Significance
Conservation outreach has long depended on an intuitive sense of which species are more “charismatic” or engaging, for example, placing focus on certain charismatic megafauna in advertising materials. Online community science databases like eBird and iNaturalist provide records of how people engage with different birds under differing data collection protocols. Comparisons between the two databases reveal biases in bird reporting rates. Larger, more colorful, and rarer birds are preferentially engaged with opportunistically in iNaturalist records compared to more systematic eBird records. These relationships and the species-specific engagement indexes determined from these data can be applied to conservation and outreach efforts to help foster a public relationship with nature and can be used to improve models using these two databases.
Keywords: animal charisma, birds, eBird, iNaturalist, generalized additive models
Abstract
Identifying rates at which birders engage with different species can inform the impact and efficacy of conservation outreach and the scientific use of community-collected biodiversity data. Species that are thought to be “charismatic” are often prioritized in conservation, and previous researchers have used sociological experiments and digital records to estimate charisma indirectly. In this study, we take advantage of community science efforts as another record of human engagement with animals that can reveal observer biases directly, which are in part driven by observer preference. We apply a multistage analysis to ask whether opportunistic birders contributing to iNaturalist engage more with larger, more colorful, and rarer birds relative to a baseline approximated from eBird contributors. We find that body mass, color contrast, and range size all predict overrepresentation in the opportunistic dataset. We also find evidence that, across 472 modeled species, 52 species are significantly overreported and 158 are significantly underreported, indicating a wide variety of species-specific effects. Understanding which birds are highly engaging can aid conservationists in creating impactful outreach materials and engaging new naturalists. The quantified differences between two prominent community science efforts may also be of use for researchers leveraging the data from one or both of them to answer scientific questions of interest.
Birds have received special attention in conservation (1), and investigations into stated preferences for birds have found that species traits—color, pattern, and shape—influence their perceived charisma (2–4). Sociological experiments studying animal charisma have relied on stated preferences to find correlations between hypothetical “willingness to pay” or “empathy” for a species’ conservation and species’ size, color, and aesthetic appeal (5–7). Recognizing the increasing availability of digital records of public engagement with animals that reveal preferences, an emerging field of “culturomics” uses Google search results, Wikipedia article activity, and records of digital engagement to identify charismatic species and traits (8–10). Others have taken advantage of revealed-preference data for birds from Google search results and the Common Breeding Bird Monitoring Scheme (11) and eBird data (12) to similarly investigate the public’s perception of different birds.
Online community science platforms, which collect data contributed by volunteers, provide a more direct way to study people’s engagement with species in the wild. Community scientists, sometimes called “citizen scientists,” volunteer contributions to scientific databases as self-guided nonprofessionals. Two biodiversity platforms in particular are of great interest for investigating bird charisma and engagement on a large scale. eBird, an app for hobbyist birders that has generated one of the world’s largest biodiversity databases, has recorded over 550 million records in North America to date (13). Many of these records come from over 46 million “complete checklists” and thus represent a rigorous reporting protocol, with reliable information on when species were not detected alongside when they were detected as well as the inclusion of sampling metadata. Another popular platform is iNaturalist, a nature app designed to encourage public engagement with all species. The primary goal of iNaturalist is to “connect people to nature” (14). The app allows an observation of any species at any time or place to be entered, so reporting rates depend on relative interest in different species among other factors.
Biodiversity records such as those aggregated on online community science platforms are important for informing species distribution models (15–17). Complete checklists from eBird are lauded as appropriate for species distribution modeling (18, 19), whereas opportunistic records such as from iNaturalist are known to contain particular biases (20, 21). These biases are often characterized as noise, but they may actually contain a strong signal of the habits and preferences of opportunistic naturalists. In this paper, we estimate those biases in relation to eBird and thereby analyze public engagement with species traits such as size and color contrast while controlling for phylogenetic relatedness, taxonomic order, and distribution characteristics such as abundance and range size. By harnessing two huge but very different community science datasets, we gain insight into how humans engage with biodiversity they encounter in the wild.
