Significance
G protein-coupled receptor (GPCR) structures determined by X-ray crystallography or cryo-electron microscopy include 28 receptors for which complexes with agonists and antagonists can be compared. In all these comparisons, an interatomic distance representing the size of the orthosteric ligand binding groove differs by less than 2.9 Å. In this report, 19F-NMR observations of the NK1R-bound drug molecule aprepitant show that the orthosteric binding groove undergoes transient fluctuations with amplitudes 6 to 8 Å. We propose that this large-amplitude plasticity enables a multistep selection of functional ligands with variable efficacies. These insights into structural dynamics also provide a rationale for the observation that diffracting crystals are obtained for GPCR complexes with only few of the ligands that bind to the receptors.
Keywords: aromatic ring flips; NMR saturation transfer; nanodiscs; protein dynamics; 2D [19F,19F]-EXSY
Abstract
Comparisons of G protein-coupled receptor (GPCR) complexes with agonists and antagonists based on X-ray crystallography and cryo-electron microscopy structure determinations show differences in the width of the orthosteric ligand binding groove over the range from 0.3 to 2.9 Å. Here, we show that there are transient structure fluctuations with amplitudes up to at least 6 Å. The experiments were performed with the neurokinin 1 receptor (NK1R), a GPCR of class A that is involved in inflammation, pain, and cancer. We used 19F-NMR observation of aprepitant, which is an approved drug that targets NK1R for the treatment of chemotherapy-induced nausea and vomiting. Aprepitant includes a bis-trifluoromethyl-phenyl ring attached with a single bond to the core of the molecule; 19F-NMR revealed 180° flipping motions of this ring about this bond. In the picture emerging from the 19F-NMR data, the GPCR transmembrane helices undergo large-scale floating motions in the lipid bilayer. The functional implication is of extensive promiscuity of initial ligand binding, primarily determined by size and shape of the ligand, with subsequent selection by unique interactions between atom groups of the ligand and the GPCR within the binding groove. This second step ensures the wide range of different efficacies documented for GPCR-targeting drugs. The NK1R data also provide a rationale for the observation that diffracting GPCR crystals are obtained for complexes with only very few of the ligands from libraries of approved drugs and lead compounds that bind to the receptors.
The neurokinin 1 receptor (NK1R) is activated by an indigenous polypeptide agonist, substance P (SP) (1–3). NK1R is a G protein-coupled receptor (GPCR) of class A that is found in central and peripheral regions of the human nervous system, where activation of the receptor by SP affects key physiological processes, such as inflammation, nausea, pain, and depression (4). The drug aprepitant is an NK1R antagonist, which was approved by the US Food and Drug Administration for the treatment of chemotherapy-induced nausea and vomiting (5, 6). Crystal structures of NK1R bound to aprepitant and other antagonists (i.e., CP-99994, netupitant, and L760735), provided structural information about the location of these ligands in the orthosteric binding site (Fig. 1 A and B) (7–9), and cryo-electron microscopy (cryo-EM) structures of NK1R bound to SP and intracellular partner proteins are also available, representing activated states of the receptor (3). 19F-NMR experiments were performed with a thermostabilized fusion construct of NK1R used for crystallography; they showed that the binding mode of aprepitant involves multiple simultaneously populated orientations of the drug molecule in the orthosteric binding site (9). Here, we present a spatiotemporal characterization of the NK1R drug binding site by 19F-NMR observation (10) of the two trifluoromethyl groups of bound and free aprepitant (Fig. 1), using a truncated wild-type NK1R construct, NK1R[2–335], reconstituted in different detergents and in nanodiscs.
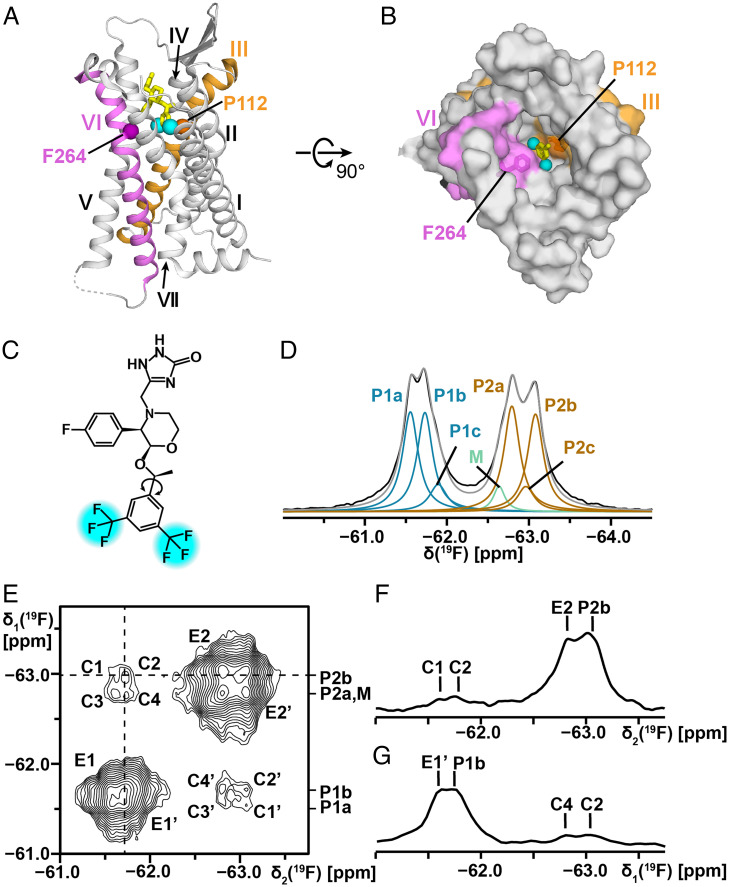
Fig. 1.
