. Neurosci. 34, 8164–8174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang L., et al. , Heterotrimeric kinesin-2 (KIF3) mediates transition zone and axoneme formation of mouse photoreceptors. J. Biol. Chem. 290, 12765–12778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X., Udovichenko I. P., Brown S. D., Steel K. P., Williams D. S., Myosin VIIa participates in opsin transport through the photoreceptor cilium. J. Neurosci. 19, 6267–6274 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfrum U., Schmitt A., Rhodopsin transport in the membrane of the connecting cilium of mammalian photoreceptor cells. Cell Motil. Cytoskeleton 46, 95–107 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Corral-Serrano J. C., et al. , PCARE and WASF3 regulate ciliary F-actin assembly that is required for the initiation of photoreceptor outer segment disk formation. Proc. Natl. Acad. Sci. U.S.A. 117, 9922–9931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arikawa K., Williams D. S., Organization of actin filaments and immunocolocalization of alpha-actinin in the connecting cilium of rat photoreceptors. J. Comp. Neurol. 288, 640–646 (1989). [DOI] [PubMed] [Google Scholar]
- 47.Chakraborty D., Conley S. M., Al-Ubaidi M. R., Naash M. I., Initiation of rod outer segment disc formation requires RDS. PLoS One 9, e98939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian G., et al. , An unconventional secretory pathway mediates the cilia targeting of peripherin/rds. J. Neurosci. 34, 992–1006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fariss R. N., Molday R. S., Fisher S. K., Matsumoto B., Evidence from normal and degenerating photoreceptors that two outer segment integral membrane proteins have separate transport pathways. J. Comp. Neurol. 387, 148–156 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Lem J., et al. , Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc. Natl. Acad. Sci. U.S.A. 96, 736–741 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humphries M. M., et al. , Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat. Genet. 15, 216–219 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Chih B., et al. , A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 14, 61–72 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Steinberg R. H., Fisher S. K., Anderson D. H., Disc morphogenesis in vertebrate photoreceptors. J. Comp. Neurol. 190, 501–508 (1980). [DOI] [PubMed] [Google Scholar]
- 54.Chuang J. Z., Hsu Y. C., Sung C. H., Ultrastructural visualization of trans-ciliary rhodopsin cargoes in mammalian rods. Cilia 4, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chuang J. Z., Zhao Y., Sung C. H., SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell 130, 535–547 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carter-Dawson L. D., LaVail M. M., Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J. Comp. Neurol. 188, 263–272 (1979). [DOI] [PubMed] [Google Scholar]
- 57.Miyaguchi K., Hashimoto P. H., Evidence for the transport of opsin in the connecting cilium and basal rod outer segment in rat retina: Rapid-freeze, deep-etch and horseradish peroxidase labelling studies. J. Neurocytol. 21, 449–457 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.