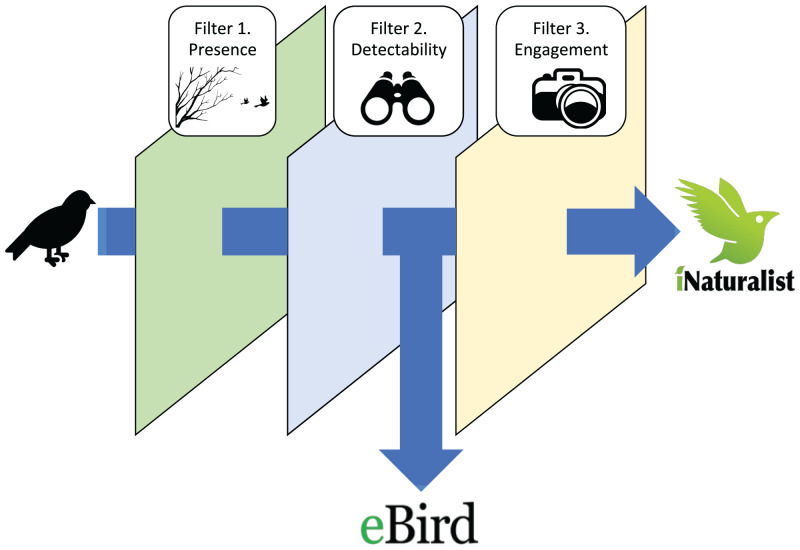
We construct a conceptual model to relate eBird and iNaturalist’s data-generating processes and show how they can be studied to characterize observer biases (Fig. 1). Imagine an iNaturalist user who notices a bird, takes a picture of it, and submits the photo to iNaturalist. For this event to occur, three separate conditions must be satisfied. First, the species must be present in the environment. We call this condition the “presence” filter, and characterizing this process is the main goal of most species distribution models that use community science data. Second, the bird itself must be visible or audible to a skilled observer—this is the “detectability” filter, which is controlled for in ecological studies as imperfect detection and includes factors like the species’ loudness, visibility, and distinctiveness. Were this a complete checklist in eBird, the process would stop here, because all detected birds must be reported under the complete-checklist protocol. However, in iNaturalist, a third event must occur: The observer must record an audio clip or, far more commonly, take a photo and upload it to iNaturalist. In practice, an observer might ultimately not report a species for a number of reasons: The observer may fail to notice or identify the bird due to skill or experience level; documenting the observation may be logistically challenging; or, the observer may find it uninteresting or otherwise not worth documenting. We call the factors that lead an observer to engage with one bird over another the “engagement” filter.
Fig. 1.
We conceptualize community science reporting as a filtering process. eBird complete checklist observations pass through two conceptual filters: 1) presence and 2) detectability. In iNaturalist, a hypothetical report must pass through an additional filter, 3) engagement, which represents the factors that lead an observer to report one bird over another, encompassing human-driven differences in these data, including the bird’s perceived charisma, the birder’s skill level, logistical obstacles to recording the detection, and spatial sampling effort.
We hypothesized two main types of bias that could drive variation in the engagement filter. The first, photogenic bias, refers to sampling differences due to aesthetic preferences of observers. One component of photogenic bias relates to relevant aspects of species charisma, such as size and color. Another component of photogenic bias is logistical: Particular bird species might be overreported by virtue of being easier to document rather than more charismatic. Since iNaturalist observations are almost always associated with photos, this could lead to a difference between the datasets. The second type of bias, novelty bias, may occur where users in iNaturalist preferentially report species that are new to them or that they observe infrequently. Other biases may also occur. For example, observers might engage differently with species based on what habitats or locations the observer engages with, which may vary between the two datasets at a scale finer than was feasible to analyze in this study. Characterizing the engagement filter, visible as deviations in iNaturalist reporting rates relative to eBird, is the main goal of this study.
Results
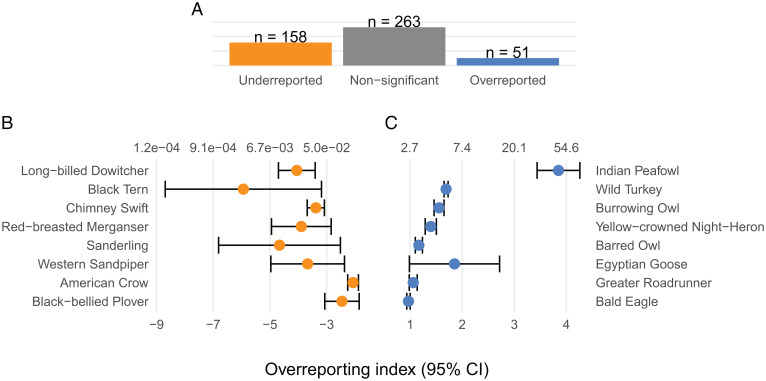
We estimated an overreporting index with uncertainty for each of 472 species of interest. This index is interpreted as the typical deviation across space in iNaturalist relative to eBird reporting rates, and its value is the difference in the typical log odds-scale reporting rate of a species in each dataset. For example, an overreporting index of ln(2) (∼0.693) means that the odds that a new observation in iNaturalist is the species are twice the odds that a new observation in eBird is the species. After controlling the false discovery rate, 210 species’ overreporting indexes were significantly different from zero, giving evidence that iNaturalist and eBird reporting rates were meaningfully different for many species. A significantly negative overreporting index means iNaturalist observers engaged with the bird at a lower rate than eBird observers, while a significantly positive one means they engaged with the bird at a higher rate. Fig. 2 shows the most extreme over- and underreported birds. The most overreported birds are each some combination of large (wild turkey), well known for their appeal (burrowing owl), or considered especially beautiful (Indian peafowl). The Indian peafowl may be particularly overreported in iNaturalist due to a technical rule that some eBirders may be following, namely that captive birds are not countable. Examining the least overreported birds, we notice that they are smaller birds, waders and gulls, or have large ranges (crow), characteristics we investigate in a more formal analysis next.
Fig. 2.
