Abstract
Abrupt aggregation of misfolded proteins is a hallmark of a large number of severe pathologies, including diabetes types 1 and 2, Alzheimer, and Parkinson diseases. A growing body of evidence suggests that lipids can uniquely change rates of amyloid-associated proteins as well as modify the structure of formed oligomers and fibrils. In this study, we utilize atomic force microscopy infrared (AFM-IR) spectroscopy, also known as nano-IR spectroscopy, to examine the structure of individual insulin oligomers, protofilaments, and fibrils grown in the presence of phospholipids. Our findings show that AFM-IR spectra of insulin oligomers have strong signals of C–H and PO2− vibrations, which points on the presence of lipids in the oligomer structure. Furthermore, substantial shifts in lipid vibrations in AFM-IR spectra of the oligomers relative to the corresponding bands of pure lipids have been observed. This points on strong interactions between a lipid and a protein that are developed at the stage of the oligomer formation.
Graphical Abstract
A growing body of evidence suggests that more than 20 different peptides and proteins can aggregate forming β-sheet-rich oligomers and fibrils.1–3 Some of these protein aggregates are a hallmark of Alzheimer, Huntington, and many other amyloid diseases. In vitro experiments shed light on the kinetic and thermodynamic nature of protein aggregation.4,5 It has been found that protein aggregation has a lag phase that is followed by a rapid growth of fibrils. These findings suggest that a critical amount of oligomers, which are formed at the lag-phase, is required to catalyze their growth.4,6–11 Utilization of cryo-electron microscopy and solid-state nuclear magnetic resonance (ss-NMR) allowed to resolve the secondary structure of amyloid fibrils.12–14 It has been found that amyloid fibrils possessed a very stable cross-β-sheet secondary structure that had the lowest thermodynamic minimum from all possible protein secondary structures. Thus, minimization of the free energy is considered to be the driving force of protein aggregation.
Microscopic analysis of Lewy bodies localized in a midbrain of patients diagnosed with Parkinson disease (PD) revealed presence of fragments of lipid-rich membranes, organelles, and vesicles.15–17 This microscopic evidence suggests that aggregation of α-synuclein (α-Syn), a protein that is directly linked to PD, occurs in the presence of lipids. This hypothesis is supported by experimental results reported by Galvagnion and co-workers. It has been found that lipids can alter the rate of α-Syn aggregation. The acceleration or deceleration effect is determined by the chemical structure of the lipid and lipid-to-protein ratio.15–17
Our own experimental findings show that lipids also uniquely alter the secondary structure of α-Syn oligomers.5 This important information was revealed by using atomic force microscopy infrared (AFM-IR) spectroscopy.18–22 In AFM-IR, a metalized scanning probe can be positioned at individual oligomers that are illuminated by pulsed tunable IR light.23–25 IR radiation induces expansions in protein aggregates that are recorded by the scanning probe.26,27 If the frequency of the laser is tuned to the resonance frequency of the scanning probe, the induced resonance effect allows for reaching the single-monolayer and even single-molecule sensitivity.28,29 This high sensitivity and nanometer spatial resolution made AFM-IR highly attractive for structural analysis of amyloid fibrils,18,20,21,30–32 plant epicuticular waxes,33,34 polymers,35 malaria blood cells,36 meteorites,37 bacteria,38–40 liposomes,41 and polycrystalline perovskite films.42 Using AFM-IR, our group found that α-Syn yields structurally different oligomers if aggregates in the presence of dimyristoylphosphatidylcholine (PC) and dimyristoylphosphatidylserine (PS).5 Furthermore, the structure of these oligomers is drastically different from α-Syn aggregates grown in the lipid-free environment. The question to ask is: do lipids alter the structure of α-Syn, or is this a general phenomenon that can be applied to a large group of amyloid-associated proteins? Also, one may wonder: are lipids present only in amyloid oligomers, or are they also present in fibrils?
To answer these questions, we aggregated insulin in the presence of PC and cardiolipin (CL), a lipid that is uniquely present in mitochondria. Our findings show that insulin oligomers grown in the presence of both PC and CL possess lipid molecules in their structure. This finding suggests that lipids template insulin aggregation. Our results also show that insulin filaments and fibrils grown in the presence of PC and CL possess very little of any lipids. We have also questioned whether insulin has any presences in lipid binding if more than one lipid is available. To answer this question, we aggregated insulin in an equimolar mixture of PC and CL. Our findings show that AFM-IR spectra collected from the corresponding insulin oligomers exhibit PC vibrations with no defined CL bands.
These findings are significant because insulin aggregation is linked to injection amyloidosis.43,44 This severe pathology is caused by high local concentration of insulin in the skin dermis and dermal fat. The increase in the insulin concentration is caused by constant hormone injections, which are required upon diabetes type 1. The presence of insulin at high concentrations catalyzes its aggregation into oligomers and fibrils that, in turn, can trigger the aggregation of other proteins present in cell media which may result in systemic amyloidosis.45 Our results show that insulin can interact with lipids present in the dermal fat making lipid–protein oligomers that are structurally different from oligomers formed by insulin itself. Furthermore, such oligomers may or may not yield fibrils. Finally, insulin fibrils grown in the presence of lipids and fibrils grown in the lipid-free environment will exert different cell toxicities.
Microscopic examination of insulin aggregation in the equimolar concentration of large unilamellar vesicles (LUVs) of PC (Ins:PC) revealed the presence of small oligomers that are 3–8 nm in height (Figure 1). AFM-IR spectra collected from these aggregates exhibited vibrational bands that can be assigned to both proteins and lipids. Specifically, we observed both amide I and amide II at 1620–1700 and 1480–1580 cm−1, respectively (Figure 2). The position of amide I band can be used to determine the secondary structure of protein aggregates. If the amide I is centered around 1620 cm−1, the aggregates exhibit primarily parallel β-sheet secondary structure, whereas the shift of this vibration to ~1695 cm−1 is indicative of the antiparallel β-sheet.5,10 Proteins with unordered secondary structure exhibit amide I around 1660 cm−1, whereas α-helical proteins possess amide I around 1640–1650 cm−1.5,10 On the basis of the acquired spectra, we can conclude that Ins:PC aggregates have predominantly unordered protein secondary structure.
