Significance
We report on a deregulatory activity on microRNA (miRNA) biogenesis by the FMS-like tyrosine kinase 3 (FLT3)-internal tandem duplication (ITD) in acute myeloid leukemia. FLT3-ITD provides a divergent signal for concurrent and aberrant miR-155 up-regulation and miR-126 down-regulation via a series of miRNA–protein regulatory loops interconnected through SH2-containing inositol phosphatase 1 (SHIP1)/phosphor-protein kinase B (AKT)/Sprouty related EVH1 domain containing 1 (SPRED1) signaling. This divergent input signal eventually converges and amplifies an output signal for leukemic growth.
Keywords: acute myeloid leukemia, FLT3-ITD, miR-126, miR-155, AKT
Abstract
MiR-126 and miR-155 are key microRNAs (miRNAs) that regulate, respectively, hematopoietic cell quiescence and proliferation. Herein we showed that in acute myeloid leukemia (AML), the biogenesis of these two miRNAs is interconnected through a network of regulatory loops driven by the FMS-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD). In fact, FLT3-ITD induces the expression of miR-155 through a noncanonical mechanism of miRNA biogenesis that implicates cytoplasmic Drosha ribonuclease III (DROSHA). In turn, miR-155 down-regulates SH2-containing inositol phosphatase 1 (SHIP1), thereby increasing phosphor-protein kinase B (AKT) that in turn serine-phosphorylates, stabilizes, and activates Sprouty related EVH1 domain containing 1 (SPRED1). Activated SPRED1 inhibits the RAN/XPO5 complex and blocks the nucleus-to-cytoplasm transport of pre-miR-126, which cannot then complete the last steps of biogenesis. The net result is aberrantly low levels of mature miR-126 that allow quiescent leukemia blasts to be recruited into the cell cycle and proliferate. Thus, miR-126 down-regulation in proliferating AML blasts is downstream of FLT3-ITD–dependent miR-155 expression that initiates a complex circuit of concatenated regulatory feedback (i.e., miR-126/SPRED1, miR-155/human dead-box protein 3 [DDX3X]) and feed-forward (i.e., miR-155/SHIP1/AKT/miR-126) regulatory loops that eventually converge into an output signal for leukemic growth.
MicroRNAs (miRNAs) are small noncoding RNAs that concurrently control the expression of multiple target genes directly and through networks that integrate the functions of their respective targets and other noncoding RNA species to regulate cellular homeostasis and functions (1). MiRNAs are initially transcribed from their genomic loci as pri-miRNAs and are converted by Drosha ribonuclease III (DROSHA) into pre-miRNAs that once transported from the nucleus to the cytoplasm are processed by endoribonuclease Dicer (DICER) into mature miRNA. MiRNAs have been found to be deregulated in cancer and leukemia and to contribute to disease initiation and maintenance.
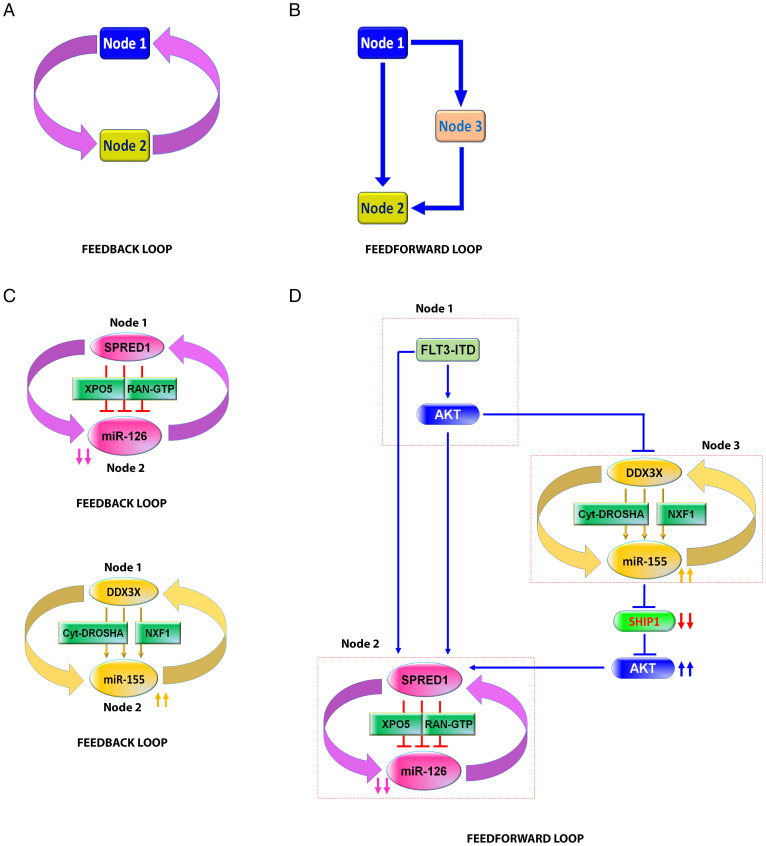
Acute myeloid leukemia (AML) is a clinically and genetically heterogeneous myeloid malignancy characterized by differentiation arrest and the proliferation of transformed hematopoietic cells that leads to rapid bone marrow (BM) failure. The biologic, prognostic, and therapeutic roles of aberrantly expressed miRNAs in AML have been previously reported (2–4). However, while most of the studies have focused on the mechanistic contributions of individual miRNAs to leukemia initiation and growth, miRNAs generate a much more complex array of interactions that involve their own multiple messenger RNA (mRNA) targets and other species of noncoding RNAs (2–4). To this end, Anastasiadou et al. recently reviewed the principles of noncoding RNA networks in cancer and described elements of the network theory that are applicable to the circuitous nature of the normal and aberrant miRNome dynamics (5). In this context, a feedback loop is defined as the interaction through which an miRNA (e.g., node 1) controls the expression of target genes (e.g., node 2), which in return control the expression of their own targeting miRNA (schematic model in Fig. 1A); a feed-forward loop instead exemplifies an interaction where an miRNA directly (nodes 1 and 2) and indirectly controls the expression of a target through an intermediate node (e.g., node 3), in which other miRNAs or proteins also participate (Fig. 1B). Herein, we applied these concepts to dissect the interplay of two miRNAs (i.e., miR-126 and miR-155) that have been found to be consistently altered in AML blasts carrying the FMS-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD) (Fig. 1 C and D) and that seems to play a pivotal role for FLT3-ITD AML growth (6–12).
Fig. 1.
Schematic models of FLT3-ITD–regulated miR-126 and miR-155 expression through feedback and feed-forward loops network. (A and B) Examples of feedback (A) and feed-forward (B) loops. (C) Top, a feedback loop of miR-126 and SPRED1. Bottom, a feedback loop of miR-155 and DDX3X. (D) Feed-forward loop of FLT3-ITD–regulated miR-126 expression through miR-155/SHIP1/AKT axis. Node 1, FLT3-ITD/AKT; node 2, SPRED1/miR-126; node 3, DDX3X/miR-155. GTP, guanosine-5'-triphosphate.