Chemical structure of the drug aprepitant, its location in the crystal structure of the complex with NK1R, and its 19F-NMR spectrum when bound to NK1R[2–335]. (A and B) Crystal structure of an NK1R complex with aprepitant (PDB ID code 6J20). (A) Side view in ribbon presentation. Aprepitant is shown in yellow stick presentation, with cyan spheres representing the trifluoromethyl groups. The TM helices are identified with Roman numerals. On the gray background the TM III and VI are highlighted in orange and purple, respectively. The locations of the α-carbon atoms of P1123.32 in TM III and F2646.51 in TM VI are represented by spheres. (B) View onto the extracellular surface in a space-filling atom presentation, same color code as in A. (C) Chemical structure of aprepitant. The two –CF3 groups are highlighted in cyan. The arrow indicates the single bond about which the bis-trifluoromethyl-phenyl ring undergoes 180° rotational flipping motions. (D) One-dimensional 19F-NMR spectrum of aprepitant bound to NK1R[2–335] in mixed micelles of LMNG and CHS at 298 K. The black line represents the experimental 19F-NMR signals of the two –CF3 groups. Lorentzian deconvolution revealed the following substates: P1a, P1b, and P1c (blue) represent one of the two –CF3 groups; M (green) represents micelle-associated “free” aprepitant; P2a, P2b, and P2c (brown) represent the second –CF3 group; the sum of P1, M, and P2 is shown in gray (SI Appendix, Table S1 for the relative integrals of the individual components). (E) Two-dimensional [19F,19F]-EXSY spectrum of NK1R[2–335] in LMNG/CHS micelles at 298 K, protein concentration ∼200 μM, mixing time = 600 ms. The cross-peaks C and C′ represent the exchange between P1 and P2, where exchange between pairs of substates is manifested by the fine structures of C and C′, with the components C1 to C4 and C1′ to C4′ (see text). The cross peaks E1, E1′, E2, and E2′ represent exchange among substates within the manifolds of P1 and P2. (F) One-dimensional cross-section along the dashed horizontal line in E. (G) One-dimensional cross-section along the dashed vertical line in E.
For 28 class A GRCRs, structures of complexes with both agonists and antagonists have been determined by X-ray crystallography or cryo-EM; these data reveal distinct structural differences between complexes with activating and inactivating ligands. Taking the distance between the α-carbon atoms of the residues 3.32 and 6.51 (Ballesteros–Weinstein notation) (11) as a measure for comparison of the dimensions of the orthosteric binding grooves in the active-like and inactive states, we find that the differences are in the range from 0.3 Å to 2.9 Å in these 28 GPCRs (Table 1). We now use the distance between the two trifluoromethyl groups of aprepitant (Fig. 1C) as a reference for estimating the amplitudes of transient fluctuations of the orthosteric binding site.
Table 1.
Comparison of the distance between the TM helices III and VI in X-ray crystal and cryo-EM structures of GPCR complexes with agonists and antagonists
| GPCR | PDB ID | Ligand* | TM III3.32† | TM VI6.51† | d (Å)‡ | Δd (Å)§ | Bound protein¶ |
|---|---|---|---|---|---|---|---|
| A1R | 6D9H | Adenosine | V87 | L250 | 14.1 | 1.2 | Gi |
| 5UEN | DU172 | 12.9 | — | ||||
| A2AAR | 6GDG | NECA | V84 | L249 | 14.5 | 1.4 | Nb35 |
| 4EIY | ZM241385 | 13.1 | — | ||||
| AT1R | 6DO1 | S1I8 | V108 | H256 | 13.3 | −0.6 | NbAT110i1 |
| 4YAY | ZD7155 | 13.9 | — | ||||
| AT2R | 5XJM | S1I8 | L124 | F272 | 15.3 | −0.3 | Fab |
| 5UNG | 8ES | 15.6 | — | ||||
| β1AR | 6H7J | Isoprenaline | D138 | F340 | 13.8 | 0.4 | Nb80 |
| 2VT4 | (S)-Cyanopindolol | 13.4 | — | ||||
| β2AR | 3SN6 | Bl-167107 | D113 | F289 | 14.2 | 0.5 | Gs |
| 2RH1 | Carazolol | 13.7 | — | ||||
| CB1 | 6N4B | MDMB-FUB | V196 | L359 | 14.4 | 1.9 | Gi |
| 5TGZ | AM6538 | 12.5 | — | ||||
| CB2 | 6KPF | AM841 | V113 | V261 | 14.0 | −0.7 | Gi |
| 5ZTY | AM10257 | 14.7 | — | ||||
| CCR5 | 7F1R | CCL5 | Y108 | Y251 | 15.4 | 0.3 | Gi |
| 4MBS | Maraviroc | 15.1 | — | ||||
| D2R | 6VMS | Bromocriptine | D114 | F389 | 14.4 | 0.5 | Gi |
| 6LUQ | Haloperidol | 13.9 | — | ||||
| D3R | 7CMU | Pramipexole | D110 | F345 | 14.1 | 0.2 | Gi |
| 3PBL | Eticlopride | 13.9 | — | ||||
| δOR | 6PT2 | KGCHM07 | D128 | I277 | 14.5 | 1.2 | — |
| 4N6H | Naltrindole | 13.3 | — | ||||
| ETBR | 5GLH | Endothelin-1 | Q181 | L339 | 13.8 | −1.0 | — |
| 5X93 | K-8794 | 14.8 | — | ||||
| EP4R | 7D7M | Prostaglandin E2 | L99 | L288 | 12.6 | 2.2 | Gs |
| 5YWY | ONO-AE3-208 | 10.4 | — | ||||
| FFAR1 | 4PHU | TAK-875 | H86 | Y240 | 12.9 | −0.4 | — |
| 5KW2 | Propanoic acid | 13.3 | — | ||||
| H1R | 7DFL | Histamine | D107 | Y431 | 13.5 | −0.7 | Gq |