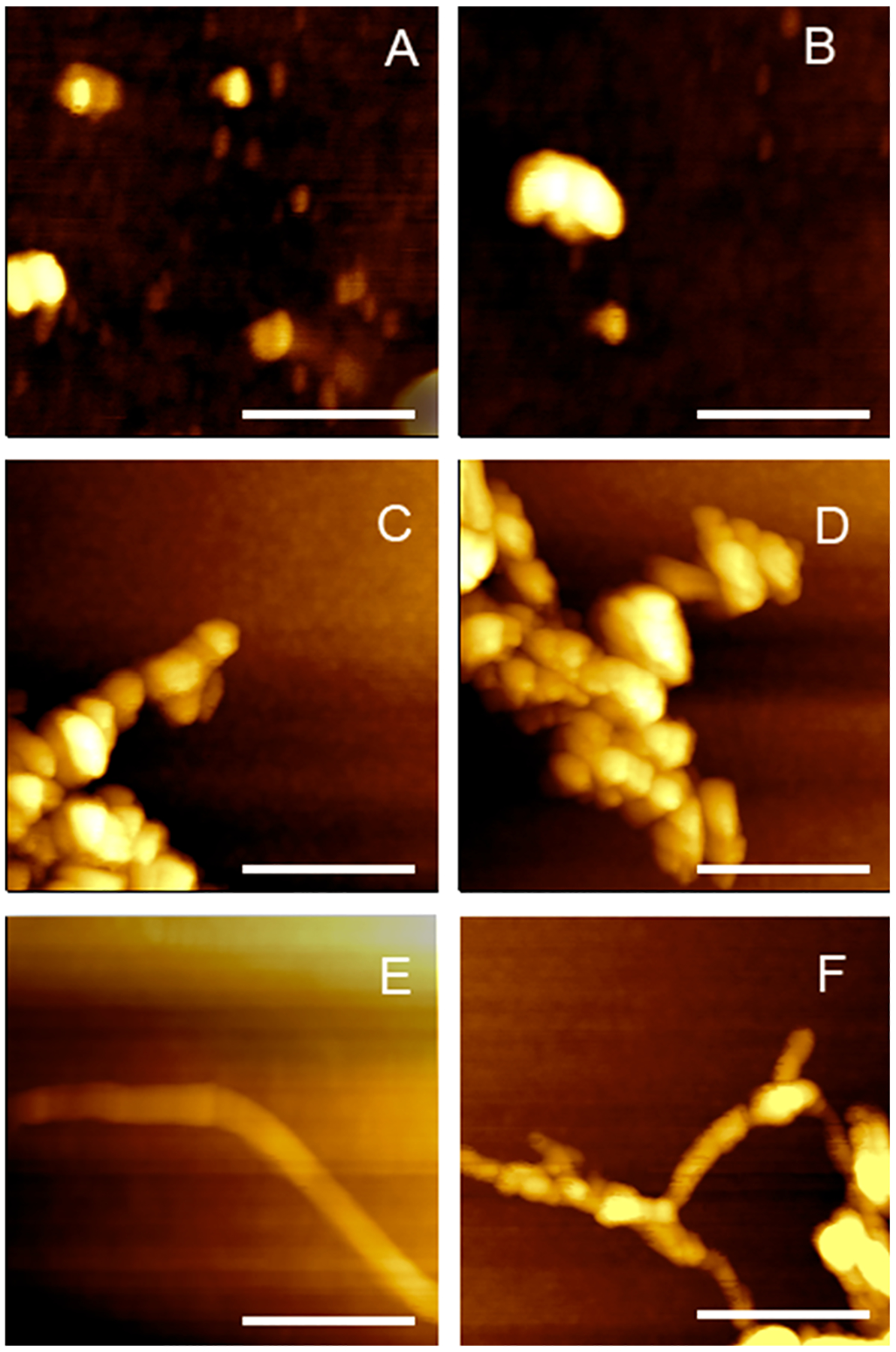
Figure 1.
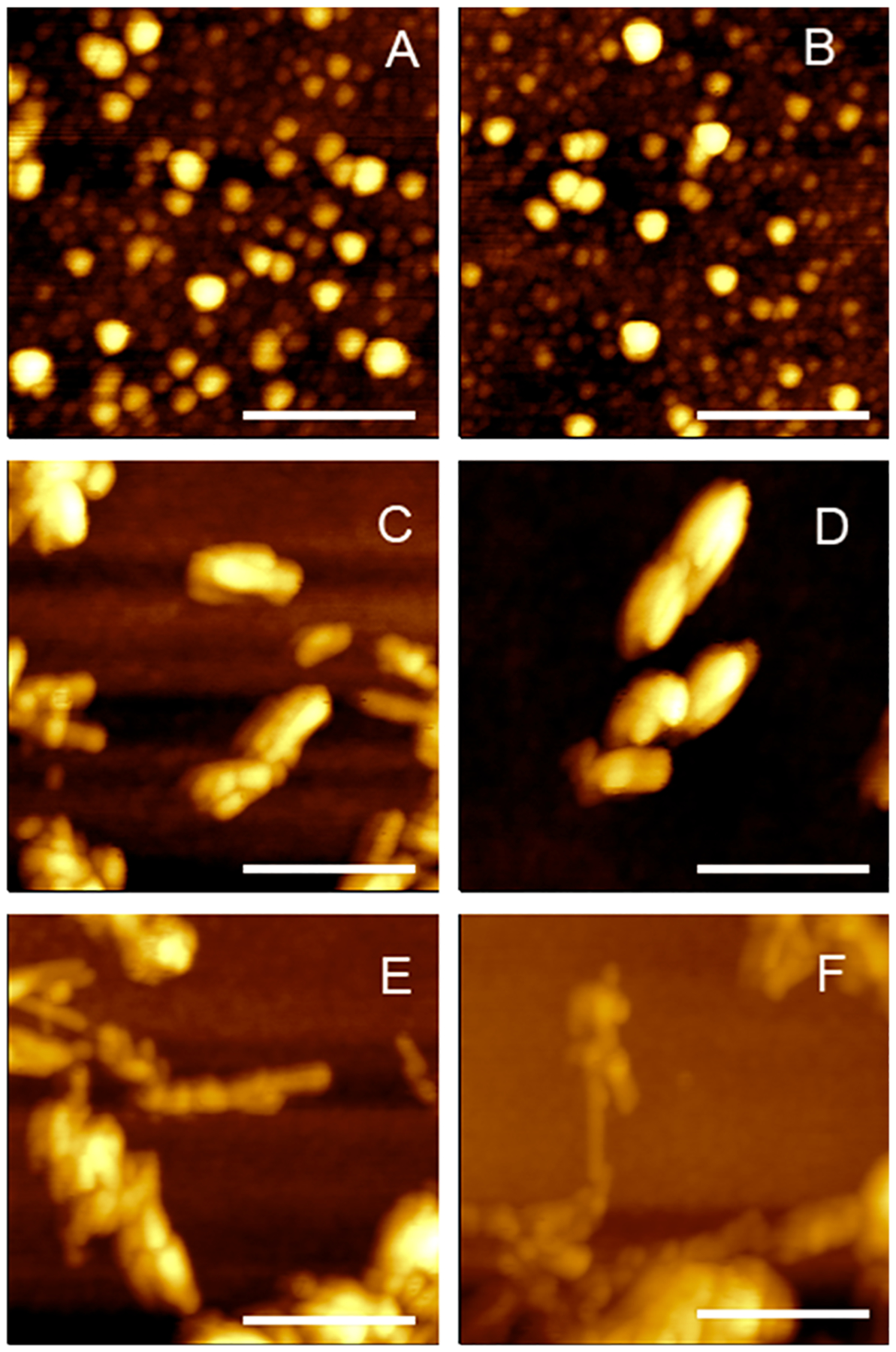
AFM images of Ins:PC oligomers (A, B), Ins:CL oligomers (C, D), protofibrils (E), and fibrils (F).
Figure 2.