FLT3-ITD is one of the most common mutations in AML that confers growth and survival advantages to leukemic blasts. The mutant protein has been associated with a high expression of miR-155 to produce a highly proliferative leukemic phenotype (12–15). Recently, we showed that FLT3-ITD also associates with low-expression miR-126, which reportedly controls the quiescence and self-renewal of normal stem cells and leukemia stem cells (10, 11). FLT3-ITD decreases miR-126 levels by interfering with pre-miR-126 processing via the phosphorylation of Sprouty related EVH1 domain containing 1 (SPRED1), itself a miR-126 target (10, 11). In fact, FLT3-ITD phosphorylates SPRED1, which in turn binds and inhibits the RAs-related nuclear protein (RAN)/Exportin-5 (XPO5) complex. The latter regulates the transport of pre-miRNAs from the nucleus to the cytoplasm for the last step of miRNA maturation. Thus, the FLT3-ITD-dependent SPRED1 phosphorylation decreases the production of mature miR-126 (10, 11) by interrupting the canonical transport mechanisms of pre-miR-126 to the cytoplasm (Fig. 1 C, Top). Of note, as RAN/XPO5 also controls miR-155 biogenesis, we postulated and proved noncanonical mechanisms of miRNA biogenesis that circumvent the FLT3-ITD self-imposed block of the RAN/XPO5 complex and lead to the high levels of mature miR-155 observed in FLT3-ITD AML blasts. To this end, we showed that in FLT3-ITD AML blasts, miR-155 follows a different maturation path with respect to other miRNAs (i.e., noncanonical miRNA biogenesis) by virtue of being initially transcribed as a long noncoding RNA (i.e., lnc-RNA BIC-155; hereafter called BIC-155) (11). In fact, FLT3-ITD enhances BIC-155 transcription while also inhibiting the human dead-box protein 3 (DDX3X)-dependent splicing of BIC-155. The net result is an abundant unspliced BIC-155 that binds to nuclear RNA export factor 1 (NXF1) and is transported to the cytoplasm, where it is processed by the recently discovered cytoplasmic DROSHA into pre-miR155 and in turn by Dicer into mature miR-155 (11). Thus, high levels of miR-155 occur in FLT3-ITD AML blasts despite the RAN/XPO5 complex blockage (Fig. 1 C, Bottom).
Given the emerging concept of the circuitous nature of the miRNome (5), it is then logical to ask how these two different “paths” of miRNA biogenesis are interconnected to ensure that a seemingly divergent FLT3-ITD input signal for concurrent up- and down-regulation of miRNAs eventually converges into a unified output signal for leukemia growth. To this end, we show that the convergence of the FLT3-ITD split signal is assured by the upstream and prioritized regulatory activity of miR-155 over the miR-126 biogenesis via a complex circuit of miRNA–protein interactions that is regulated by the SH2-containing inositol phosphatase 1(SHIP1)/phosphor-protein kinase B (AKT) (p-AKT)/SPRED1 axis (Fig. 1D). Interestingly, this functional network is not unique to leukemogenesis but is rather hijacked from normal hematopoietic cells, which under physiologic conditions respond similarly to FLT3-ligand (FLT3-L) stimulation and/or inflammation.
Results
FLT3-ITD Splits Signal for Concurrent Up-Regulation of miR-155 and Down-Regulation of miR-126.
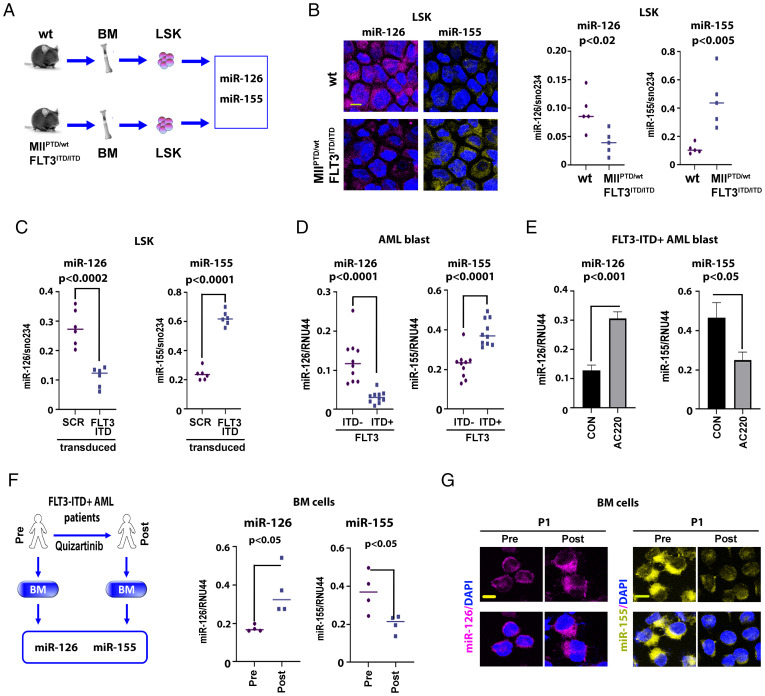
Using the MllPTD/wt/Flt3ITD/ITD AML mouse that is similar to its human counterpart disease (16) (Fig. 2A), we showed in vivo that miR-126 and miR-155 were respectively down- and up-regulated in Lin−Sca-1+c-Kit+ (LSK) cells compared to control normal wild-type (wt) mice, as demonstrated by live miRNA staining with SmartFlare probes and qPCR (Fig. 2B and SI Appendix, Fig. S1). These observations were recapitulated with the transduction of normal murine LSK cells with lentiviral vectors carrying FLT3-ITD, which decreased miR-126 levels and increased miR-155 levels compared with control vector-transduced cells (Fig. 2C). We further corroborated these results in human blasts by comparing miR-126 and miR-155 expression levels in primary BM CD34+ cells from FLT3-ITD+ (n = 10) and FLT3-ITD− AML (n = 10) patient samples (Fig. 2D). Levels of miR-126 and miR-155 were respectively and significantly lower and higher in FLT3-ITD+ samples compared with FLT3-ITD− samples. Conversely, pharmacologic ex vivo inhibition of FLT3-ITD with the tyrosine kinase inhibitor (TKI) AC220 (quizartininb; [17]) increased miR-126 expression and decreased miR-155 expression in FLT3-ITD+ (human) AML blasts (Fig. 2E). These results were also corroborated in vivo by the observation of increased miR-126 and decreased miR-155 in samples from FLT3-ITD+ AML patients treated with AC220 (Fig. 2 F and G and SI Appendix, Fig. S2). The discordant expression levels between miR-126 and miR-155 in FLT3-ITD AML prompted us to hypothesize that the biogenesis of these two miRNAs was coregulated to provide a unified input signal for leukemia growth.
Fig. 2.