| 3RZE | Doxepin | 14.2 | — | ||||
| κOR | 6B73 | MP1104 | D138 | I290 | 14.1 | 1.4 | Nb39 |
| 4DJH | JDTic | 12.7 | — | ||||
| M2R | 4MQS | Iperoxo | D103 | Y403 | 14.0 | −0.6 | Nb9-8 |
| 3UON | (R)-QNB | 14.6 | — | ||||
| MC4R | 7AUE | Aminoserine | I129 | F261 | 13.7 | 0.6 | Gs |
| 6W25 | SHU9119 | 13.1 | — | ||||
| MT1AR | 7DB6 | Ramelteon | M107 | L254 | 15.2 | 1.1 | Gi |
| 6PS8 | 2-phenylmelatonin | 14.1 | — | ||||
| μOR | 6DDF | DAMGO | D149 | I296 | 14.1 | 0.7 | Gi |
| 4DKL | BF0 | 13.4 | — | ||||
| NK1R | 7RMH | SP | P112 | F264 | 15.6 | 2.9 | Gs |
| 6J20 | aprepitant | 12.7 | — | ||||
| OX2R | 7L1U | Orexin-A | Q134 | I320 | 13.7 | 0.8 | Gs |
| 5WQC | EMPA | 12.9 | — | ||||
| P2Y12R | 4PXZ | 2-MeSADP | F104 | F252 | 11.8 | −0.3 | — |
| 4NTJ | AZD1283 | 12.1 | — | ||||
| 5-HT1BR | 6G79 | Donitriptan | D129 | F330 | 13.2 | −0.9 | Go |
| 5V54 | Methiothepin | 14.1 | — | ||||
| 5-HT2AR | 6WHA | 25-CN-NBOH | D155 | F339 | 15.0 | 1.1 | Gq |
| 6A94 | Zotepine | 13.9 | — | ||||
| 5-HT2BR | 5TUD | Ergotamine | D135 | F340 | 13.4 | −0.5 | — |
| 6DRZ | Methysergide | 13.9 | — | ||||
| 5-HT2CR | 6BQG | Ergotamine | D134 | F330 | 13.6 | −0.7 | — |
| 6BQH | Ritanserin | 14.3 | — |
*Orthosteric ligand bound; for each receptor the agonist is listed first.
†Ballesteros–Weinstein notation of the residues in TM III and TM VI between which the Cα–Cα distance was evaluated.
‡Distance between the α-carbons of residues 3.32 and 6.51.
§Difference between the two structures of each receptor; negative numbers represent that the distance in the agonist-bound structure is smaller than in the antagonist-bound structure.
¶Intracellular partner protein bound in a tertiary complex with GPCR and an orthosteric ligand; the structures of all but one of the tertiary complexes resulted from cryo-EM studies.
The experiments used are based on the absence of periodicity in the distribution of atom groups across the interior of folded proteins, which results in nonuniform magnetic microsusceptibility experienced by pairs of chemically identical substituents across aromatic rings (12, 13); for NK1R-bound aprepitant, this results in a 1.5-ppm chemical-shift difference between the signals of the two trifluoromethyl groups (P1 and P2 in Fig. 1D) (9), whereas these trifluoromethyl groups are not distinguishable in solution or when bound to detergent micelles (M in Fig. 1D). Exchange of symmetry-related substituents of aromatic rings by 180° rotational flipping motions about the single bond, by which they are attached to the remainder of the chemical structure (Fig. 1C), connects two nondistinguishable conformational states of the rings; depending on the frequency of these “ring-flipping” events, the line shapes of the NMR signals of the ring substituents can be widely different. In soluble globular proteins, such flipping motions have been observed for phenylalanine and tyrosine rings, with frequencies over the range from about 1 s−1 to larger than 108 s−1 (14–17). Ring-flipping motions of phenylalanine and tyrosine manifest transient structure fluctuations with an amplitude of about 1.5 Å (12). For the trifluoromethyl-substituted aromatic ring in aprepitant (Fig. 1C), similar ring-flipping would manifest transient structure fluctuations with an amplitude of 6 to 8 Å. Using 19F-NMR (10), we explored the possibility that such large-amplitude structural fluctuations occur in the orthosteric binding groove of NK1R in solution at ambient temperature.
Results and Discussion
The main line of 19F-NMR experiments was pursued with aprepitant complexes of a truncated wild-type NK1R, NK1R[2–335], reconstituted either in lauryl-maltose-neopentyl-glycol (LMNG)/cholesteryl hemisuccinate (CHS) mixed micelles, or in MSP1D1 nanodiscs. The construct NK1R[2–335] contains the additional N-terminal residues GP and the additional C-terminal residues ENLYFQ, due to the engineered PPase and tobacco etch virus cleavage sites, respectively (SI Appendix, Fig. S1A). Most of the protein samples were expressed in Pichia pastoris, which provided higher yields than the more commonly used insect cell Sf9 expression system. For comparison, we also expressed the variant protein NK1R[2–335, E782.50N], which includes the single mutation E782.50N, and a fusion protein with modified T4 lysozyme in the intracellular loop 3, NK1R[2–335, E782.50N]–mT4L, which was previously used for crystal structure determination and exploratory 19F-NMR studies (9). SDS/PAGE and analytical size-exclusion chromatography were used to check that the NMR samples were monodisperse and homogenous (SI Appendix, Figs. S1 and S2).
Conformational Polymorphisms in the NK1R–Aprepitant Complexes.