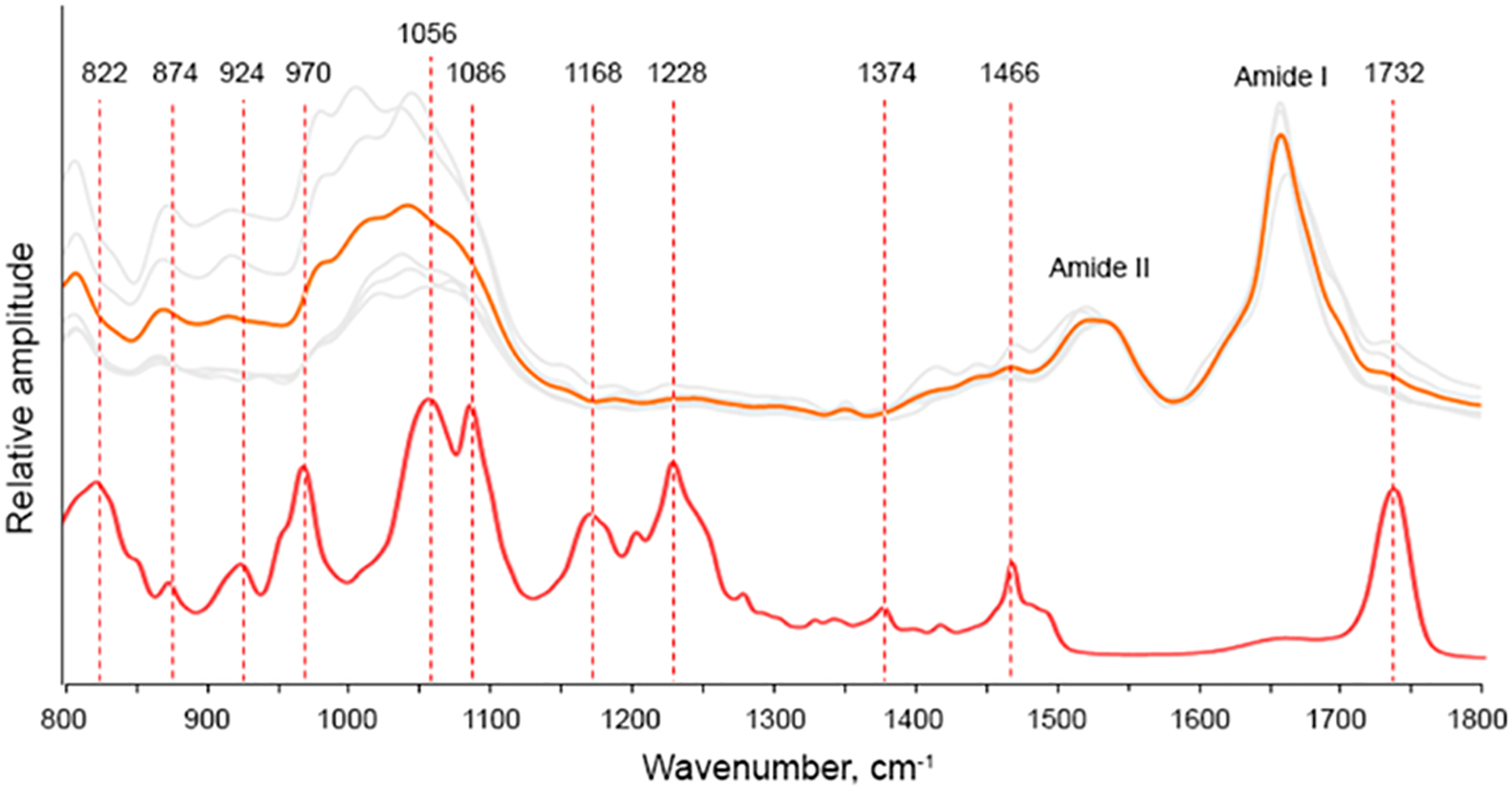
Nanoscale analysis of lipid content of insulin aggregates grown in the presence of PC. AFM-IR spectra of Ins:PC (gray); the average spectrum is shown in orange. FTIR spectrum of PC (red).
AFM-IR spectra acquired form Ins:PC also possess vibrations that can be assigned to PC itself (Figure 2 and Figure S1). Specifically, we observed a weak band at 1732 cm−1, which corresponds to the ester vibration of PC as well as a set of bands in the range 822–1086 cm−1 (Figure 2 and Table 1). These set of bands includes ν(C–N+–C), ν(PO2−), and C–O vibrations. We have also observed weak P═O, ν(PO2−), and C–O–P vibrations of PC at 1168–1228 cm−1 as well as the C–O stretching vibration of the lipid at 1374 cm−1. It should be noted that these vibrational bands are not present in the insulin aggregates grown in a lipid-free environment (Figure 3). To demonstrate this, we collected more than 10 spectra from different insulin aggregates shown in Figure 3A–D. These spectra (Figure 3E) do not possess vibrations that can be assigned to lipids (Table 1).
Table 1.
Vibrational Bands and Their Assignments for Phospholipids and Proteins
| band wavenumber (cm−1) | vibration mode | assignment |
|---|---|---|
| 820–880 | ν(C–N+–C) | lipid49 |
| 920–950 | ν(C–N+–C) | lipid49 |
| 970 | ν(C–N+–C) | lipid49 |
| 1020–1028 | C–O stretch | lipid50,51 |
| 1082–1090 | ν(PO2−) | lipid49 |
| 1168–1200 | P═O C–O–P |
lipid52 |
| 1220–1260 | ν(PO2−) | lipid53 |
| 1350 | stretching C–O | lipid54 |
| 1374 | CH3, C–O stretching | lipid49,53,54 |
| 1466 | CH2 scissoring | proteins and lipids10 |
| 1480–1580 | C–N stretching and N–H bending (amide II) | proteins10 |
| 1620–1700 | C═O (amide I) | proteins10 |
| 1730–1745 | ν(C═O) | lipid54 |
Figure 3.
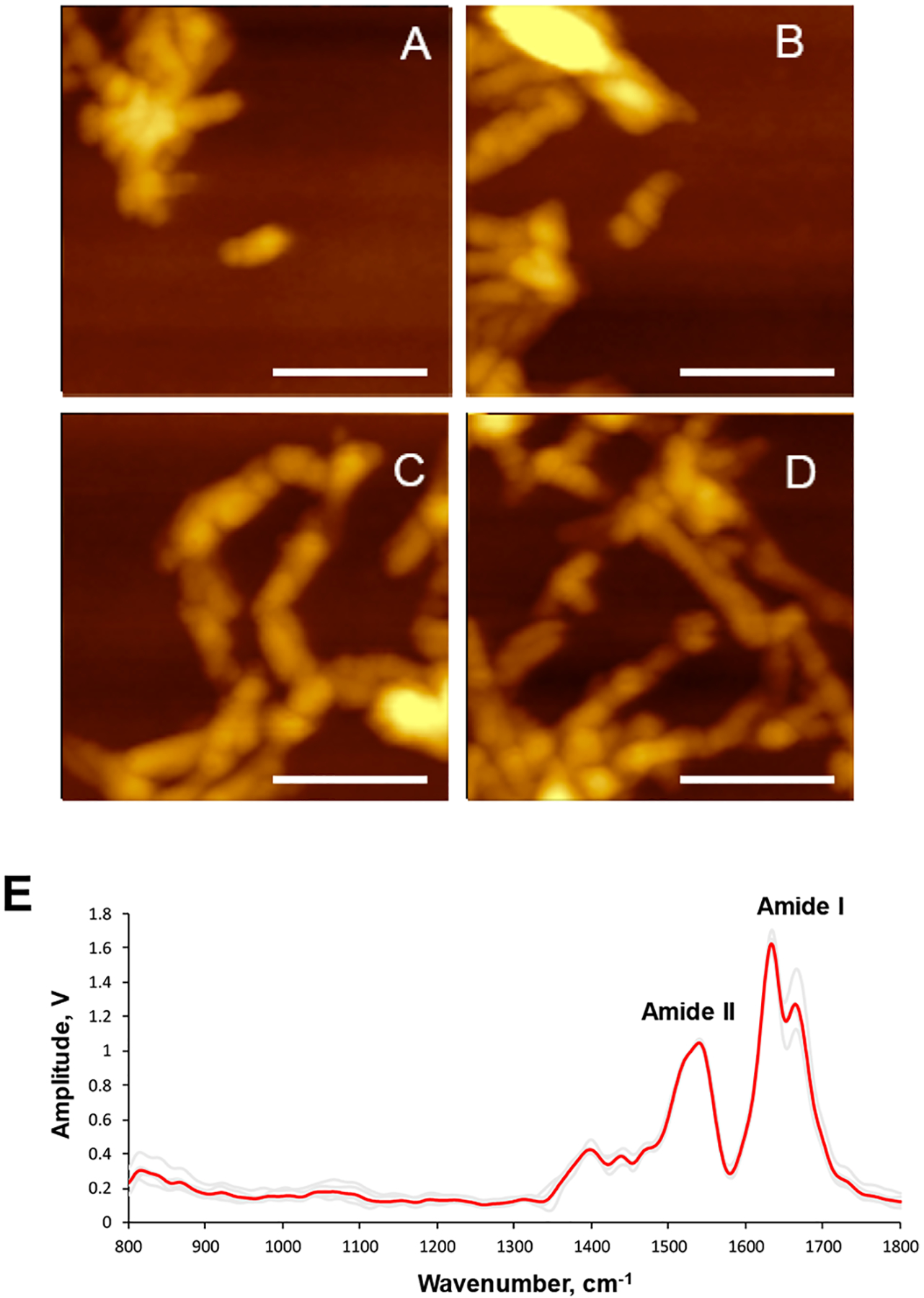
AFM images (A–D) of insulin aggregates grown in the lipid-free environment; AFM-IR spectra collected from these aggregates (E). Individual spectra are in gray, and the average spectrum is in red.