Species overreporting indexes. (A) Counts of bird overreporting indexes by significance level, controlling the false discovery rate (FDR) to account for multiple comparisons. The odds of appearing in iNaturalist for the most underreported species (B) were between 0.002 and 0.13 as small as in eBird, while the odds for the most overreported species (C) were roughly 1.6 to 45 as large as in eBird. The bottom axis shows the overreporting index on the log scale as used elsewhere in this paper. The top axis shows the index on the real scale. For each species, the index is plotted with its 95% CI.
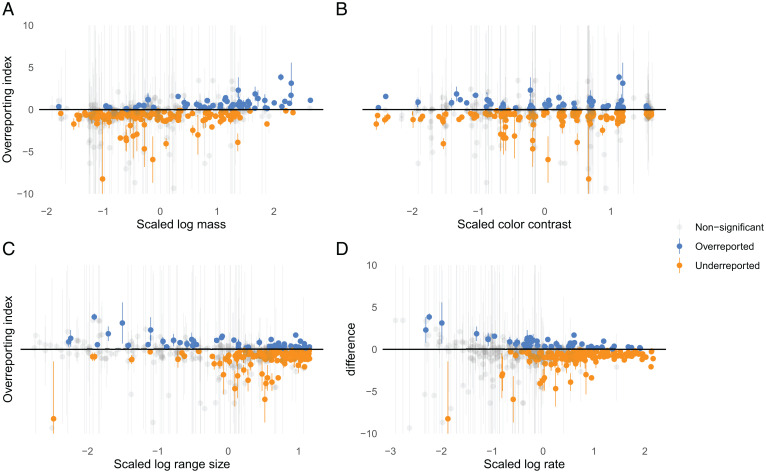
We hypothesized that differences at the species level, represented by the third filter layer (Fig. 1), could be driven by a variety of mechanisms. Fig. 3 shows the relationship between overreporting indexes and the species traits that relate to the proposed biases. We found a statistically significant relationship between the overreporting index and size, color, and number of hexagons (hexes ) containing observations of the species (a proxy for range size) in a meta-analysis of species-level effects incorporating uncertainties. We did not find a statistically significant relationship (in the presence of other covariates) between the overreporting index and reporting rate (a proxy for overall prevalence). This means that opportunistic birders in the contiguous United States engaged more with larger, more colorful, and more range-limited birds more often than would be expected based on the corresponding species’ detection rates in eBird.
Fig. 3.
Species overreporting indexes plotted against four traits—(A) log mass, (B) color contrast (higher values indicate greater color contrast), (C) log range size, and (D) log reporting rate (prevalence)—whose associations were studied in a meta-analysis. Vertical error bars show 95% CIs of indexes not adjusted for multiple testing.
While accounting for phylogenetic structure across species we found a 0.31 effect size on scaled log mass with a 95% CI of (0.15, 0.47), 0.12 for scaled color (0.04, 0.19), –0.13 for scaled log number of hexes where reported (–0.25, –0.03), and –0.08 for scaled log proportion of eBird checklists where reported (–0.19, 0.03). See SI Appendix, Table S1 for more interpretation. The independent random effect dominates the phylogenetically structured random effect in magnitude, suggesting there is not much phylogenetic structure in the residual variance. If one is not interested in relationships to the phylogeny, one might fit the same models without accounting for phylogenetic structure. We present the results of this analysis ignoring phylogeny, which are similar but slightly less conservative, in SI Appendix, Table S5.
We tested sensitivity of results to the choice to exclude low information species associated with extreme overreporting indexes. Including these indexes meaningfully changed the results of the meta-analysis, indicating that the choice to exclude low information species was well motivated (SI Appendix, Tables S2 and S6).
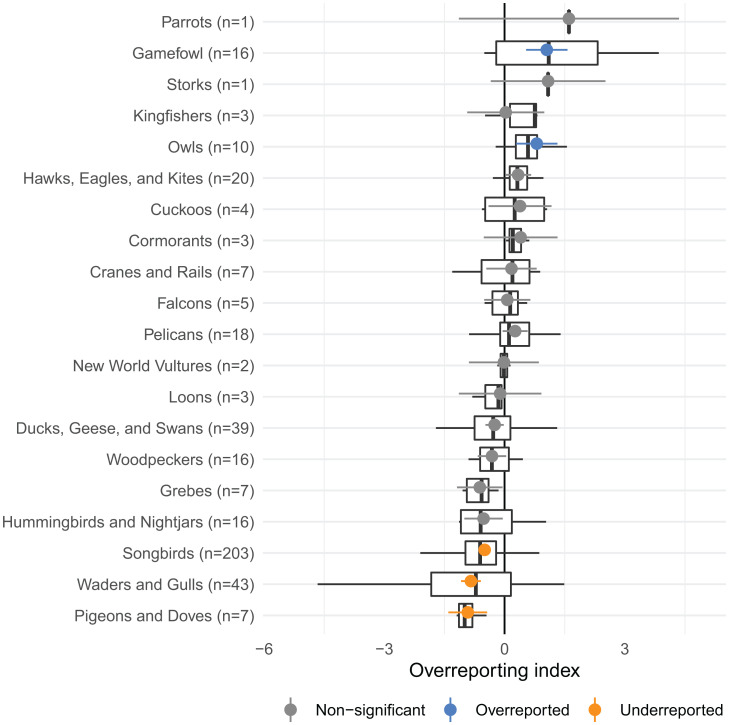
We accounted for phylogeny at the species level in the meta-analysis, but it can also be helpful to visualize relationships at the order level for improved interpretation. We identified five taxonomic orders whose typical overreporting indexes were significantly different from zero after correcting for multiple testing (Fig. 4). Owls and gamefowl tend to be relatively large, which may explain their overreporting. However, many waders and gulls are large, and this order is underreported, along with songbirds and doves.