The 19F-NMR spectrum of the NK1R[2–335]–aprepitant complex in mixed micelles of LMNG and CHS at 298 K (Fig. 1D) shows two well-separated signals centered at −61.6 ppm and −62.9 ppm, which correspond to the two trifluoromethyl groups of the bound aprepitant (Fig. 1C). The 4-fluoro-phenyl group shows a somewhat broadened resonance at −113.2 ppm (SI Appendix, Fig. S3), which was not further analyzed here because the structure of aprepitant (Fig. 1C) implies that this fluorine atom is not affected by flipping motions of the di-trifluoromethyl-substituted ring. Ring current shifts of the trifluoromethyl groups were calculated with the program MOLMOL (18) from the atomic coordinates of the crystal structure in Fig. 1 A and B (9); they showed that a large shift for one of the trifluoromethyl groups is due to the aromatic residue W261 in the binding groove. The 19F-NMR signal P1 (Fig. 1D) was thus assigned to the trifluoromethyl group located near the transmembrane helix (TM) II, the signal P2 to the trifluoromethyl group located near TM V (Fig. 1 A and B), and the signal M to “free” aprepitant in the detergent micelles; these assignments coincide with earlier studies of the variant fusion protein NK1R[2–335, E782.50N]–mT4L (9).
So far, the analysis of the 19F-NMR spectrum of Fig. 1D confirms conclusions that were previously drawn from studies with the fusion protein NK1R[2–335, E782.50N]–mT4L (8). The earlier investigation had further provided evidence for the occurrence of conformational substates in the NK1R complex with aprepitant. Based on the greatly improved quality of the NMR spectra obtained in the present study, which resulted from the lower molecular weight of the NK1R construct used as well as from the high expression yield in P. pastoris, the analysis of the NMR data can now be reliably extended. The deconvolution of the spectrum in Fig. 1D reveals that P1 and P2 each include two main substates, a and b, and a less-populated substate c, with relative populations of about 45%, 45%, and 10% (SI Appendix, Table S1). The observation of two separate signals for P1 and P2, and of separate, quite narrow signals for the three substates in each of P1 and P2, shows that the exchange among these locally different structures is slow on the chemical-shift time scale (i.e., exchange rates must be slower than 103 s−1). While the chemical-shift difference between the signals P1 and P2 is due to different microsusceptibilities at the positions of the two trifluoromethyl groups of aprepitant in the NK1R orthosteric ligand binding groove, the substates within each of P1 and P2 most likely correspond to different rotational states about one or multiple single bonds preceding the bond which links the trifluoromethylphenyl ring to the skeleton of aprepitant (Fig. 1C).
Ring Flips of the Aprepitant Di-trifluoromethylphenyl Ring Reveal Large-Amplitude Structure Fluctuations of NK1R.
In the two-dimensional (2D) [19F,19F]-EXSY experiment with the NK1R[2–335]–aprepitant complex (Fig. 1E), the mixing time was set to 0.6 s in order to generate strong cross-peaks (19). Cross-peaks between P1 and P2 reveal exchange of the two trifluoromethyl groups of aprepitant. The fine structures of the cross-peaks further manifest exchange between the major substates a and b of P1 and P2 (Fig. 1E). The cross-section along the horizontal dashed line represents the exchange peaks of P2b with P1a and P1b (C1 and C2 in Fig. 1F). Similarly, the cross-peaks of P1b with P2b and P2a are seen along the vertical cross-section (C4 and C2 in Fig. 1G). The cross-sections further show the exchange peaks E2 and E1′ between substates within P1 and P2.
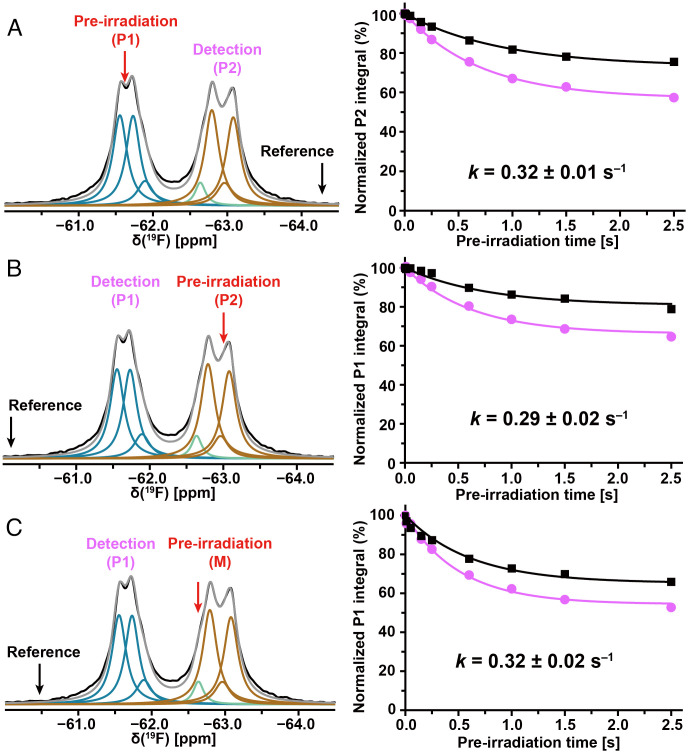
Additional information on the exchange between P1 and P2 was obtained with one-dimensional (1D) 19F saturation transfer NMR experiments (Fig. 2 A and B). In the experiment of Fig. 2A, the preirradiation was at −61.5 ppm (P1, red arrow in Fig. 2A), detection at −62.8 ppm (P2), and a reference was recorded with preirradiation at −64.1 ppm (black arrow in Fig. 2A). The preirradiation times were varied over the range 0.01 to 2.50 s. Plots of the normalized peak integrals of P1 versus the preirradiation time (Fig. 2A) were used for a quantitative analysis with the Bloch–McConnell formalism (20, 21), based on the longitudinal spin relaxation times (T1) of 0.94 ± 0.01 s and 1.23 ± 0.01 s for P1 and P2, respectively (SI Appendix, Fig. S4). An exchange rate of 0.32 ± 0.01 s−1 between P1 and P2 was obtained (Fig. 2A), and the reverse experiment yielded an exchange rate of 0.29 ± 0.02 s−1 (Fig. 2B).
Fig. 2.