These findings show that Ins:PC aggregates possess lipids in their structure. Finally, in the AFM-IR spectra acquired form Ins:PC, we observed the CH2 vibration at 1466 cm−1. However, this vibration cannot be exclusively assigned to a particular class of biological molecules because CH2 groups present in both lipids and protein.
We also found that frequencies and intensities of a vast majority of lipid vibrations in the AFM-IR spectra collected from Ins:PC have been changed relative to the reference IR spectrum of PC (Figure 2). Specifically, we observed a drastic decrease in the intensity of the C═O ester vibration (1730 cm−1) and the C–O stretching vibration (1380 cm−1) in the AFM-IR spectrum relative to the intensities of these bands in the IR spectrum of PC. We have also observed a shift of most of v(C–N+–C) vibrations relative to the reference vibrations of these bands in the IR spectrum of PC. These spectral changes show that polar PC groups interact with insulin molecules upon protein aggregation.
Previously reported NMR studies shed light on the nature of such lipid–protein interactions. Specifically, it has been demonstrated that polar head groups of lipids build electrostatic interactions with charged amino acid residues of α-Syn.46 NMR and fluorescence methods also revealed that lipid–protein interactions are facilitated by hydrophobic interactions between nonpolar amino acid residues of the protein and fatty acid tails of lipids.47,48 These interactions alter the structure of Ins:PC aggregates, preventing their propagation into mature fibrils. Thus, PC can have an inhibitory effect on insulin aggregation.
CL is a phospholipid that is uniquely present in the inner membranes of mitochondria.55,56 Changes in the concentration of CL are directly linked to a large group of diseases, including Parkinson disease.57,58 Therefore, we examined the extent to which insulin aggregation can be altered by CL. We found that in the presence of CL insulin formed oligomers that propagated into protofibrils and fibrils (Figure 1C–F). Ins:CL oligomers exhibit an amide I band centered at 1660 cm−1 with a shoulder around 1624 cm−1 (Figure 4). This suggests the dominance of the unordered protein secondary structure in Ins:CL oligomers with a small contribution of parallel β-sheet. Similar to Ins:PC, AFM-IR spectra collected from Ins:CL oligomers exhibit bands that can be assigned to the C═O ester vibration of CL (1730 cm−1) as well as a set ν(C–N+–C), ν(PO2−), and C–O vibrations at 800–1098 cm−1 (Figure 4 and Figure S2). We have also observed a peak at 1466 cm−1 which could be assigned to CH2 vibrations. These data show that CL is present in Ins:CL oligomers. Similar to Ins:PC, the intensity of the C═O ester vibration was found to be significantly lower in the AFM-IR spectra of Ins:CL compared to the intensity of this vibration in the reference spectrum of CL. This conclusion can be also made about the ~840 cm−1 vibration. Finally, we found that most of v(C–N+–C) vibrations were shifted in the AFM-IR spectrum of Ins:CL relative to the reference vibrations of these bands in the IR spectrum of CL. It should be noted that vibrational bands that can be assigned to CL are not present in the insulin aggregates grown in the absence of lipids (Figure 3). These findings show that insulin develops strong charge interactions with the headgroup of CL. This Ins:CL complex templates protein aggregation that yields oligomers, protofibrils, and fibrils.
Figure 4.
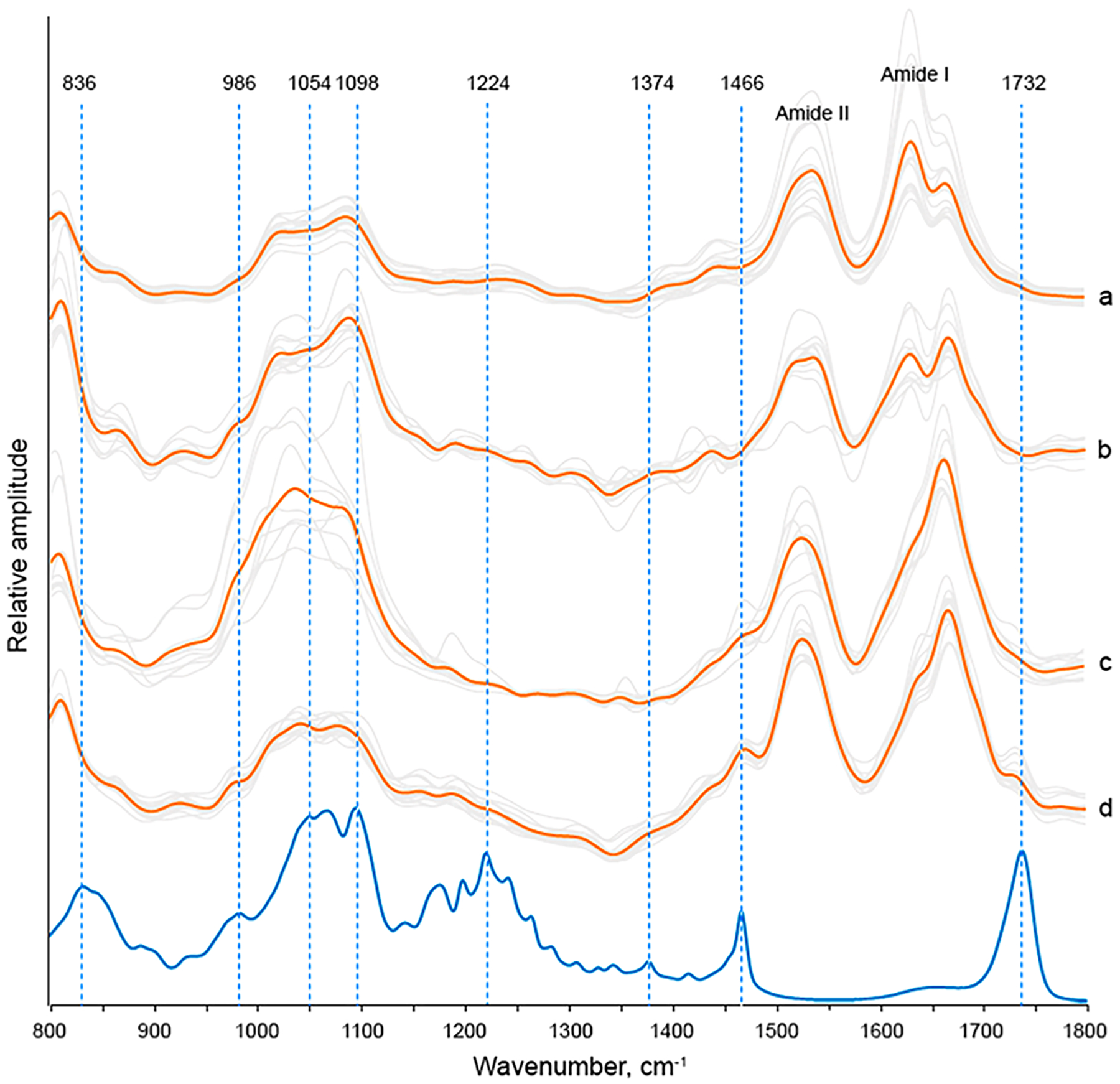
AFM IR spectra of Ins:CL fibrils (a), protofibrils (b), and oligomers (c, d) (gray) with corresponding average spectra shown in orange. FTIR spectrum of CL (blue).