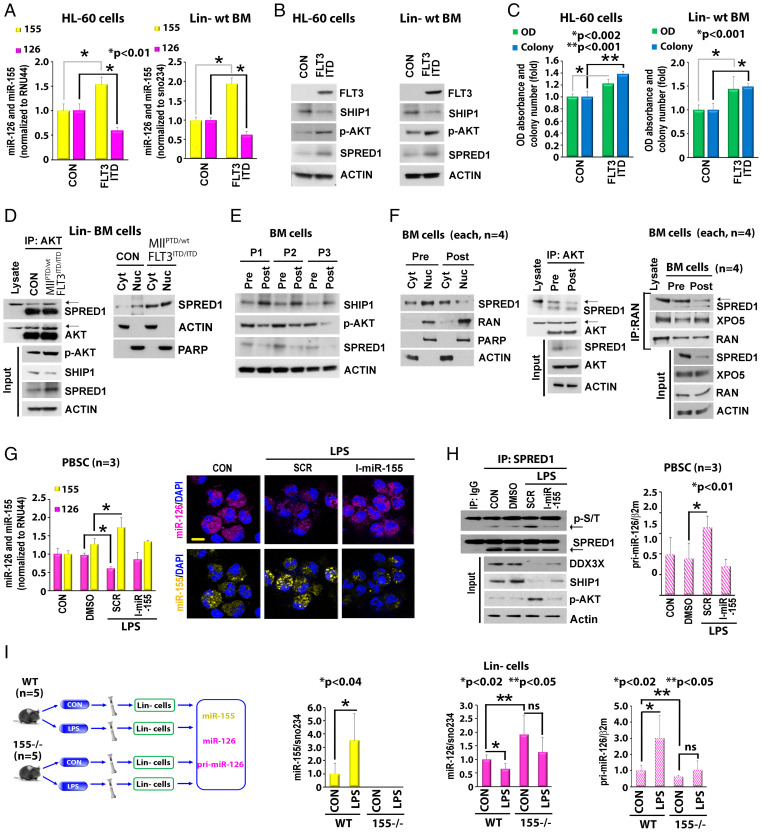
FLT3-ITD concurrently regulates miR-126 and miR-155 expression. (A and B) MiR-126 and miR-155 expression in LSK population from wt vs. MllPTD/wt/Flt3ITD/ITD AML mice. (A) Experimental design. (B, Left), representative images of cell staining with miR-126 and miR-155 SmartFlare probes. Right, miRNA expression was measured by qPCR (each group, n = 5). (C) LSK cells from wt mouse (n = 6) transduced with scramble control (SCR) or FLT3-ITD (FLT3) lentivirus vectors. Mir-126 (Left) and miR-155 (Right) expression levels were measured by qPCR at 48 h. (D) miR-126 (Left) and miR-155 (Right) expression in FLT3-ITD+ vs. FLT3-ITD− AML patients in BM primary CD34+ blasts. (E) miR-126 (Left) and miR-155 (Right) expression in CD34+ blasts from FLT3-ITD+ AML patients (n = 3) treated ex vivo with DMSO (CON) or the tyrosine kinase inhibitor AC220 (20 nM) measured by qPCR. (F and G) Effects of AC220 on miR-126 and miR-155 levels in vivo. (F, Left) In vivo experimental design. The BM cells from FLT3-ITD+ AML patients pre- or posttreated with AC220 (n = 4) were collected to analyze as indicated. Right, miR-126 and miR-155 expression levels were measured by qPCR. Individual value of miR-126 and miR-155 levels is shown in SI Appendix, Fig. S2. (G) Representative images of cell staining with miR-126 and miR-155 SmartFlare probes. More staining images of different samples are shown in SI Appendix, Fig. S2 (Scale bar, 10 μm).
miR-155 Is Upstream and Controls miR-126 Biogenesis.
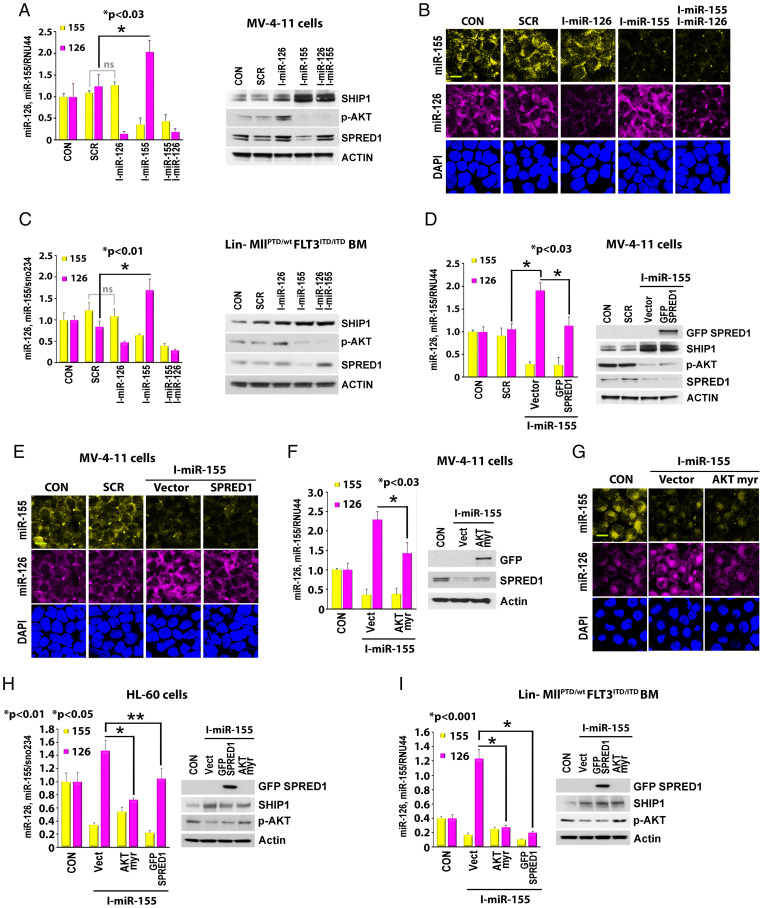
To dissect the coregulatory mechanisms of miR-155 and miR-126 biogenesis in FLT3-ITD AML blasts, we knocked down each of two miRNAs in FLT3-ITD+ FLT3-ITD+ MV-4-11 leukemic cells (MV-4–11) and assessed the reciprocal effect on their expression levels. Using qPCR assay and live miR-126 and miR-155 staining with SmartFlare probes (Fig. 3 A, Left, and Fig. 3B), we showed that miR-126 knockdown (KD) (10) produced no effect on miR-155 levels in MV-4–11 AML cells; conversely, miR-155 KD (18) increased the levels of miR-126. Similar results were observed when we analyzed the reciprocal changes of miR-126 and miR-155 after knocking down each individual miRNA in lineage-negative (Lin−) BM cells from MllPTD/wt/Flt3ITD/ITD AML mice (Fig. 3 C, Left), normal wt mice, and FLT3-ITD-HL-60 leukemic cells (HL-60) (SI Appendix, Fig. S3 A and B, Left).
Fig. 3.
Regulation of miR-126 by miR-155. (A–C) Effects of miR-155 inhibition on SHIP1/p-AKT/SPRED1 signaling and miR-126 expression. MV-4–11 cells (A and B) or Lin− BM cells from MllPTD/wt/Flt3ITD/ITD AML mice (C) were treated with either miR-126 inhibitor, miR-155 inhibitor alone, or combination for 24 h. (A and C) Left, expression levels of miR-126 and miR-155 by qPCR. Right, cell lysate was immunoblotted with indicated antibodies. (*, P value.) (B) Representative images of cell staining with miR-126 and miR-155 SmartFlare probes. (D–I) Effects of SPRED1 and AKT-myr overexpression (OE) on miR-155–regulated miR-126 expression. MV-4–11 cells (D–G), HL-60 cells (H), and Lin− BM cells from MllPTD/wt/Flt3ITD/ITD AML mice (I) were treated with miR-155 inhibitor in the presence or absence of SPRED1 or AKT-myr OE. (D, F, H, and I) Left, expression levels of miR-126 and miR-155 by qPCR. Right, cell lysate was immunoblotted with indicated antibodies. (*, P value.) (E and G) Representative images of cell staining with miR-126 and miR-155 SmartFlare probes (Scale bar, 10 μm).