Large-amplitude structure fluctuations of NK1R[2–335] in mixed micelles of LMNG and CHS observed at 298 K by 19F saturation transfer between the two trifluoromethyl groups of bound aprepitant. (A–C) In each experiment, the red arrow indicates the carrier position for the preirradiation, and the black arrow the position of the reference irradiation; the detection position is indicated with purple lettering. Lorentzian deconvolution of the 1D 19F-NMR spectra is used to identify a minimal number of overlapping signals that provide a quantitative fit of the experimental data. The plots on the right show the normalized integrals of the observed peak at different preirradiation times on-resonance and at the reference position. The Bloch–McConnell equation was used to fit the experimental data and derive the exchange rates.
Exchange rates among the substates a and b of P1 and P2 (Fig. 1D) were investigated using the 2D [19F,19F]-EXSY data (Fig. 1 E–G). The observation of separate subpeaks within P1 and P2 establishes an upper limit for the exchange among the corresponding substates within P1 and P2 of k ≤ 6 × 102 s−1. The analysis of the EXSY spectrum (Fig. 1 E–G) further showed that the exchange rates between P1a and P1b (E1, E1′) and between P2a and P2b (E2, E2′) at 298 K are 6.2 s−1 and 7.0 s−1 (SI Appendix, Eq. S1), respectively. Compared with E1 and E2, the cross-peaks C1 to C4 have lower intensities (Fig. 1 F and G), showing that the ring flips occur at lower frequencies than the exchange between the substates. Exchange rates between P1a and P2a (C3, C3′), P1a and P2b (C1, C1′), P1b and P2a (C4, C4′), and P1b and P2b (C2, C2′) were evaluated as 0.5, 0.4, 0.5, and 0.6 s−1, respectively. Exchange with the substate c (Fig. 1D) was not evaluated due to the low population of these substates.
Replacement of residue E782.50 by asparagine results in a nonfunctional variant of NK1R (22), but this variant provides higher expression yields and stability when compared to wild-type NK1R. The 19F-NMR spectra of NK1R[2–335, E782.50N] show similar substates a to c in P1 and P2 as NK1R[2–335] (Fig. 1D), but the population of the substate a is significantly smaller (SI Appendix, Fig. S5A and Table S1). One-dimensional 19F saturation transfer experiments with NK1R[2–335, E782.50N] in LMNG/CHS micelles were performed at a concentration of 260 μM and with similar preirradiation times to those in Fig. 2 A and B. The exchange rates of 0.36 ± 0.02 s−1 between P1 and P2 (SI Appendix, Fig. S5A) and 0.37 ± 0.04 s−1 for the reverse experiment (SI Appendix, Fig. S5B) are about 20% faster than for NK1R[2–335]. The 2D [19F,19F]-EXSY experiment with NK1R[2–335, E782.50N] in LMNG/CHS micelles (SI Appendix, Fig. S5 C–E) contains cross-peaks between P1 and P2 with more clearly resolved fine structures than in NK1R[2–335] (Fig. 1 E–G). The quantitative data analysis based on T1 measurements (SI Appendix, Fig. S6) yielded exchange frequencies between P1a and P2a (C3, C3′ in SI Appendix, Fig. S5), P1a and P2b (C1, C1′), P1b and P2a (C4, C4′), and P1b and P2b (C2, C2′) of 1.1, 1.3, 0.9, and 1.2 s−1, respectively, which corresponds to about twice the corresponding values for NK1R[2–335].
Exchange between Free and Protein-Bound Aprepitant Did Not Affect the 19F-NMR Measurements of Intramolecular Rate Processes in NK1R.
Because of the spectral overlap of the signals M and P2 (Fig. 1D), exchange between free and protein-bound aprepitant might contribute to the cross-peak intensities in 2D [19F,19F]-EXSY spectra (Fig. 1E) and invalidate the results of saturation transfer experiments (Fig. 2). In an initial check on possible effects from aprepitant exchange, the saturation transfer measurement of Fig. 2B was repeated with the preirradiation centered on the signal M at −62.6 ppm (Fig. 2C). The ring flip rate between P1 and P2 thus obtained, 0.32 ± 0.02 s−1, coincides with the result of Fig. 2A. The preirradiation of M in concert with P2 in the experiment of Fig. 2B thus did not interfere with the measurement of the ring-flip rates. This observation was confirmed by two different experimental approaches. First, we noticed that the temperature dependence of the chemical shift for M is larger than that for P2 (SI Appendix, Fig. S7). Therefore, the signals M and P2 are better resolved at lower temperature, where the 2D [19F,19F]-EXSY spectrum of the NK1R[2–335]–aprepitant complex showed no cross-peak between P1 and M (SI Appendix, Fig. S8 A and B). The micelle-bound “free” aprepitant did thus not interfere with the 2D [19F,19F]-EXSY measurements of the ring-flip rates between P1 and P2. Second, we compared 19F saturation transfer experiments with preirradiation on M and detection at P1 performed at different concentrations of “free” aprepitant (SI Appendix, Fig. S8 C and D). A twofold increase of the concentration of aprepitant in a solution of the NK1R[2–335]–aprepitant complex did not result in an intensity change at position P1, showing again that the micelle-bound “free” aprepitant did not affect the measurements of the ring-flip frequencies.
Large-Amplitude NK1R Dynamics Is Maintained in Different Detergent Micelles and in Nanodiscs.
One-dimensional 19F-NMR spectra of NK1R[2–335, E782.50N] in n-dodecyl-β-D maltopyranoside (DDM)/CHS showed similar chemical-shift dispersion as in LMNG/CHS micelles (SI Appendix, Fig. S9 and Table S1). The average line widths for the P1 and P2 substates of NK1R[2–335, E782.50N] in DDM/CHS or LMNG/CHS micelles are 170 Hz and 160 Hz, respectively, and thus not significantly different. The exchange rates between P1 and P2 of 0.25 ± 0.04 s−1 and 0.46 ± 0.05 s−1 measured by 1D 19F saturation transfer experiments (SI Appendix, Fig. S9) differ only slightly from those in LMNG/CHS (SI Appendix, Fig. S5); the smaller value measured with P2 irradiation was possibly due to lesser irradiation efficiency caused by the wider dispersion of the chemical shifts among the P2 substates (SI Appendix, Fig. S9).
