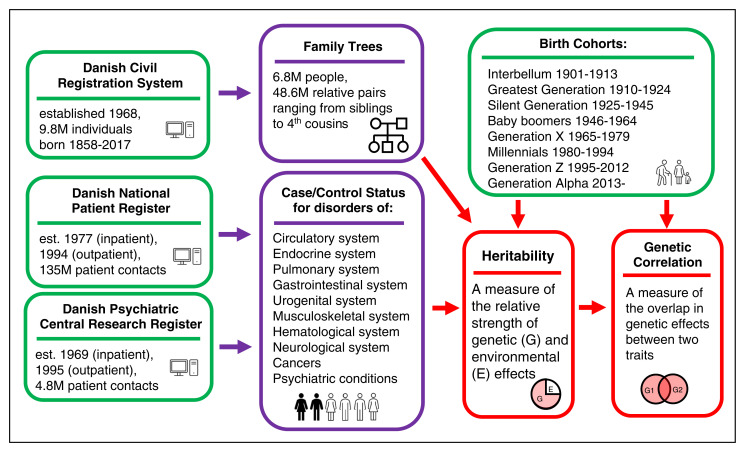
Family studies are a powerful tool for quantifying the extent to which genetic factors contribute to variation among individuals and for exploring the interaction of genetic and environmental factors. They can also be used to study shared genetic effects across traits, providing clues about etiological pathways. In PNAS, Athanasiadis et al. (1) describe what is one of the largest human family studies to date, including over 6 million individuals who form over 48 million pairs of relatives spanning as many as six generations. They achieved this massive feat by drawing parent–child relationships from the Danish Civil Registration System and connecting overlapping sets of parents and children to identify siblings, aunts, uncles, grandparents, and cousins to build large family trees. This pedigree information was then combined with records from the national health care system to score each individual for the presence or absence of medical disorders in 10 major categories (Fig. 1).
Fig. 1.
Overview of the Athanasiadis et al. (1) study. Green boxes represent study inputs, purple boxes represent analytical products, and red boxes represent study outcomes. E, environmental; est., established; M, million; G, genetic.
The resource built by Athanasiadis et al. (1) is extremely flexible and will facilitate numerous future genetic studies of both rare and common disorders. In this introductory paper, the authors have used it to examine the relative strength of genetic effects (i.e., heritability) on general categories of disorders and the overlap in genetic effects between disorders (i.e., genetic correlation). They also explored how these effects changed over time. This analysis had several uncommon features that complement what we know from other studies. First, we typically estimate heritability for a single disorder at a time. It is less common to estimate heritability for general categories of disorders, such as any urogenital disorder or any gastrointestinal disorder. Often, we are concerned with maximizing the genetic signal for gene localization by ensuring that we have a single diagnostic entity, which will hopefully reduce the underlying genetic and etiological heterogeneity. In contrast, Athanasiadis et al. (1) are interested in general liability or the idea that heritable factors may affect an entire body system and predispose an individual to multiple disorders within or across body systems. Identifying such cross-system connections may provide valuable clues into the pathophysiology of the associated disorders.
In PNAS, Athanasiadis et al. describe what is one of the largest human family studies to date, including over 6 million individuals who form over 48 million pairs of relatives spanning as many as six generations.
Another unusual aspect of this work is the examination of the change in heritability and genetic correlations across birth cohorts. It is rare for a family study to include individuals spanning more than three generations with large sample sizes in both earlier and more recent birth cohorts. Health information from the Danish National Patient Register and the Danish Psychiatric Central Research Register was available for people born over a period of more than a century, from 1901 to 2017. This led to the intriguing observation that heritability for all disorders increased in more recent generations. Note, however, that if we divide the sources of variation between individuals into genetic (G) and environmental (E) components, heritability (h2) is defined as the proportion of the overall variance that is attributable to genetic factors: h2 = G/(G + E). This means that heritability can change through alterations in the strength of either genetic or environmental factors. If the contribution of environmental factors toward variation in disease risk is decreased through improved nutrition or improved health care, the total variance, which is the denominator in the heritability equation, would decrease, and thus, the heritability could increase without the genetic component changing. This complicates the interpretation of increasing heritability in more recent cohorts. In contrast, for pairs of body systems showing shared genetic effects, Athanasiadis et al. (1) find that the genetic correlations are generally stronger in older generations and decreased in more recent cohorts, although there are some exceptions to this pattern.
Family studies can be difficult to conduct as they place additional limitations on subject recruitment and require specialized approaches to data analysis. Many large-scale gene localization studies these days rely on unrelated individuals either recruited for the presence or absence of a particular disorder or chosen without regard to phenotype in the hopes of capturing the range of variation within a population. In this study design, family relationships are treated as a nuisance, and often, study participants are filtered to remove closely related individuals. Recently, however, investigators have sought to utilize the information in these family relationships and have begun to assemble families from what were originally large population-based studies, such as the UK Biobank, or from registry-based studies.
The Athanasiadis et al. (1) Danish registry cohort joins similar extended genealogy family studies assembled from national population registries or health insurance databases in Taiwan (2–4); Sweden (5–10); Manitoba, Canada (11, 12); Western Australia (13); Norway (14, 15); and Iceland (16). The availability of multiple such cohorts from around the world is important as comparisons between results from different studies will help to control for and disentangle sources of variation that differ between populations, such as cultural or environmental factors and genetic ancestry. These cohorts have been used to study familial risk in heart disease (3, 8), renal disease (2), inflammatory bowel disease (17), cancer (4, 5, 15), fracture risk (11, 12), cardiorespiratory conditions (12), and psychiatric disorders (6, 7, 9, 10, 13, 18).