Fig. 4.
The predicted effect of order, representing the typical overreporting index for that group of species, excluding low-information outliers. Colors indicate the results of a false discovery rate-controlling significance test. Box plots show the range of estimated overreporting indexes in each group.
Discussion
When using community science data to study biodiversity, the inescapable fact of variation between users and across platforms is often ignored or treated as noise that at best adds uncertainty and at worst causes biased estimation. However, when studying people’s relationships to animals rather than the animals themselves, with an eye toward using this knowledge to inform conservation efforts (22), the paradigm flips, and variation across platforms becomes the data-generating process of interest. For this task, community science is not merely a source of noisy data, but also a unique and invaluable source of information about how members of the public engage with the natural world.
The iNaturalist dataset in particular is ideal for characterizing naturalists’ biases. Because iNaturalist is open to all living organisms, anything like eBird’s complete checklist protocol is impossible, as an observer will likely detect hundreds of species of plants, animals, and fungi for every individual they choose to report. iNaturalist therefore forces its users to constantly discern which organisms they consider noteworthy and which to ignore. Therefore, differences between iNaturalist and eBird data are partially driven by the human element of choice.
To obtain estimates of birder engagement with 472 bird species, we modeled variation in iNaturalist reports across species relative to the eBird baseline. To increase confidence that any findings reflect true differences in reporting behaviors, we included four critical layers of statistical rigor intended to estimate unbiased reporting rates and accurately estimate uncertainty: spatially explicit models for each dataset so neither one serves as a noisy predictor of the other, a quasi-binomial error distribution to account for remaining variation, a parametric bootstrap on the overreporting index to accurately quantify uncertainty to propagate forward, and a phylogenetically explicit meta-analysis to account for potential lack of independence between species. Callaghan et al. (23) give a simpler analysis from regressing one dataset against the other.
In the wild, opportunistic birders engage more with species whose traits have been identified as charismatic in artificial contexts such as surveys (5–7). In particular, this study reinforces the concept of the charismatic megafauna, the idea that larger species are more interesting, sympathetic, or accessible to members of the public (24–26). The finding that more range-restricted birds are overrepresented is also consistent with the hypothesis that iNaturalist users may optimize for lengthening their “life list,” the list of unique species they have ever observed on the app (27), or want assistance identifying species that are new to them. The strong relationship we find between size and overreporting is also consistent with the hypothesis that iNaturalist users engage more with birds that are easier to photograph. Logistical constraints around photography are likely not the only drivers of variation, as hinted by the fact that the American crow, a common, large, and relatively bold bird, is quite easy to photograph but is strongly underreported.
Three taxonomic orders—songbirds, waders and gulls, and pigeons and doves—were associated with underreporting. Songbirds, a group that comprised more than 40% of the species studied, are a phenotypically diverse group, so it is difficult to speculate why they may be overall underreported, but their general small size, ubiquity, and difficulty to identify may all contribute to their underreporting. Traits that are harder to quantify than those we studied might contribute to these three taxa being underreported: Gulls, pigeons, and some songbirds (such as American crows, European starlings, and chimney swifts) might be considered nuisances in urban or agricultural settings, and opportunistic observers might ignore nuisance species; many species in these three groups flock, which could drive engagement; and species may use space differently; for example, shorebirds may congregate in locations that contain few iNaturalist observations.
Nature photography and film making have played a large role in conservation by bringing biodiversity to the attention of the public (28, 29). As camera lenses and photos have become ubiquitous in our culture, conservation sites and museums have leveraged this fact, using Instagram and other social media platforms to understand and further engage visitors, both by producing aesthetically compelling imagery and by encouraging visitors to take and share their own photographs (30, 31). iNaturalist takes advantage of commonplace camera phones to provide users with automatic identifications of their observations (32) and a platform on which to document and share their experiences. The logistics and charisma dimensions of this engagement bias, seen through a camera lens, are likely correlated, and we were not able to disentangle them within this framework. However, because photographs and imagery play a large role in modern communication and social media, logistical obstacles to photographing a bird may play a large role in how that bird is known or perceived by community members and therefore patterns in logistical bias may themselves be of interest.