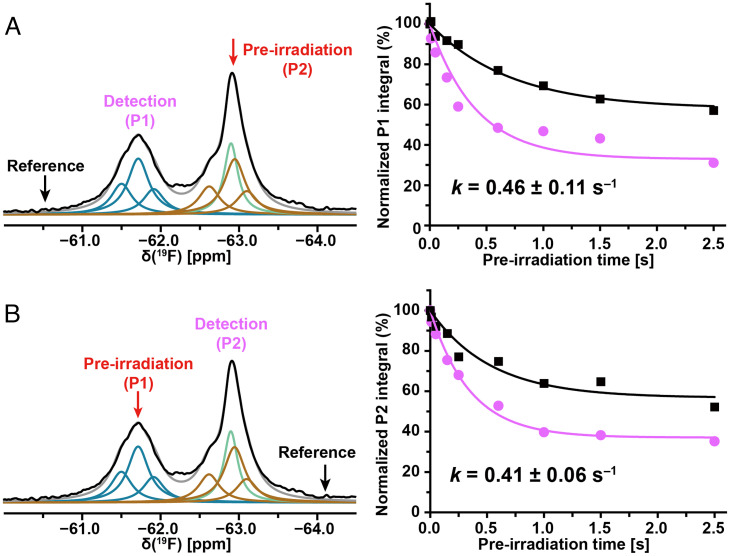
The 19F-NMR spectra of the aprepitant–NK1R[2–335] complex in nanodiscs showed similar chemical-shift dispersion to that in detergent micelles (Fig. 3 and SI Appendix, Table S1). The line widths of the substates in the nanodiscs sample are about 180 Hz, as compared to 130 Hz in the detergent micelles. This difference may reflect the larger size of the nanodiscs, but this was not confirmed with NK1R[2–335, E782.50N] (SI Appendix, Table S1). Exchange rates for the aprepitant complex of NK1R[2–335] in nanodiscs, as measured with 19F saturation transfer experiments, are about 40% faster than those in detergent micelles (Figs. 2 and 3). For the NK1R[2–335, E782.50N]–aprepitant complex in nanodiscs, the exchange rates between P1 and P2, 0.36 ± 0.03 s−1 and 0.33 ± 0.05 s−1 for preirradiation at P2 and P1, respectively, are nearly identical to those in LMNG/CHS micelles (SI Appendix, Fig. S5).
Fig. 3.
Large-amplitude structure fluctuations of NK1R[2–335] in nanodiscs at 298 K detected by 19F-NMR observation of the bound drug aprepitant. The color code used for the Lorentzian deconvolution is presented the same as in Fig. 1D. A green line represents nanodiscs-associated “free” aprepitant. (A and B) 19F-NMR saturation transfer between the trifluoromethyl groups of aprepitant bound to NK1R[2–335]. Same presentation as in Fig. 2.
Conclusions
Structure determinations of GPCRs, and in particular class A GPCRs, have long implicated function-related structural plasticity in this large-membrane protein family. Differences in the orientation of individual TM helices between complexes with agonists and antagonists have been widely discussed; systematic examination of the structures shows that these conformational changes are associated with rather small variations in the dimensions of the orthosteric ligand binding groove (Table 1). In the crystal structures, the hand-shaped binding groove has a thickness of about 3 Å along a height of 14 Å (SI Appendix, Fig. S11). In addition to the information obtained from comparison of crystal and cryo-EM structures at different stages of activation, spectroscopy techniques and molecular dynamics simulations revealed local conformational polymorphisms and rate processes associated with conformational exchange between two or multiple simultaneously populated structural states (23–38). The present paper adds insights, showing that infrequent large-amplitude fluctuations are part of the NK1R structure, and indicating that this feature might be common to class A GPCRs.
In the complex with NK1R, the two trifluoromethyl groups of aprepitant (Fig. 1C) are in van der Waals contact with the residues F2646.51 and P1123.32 (Fig. 1 A and B). During a ring-flipping motion, the distance between these two residues must transiently expand by 6 to 8 Å. These large-amplitude structure fluctuations are observed in the complexes of aprepitant with NK1R[2–335], as well as in the complexes with the NK1R variants NK1R[2–335, E782.50N] and NKIR[2–335, E782.50N]–mT4L in solution at ambient temperature (Figs. 1–3 and SI Appendix, Figs. S5 and S10). The ring-flipping frequencies are similar for these three different proteins, and essentially identical in two different detergent micelles and in nanodiscs (Figs. 2 and 3, and SI Appendix, Figs. S9 and S10). We conclude that the observed large-amplitude structure fluctuations are an inherent feature of NK1R, which is likely to prevail also in the physiological membrane environment at ambient temperatures.
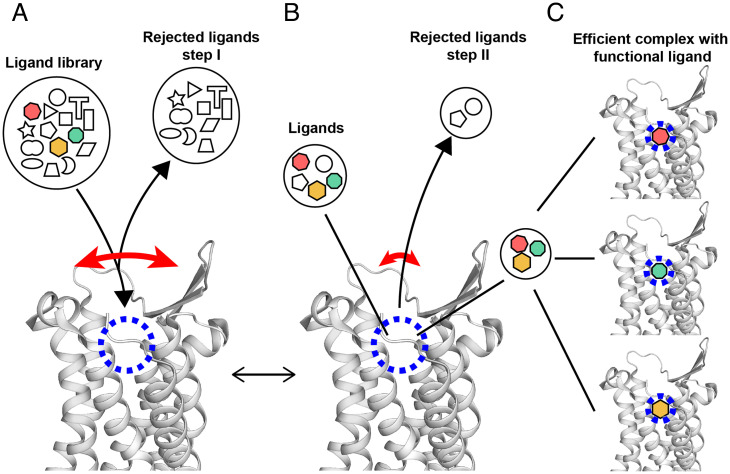
Overall, the results presented in this paper add a new perspective to the wealth of information that is already available on structural differences between different activation states of GPCRs (Table 1), and on intramolecular polymorphisms and rate processes observed by spectroscopic techniques and molecular dynamics simulations (23–38). The present spatio-temporal characterization of the orthosteric ligand-binding site reveals unprecedented large amplitudes for infrequent structure fluctuations. Model considerations indicate that the large fluctuations of the distance between the NK1R residues F2646.51 and P1123.32 go along with comparable fluctuations of the entrance to the orthosteric binding site (SI Appendix, Fig. S11). This then leads us to interpret these observations by a multistep selection mechanism of function-related ligand binding to NK1R (Fig. 4), which might be quite generally applicable to class A GPCRs.