An advantage of these registry-based studies is that they include nearly everyone born or receiving medical care in the geographic territory covered by the registry. This represents an important advantage for genetic studies. In a typical study, we are often concerned about the hidden biases in how a sample was selected and the implications of these biases for the generalizability of study findings. Participants in scientific studies may not be representative of the underlying population. They may be wealthier or more educated than average. Often, females are more likely to participate in research than males. In some cases, individuals with more severe disease may be more motivated to participate in research than individuals with milder symptoms. In other cases, individuals who are the most severely affected may not have the time or ability to participate in research. Study participants also may not represent the full racial, ethnic, and cultural diversity of the population. Registry-based cohorts with essentially complete ascertainment of an entire population avoid these biases and may provide samples in which to examine the effects of ascertainment bias through analyses of subsets of the cohort.
A little more than a century ago in 1918, R. A. Fisher (19) wrote a seminal paper in which he laid out the expected resemblance between relatives of different degrees of relationship presuming a trait was influenced by a large number of genetic factors, each subject to the laws of Mendelian inheritance. Fisher’s paper formed the theoretical basis for classical quantitative genetics and family studies, which have been widely used to establish the heritability of medical disorders and anthropometric traits as well as in agriculture and ecology (19). The work of Athanasiadis et al. (1), along with other registry-based genetic studies, brings these classical quantitative genetic approaches into the twenty-first century and the era of big data, allowing us to conduct family studies on a scale undreamt of a hundred years ago.
Acknowledgments
The research of L.A. is supported by NIH Grants U01MH119690, R01MH119219, U01MH119737, U10AA008401, and U01MH124962 as well as the University of Pennsylvania Autism Spectrum Program of Excellence.
Footnotes
The author declares no competing interest.
See companion article, “A comprehensive map of genetic relationships among diagnostic categories based on 48.6 million relative pairs from the Danish genealogy,” 10.1073/pnas.2118688119.
References
- 1.Athanasiadis G., et al. , A comprehensive map of genetic relationships among diagnostic categories based on 48.6 million relative pairs from the Danish genealogy. Proc. Natl. Acad. Sci. U.S.A. 119, 10.1073/pnas.2118688119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu H. H., et al. , Family aggregation and heritability of ESRD in Taiwan: A population-based study. Am. J. Kidney Dis. 70, 619–626 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Wang C.-L., et al. , Familial aggregation of myocardial infarction and coaggregation of myocardial infarction and autoimmune disease: A nationwide population-based cross-sectional study in Taiwan. BMJ Open 9, e023614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin H.-T., et al. , Familial aggregation and heritability of nonmedullary thyroid cancer in an Asian population: A nationwide cohort study. J. Clin. Endocrinol. Metab. 105, e2521–e2530 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Hemminki K., Czene K., Attributable risks of familial cancer from the Family-Cancer Database. Cancer Epidemiol. Biomarkers Prev. 11, 1638–1644 (2002). [PubMed] [Google Scholar]
- 6.Yao S., et al. , Familial liability for eating disorders and suicide attempts: Evidence from a population registry in Sweden. JAMA Psychiatry 73, 284–291 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Nordsletten A. E., et al. , Patterns of nonrandom mating within and across 11 major psychiatric disorders. JAMA Psychiatry 73, 354–361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindgren M. P., et al. , Mortality risks associated with sibling heart failure. Int. J. Cardiol. 307, 114–118 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Taylor M. J., et al. , Etiology of autism spectrum disorders and autistic traits over time. JAMA Psychiatry 77, 936–943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang R., et al. , Familial co-aggregation of schizophrenia and eating disorders in Sweden and Denmark. Mol. Psychiatry 26, 5389–5397 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Yang S., et al. , Objectively-verified parental non-hip major osteoporotic fractures and offspring osteoporotic fracture risk: A population-based familial linkage study. J. Bone Miner. Res. 32, 716–721 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Yang S., et al. , Parental cardiorespiratory conditions and offspring fracture: A population-based familial linkage study. Bone 139, 115557 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Morgan V. A., et al. , Are familial liability for schizophrenia and obstetric complications independently associated with risk of psychotic illness, after adjusting for other environmental stressors in childhood? Aust. N. Z. J. Psychiatry 53, 1105–1115 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Næss Ø., Hoff D. A., The Norwegian Family Based Life Course (NFLC) study: Data structure and potential for public health research. Int. J. Public Health 58, 57–64 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Del Risco Kollerud R., et al. , Family history of cancer and risk of paediatric and young adult’s testicular cancer: A Norwegian cohort study. Br. J. Cancer 120, 1007–1014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaitlen N., et al. , Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 9, e1003520 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orholm M., Fonager K., Sørensen H. T., Risk of ulcerative colitis and Crohn’s disease among offspring of patients with chronic inflammatory bowel disease. Am. J. Gastroenterol. 94, 3236–3238 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Bai D., et al. , Association of genetic and environmental factors with autism in a 5-country cohort. JAMA Psychiatry 76, 1035–1043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher R. A., The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 52, 399–433 (1918). [Google Scholar]