We have argued that these engagement results are interesting in part because of their implications for birders’ preferences, i.e., the perceived charisma of the species we studied. However, the role of perceived charisma in effective conservation is complicated. While charismatic species are often used for public outreach as “flagship species,” this may be associated with the misallocation of conservation resources as the species in greatest need are not always those deemed charismatic (6). A similar imbalance is present in community science data; the “gotta catch ’em all” commodification of nature associated with birding life lists can be at odds with the scientific pursuits that these data inform (33). As our overreporting indexes show, people do engage differently with different species. These differences can inform effective outreach and fundraising even while conservationists maintain preference-blind standards in prioritizing species for conservation management (34). Conservationists may also tailor outreach materials, arguments, and experiences to best align with people’s levels of engagement, whether by focusing on high engagement or by identifying engagement gaps where species are ignored.
It is important to note that neither eBird nor iNaturalist is uniformly used or accessible across user demographics, so these results can only reveal the behaviors of people who are already engaging with nature through these community science platforms, who have self-selected based on a general interest in birds, and who are not blocked from engaging with nature due to economic disadvantage or other systemic barriers. Future work could investigate the engagement filter in further subpopulation analyses (e.g., children, urban areas) (35, 36). Since bird plumage varies seasonally, an investigation of the engagement filter variation across seasons may also be of interest to clarify the relationship between color and charisma (37).
Quantifying the difference between eBird and iNaturalist may also be of interest to scientists using the data from these sources in downstream analyses of species abundance. Future work could include evaluating and improving modeling with community data using these quantified differences between the two datasets. For example, many hierarchical ecological models have a latent abundance or occurrence layer and an imperfect detection layer. Our overreporting index could be used directly in a model informing the imperfect detection layer. For statistical methods that do not explicitly model the detection process, our relative reporting rates can play a role similar to a different sampling effort for the two datasets: The indexes we estimated could help calibrate iNaturalist detection rates across species. Our methods could be repurposed to create a difference index to calibrate detection or reporting between any two data sources.
We analyzed the differences in these two datasets with data aggregated over time, at a large spatiotemporal scale. Studies on smaller spatiotemporal scales could obtain more tailored estimates using the methods we describe. High-engagement species may differ across regions or communities. Similarly, downstream analyses may call for detection or sampling effort corrections estimated at more local scales. By tuning the methods with select hex size and data filters, a set of tailored overreporting indexes could be obtained for a variety of contexts.
By treating observer biases as signal rather than noise, we estimated overreporting indexes characterizing community scientists’ rates of engagement with over 400 species of US birds. We identified individual species that birders either engage with preferentially or ignore as well as species traits and taxonomic groups associated with these patterns. This work has the potential to inform conservation decision making and improve models of bird species distributions.
Materials and Methods
eBird and iNaturalist Preprocessing and Data Structures.
In August 2020 we downloaded the eBird Basic Dataset and extracted all complete checklists (13). We also obtained all iNaturalist research grade observations of bird species from the Global Biodiversity Information Facility (38). For both datasets, we considered only observations made in the contiguous 48 United States and Washington, DC on or before 31 December 2019. We excluded checklists obtained in 2020 or later to avoid accidentally capturing changes in observer behavior related to the COVID-19 pandemic (39, 40). We associated species across the two datasets using the R package taxalight (41).
We aggregated observations to a spatial grid of regular hexagonal cells covering the contiguous United States such that spatial grid cells had a long radius (center to vertex) of 20 km. In each “hex,” a count was obtained for the number of times each species was detected. The total number of observations in each hex was calculated as the sum of the species observation events in that hex. For consistency with iNaturalist, an eBird checklist that reported more than one species was counted as a series of separate species observation events (a checklist with three species reported constituted three observations). If a species was observed multiple times on a checklist, this counted as only one species observation event. This framework ignores species counts on each checklist to focus on species encounter rates in the creation of the overreporting index and is intended to homogenize procedures across the two datasets as iNaturalist does not report species counts. By aggregating over time we assumed that the primary variation is spatial. We accounted for secondary variation elsewhere in the modeling approach.
We subselected species according to the following criteria: First, to capture spatial variability in sampling, we considered only species that were observed one or more times in at least 100 different hexes in eBird in the contiguous United States, ensuring a minimally well-informed baseline of detection across space for all species. Second, we considered only species in the EltonTraits database (42). We eliminated “pelagic specialists” according to EltonTraits, expecting the sampling process generating these data to be fundamentally different from that of terrestrial birds.
Species-Level Spatial Analysis.
We first estimated a typical overreporting index characterizing each species. The unit of analysis for the first stage was yijk, the count of reports of species j reported at the ith spatial hex in dataset k (either eBird or iNaturalist). We modeled the number of “successes” in a binomial random draw yijk as Here Cik is the known number of “attempts,” which we define for the presence-only iNaturalist data to be the total number of iNaturalist observations across all bird species in spatial hex i and for the eBird checklist data to be the total number of species observation events on all of the eBird checklists in spatial hex i. (Note that for eBird, a single checklist on which five different species are observed would be considered five species observation events, rather than one, to match the iNaturalist sampling schema.) Rijk is the reporting rate that we model as a function of location i for each species j and dataset k combination.