AFM-IR analysis of Ins:CL protofibrils and fibrils showed graduate changes in the relative intensities of 1660 and 1624 cm−1 in their spectra. We found that, similar to the oligomers, Ins:CL protofibrils have a more intense 1660 cm−1 vibration, whereas in the AFM-IR spectrum of Ins:CL fibrils, the 1624 cm−1 bond dominates. These spectral changes indicate a graduate increase in the amount of parallel β-sheet with a reversed decrease in the amount of unordered protein secondary structure as Ins:CL oligomers propagate into fibrils.
However, we have not observed the C═O ester vibration of PC (1730 cm−1) either in the AFM-IR spectrum of Ins:CL protofibrils or in the spectrum of Ins:CL fibrils. Furthermore, we have found a change in the relative intensities of v(C–N+–C) and amide vibrations in the spectra of protofibrils and fibrils. If in the AFM-IR spectrum of Ins:CL protofibril vibrations within 986–1098 cm−1 were more intense than both amide I and II bands, in the spectrum of Ins:CL these ratios were reversed. These findings show that the amounts of lipids relative to the amount of protein decreases as protofibrils are developed into fibrils. Thus, although some residual CL is present in Ins:CL fibrils, these aggregates are modestly lipid-free compared to their oligomers.
The question to ask is whether insulin has certain lipid preferences if aggregated in the presence of both CL and PC. To answer this question, we aggregated insulin in the equimolar rations of premade LUVs of 1:1 PC:CL. Microscopic examination of Ins:PC:CL aggregates revealed the presence of oligomers, protofibrils, and fibrils (Figure 5).
Figure 5.
AFM images of Ins:PC:CL oligomers (A, B), protofibrils (C, D), and fibrils (E, F).
AFM-IR spectra collected from the oligomers exhibit vibrational bands that can be assigned to the protein (amide I and II) (Figure 6). In the spectra collected from the oligomers, the amide I is centered around 1660 cm−1, indicating the dominance of unordered protein secondary structure, whereas in the spectra collected from protofibrils, the presence of both 1624 and 1660 cm−1 bands is evident. In the spectra collected from the oligomers, we have also observed vibrations that can be assigned to PC. Specifically, we have observed the C═O ester vibration around 1730 cm−1 as well as the ν(C–N+–C), ν(PO2−), and C–O vibrations at 822–1068 cm−1. However, we have not observed the CL marker vibrational bands at 840 and 1130 cm−1 in the spectra collected from Ins:PC:CL oligomers. This finding shows that in the presence of equimolar concentrations of PC and CL insulin exclusively interacts with PC. It should be noted that all analyzed aggregates exhibited similar if not identical lipid vibrations (Figure 6). This finding shows that Ins:PC:CL aggregates are homogeneous rather than heterogeneous from the perspective of their structural organization.
Figure 6.
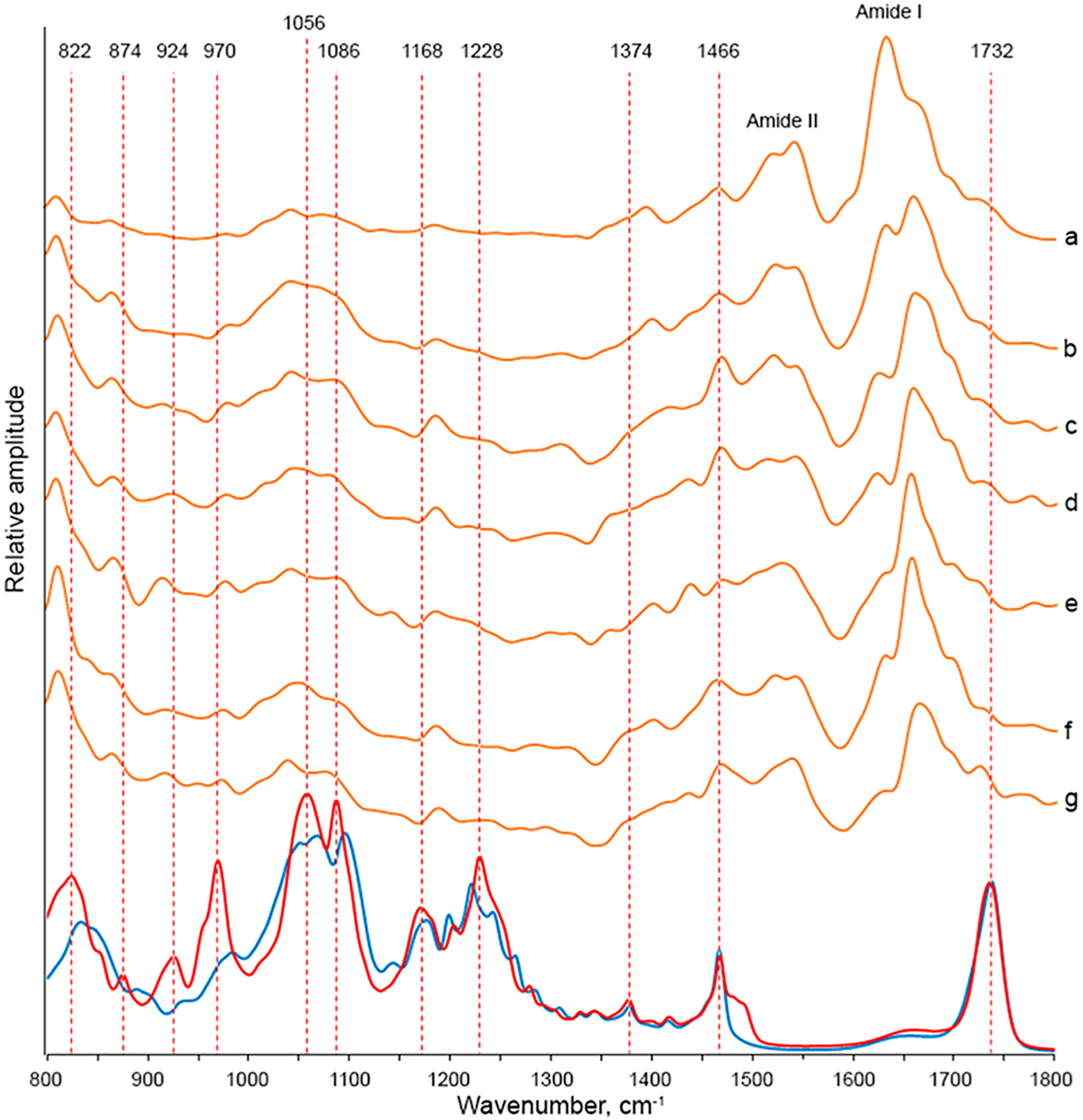
AFM-IR spectra of Ins:CL collected from individual fibrils (a), protofibrils (b), and oligomers (c–h). FTIR spectra of CL (blue) of PC (red).