miR-155 reportedly targets SH2-containing inositol phosphatase 1 (SHIP1) (19) (Fig. 3 A, C, and D and SI Appendix, Fig. S3 A and B, Right), a negative phosphatase that regulates the serine-threonine AKT kinase (20, 21). Of note, we observed that treatment with an miR-155 inhibitor (i.e., anti-miR-155) up-regulates SHIP1 and in turn down-regulates p-AKT and SPRED1 (Fig. 3 A, C, and D and SI Appendix, Fig. S3 A and B, Right). Since we have previously demonstrated that SPRED1 inhibits the RAN/XPO5 complex and significantly decreases miR-126 biogenesis (10, 11), we then postulated that the aberrant activation of AKT/SPRED1 signaling via SHIP1 down-regulation mediates miR-155–driven mechanisms of miR-126 down-regulation. To this end, we observed that an miR-155 inhibitor led to SHIP1 up-regulation, p-AKT and SPRED1 decrease (Fig. 3 A, C, and D, Right, and SI Appendix, Fig. S3 A and B, Right), and miR-126 increase (Fig. 3 A–E and SI Appendix, Fig. S3 A and B). Conversely, constitutively activated protein kinase B-myristoylated (AKT-myr) or forced overexpression of SPRED1 reversed the effects of the miR-155 inhibitor on miR-126 (Fig. 3 D–I). Furthermore, SHIP1 KD or the pharmacologic inhibition of SHIP1 by K118 (22) decreased miR-126 levels in FLT3-ITD+ MV-4–11 cells (SI Appendix, Fig. S3C, Left). We previously showed that the aberrant tyrosine kinases (TK; e.g., BCR/ABL and FLT3-ITD) down-regulate miR-126 and conversely that TKIs (nilotinib and AC220) increase miR-126 expression (10, 11). Herein, we observed that a combination of AC220 and K118 attenuated the AC220-dependent increase in miR-126 expression and enhanced apoptosis compared with either of the compounds used as single agents at concentrations lower than their respective half maximal inhibitory concentrations (IC50s) (SI Appendix, Fig. S3C, Middle and Right) (22–25). These results support the notion that the miR-155–mediated SHIP1 down-regulation is likely to contribute to lower levels of miR-126 in FLT3-ITD+ AML via AKT/SPRED1 axis activation. But how do these observations reconcile with the phosphorylated SPRED1–driven RAN/XPO5 inhibition (Fig. 1D) (11)?
AKT Phosphorylates SPRED1 and Regulates Its Protein Stability and Activity.
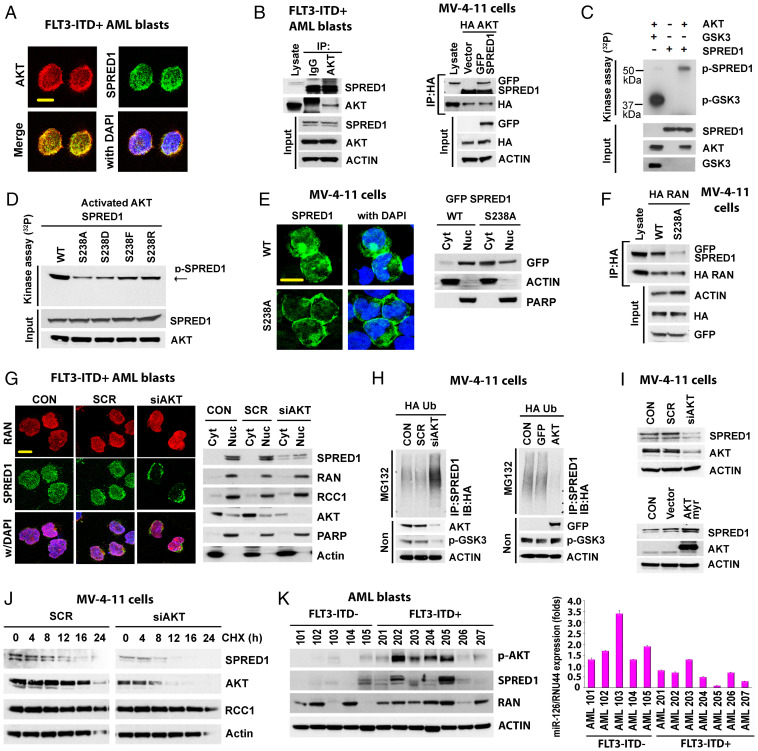
To this end, while SPRED1 is a reported TK substrate (10, 11), we observed that this protein also harbors a serine/threonine AKT phosphorylation motif at the serine 238 residue. Thus, we postulated that SPRED1 could be phosphorylated and activated at this site by the AKT kinase that, as described above, increases via miR-155–mediated SHIP1 down-regulation. Serine-phosphorylated SPRED1 could in turn inhibit the RAN/XPO5 complex (Fig. 1D).
To validate this hypothesis, first we proved that AKT and SPRED1 colocalized and physically interacted in FLT3-ITD+ AML cells (Fig. 4 A and B). The interaction of SPRED1 and AKT was also observed in Lin− BM cells from MllPTD/wt/Flt3ITD/ITD AML mice (SI Appendix, Fig. S4A) and in AKT and SPRED1-overexpressed FLT3-ITD− HL60 cells (SI Appendix, Fig. S4B). Second, using in vitro 32P labeling phosphorylation assays (26), we showed that AKT directly phosphorylated SPRED1 wt (Fig. 4C) but not the SPRED1 S238 mutant (Fig. 4D), thereby suggesting that SPRED1 S238 is a main substrate for p-AKT. Of note, in contrast to the wt protein that localized mainly in the perinuclear region, the ectopically expressed SPRED1 S238A mutant localized preferentially to the cytoplasm in FLT3-ITD+ MV-4–11 cells (Fig. 4E), Lin− BM cells from MllPTD/wt/Flt3ITD/ITD AML mice, and FLT3-ITD− HL-60 cells (SI Appendix, Fig. S4C). SPRED1 S238A also interacted less with RAN (Fig. 4F and SI Appendix, Fig. S4D) and had a decreased half-life compared to SPRED1 wt (SI Appendix, Fig. S4E). These results suggested that the serine phosphorylation of SPRED1 by p-AKT contributes to protein stabilization and activation. In fact, AKT KD or AKT pharmacologic inhibition by AZD8055, AKTi viii, or MK2206 inhibitors resulted in preferential SPRED1 cytoplasmic relocalization (Fig. 4G and SI Appendix, Fig. S4 F and G) and increased SPRED1 ubiquitination (Fig. 4H and SI Appendix, Fig. S4 H, Left) and degradation (Fig. 4 I, Top, and Fig. 4J and SI Appendix, Fig. S4 I, Top, and J); these effects were promptly reversed by the ectopic expression of the activated AKT-myr (Fig. 4H and SI Appendix, Fig. S4 H, Right, and Fig. 4I and SI Appendix, Fig. S4 I, Bottom). Of note, higher levels of p-AKT and SPRED1 were also observed in FLT3-ITD+ compared with FLT3-ITD− AML blasts (Fig. 4K).
Fig. 4.