Fig. 4.
Hypothetical multistep selection of functional orthosteric ligands by GPCRs based on the NMR observation of transient large-amplitude structure fluctuations and observations with crystallization assays (39). A side-view of the extracellular part of the NK1R crystal structure (PDB ID code 6J20) is shown. The long curved red arrow indicates large transient openings of the ligand-binding groove. The short red arrow indicates a closed conformation of the GPCR, which is populated most of the time. The blue dashed circle indicates the location in the binding groove where ligands interact with specific amino acid residues of the GPCR. The black lines refer symbols to locations in the NK1R structure. Black arrowheads indicate flows of ligands leading to equilibrium at given concentrations of NK1R and ligands, solution conditions and temperature. (A) Initial screening of ligands by size and shape. (B) Selection of high-affinity ligands within the binding groove. (C) Different functional ligands are specifically bound to the receptor, yielding different functional states. Depending on the availability of such, ligands in the solution milieu (colored symbols in A) and their relative binding affinities, one of the ligands may be predominantly bound and imprint its efficacy on GPCR.
The proposed multistep selection of functional orthosteric ligands by GPCRs is outlined in Fig. 4. In a first step, transient openings of the ligand binding groove provide a basis for selecting potential functional ligands on the basis of size and shape. In a second step, these initially accepted ligands are screened within the binding groove for specific interactions with amino acid residues of the GPCR. The nonfunctional ligands are released during subsequent large-amplitude openings of the ligand binding groove, and the functional ligands are stabilized in the binding groove.
In proposing the model of Fig. 4, we considered the experiences of others when pursuing crystallization trials with GPCRs, as surveyed in Zhang et al. (39). Success of a crystal structure determination has been achieved for complexes with a very limited selection of ligands, which were typically taken from libraries of drug candidates. In our model, all ligands bound to a given GPCR would be in the state characterized by Fig. 4B. Only the targeted functional ligands would interact in specific ways with the protein and proceed to a state corresponding to Fig. 4C. In the practice of GPCR crystallography, the bound ligands are evaluated by the extent to which they increase the thermal stability of the receptor when compared to the apo-form; thereby, the fact that “poor” ligands also increase the thermal stability, although to a lesser extent, indicates that they bind in a related but less-specific manner than the successful ligands. Considering that the structure fluctuations manifested by the flipping motions of the di-trifluoromethyl–substituted ring of aprepitant (Fig. 1C) are an intrinsic property of NK1R and are not induced by the presence of the ligand, the considerations of the model in Fig. 4 should apply generally to class A GPCRs, and possibly even to GPCRs of all classes.
Materials and Methods
NK1R and variants thereof were expressed in P. pastoris Bg12 or in Sf9 insect cells, purified by immobilized metal-affinity chromatography, and analyzed by analytical size-exclusion chromatography and SDS/PAGE. NMR data were collected on a Bruker Avance 600 MHz spectrometer equipped with a cryoprobe. 19F-NMR spectra were Lorentzian deconvoluted by MestReNova, and resulting data were analyzed by Origin Pro-2019. Details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Shuanghong Chen, Prof. Qiang Zhao, and Prof. Beili Wu from the Shanghai Institute of Materia Medica for providing the starting insect cell expression plasmid; and the Cloning, Insect Cell Expression, Protein Purification, and NMR Core Facilities of the iHuman Institute for their support. D.L. and L.Y. acknowledge funding from the National Natural Science Foundation of China (31971153 and 31670733 to D.L.; 21904088 to L.Y.). K.W. is the Cecil H. and Ida M. Green Professor of Structural Biology at Scripps Research.
Footnotes
Reviewers: M.S., University of Maryland; G.W., Harvard Medical School; and D.W., University of California, Berkeley.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122682119/-/DCSupplemental.
Data Availability
All study data are included in the main text and SI Appendix.