Although CL was not detected in the structure of Ins:PC:CL aggregates, AFM results show that a presence of CL in the lipid mixture suppresses the inhibitory activity of PC on fibril formation. Specifically, in the presence of PC, insulin forms only oligomers, whereas both protofibrils and fibrils are observed for PC:CL. Thus, although CL does not bind directly to insulin, it modifies the structure of the aggregates that were grown in its presence. Consequently, one can expect that such aggregates would exert distinctly different toxicities compared to both Ins:CL and Ins:PC aggregates.
To answer this question, we performed lactate dehydrogenase test (LDH) in which we examined the toxicity of Ins:PC, Ins:CL, and Ins:PC:CL. We have also compared toxicities of these aggregates to the toxicities exerted by lipids themselves and insulin aggregates grown in the lipid-free environment (Figure 3).
Our results show that Ins:CL:PC oligomers exerted very similar toxicities to Ins:PC, which confirms discussed above structural similarities between these two types of oligomeric species (Figure 7). It should be noted that toxicities of both Ins:CL:PC and Ins:PC oligomers are greater than the toxicities of Ins and Ins:CL. However, toxicity exerted by Ins:CL:PC is more similar to Ins:CL, which confirms structural similarities between these two types of insulin aggregates. It should be noted that Ins:PC exerted significantly lower cell toxicity than Ins, Ins:CL, and Ins:CL:PC. The toxicities of Ins, Ins:PC, Ins:CL, and Ins:CL:PC fibrils showed that Ins:CL:PC exerted a higher cell toxicity than Ins:PC and a lower toxicity than Ins:CL. At the same time, all aggregates grown in the presence of lipids exerted lower cell toxicities compared to insulin aggregates grown in the lipid-free environment. The LDH assay also revealed that insulin protofibrils and fibrils exert greater toxicities than oligomers. It should be noted that lipids themselves did not exert any significant cell toxicity.
Figure 7.
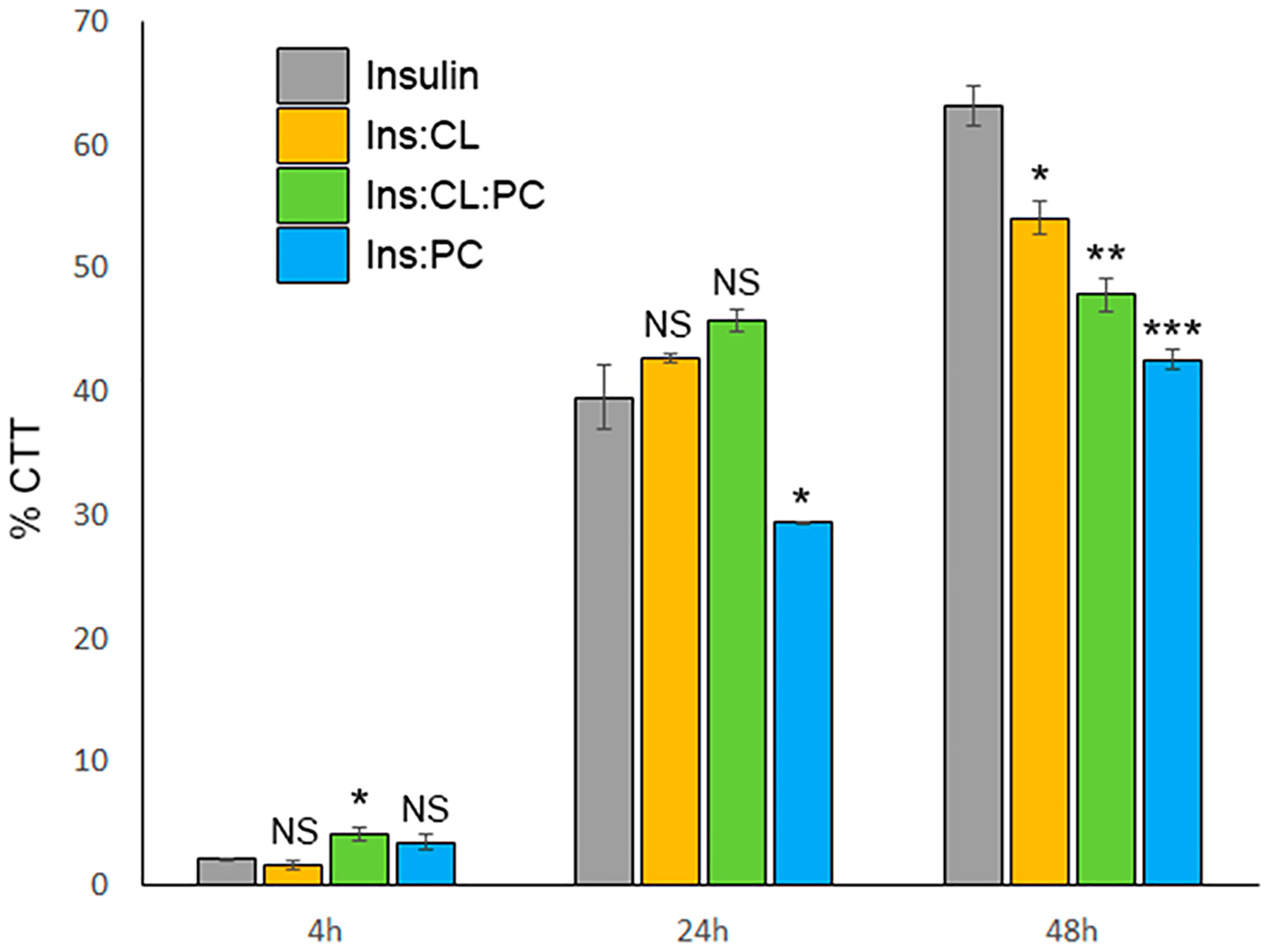
LDH-determined toxicity of oligomers (4 h), protofibrils (24 h), and fibrils (48 h) of Ins, Ins:PC, Ins:CL, and Ins:CL:PC.
Summarizing, our experimental findings show that phospholipids interact with insulin drastically changing the structure and toxicity of the resulting protein aggregates. Insulin oligomers possess a high amount of lipids and exert low cell toxicities, whereas protofibrils and fibrils exhibit very low lipid content simultaneously exerting high cell toxicity. Our findings also show that insulin has higher affinity toward PC than CL if aggregated in their equimolar mixture. However, although presence of CL in this case has very little if any effect on insulin oligomers, CL presence strongly suppresses the inhibitory effect of PC on insulin aggregation. The resulting Ins:PC:CL protofibrils and fibrils exert a similar toxicity to Ins:CL aggregates.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the National Institutes of Health for the provided financial support (R35GM142869).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpclett.1c04012.
Detailed description of spectra acquisition methods and instrumentation; FITR and corresponding AFM-IR spectra of PC and CL shown in Figures S1 and S2, respectively (PDF)
Transparent Peer Review report available (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jpclett.1c04012
The authors declare no competing financial interest.
Contributor Information
Stanislav Rizevsky, Department of Biochemistry and Biophysics, Texas A&M University, College Station, Texas 77843, United States; Department of Biotechnology, Binh Duong University, Thu Dau Mot 820000, Vietnam.