AKT regulates SPRED1 phosphorylation and stability. (A and B) Colocalization and interaction of AKT and SPRED1. (A) FLT3-ITD+ AML blasts were stained with anti-AKT and anti-SPRED1 antibodies (Scale bar, 10 μm). (B) Left, protein lysate from FLT3-ITD+ AML blasts (n = 3) was immunoprecipitated with anti-IgG or anti-AKT antibody and immunoblotted with anti-SPRED1 antibody. Right, MV-4–11 cells were transfected with hemagglutinin (HA)-AKT and GFP-SPRED1 and immunoprecipitation and immunoblotting were performed as shown. (C) SPRED1 phosphorylation by AKT. Cell-free in vitro kinase phosphorylation assay using recombinant proteins of AKT, glycogen synthase kinase-3 (GSK3), and SPRED1 protein. AKT-phosphorylated GSK3 was used as a positive control for the phosphorylation assay. (D) AKT-phosphorylated SPRED1 at S238. Cell-free in vitro phosphorylation using recombinant proteins of AKT with SPRED1 wt or indicated mutation of SPRED1 at S238. (E) Cellular distribution of SPRED1 WT and SPRED1 S238A (non-AKT-phosphorylated mutation) in MV-4–11 cells. Left, representative images of cells overexpressed with SPRED1 wt or S238A (Scale bar, 10 μm). Right, indicated transfected cells were fractionated and immunoblotted with indicated antibodies. (F) Interaction between SPRED1 wt and SPRED1 S238A with RAN. MV-4–11 cells were transfected with HA-RAN and GFP-SPRED1 wt or GFP-SPRED1 S238A. Immunoprecipitation and immunoblotting as shown. (G) Effects of AKT KD on SPRED1 localization. MV-4–11 cells were transfected with scrambled control siRNA (siSCR) or AKT siRNA (siAKT) for 24 h. Left, transfected cells were stained with anti-RAN and anti-SPRED1 antibodies; representative images are shown (Scale bar, 10 μm). Right, fractionated lysate of transfected cells was immunoblotted with indicated antibody. PARP, Poly (ADP-ribose) polymerase; Cyt, Cytoplasm; Nuc, Nucleus; RCC1, Regulator Of Chromosome Condensation 1; (H) Effect of AKT KD and OE on SPRED1 ubiquitination. MV-4–11 cells were cotransfected with HA-ubiquitin and siSCR or siAKT (Left) and green fluorescent protein (GFP) vector or GFP-AKT (Right) for 24 h. Ubiquitination assay was performed with anti-SPRED1, anti-GFP, and anti-HA-ubiquitin antibodies. (I) Effect of AKT KD and OE on protein stability of SPRED1. MV-4–11 cells were transfected with siSCR and siAKT (Top) or HA vector control and HA-AKT (Bottom) for 24 h. Lysate from indicated transfected cells was immunoblotted with anti-SPRED1, anti-AKT, anti-HA, and anti-actin antibodies. (J) Effect of AKT KD on SPRED1 protein stability. MV-4–11 cells were transfected with siSCR or siAKT for 24 h. The cells were then treated with cycloheximide (CHX) (10 μM) for the indicated times and lysate was immunoblotted with indicated antibodies. (K) Expression levels of miR-126, p-AKT, and SPRED1 protein in FLT3-ITD- and FLT3-ITD+ primary AML blasts. Left, the cell lysate of each sample was immunoblotted with anti-SPRED1, anti-p-AKT, anti-RAN, and anti-actin antibodies. Right, levels of miR-126 as measured by qPCR. MG132, proteasome inhibitor MG132.
In order to demonstrate a direct effect of FLT3-ITD on the miR-155/miR-126 network through the SHIP1/p-AKT/SPRED1 axis, we then transduced FLT3-wt− HL-60 cells and Lin−BM cells from a normal wt mouse with an FLT3-ITD lentivirus. Forced expression of FLT3-ITD in these cells resulted in up-regulated miR-155 (Fig. 5A), decreased SHIP1, and increased p-AKT and SPRED1 levels and AKT/SPRED1 interaction (Fig. 5B and SI Appendix, Fig. S5A), which led to lower miR-126 levels (Fig. 5A) and increased proliferation and colony formation activities (Fig. 5C). Next, we examined whether this miRNA coregulation also occurred in vivo. To this end, we compared Lin− BM cells isolated from MllPTD/wt/Flt3ITD/ITD AML mice with those from normal wt mice. A decrease in SHIP1 and an increase in p-AKT, AKT/SPRED1 interaction, and levels of nuclear SPRED1 were observed in MllPTD/wt/Flt3ITD/ITD Lin− BM cells compared to the normal, wt counterpart cells (Fig. 5D). Furthermore, in FLT3-ITD+ AML patients receiving AC220, CD34+ cells showed higher levels of SHIP1 and lower levels of p-AKT and SPRED1 in posttreatment vs. pretreatment human samples (Fig. 5E). We also observed the relocalization of (peri)-nuclear SPRED1 to cytoplasm (Fig. 5 F, Left, and SI Appendix, Fig. S5B), decreased SPRED1-AKT interaction (Fig. 5 F, Middle), decreased SPRED1-RAN binding, and increased RAN/XPO5 binding in posttreatment vs. pretreatment samples from FLT3-ITD+ AML patients receiving AC220 (Fig. 5 F, Right).
Fig. 5.
Regulation of miR-126 by miR-155 in Lin− BM cells from wt vs. MllPTD/wt/Flt3ITD/ITD AML mice and in normal cells. (A–C) Effects of FLT3-ITD overexpression on miR-155–regulated miR-126 through SHIP1/p-AKT/SPRED1 axis in FLT3-ITD− HL-60 cells and Lin− BM cells from wt mice. HL-60 cells (Left) and Lin− BM cells from normal wt mice (Right) were infected with FLT3-ITD lentiviruses for 48 h. (A) Levels of miR-126 and miR-155 by qPCR. (*, P value.) (B) Levels of SHIP1, p-AKT, and SPRED1 by immunoblotting. (C) Levels of proliferation and colony formation. (*, P value.) (D) Different levels of AKT/SPRED1 interaction and SPRED1 distribution in Lin− BM cells isolated from wt vs. MllPTD/wt/Flt3ITD/ITD AML mice. Left, interaction between AKT and SPRED1. Right, cellular distribution of SPRED1. (E and F) Regulation of SHIP1/p-AKT/SPRED1 and SPRED1/RAN/XPO5 signaling in AC220 pre- and posttreated cells. The BM cells from FLT3-ITD+ AML patients pre- or posttreated with AC220 (n = 4) were collected to analyze as indicated. Left, levels of SHIP1, p-AKT, and SPRED1 by immunoblotting. Middle, interaction between AKT and SPRED1. Right, interaction of SPRED1/RAN and RAN/XPO5. (G and H) Effects of LPS on miR-155 and miR-126 expression in normal peripheral blood stem cells (PBSC). The cells were treated with LPS in the presence of SCR control or miR-155 inhibitor for 24 h. (G) Levels of miR-126 and miR-155 expression (Left) and miR-126 and miR-155 staining (Right) (Scale bar, 10 μm). (*, P value.) (H) Left, levels of SPRED1 phosphorylation and expression of DDX3X, SHIP1, and p-AKT protein. Right, levels of pri-miR-126 expression. (*, P value.) (I) Left, in vivo experimental design. The wt or miR-155 KO mice were treated with vehicle or LPS for 24 h (each group, n = 5). The Lin− stem-progenitor cell population from BM was isolated to analyze as indicated. Middle and Right, effects of LPS on miR-155 and miR-126 expression in Lin− cells. Middle, levels of miR-155. Right, levels of miR-126 and pri-miR-126. Ns indicates nonsignificance. (*, P value.) RNU44, small-nucleolar RNA RNU44; sno234, small control RNA sno234; OD, optical density.
Next, we treated FLT3 wild type (wt) THP1 leukemic cells (THP1) and FLT3-ITD MV-4–11 cells, CD34+ mononuclear cells (MNCs) from normal donors, and CD34+ FLT3 wt and FLT3-ITD AML blasts with the FLT3 ligand (100 ng/mL) for 24 h (SI Appendix, Fig. S5 C and D). Exposure to the FLT3-L resulted respectively in an increase and decrease of miR-155 and miR-126 and a decrease of SHIP1 and increase of p-AKT and SPRED1 in FLT3 wt THP1 cells, FLT3 wt AML blasts, and normal CD34 MNCs. In contrast, the FLT3-L did not significantly affect levels of miR-155, miR-126, and SHIP1/p-AKT/SPRED1 signaling in FLT3-ITD MV-4–11 cells and AML blasts, in which the downstream FLT3 signaling was already constitutively activated by the presence of ITD.