We used a quasi-binomial generalized additive model (GAM) with a logit link to capture spatial variability via a multidimensional tensor-product smooth of the longitude and latitude coordinates of the hexes (43). The motivation for this approach was twofold. First, we anticipated that many differences between the datasets could be due to spatial heterogeneity in sampling. Both eBird and iNaturalist are highly spatially variable with different hotspots, meaning that sampling intensity differs by dataset and a spatially explicit analysis is called for. We chose a pragmatic resolution (20-km hexagons) that addressed spatial variation while remaining computationally feasible to estimate, and as a consequence smaller-scale variation in sampling is in the engagement filter of interest. A spatially explicit approach is also necessary since user habits may themselves be spatially nonindependent. To obtain accurate CIs on parameter estimates, spatial autocorrelation in the data-generating process must be accounted for. Second, we anticipated extrabinomial, nonspatial variability across units (e.g., temporal, weather conditions, observer variability, etc.). We chose a quasi-binomial approach as a way to account for this in the uncertainty quantification.
A GAM was fitted for each species and dataset combination. The basis dimensions were chosen to use 20 × 20 knots. We fitted all models and then iteratively increased knots by 5 in each axis until the model passed a hypothesis test of whether the basis dimension for a smooth was adequate using a P-value cutoff (given in the R function mgcv::gam.check).
From the quasi-binomial GAMs, we obtained estimates of the spatially smooth surface of the reporting rate at each hex, in each dataset. We calculated the overreporting index as the median predicted difference in log-scale reporting rates across hexes for each species. We used the median predicted difference rather than the mean as the median is a more robust summary across all hexes when a few hexes have large and uncertain differences that would distort the mean. To obtain accurate CIs on the overreporting index, we used a parametric bootstrap approach, making random draws of the spatial surface and recomputing the index each time, to obtain an estimate of uncertainty for each index. Each species’ overreporting index represents the typical deviation in the iNaturalist reporting rate relative to the eBird baseline for that species.
To assess which overreporting indexes were significantly different from zero, we used a P-value threshold that was adjusted to account for the fact that we made multiple comparisons (one for each species). We used a false discovery rate controlling method to ensure that across comparisons the false positive rate was no more than 0.05 (44).
Cross-Species Meta-Analysis.
We used a meta-analysis to ask whether species traits can help explain differences in birder engagement measured by overreporting indexes. A meta-analysis allowed us to propagate the uncertainty estimated in the first stage of the analysis (45). The median differences in reporting rates for all of the species, along with their SEs, became the response in this stage of the analysis.
To investigate the effect of size, we retrieved species’ log mass from the EltonTraits dataset (42). To represent how colorful or striking a bird is, we used an index of maximum color contrast originally developed by Schuetz and Johnston (12). We used two covariates as proxies of different aspects of rarity: the number of hexes a species is reported in (a proxy for size of effective range) and the proportion of all eBird checklists where the species was found (a proxy for overall prevalence). We centered and scaled these covariates for log mass, maximum color contrast, log range size, and log prevalence.
We also considered species’ level of risk for extinction as a potential covariate (SI Appendix, Tables S3 and S4). However, many of the birds most at risk fail to appear in our dataset due to the sample size filters we put in place, mentioned above. Therefore, the results presented in this paper reveal conservation insight at medium levels of risk rather than the most extreme.
In the meta-analysis we also incorporated phylogenetic structure to account for the possibility that phylogenetically closer species have more similar reporting indexes due to evolutionary nonindependence of unmodeled but important traits (46). We obtained multiple phylogenetic trees from BirdTree.org (47). We then obtained a consensus tree including branch edges using the R package phytools (48). Finally, we computed a variance–covariance matrix based on this consensus tree using the R package ape (49). We allowed for both a random effect for species with this variance–covariance structure and an unstructured random effect for species.
We fitted a second meta-analysis including only the effect of taxonomic order and excluding phylogenetic structure to obtain estimates of each order’s mean overreporting index with properly propagated error (50).
Models for nine species failed to fit (SI Appendix) and were therefore dropped from the second-stage analysis. Three of these failed to pass a test for adequate knots with a basis dimension of 35 × 35, above which computation became infeasible. Six of these failed to converge in under 24 h, which was chosen as a practical cutoff.
We removed 49 species that had overreporting indexes outside the range –10 to 10 from the meta-analysis stage. Values less than –10 or greater than 10 arose in cases where, among the union of hexes where a species was reported in either dataset, one dataset reported no observations in over half of those hexes. Because the reporting index uses median differences in log reporting rates, it could not be reliably estimated, nor its uncertainty reliably quantified for the meta-analysis step, in these cases. After these two filters, 424 species were included in the meta-analysis. To test the sensitivity of results, we repeated the meta-analysis including the extreme overreporting indexes.