Mikhail Matveyenka, Department of Biochemistry and Biophysics, Texas A&M University, College Station, Texas 77843, United States.
Dmitry Kurouski, Department of Biochemistry and Biophysics and Department of Biomedical Engineering, Texas A&M University, College Station, Texas 77843, United States;.
REFERENCES
- (1).Chiti F; Dobson CM Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress over the Last Decade. Annu. Rev. Biochem 2017, 86, 27–68. [DOI] [PubMed] [Google Scholar]
- (2).Knowles TP; Vendruscolo M; Dobson CM The Amyloid State and Its Association with Protein Misfolding Diseases. Nat. Rev 2014, 15, 384–96. [DOI] [PubMed] [Google Scholar]
- (3).Iadanza MG; Jackson MP; Hewitt EW; Ranson NA; Radford SE A New Era for Understanding Amyloid Structures and Disease. Nat. Rev. Mol. Cell Biol 2018, 19, 755–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Chen SW; et al. Structural Characterization of Toxic Oligomers That Are Kinetically Trapped During Alpha-Synuclein Fibril Formation. Proc. Natl. Acad. Sci. U. S. A 2015, 112, E1994–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Dou T; Zhou L; Kurouski D Unravelling the Structural Organization of Individual Alpha-Synuclein Oligomers Grown in the Presence of Phospholipids. J. Phys. Chem. Lett 2021, 12, 4407–4414. [DOI] [PubMed] [Google Scholar]
- (6).Pieri L; Madiona K; Melki R Structural and Functional Properties of Prefibrillar A-Synuclein Oligomers. Sci. Rep 2016, 6, 24526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cremades N; et al. Direct Observation of the Interconversion of Normal and Toxic Forms of Alpha-Synuclein. Cell 2012, 149, 1048–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Apetri MM; Maiti NC; Zagorski MG; Carey PR; Anderson VE Secondary Structure of Alpha-Synuclein Oligomers: Characterization by Raman and Atomic Force Microscopy. J. Mol. Biol 2006, 355, 63–71. [DOI] [PubMed] [Google Scholar]
- (9).O’Leary EI; Lee JC Interplay between Alpha-Synuclein Amyloid Formation and Membrane Structure. Biochim. Biophys. Acta Proteins Proteom 2019, 1867, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kurouski D; Van Duyne RP; Lednev IK Exploring the Structure and Formation Mechanism of Amyloid Fibrils by Raman Spectroscopy: A Review. Analyst 2015, 140, 4967–80. [DOI] [PubMed] [Google Scholar]
- (11).Hong DP; Han S; Fink AL; Uversky VN Characterization of the Non-Fibrillar Alpha-Synuclein Oligomers. Prot. Pept. Lett 2011, 18, 230–40. [DOI] [PubMed] [Google Scholar]
- (12).Li B; et al. Cryo-Em of Full-Length Alpha-Synuclein Reveals Fibril Polymorphs with a Common Structural Kernel. Nat. Commun 2018, 9, 3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Guerrero-Ferreira R; Taylor NM; Mona D; Ringler P; Lauer ME; Riek R; Britschgi M; Stahlberg H Cryo-Em Structure of Alpha-Synuclein Fibrils. Elife 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tycko R Solid-State Nmr Studies of Amyloid Fibril Structure. Annu. Rev. Phys. Chem 2011, 62, 279–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Alza NP; Iglesias Gonzalez PA; Conde MA; Uranga RM; Salvador GA Lipids at the Crossroad of Alpha-Synuclein Function and Dysfunction: Biological and Pathological Implications. Front. Cell Neurosci 2019, 13, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Galvagnion C The Role of Lipids Interacting with α-Synuclein in the Pathogenesis of Parkinson’s Disease. J. Parkins. Dis 2017, 7, 433–450. [DOI] [PubMed] [Google Scholar]
- (17).Galvagnion C; Brown JW; Ouberai MM; Flagmeier P; Vendruscolo M; Buell AK; Sparr E; Dobson CM Chemical Properties of Lipids Strongly Affect the Kinetics of the Membrane-Induced Aggregation of Alpha-Synuclein. Proc. Natl. Acad. Sci. U. S. A 2016, 113, 7065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Rizevsky S; Kurouski D Nanoscale Structural Organization of Insulin Fibril Polymorphs Revealed by Atomic Force Microscopy-Infrared Spectroscopy (Afm-Ir). Chembiochem 2020, 21, 481–485. [DOI] [PubMed] [Google Scholar]
- (19).Ruggeri FS; Charmet J; Kartanas T; Peter Q; Chia S; Habchi J; Dobson CM; Vendruscolo M; Knowles TPJ Microfluidic Deposition for Resolving Single-Molecule Protein Architecture and Heterogeneity. Nat. Commun 2018, 9, 3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ruggeri FS; Flagmeier P; Kumita JR; Meisl G; Chirgadze DY; Bongiovanni MN; Knowles TPJ; Dobson CM The Influence of Pathogenic Mutations in Alpha-Synuclein on Biophysical and Structural Characteristics of Amyloid Fibrils. ACS Nano 2020, 14, 5213–5222. [DOI] [PubMed] [Google Scholar]
- (21).Ruggeri FS; Longo G; Faggiano S; Lipiec E; Pastore A; Dietler G Infrared Nanospectroscopy Characterization of Oligomeric and Fibrillar Aggregates During Amyloid Formation. Nat. Commun 2015, 6, 7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhou L; Kurouski D Structural Characterization of Individual Alpha-Synuclein Oligomers Formed at Different Stages of Protein Aggregation by Atomic Force Microscopy-Infrared Spectroscopy. Anal. Chem 2020, 92, 6806–6810. [DOI] [PubMed] [Google Scholar]
- (23).Dazzi A; Glotin F; Carminati R Theory of Infrared Nanospectroscopy by Photothermal Induced Resonance. J. Appl. Phys 2010, 107, 124519. [Google Scholar]
- (24).Dazzi A; Prater CB Afm-Ir: Technology and Applications in Nanoscale Infrared Spectroscopy and Chemical Imaging. Chem. Rev 2017, 117, 5146–5173. [DOI] [PubMed] [Google Scholar]
- (25).Kurouski D; Dazzi A; Zenobi R; Centrone A Infrared and Raman Chemical Imaging and Spectroscopy at the Nanoscale. Chem. Soc. Rev 2020, 49, 3315–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Katzenmeyer AM; Aksyuk V; Centrone A Nanoscale Infrared Spectroscopy: Improving the Spectral Range of the Photothermal Induced Resonance Technique. Anal. Chem 2013, 85, 1972–1979. [DOI] [PubMed] [Google Scholar]
- (27).Katzenmeyer AM; Holland G; Kjoller K; Centrone A Absorption Spectroscopy and Imaging from the Visible through Mid-Infrared with 20 Nm Resolution. Anal. Chem 2015, 87, 3154–3159. [DOI] [PubMed] [Google Scholar]
- (28).Ruggeri FS; Mannini B; Schmid R; Vendruscolo M; Knowles TPJ Single Molecule Secondary Structure Determination of Proteins through Infrared Absorption Nanospectroscopy. Nat. Commun 2020, 11, 2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lu F; Jin MZ; Belkin MA Tip-Enhanced Infrared Nanospectroscopy Via Molecular Expansion Force Detection. Nat. Photonics 2014, 8, 307–312. [Google Scholar]