Taken together, these results support the notion of a twofold phosphorylated SPRED1–dependent XPO5/RAN inhibition by FLT3-ITD (or FLT3-L–FLT3 wt), directly via an SPRED1 tyrosine phosphorylation (11) and indirectly via p-AKT–dependent SPRED1 serine phosphorylation.
miR-155 Also Controls miR-126 Biogenesis in Normal Cells.
While we discovered the miR-155 controls over miR-126 biogenesis in FLT3-ITD+ AML blasts, the interconnected coregulation of these two miRNAs is unlikely to be unique to leukemogenesis; rather, it may represent a mechanism of leukemia growth that is hijacked from normal cells, which utilize it in situations demanding increased hematopoietic output. To this end, we showed an increase in miR-155 and a decrease in miR-126 when CD34+ MNCs from normal individuals are exposed to the FLT3-L (SI Appendix, Fig. S5D). Furthermore, as lipopolysaccharide (LPS) activates nuclear factor kappa B (NFκB) and the enhancement of NFκB binding to the BIC-155 promoter during inflammation (11, 27), we also showed that exposure of normal CD34+ cells to LPS resulted in higher levels of miR-155 (Fig. 5G) that were prevented by pretreatment with an miR-155 inhibitor (Fig. 5G). LPS exposure also resulted in an miR-126 decrease, via down-regulation of SHIP1, increase in p-AKT and p-SPRED1 (Fig. 5 G and H). We validated these results in vivo by injecting LPS into miR-155 wt (n = 5) and miR-155 knockout (KO) (n = 5) B6 mice (Fig. 5 I, Left). LPS treatment resulted in a twofold increase in miR-155 expression in BM Lin− MNCs from miR-155 wt mice compared to vehicle-treated controls and had no effect on BM Lin− MNCs from miR-155 KO mice (Fig. 5 I, Middle). LPS treatment also blocked the RAN/XPO5 complex as supported by an increase in pri-miR-126 levels and ultimately decreased mature miR-126 in BM Lin− MNCs from miR-155 wt mice compared to vehicle-treated controls (Fig. 5 I, Right). These effects, however, were not observed in BM Lin− MNCs from miR-155 KO mice (Fig. 5 I, Right), supporting the notion that miR-155 is required for miR-126 down-regulation.
Modeling the FLT3-ITD “Reshaped” miRNome.
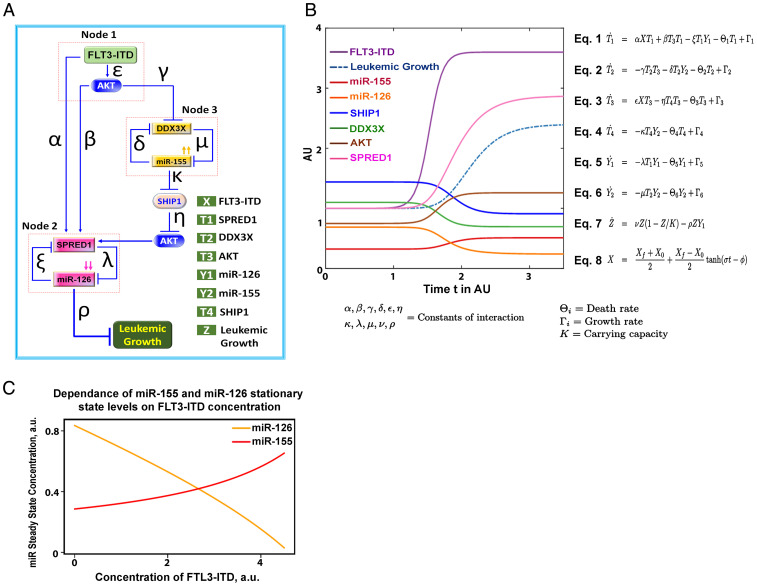
Given the circuitous nature and the complexity of the biological interactions of miR-155 and miR-126, we then studied a mathematical representation of this system to describe how the relationships among the variables was compatible with our experimental results (Fig. 6 A–C). Model simulations were based on a particular set of parameter values estimated from the rates of reactions relative to the timescale of leukemia growth and from the relative proportions of concentrations of the variables at steady-state in the presence and absence of FLT3-ITD (SI Appendix, Table S6). We tested the effect on increasing levels of FLT3-ITD on stationary states of miR-155 and miR-126 expression by performing numerical simulations of the mathematical model for different initial values of FLT3-ITD. We modeled the dynamics of the FLT3-ITD–dependent miRNA–protein network with a set of coupled ordinary differential equations under the assumption of the law of mass action, which states that the rate of change is proportional to the chemical concentration. In particular, the constant rates of interactions between the variables involved in the different nodes of the FLT-ITD–dependent miRNA network were given by the parameters α, β, γ, δ, ε, η, κ, λ, μ, ν, and ρ. We assumed that all variables have an intrinsic production/synthesis rate Γi and an intrinsic degradation rate Θi (where the index i = 1,2,...,6 represents the rate for variable i). The time evolution of this system was investigated assuming a rapid change in the level of FLT3-ITD from the initial value X0 to a final value Xf. The variation of FLT3 was modeled by equation (Eq. 8, Fig. 6B), where the parameters σ and ϕ determined the velocity and the time, respectively, at which the change of FLT3-ITD level occurs. We numerically solved this system of differential equations and showed that an increase of FLT3-ITD was expected to drive higher levels of miR-155 and lower levels of miR-126 (Fig. 6B), just as we observed in our in vitro and in vivo biological models.
Fig. 6.
Mathematical model of FLT3-ITD–regulated miR-126 and miR-155. (A) Schematic representation of a mathematical model of how FLT3-ITD controls the dynamics of miR-126/miR-155 circuit. (B) The mathematical model parameters are defined as follows: K is the carrying capacity of AML growth and Θi and Γi (where the index i = 1,2,…,6) are the degradation and production rates, respectively. The parameters α, β, γ, δ, ε, η, κ, λ, μ, ν, and ρ are constants that quantify the interaction between variables. Newton’s notation is used to indicate differentiation with respect to time (). Time and concentration are scaled and shown in arbitrary units (AU). The activation of FLT3-ITD (X) from an initial value to a final value is modeled by Eq. 8, in which represents the velocity at which activation of FLT3-ITD occurs and determines the time when the activation takes place. (C) Levels of miR-126 (yellow) and miR-155 (red) at the stationary states are obtained by numerically solving the system of ordinary differential equations for increasing levels of FLT3-ITD. Numerical integration of the mathematical model in B and C is shown with a particular set of estimated values for many unknown parameters.
Discussion
We show here that FLT3-ITD produces a divergent signal for leukemogenic miRNA deregulation as exemplified by the concurrent up-regulation of miR-155 and down-regulation of miR-126. Both miRNAs have been previously reported to play a relevant and differential role in AML growth as they control cell proliferation and quiescence, respectively (6–9, 12–15). Despite providing inputs in opposing directions, the divergent FLT3-ITD signal eventually converges and amplifies an output signal for leukemic growth through a complex network of feed-forward and feedback regulatory loops (see Anastasiadou et al. for definitions [5]) (Figs. 1 C and D and 6 and SI Appendix, Fig. S6). The key functional element of this leukemic multiloop circuit of deregulated miRNAs is the FLT3-ITD–induced SPRED1 phosphorylation that inhibits the RAN/XPO5 complex and in turn interferes with the canonical nucleus-to-cytoplasm transport of pre-miR-126 (11) (SI Appendix, Fig. S6, Left, node 2). This development results in low levels of mature miR-126 that allow quiescent leukemia cells to enter the cell cycle and sustain leukemia growth (10, 11). Since SPRED1 is also an miR-126 target (10, 11, 28), a feedback loop is also created in which progressively lower levels of miR-126 lead to progressively higher levels of SPRED1 and in turn to the amplification of the RAN/XPO5 blockage (Fig. 1 C, Top, and SI Appendix, Fig. S6, Left, node 2).