Supplementary Material
Acknowledgments
We thank S. Beissinger, C. Boettiger and his laboratory group, M. Chapman, J. Clare, and W. Fithian, for comments and support and the dedicated users of eBird and iNaturalist who provided data for this study. S.S. was supported by the Gordon and Betty Moore Foundation through Grant GBMF3834 and by the Alfred P. Sloan Foundation through Grant 2013-10-27 to the University of California, Berkeley. B.R.G. was supported by the NSF Graduate Research Fellowship under Grant 1752814. This work used the Extreme Science and Engineering Discovery Environment, which is supported by NSF Grant ACI-1548562. We thank the reviewers who helped strengthen this work.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110156119/-/DCSupplemental.
Data Availability
Phylogenetic tree data (47) and the code used to process the data, perform the analyses, and make the figures have been deposited on Github (https://github.com/sastoudt/charismatic-birds). eBird data are freely available to download from https://ebird.org/data/download or from the Global Biodiversity Information Facility (51). We used version ebd_relAug-2020 directly from eBird. Previously published data were used for this work (12, 38, 42, 52).
References
- 1.Robinson S. K., Bird niches in human culture and why they matter. Proc. Natl. Acad. Sci. U.S.A. 116, 10620–10622 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brambilla M., Gustin M., Celada C., Species appeal predicts conservation status. Biol. Conserv. 160, 209–213 (2013). [Google Scholar]
- 3.Liskova S., Frynta D., What determines bird beauty in human eyes? Anthrozoos 26, 27–41 (2013). [Google Scholar]
- 4.Garnett S. T., Ainsworth G. B., Zander K. K., Are we choosing the right flagships? The bird species and traits Australians find most attractive. PLoS One 13, e0199253 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serpell J. A., Factors influencing human attitudes to animals and their welfare. Anim. Welf. 13, S145–S152 (2004). [Google Scholar]
- 6.Colléony A., Clayton S., Couvet D., Saint Jalme M., Prevot A., Human preferences for species conservation: Animal charisma trumps endangered status. Biol. Conserv. 206, 263–269 (2016). [Google Scholar]
- 7.Curtin P., Papworth S., Coloring and size influence preferences for imaginary animals, and can predict actual donations to species-specific conservation charities. Conserv. Lett. 13, e12723 (2020). [Google Scholar]
- 8.Kim J. Y., Do Y., Im R., Kim G., Joo G., Use of large web-based data to identify public interest and trends related to endangered species. Biodivers. Conserv. 23, 2961–2984 (2014). [Google Scholar]
- 9.Burivalova Z., Butler R. A., Wilcove D. S., Analyzing Google search data to debunk myths about the public’s interest in conservation. Front. Ecol. Environ. 16, 509–514 (2018). [Google Scholar]
- 10.Mittermeier J. C., Correia R., Grenyer R., Toivonen T., Roll U., Using Wikipedia to measure public interest in biodiversity and conservation. Conserv. Biol. 35, 412–423 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Zmihorski M., Dziarska-Palac J., Sparks T. H., Tryjanowski P., Ecological correlates of the popularity of birds and butterflies in Internet information resources. Oikos 122, 183–190 (2013). [Google Scholar]
- 12.Schuetz J. G., Johnston A., Characterizing the cultural niches of North American birds. Proc. Natl. Acad. Sci. U.S.A. 116, 10868–10873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.eBird; Cornell Lab of Ornithology, eBird: An online database of bird distribution and abundance. http://www.ebird.org. Accessed 28 May 2021.
- 14.iNaturalist, What is it. https://www.inaturalist.org/pages/what+is+it. Accessed 28 May 2021.
- 15.Supp S. R., et al., Citizen-science data provides new insight into annual and seasonal variation in migration patterns. Ecosphere 6, 1–19 (2015). [Google Scholar]
- 16.Pacifici K., et al., Integrating multiple data sources in species distribution modeling: A framework for data fusion. Ecology 98, 840–850 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Van Eupen C., et al., The impact of data quality filtering of opportunistic citizen science data on species distribuiton model performance. Ecol. Modell. 444, 109453 (2021). [Google Scholar]
- 18.Callaghan C. T., Gawlik D. E., Efficacy of eBird data as an aid in conservation planning and monitoring. J. Field Ornithol. 86, 298–304 (2015). [Google Scholar]
- 19.Steen V. A., Elphick C. S., Tingley M. W., An evaluation of stringent filtering to improve species distribution models from citizen science data. Divers. Distrib. 25, 1857–1869 (2019). [Google Scholar]
- 20.Isaac N. J. B., Pocock M. J. O., Bias and information in biological records. Biol. J. Linn. Soc. Lond. 115, 522–531 (2015). [Google Scholar]
- 21.Meyer C., Weigelt P., Kreft H., Multidimensional biases, gaps and uncertainties in global plant occurrence information. Ecol. Lett. 19, 992–1006 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Ladle R. J., et al., Conservation culturomics. Front. Ecol. Environ. 14, 269–275 (2016). [Google Scholar]
- 23.Callaghan C. T., Poore A. G. B., Hofmann M., Roberts C. J., Pereira H. M., Large-bodied birds are over-represented in unstructured citizen science data. Sci. Rep. 11, 19073 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knegtering E., Van der Windt H. J., Schoot Uiterkamp A. J. M., Public decisions on animal species: Does body size matter? Environ. Conserv. 38, 28–36 (2010). [Google Scholar]
- 25.Ducarme F., Luque G. M., Courchamp F., What are “charismatic species” for conservation biologists ? BioSciences Master Rev. 10, 1–8 (2013). [Google Scholar]
- 26.Krause M. l, Charismatic species and beyond: How cultural schemas and organisational routines shape conservation. Conserv. Soc. 15, 313–321 (2017). [Google Scholar]
- 27.Loarie S., A new kind of life list. https://www.inaturalist.org/blog/42454-a-new-kind-of-life-list. Accessed 28 May 2021.