References
- 1.Kramer M. S., et al. , Distinct mechanism for antidepressant activity by blockade of central substance P receptors. Science 281, 1640–1645 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Mantyh P. W., Substance P and the inflammatory and immune response. Ann. N. Y. Acad. Sci. 632, 263–271 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Harris J. A., et al. , Selective G protein signaling driven by substance P-neurokinin receptor dynamics. Nat. Chem. Biol. 18, 109–115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinhoff M. S., von Mentzer B., Geppetti P., Pothoulakis C., Bunnett N. W., Tachykinins and their receptors: Contributions to physiological control and the mechanisms of disease. Physiol. Rev. 94, 265–301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hale J. J., et al. , Structural optimization affording 2-(R)-(1-(R)-3, 5-bis(trifluoromethyl)phenylethoxy)-3-(S)-(4-fluoro)phenyl-4- (3-oxo-1,2,4-triazol-5-yl)methylmorpholine, a potent, orally active, long-acting morpholine acetal human NK-1 receptor antagonist. J. Med. Chem. 41, 4607–4614 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Hesketh P. J., et al. ; Aprepitant Protocol 052 Study Group, The oral neurokinin-1 antagonist aprepitant for the prevention of chemotherapy-induced nausea and vomiting: A multinational, randomized, double-blind, placebo-controlled trial in patients receiving high-dose cisplatin—The Aprepitant Protocol 052 Study Group. J. Clin. Oncol. 21, 4112–4119 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Yin J., et al. , Crystal structure of the human NK1 tachykinin receptor. Proc. Natl. Acad. Sci. U.S.A. 115, 13264–13269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schöppe J., et al. , Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists. Nat. Commun. 10, 17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S., et al. , Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography. Nat. Commun. 10, 638 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didenko T., Liu J. J., Horst R., Stevens R. C., Wüthrich K., Fluorine-19 NMR of integral membrane proteins illustrated with studies of GPCRs. Curr. Opin. Struct. Biol. 23, 740–747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballesteros J. A., Weinstein H., Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in Neurosci. 25, 366–428 (1995). [Google Scholar]
- 12.Wüthrich K., NMR of Proteins and Nucleic Acids (Wiley, New York, 1986). [Google Scholar]
- 13.Liu D., Wüthrich K., Ring current shifts in (19)F-NMR of membrane proteins. J. Biomol. NMR 65, 1–5 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Wüthrich K., Wagner G., NMR investigations of the dynamics of the aromatic amino acid residues in the basic pancreatic trypsin inhibitor. FEBS Lett. 50, 265–268 (1975). [DOI] [PubMed] [Google Scholar]
- 15.Wagner G., DeMarco A., Wüthrich K., Dynamics of the aromatic amino acid residues in the globular conformation of the basic pancreatic trypsin inhibitor (BPTI). I. 1H NMR studies. Biophys. Struct. Mech. 2, 139–158 (1976). [DOI] [PubMed] [Google Scholar]
- 16.Weininger U., Modig K., Akke M., Ring flips revisited: (13)C relaxation dispersion measurements of aromatic side chain dynamics and activation barriers in basic pancreatic trypsin inhibitor. Biochemistry 53, 4519–4525 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Martin B. T., Malmstrom R. D., Amaro R. E., Wüthrich K., OCRE domains of splicing factors RBM5 and RBM10: Tyrosine ring-flip frequencies determined by integrated use of 1H NMR spectroscopy and molecular dynamics simulations. ChemBioChem 22, 565–570 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Koradi R., Billeter M., Wüthrich K., MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Perrin C. L., Dwyer T. J., Application of two-dimensional NMR to kinetics of chemical exchange. Chem. Rev. 90, 935–967 (1990). [Google Scholar]
- 20.Forsén S., Hoffman R. A., Exchange rates by nuclear magnetic multiple resonance. III. Exchange reactions in systems with several nonequivalent sites. J. Chem. Phys. 40, 1189–1196 (1964). [Google Scholar]
- 21.Forsén S., Hoffman R. A., Study of moderately rapid chemical exchange reactions by means of nuclear magnetic double resonance. J. Chem. Phys. 39, 2892–2901 (1963). [Google Scholar]
- 22.Huang R. R., Yu H., Strader C. D., Fong T. M., Interaction of substance P with the second and seventh transmembrane domains of the neurokinin-1 receptor. Biochemistry 33, 3007–3013 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Liu J. J., Horst R., Katritch V., Stevens R. C., Wüthrich K., Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335, 1106–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalton J. A. R., Lans I., Giraldo J., Quantifying conformational changes in GPCRs: Glimpse of a common functional mechanism. BMC Bioinformatics 16, 124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sounier R., et al. , Propagation of conformational changes during μ-opioid receptor activation. Nature 524, 375–378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregorio G. G., et al. , Single-molecule analysis of ligand efficacy in β2AR-G-protein activation. Nature 547, 68–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solt A. S., et al. , Insight into partial agonism by observing multiple equilibria for ligand-bound and Gs-mimetic nanobody-bound β1-adrenergic receptor. Nat. Commun. 8, 1795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eddy M. T., et al. , Allosteric coupling of drug binding and intracellular signaling in the A2A adenosine receptor. Cell 172, 68–80.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sušac L., Eddy M. T., Didenko T., Stevens R. C., Wüthrich K., A2A adenosine receptor functional states characterized by 19F-NMR. Proc. Natl. Acad. Sci. U.S.A. 115, 12733–12738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frei J. N., et al. , Conformational plasticity of ligand-bound and ternary GPCR complexes studied by 19F NMR of the β1-adrenergic receptor. Nat. Commun. 11, 669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamichhane R., et al. , Biased signaling of the G-protein-coupled receptor β2AR is governed by conformational exchange kinetics. Structure 28, 371–377.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizumura T., et al. , Activation of adenosine A2A receptor by lipids from docosahexaenoic acid revealed by NMR. Sci. Adv. 6, eaay8544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imai S., et al. , Structural equilibrium underlying ligand-dependent activation of β2-adrenoreceptor. Nat. Chem. Biol. 16, 430–439 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Clark L. J., et al. , Allosteric interactions in the parathyroid hormone GPCR-arrestin complex formation. Nat. Chem. Biol. 16, 1096–1104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krug U., et al. , The conformational equilibrium of the neuropeptide Y2 receptor in bilayer membranes. Angew. Chem. Int. Ed. Engl. 59, 23854–23861 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang S. K., et al. , Delineating the conformational landscape of the adenosine A2A receptor during G protein coupling. Cell 184, 1884–1894.e1814 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Josephs T. M., et al. , Structure and dynamics of the CGRP receptor in apo and peptide-bound forms. Science 372, eabf7258 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Wang X., et al. , A genetically encoded F-19 NMR probe reveals the allosteric modulation mechanism of cannabinoid receptor 1. J. Am. Chem. Soc. 143, 16320–16325 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Zhang X., Stevens R. C., Xu F., The importance of ligands for G protein-coupled receptor stability. Trends Biochem. Sci. 40, 79–87 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.