The FLT3-ITD–induced SPRED1 phosphorylation is the result of a direct FLT3-dependent tyrosine residue phosphorylation and an indirect AKT-dependent serine residue phosphorylation via a feed-forward loop with an operative “node 3” that constitutes the DDX3X/miR-155/SHIP1/p-AKT axis. In fact, FLT3-ITD/AKT inhibits DDX3X that participates in the splicing of BIC-155 and in turn in the production of pri-miR-155 for the canonical miRNA biogenesis path. The FLT3-ITD–induced DDX3X inhibition results in an aberrant increase of unspliced BIC-155 that is then routed toward cytoplasmic DROSHA via NXF1 transportation, thereby bypassing the RAN/XPO5 blockage and allowing the production of high levels of mature miR-155 (11). miR-155 then down-regulates the SHIP1 and in turn activates AKT that serine-phosphorylates SPRED1. Thus, taken together, this miRNA–protein interaction creates a recognizable feed-forward loop motif where the FLT3-ITD–dependent miR-126 down-regulation is amplified by the miR-155/SHIP1/p-AKT axis (Fig. 1D and SI Appendix, Fig. S6, Bottom). Interestingly, since DDX3X is also a target of miR-155 (11, 29), progressively higher levels of miR-155 lead to progressively lower levels of DDX3X, thereby amplifying the activity of the miR-155/SHIP1/p-AKT axis through a distinct feedback loop motif (Fig. 1 C, Bottom, and SI Appendix, Fig. S6, Right, node 3) contained within the broader FLT3-ITD feed-forward loop (Fig. 1D and SI Appendix, Fig. S6, Bottom).
Of note, the altered miRNA regulatory circuit described here is not unique to leukemogenesis, and it may represent a normal cellular response that shifts the balance between cell quiescence and proliferation, as observed by challenging normal wt mice with LPS (Fig. 5) and normal CD34+ cells with FLT3-L (SI Appendix, Fig. S5D). While under physiological conditions, the system returns at baseline once the input signal (e.g., FLT3-L stimulation or infection) attenuates, in AML the input provided by FLT3-ITD persists and support leukemia growth indefinitely.
The individual leukemogenic relevance of miR-126 and miR-155 to FLT3-ITD AML has been previously reported (6–9, 12–15). We demonstrated that FLT3 blocks miR-126 biogenesis and decreases leukemic cell quiescence, thereby favoring leukemia growth (11, 30). Others have reported on the key leukemogenic role of the FLT3-ITD–induced miR-155 via aberrant activation of downstream NFκB and interferon signaling pathways (13, 15, 31). Herein, we show how these two miRNAs are interconnected, support FLT3-ITD–driven leukemia growth, and provide the rationale for dual miR-155/miR-126 therapeutic targeting (30). Of note, whether our results are extendable to other receptor tyrosine kinase (RTK) remains to be fully demonstrated as these two miRNAs may participate on their own in other leukemogenic mechanisms. For examples, constitutively high levels of miR-155 expression have been shown to be mutagenic for KIT, a gene encoding another member of the RTK family (32), and low levels of miR-126 are also reportedly induced by protein kinases other than RTKs to sustain leukemia growth (10, 11, 33). Nevertheless, the interdependent miRNA–miRNA regulation is an emerging concept, and relatively little is known regarding its impact on leukemia initiation and growth. It may occur through several potential mechanisms, from a direct miRNA–miRNA interaction and “sponging” to an interdependent and reciprocal targeting of the respective transcription activators or repressors (34). It has been reported, for example, that miR-122 negatively deregulates miR-21 by binding to the miR-21 primary transcript and blocking Drosha cleavage (35) and that miR-709 regulates miR-15a/16–1 production by binding to its primary transcript (36). On the other hand, miR-660–5p can alter the expression of miR-486–5p by targeting mouse double minute 2 homolog (MDM2) and p53 (37). With regard to AML, relatively little is known regarding miRNA–miRNA interactions and to our knowledge, the miR-155–miR-126 interaction as reported here is an example of a hierarchical miRNA–miRNA interdependence that contributes to disease growth.
Of note, given the circuitous nature and complexity of the miRNA–miRNA interaction, we believe it may be helpful to describe it through mathematical models. To this end, we proposed a model that predicted how a steady increase in FLT3-ITD levels can drive miR-155 up-regulation and in turn miR-126 down-regulation (Fig. 6), just as we observed experimentally in vitro and in vivo. For this model, we assumed a linear relationship among the different variables, but it is very likely that given the complexity of the network, other nonlinear models may approximate and predict even better the observed miR-155/miR-126 coregulation and may eventually be used to guide therapy.
In conclusion, our data support the notion of a kinase-dependent deregulated architecture of the miRNome in AML and provide evidence of a feed-forward loop that allows an FLT3-ITD–generated input signal for up- and down-regulation of distinct miRNAs to converge into a common output signal for leukemia growth. We anticipate future discoveries of additional, more complex regulatory loops as part of leukemogenic networks of aberrantly expressed miRNAs. Given the complexity and the extension of these interplays, one could say that during leukemogenesis, the “circuitous architecture” of the miRNome as a whole is deregulated in AML to sustain leukemia growth (5).
Materials and Methods
An extensive description of the methods can be found in the SI Appendix, Materials and Methods. Target sequences for siRNAs are shown in the SI Appendix, Table S1. TaqMan gene expression assays used for qPCR analysis are shown in the SI Appendix, Table S2. Antibodies used for immunoprecipitation and immunoblotting analysis are shown in the SI Appendix, Table S3. Proteins used for in vitro kinase assay are shown in the SI Appendix, Table S4. Characteristics of patient samples are shown in the SI Appendix, Table S5. Mathematical model parameters and initial conditions are shown in the SI Appendix, Table S6.
Human and Mouse Samples.
Normal and AML peripheral blood and BM samples were obtained, respectively, from donors and patients at the City of Hope National Medical Center, and their use for this study was Institutional Review Board–approved under the following protocols—#06229, #03162, #07047, or #18067—in accordance with an assurance filed with and approved by the Department of Health and Human Services and met all requirements of the Declaration of Helsinki. Written informed consent was obtained from donors (#06229) or patients (#03162, #07047, or #18067) prior to specimen acquisition.
MNCs were isolated using Ficoll separation. CD34+ cells were then isolated using a positive magnetic bead selection protocol (Miltenyi Biotech, Germany). To normalize for FLT3-ITD levels in our experiments, we selected only patient samples with high FLT3-ITD allelic ratios (≥ 0.7). All FLT3-ITD–negative samples were also TKD-negative.
The MllPTD/wt/Flt3ITDITD AML mouse model was used in this study (16). Lin−Sca-1−c-Kit− (L−S−K−), Lin−Sca-1+c-Kit- (L−S+K-), Lin−Sca-1−c-Kit+ (L−S−K+), and LSK (Lin-Sca-1+c-Kit+) cells were sorted and used for in vitro studies. Mouse care and experimental procedures were performed in accordance with federal guidelines and protocols and were approved by the Institutional Animal Care and Use Committee at the City of Hope.