- (30).Ruggeri FS; Benedetti F; Knowles TPJ; Lashuel HA; Sekatskii S; Dietler G Identification and Nanomechanical Characterization of the Fundamental Single-Strand Protofilaments of Amyloid Alpha-Synuclein Fibrils. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 7230–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ruggeri FS; Vieweg S; Cendrowska U; Longo G; Chiki A; Lashuel HA; Dietler G Nanoscale Studies Link Amyloid Maturity with Polyglutamine Diseases Onset. Sci. Rep 2016, 6, 31155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ramer G; Ruggeri FS; Levin A; Knowles TPJ; Centrone A Determination of Polypeptide Conformation with Nanoscale Resolution in Water. ACS Nano 2018, 12, 6612–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Farber C; Li J; Hager E; Chemelewski R; Mullet J; Rogachev AY; Kurouski D Complementarity of Raman and Infrared Spectroscopy for Structural Characterization of Plant Epicuticular Waxes. ACS Omega 2019, 4, 3700–3707. [Google Scholar]
- (34).Farber C; Wang R; Chemelewski R; Mullet J; Kurouski D Nanoscale Structural Organization of Plant Epicuticular Wax Probed by Atomic Force Microscope Infrared Spectroscopy. Anal. Chem 2019, 91, 2472–2479. [DOI] [PubMed] [Google Scholar]
- (35).Dazzi A Photothermal Induced Resonance. Application to Infrared Spectromicroscopy. In Thermal Nanosystems and Nanomaterials; Volz S, Ed.; Springer: Berlin, 2009; Vol. 118, pp 469–503. [Google Scholar]
- (36).Perez-Guaita D; Kochan K; Batty M; Doerig C; Garcia-Bustos J; Espinoza S; McNaughton D; Heraud P; Wood BR Multispectral Atomic Force Microscopy-Infrared Nano-Imaging of Malaria Infected Red Blood Cells. Anal. Chem 2018, 90, 3140–3148. [DOI] [PubMed] [Google Scholar]
- (37).Mathurin J; Dartois E; Pino T; Engrand C; Duprat J; Deniset-Besseau A; Borondics F; Sandt C; Dazzi A Nanometer Scale Infrared Chemical Imaging of Organic Matter in Ultracarbonaceous Antarctic Micrometeorites (Ucamm). Astron. Astrophys 2019, 622, A160. [Google Scholar]
- (38).Dazzi A; Prazeres R; Glotin F; Ortega JM; Al-Sawaftah M; de Frutos M Chemical Mapping of the Distribution of Viruses into Infected Bacteria with a Photothermal Method. Ultramicroscopy 2008, 108, 635–641. [DOI] [PubMed] [Google Scholar]
- (39).Mayet C; Deniset-Besseau A; Prazeres R; Ortega JM; Dazzi A Analysis of Bacterial Polyhydroxybutyrate Production by Multimodal Nanoimaging. Biotechnol. Adv 2013, 31, 369–374. [DOI] [PubMed] [Google Scholar]
- (40).Kochan K; Perez-Guaita D; Pissang J; Jiang JH; Peleg AY; McNaughton D; Heraud P; Wood BR In Vivo Atomic Force Microscopy-Infrared Spectroscopy of Bacteria. J. Royal Soc. Interface 2018, 15, 20180115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wieland K; Ramer G; Weiss VU; Allmaier G; Lendl B; Centrone A Nanoscale Chemical Imaging of Individual Chemotherapeutic Cytarabine-Loaded Liposomal Nanocarriers. Nano Res. 2019, 12, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Strelcov E; Dong Q; Li T; Chae J; Shao Y; Deng Y; Gruverman A; Huang J; Centrone A Ch3nh3pbi3 Perovskites: Ferroelasticity Revealed. Sci. Adv 2017, 3, No. e1602165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Gupta Y; Singla G; Singla R Insulin-Derived Amyloidosis. Indian J. Endocrinol. Metab 2015, 19, 174–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Shikama Y; Kitazawa J; Yagihashi N; Uehara O; Murata Y; Yajima N; Wada R; Yagihashi S Localized Amyloidosis at the Site of Repeated Insulin Injection in a Diabetic Patient. Int. Med 2010, 49, 397–401. [DOI] [PubMed] [Google Scholar]
- (45).Iwaya K; et al. Toxicity of Insulin-Derived Amyloidosis: A Case Report. BMC Endocr. Dis 2019, 19, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Viennet T; et al. Structural Insights from Lipid-Bilayer Nanodiscs Link Alpha-Synuclein Membrane-Binding Modes to Amyloid Fibril Formation. Commun. Biol 2018, 1, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Giasson BI; Murray IV; Trojanowski JQ; Lee VM A Hydrophobic Stretch of 12 Amino Acid Residues in the Middle of Alpha-Synuclein Is Essential for Filament Assembly. J. Biol. Chem 2001, 276, 2380–6. [DOI] [PubMed] [Google Scholar]
- (48).Ueda K; Fukushima H; Masliah E; Xia Y; Iwai A; Yoshimoto M; Otero DA; Kondo J; Ihara Y; Saitoh T Molecular Cloning of Cdna Encoding an Unrecognized Component of Amyloid in Alzheimer Disease. Proc. Natl. Acad. Sci. U. S. A 1993, 90, 11282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Blume A Properties of Lipid Vesicles: Ft-Ir Spectroscopy and Fluorescence Probe Studies. Curr. Opin. Colloid Interface Sci 1996, 1, 64–77. [Google Scholar]
- (50).Wood BR; Quinn MA; Tait B; Ashdown M; Hislop T; Romeo M; McNaughton D Ftir Microspectroscopic Study of Cell Types and Potential Confounding Variables in Screening for Cervical Malignancies. Biospectroscopy 1998, 4, 75–91. [DOI] [PubMed] [Google Scholar]
- (51).Wood BR; Quinn MA; Burden FR; McNaughton D An Investigation into Ftir Spectroscopy as a Biodiagnostic Tool for Cervical Cancer. Biospectroscopy 1996, 2, 143–153. [Google Scholar]
- (52).Yoshida S; Miyazaki M; Sakai K; Takeshita M; Yuasa S; Sato A; Kobayashi T; Watanabe S; Okuyama H Fourier Transform Infrared Spectroscopic Analysis of Rat Brain Microsomal Membranes Modified by Dietary Fatty Acids: Possible Correlation with Altered Learning Behavior. Biospectroscopy 1997, 3, 281–290. [Google Scholar]
- (53).Richter T; Steiner G; Abu-Id MH; Salzer R; Bergmann R; Rodig H; Johannsen B Identification of Tumor Tissue by Ftir Spectroscopy in Combination with Positron Emission Tomography. Vibrat. Spectrosc 2002, 28, 103–110. [Google Scholar]
- (54).Movasaghi Z; Rehman S; ur Rehman DI Fourier Transform Infrared (Ftir) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev 2008, 43, 134–179. [Google Scholar]
- (55).Surmeier DJ; Obeso JA; Halliday GM Selective Neuronal Vulnerability in Parkinson Disease. Nat. Rev. Neurosci 2017, 18, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Goedert M; Spillantini MG; Del Tredici K; Braak H 100 Years of Lewy Pathology. Nat. Rev. Neurol 2013, 9, 13–24. [DOI] [PubMed] [Google Scholar]
- (57).Braak H; Ghebremedhin E; Rub U; Bratzke H; Del Tredici K Stages in the Development of Parkinson’s Disease-Related Pathology. Cell Tissue Res. 2004, 318, 121–34. [DOI] [PubMed] [Google Scholar]
- (58).Del Tredici K; Rub U; De Vos RA; Bohl JR; Braak H Where Does Parkinson Disease Pathology Begin in the Brain? J. Neuropathol. Exp. Neurol 2002, 61, 413–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