- 28.Silk M. J., Crowley S. L., Woodhead A. J., Nuno A., Considering connections between Hollywood and biodiversity conservation. Conserv. Biol. 32, 597–606 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Hanisch E., Johnston R., Longnecker N., Cameras for conservation: Wildlife photography and emotional engagement with biodiversity and nature. Hum. Dimens. Wildl. 24, 267–284 (2019). [Google Scholar]
- 30.Weilenmann A., Hillman T., Jungselius B., “Instagram at the museum: Communicating the museum experience through social photo sharing” in Proceedings of the SIGCHI Conference on Human Factors in Computing Systems (Association for Computing Machinery, New York, NY, 2013), pp. 1843–1852. [Google Scholar]
- 31.Hughes K., Moscardo G., Connecting with new audiences: Exploring the impact of mobile communication devices on the experiences of young adults in museums. Visit. Stud. 20, 33–55 (2017). [Google Scholar]
- 32.Saari C., iNaturalist computer vision explorations. https://www.inaturalist.org/pages/computer_vision_demo. Accessed 28 May 2021.
- 33.Altrudi S., Connecting to nature through tech? The case of the iNaturalist app. Convergence 27, 124–141 (2021). [Google Scholar]
- 34.McGowan J., et al., Conservation prioritization can resolve the flagship species conundrum. Nat. Commun. 11, 994 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belaire J. A., Westphal L. M., Whelan C. J., Minor E. S., Urban residents’ perceptions of birds in the neighborhood: Biodiversity, cultural ecosystem services, and disservices. Condor 117, 192–202 (2015). [Google Scholar]
- 36.Aristeidou M., et al., Exploring the participation of young citizen scientists in scientific research: The case of iNaturalist. PLoS One 16, e0245682 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mittermeier J. C., Roll U., Matthews T. J., Grenyer R., A season for all things: Phenological imprints in Wikipedia usage and their relevance to conservation. PLoS Biol. 17, e3000146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Global Biodiversity Information Facility (GBIF), GBIF occurrence download. 10.15468/dl.7mt5mn. Accessed 9 March 2021. [DOI]
- 39.Hochachka W. M., Alonso H., Gutiérrez-Expósito C., Miller E., Johnston A., Regional variation in the impacts of the COVID-19 pandemic on the quantity and quality of data collected by the project eBird. Biol. Conserv. 254, 108974 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crimmins T. M., Posthumus E., Schaffer S., Prudic K. L., COVID-19 impacts on participation in large scale biodiversity-themed community science projects in the United States. Biol. Conserv. 256, 109017 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boettiger C., Norman K., taxalight: A lightweight and lightning-fast taxonomic naming interface. R Package Version 0.1.3. https://cran.r-project.org/web/packages/taxalight/index.html. Accessed 4 April 2021.
- 42.Wilman H., et al., EltonTraits 1.0: Species-level foraging attributes of the world’s birds and mammals: Ecological archives E095-178. Ecology 95, 2027 (2014). [Google Scholar]
- 43.Wood S. N., Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. B 73, 3–36 (2011). [Google Scholar]
- 44.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 45.Viechtbauer W., Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 36, 1–48 (2010). [Google Scholar]
- 46.Lajeunesse M. J., Meta-analysis and the comparative phylogenetic method. Am. Nat. 174, 369–381 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Jetz W., Thomas G. H., Joy J. B., Hartmann K., Mooers A. O., A global phylogeny of birds. https://birdtree.org/subsets/. Accessed 1 June 2021.
- 48.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 49.Paradis E., Schliep K., ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 50. Koricheva J., Gurevitch J., Mengersen K., Eds., Handbook of Meta-Analysis in Ecology and Evolution (Princeton University Press, Princeton, NJ, 2013). [Google Scholar]
- 51.Auer T., et al., Data from “EOD - eBird observation dataset.” Global Biodiversity Information Facility. https://www.gbif.org/dataset/4fa7b334-ce0d-4e88-aaae-2e0c138d049e. Accessed 1 August 2020.
- 52.International Union for Conservation of Nature, The IUCN Red List of Threatened Species. 10.15468/0qnb58. Accessed 26 September 2021. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Phylogenetic tree data (47) and the code used to process the data, perform the analyses, and make the figures have been deposited on Github (https://github.com/sastoudt/charismatic-birds). eBird data are freely available to download from https://ebird.org/data/download or from the Global Biodiversity Information Facility (51). We used version ebd_relAug-2020 directly from eBird. Previously published data were used for this work (12, 38, 42, 52).