CpG-anti-miR-155 and CpG-anti-miR-126 Design and Synthesis.
The deoxycytidyl-phosphate-deoxyguanosine (CpG)-anti-miRNA were synthesized in the DNA/RNA Synthesis Core (City of Hope) by linking CpG-oligodeoxynucleotides (ODNs) to anti-miR-126 or anti-miR-155 similarly as described before (18). The sequences of single-stranded constructs are listed below (x indicates a single C3 unit; m indicates 2’O methylation; asterisks indicate phosphorothioation sites):
CpG-miR-126 inhibitor:
5′-G*G*TGCATCGATGCAGG*G*G* G*Gxxxxx
mCmGmC mAmUmUmAmUmUmAmCmUmCmAmCmGmGmUmAmCm-GmA -3′;
CpG-miR-155 inhibitor:
5′-G*G*TGCATCGATGCAGG*G*G*G*Gxxxxx mAmCmCmCmCmTmAmTmCmAmCmAmAmTmTmAmGmCmAmTmTmAmA
CpG-scrRNA:
5′-G*G*TGCATCGATGCAGG*G*G*G*Gxxxxx
mGmUmAmGmAmAmCmCmGmUmAmCmUmCmGmUmCmAmCmUmUmA-3′,
miRNA Labeling and Analysis.
The cells were cultured and incubated with a miR-RNA SmartFlare RNA probe (EMD Millipore) for 16 h. To ensure that the cell types, including MV-4–11 and primary AML blasts, were able to effectively endocytose the SmartFlare probes, we examined the uptake of probes in these cells using a SmartFlare uptake control, a scrambled control, and a housekeeping 18S control (according to the manufacturer’s guidelines). Cells were then washed in 1× phosphate-buffered saline and fixed in 4% paraformaldehyde for 3 min. Nuclei were counterstained with DAPI, and the images were analyzed using a confocal microscope (Zeiss LSM880). A more detailed protocol is provided in the SI Appendix.
Statistical Analysis.
Where indicated, to compare the means of 2 groups, results were generally compared by using an unpaired, 2‐tailed Student’s t test, with values from at least 2 independent experiments with triplicate determination. Data are presented as mean ± SE, as indicated. P < 0.05 was considered statistically significant; ns indicates not significant (SI Appendix).
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH CA205247 (Y.H.K., G.M.), CA201184 (G.M.), CA248475 (B.Z. and G.M.), U01CA250046 (R.C.R., G.M., Y.H.K.), the Jerome Foundation (G.M.), and the Hoag Foundation (G.M.). Research reported in this publication included work performed in the City of Hope Hematopoietic Tissue Biorepository (Pathology Research Services Core), supported by the National Cancer Institute of the NIH under Grant Number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112482119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Ha M., Kim V. N., Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Garzon R., et al. , Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl. Acad. Sci. U.S.A. 105, 3945–3950 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcucci G., et al. , MicroRNA expression in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 358, 1919–1928 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Liao Q., Wang B., Li X., Jiang G., miRNAs in acute myeloid leukemia. Oncotarget 8, 3666–3682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anastasiadou E., Jacob L. S., Slack F. J., Non-coding RNA networks in cancer. Nat. Rev. Cancer 18, 5–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Leeuw D. C., et al. , Attenuation of microRNA-126 expression that drives CD34 + 38-stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res. 74, 2094–2105 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Dorrance A. M., et al. , Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia 29, 2143–2153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lechman E. R., et al. , Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell 11, 799–811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lechman E. R., et al. , miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell 29, 214–228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B., et al. , Bone marrow niche trafficking of miR-126 controls the self-renewal of leukemia stem cells in chronic myelogenous leukemia. Nat. Med. 24, 450–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen L. X. T., et al. , Cytoplasmic DROSHA and non-canonical mechanisms of MiR-155 biogenesis in FLT3-ITD acute myeloid leukemia. Leukemia 35, 2285–2298 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcucci G., et al. , Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J. Clin. Oncol. 31, 2086–2093 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerloff D., et al. , NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia 29, 535–547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace J. A., et al. , miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood 129, 3074–3086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narayan N., Bracken C. P., Ekert P. G., MicroRNA-155 expression and function in AML: An evolving paradigm. Exp. Hematol. 62, 1–6 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Zorko N. A., et al. , Mll partial tandem duplication and Flt3 internal tandem duplication in a double knock-in mouse recapitulates features of counterpart human acute myeloid leukemias. Blood 120, 1130–1136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zarrinkar P. P., et al. , AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML). Blood 114, 2984–2992 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kortylewski M., et al. , In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat. Biotechnol. 27, 925–932 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue H., Hua L. M., Guo M., Luo J. M., SHIP1 is targeted by miR-155 in acute myeloid leukemia. Oncol. Rep. 32, 2253–2259 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Baran C. P., et al. , The inositol 5’-phosphatase SHIP-1 and the Src kinase Lyn negatively regulate macrophage colony-stimulating factor-induced Akt activity. J. Biol. Chem. 278, 38628–38636 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Zhou P., et al. , SHIP1 negatively regulates proliferation of osteoclast precursors via Akt-dependent alterations in D-type cyclins and p27. J. Immunol. 177, 8777–8784 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Brooks R., et al. , Coordinate expansion of murine hematopoietic and mesenchymal stem cell compartments by SHIPi. Stem Cells 33, 848–858 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Li D., et al. , Combined inhibition of Notch and FLT3 produces synergistic cytotoxic effects in FLT3/ITD+ acute myeloid leukemia. Signal Transduct. Target. Ther. 5, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long J., et al. , FLT3 inhibition upregulates HDAC8 via FOXO to inactivate p53 and promote maintenance of FLT3-ITD+ acute myeloid leukemia. Blood 135, 1472–1483 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Pal Singh S., et al. , Overexpression of SH2-containing inositol phosphatase contributes to chronic lymphocytic leukemia survival. J. Immunol. 204, 360–374 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Nguyen X. T., Mitchell B. S., Akt activation enhances ribosomal RNA synthesis through casein kinase II and TIF-IA. Proc. Natl. Acad. Sci. U.S.A. 110, 20681–20686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatto G., et al. , Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 36, 6608–6619 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S., et al. , The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 15, 261–271 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lössner C., et al. , Quantitative proteomics identify novel miR-155 target proteins. PLoS One 6, e22146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B., et al. , Treatment-induced arteriolar revascularization and miR-126 enhancement in bone marrow niche protect leukemic stem cells in AML. J. Hematol. Oncol. 14, 122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Connell R. M., et al. , Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205, 585–594 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witten L. W., Cheng C. J., Slack F. J., miR-155 drives oncogenesis by promoting and cooperating with mutations in the c-Kit oncogene. Oncogene 38, 2151–2161 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Zhang L., et al. , Targeting miR-126 in inv(16) acute myeloid leukemia inhibits leukemia development and leukemia stem cell maintenance. Nat. Commun. 12, 6154 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill M., Tran N., MicroRNAs regulating microRNAs in cancer. Trends Cancer 4, 465–468 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Wang D., et al. , Nuclear miR-122 directly regulates the biogenesis of cell survival oncomiR miR-21 at the posttranscriptional level. Nucleic Acids Res. 46, 2012–2029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang R., et al. , Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: Evidence for a microRNA hierarchy system. Cell Res. 22, 504–515 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borzi C., et al. , mir-660-p53-mir-486 network: A new key regulatory pathway in lung tumorigenesis. Int. J. Mol. Sci. 18, 222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.