Abstract
Heparan sulfate is a widely expressed polysaccharide in the extracellular matrix and on the cell surface. 3-O-sulfated heparan sulfate represents only a small percentage of heparan sulfate from biological sources. However, this subpopulation is closely associated with biological functions of heparan sulfate. The 3-O-sulfated heparan sulfate is biosynthesized by heparan sulfate 3-O-sulfotransferase, which exists in seven different isoforms. This review article summarizes the recent progress in the substrate specificity studies of different 3-O-sulfotransferase isoforms involving the use of homogeneous oligosaccharide substrates and crystal structural analysis. The article also reviews a newly developed liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based method to analyze the level of 3-O-sulfated heparan sulfate with high sensitivity and quantitative capability. This newly emerged technology will provide new tools to study the structure and function relationship of heparan sulfate.
Keywords: heparan sulfate, heparin, glycosaminoglycans, sulfotransferases
INTRODUCTION
Heparan sulfate (HS) is an essential glycan in mammals and is involved in many biological processes, including blood coagulation, inflammatory responses, and embryonic development (1–3). HS achieves its function through the interaction with protein ligands and receptors. Many viruses utilize cell surface HS on host cells as a receptor to establish infection. Notably, HS is reportedly involved in the infection of SARS-CoV-2 (4). HS is a sulfated polysaccharide that contains a disaccharide-repeating unit of glucuronic acid (GlcA) or iduronic acid (IdoA) linked to glucosamine (GlcN). The sulfation commonly occurs on the 2-OH position of IdoA and less frequently on the 2-OH of GlcA. GlcN saccharides can carry sulfo groups on the N-position, 6-OH and 3-OH positions. Although some interactions between HS and proteins are nonspecific, many interactions require specific sulfation patterns, the presence of GlcA and/or IdoA units, and a particular length of the saccharide chains. One of the primary goals of HS-related research is to uncover the relationship between the biological functions and saccharide structures.
The biosynthesis of HS involves a series of specialized sulfotransferases, which install sulfo groups. Four N-deacetylase/N-sulfotransferase isoforms exist to convert N-acetyl glucosamine (GlcNAc) to an N-sulfo glucosamine (GlcNS) saccharide; 2-O-sulfotransferase transfers a sulfo group to the 2-OH position of IdoA or GlcA saccharides to form IdoA2S and the less common GlcA2S, respectively; while three isoforms of 6-O-sulfotransferase transfer sulfo groups to the 6-OH position of GlcNs to form either GlcNS6S or GlcNAc6S saccharides. The 3-O-sulfotransferases (3-OSTs) transfer a sulfo group to the 3-OH of GlcNS(6S) to form GlcNS(6S)3S saccharides. There are seven different isoforms of 3-OST, including isoforms 1, 2, 3A, 3B, 4, 5, and 6, in the human genome.
Compared with other sulfation types, the level of 3-O-sulfation is low in HS isolated from biological sources, but it is critically important for biological functions of many HS (5). 3-O-sulfation is known to be essential for the anticoagulant activity of HS. As an analog of HS, heparin is a widely prescribed anticoagulant drug in hospitals and contains the 3-O-sulfation, embedded within a specific pentasaccharide sequence. Removal of the 3-O-sulfation reduces the anticoagulant activity by 1,000-fold (6). In addition, 3-O-sulfated HS serves as entry receptors for herpes simplex virus (HSV) into host cells to establish infection (7), regulate axon guidance and growth of neurons (8), and control the progenitor cell expansion for salivary gland development (9). The 3-O-sulfated HS also involves the antithrombin-mediated anti-inflammatory effect in mice (10) and enhances tau internalization into cells (11). Effects in other model organisms were also reported, including neurogenesis in Drosophila (12), neurite branching in Caenorhabditis elegans (C. elegans) (13), and cardiac contraction in zebra fish (14). Interestingly, it was found that different saccharide sequences around the 3-O-sulfated GlcN saccharides contribute to the distinct biological functions and that the 3-OST isoform specificities are experimentally distinguishable (15). The connection between the biological functions and saccharide sequences carrying 3-O-sulfated glucosamine (GlcNS3S ± 6S) has attracted considerable interest from chemists and biologists.
In recent years, significant advancements in the development of chemical and biochemical tools to study 3-O-sulfated HS have greatly accelerated HS research. First, structurally homogeneous 3-O-sulfated oligosaccharides are now possible by either chemical synthesis (16) or a chemoenzymatic approach (17, 18). The sulfotransferases, in combination with C5-epimerase and heparosan synthase 2 from Pasteurella multocida, can be used to synthesize homogeneous HS oligosaccharides with specific sulfation patterns and are commercially available. Second, crystal structures of 3-O-sulfotransferase isoforms, including -1, -3, and -5, complexed with oligosaccharide substrates have emerged. The structures offer atomic-level detail into the interactions between enzymes and saccharides, uncovering the substrate recognition mechanisms of the sulfotransferases. Third, sensitive and quantitative methods for the analysis of 3-O-sulfated HS isolated from biological sources are emerging. The synthesis of structurally homogeneous HS oligosaccharides has been reviewed previously (19). Although the mechanism of 3-O-sulfotransferase has been previously discussed (20, 21), in the current review, we focus on reviewing the substrate specificity of different 3-O-sulfotransferase isoforms and the methods for the analysis of 3-O-sulfated HS.
Substrate Specificities of 3-O-Sulfotrasnferase Isoforms
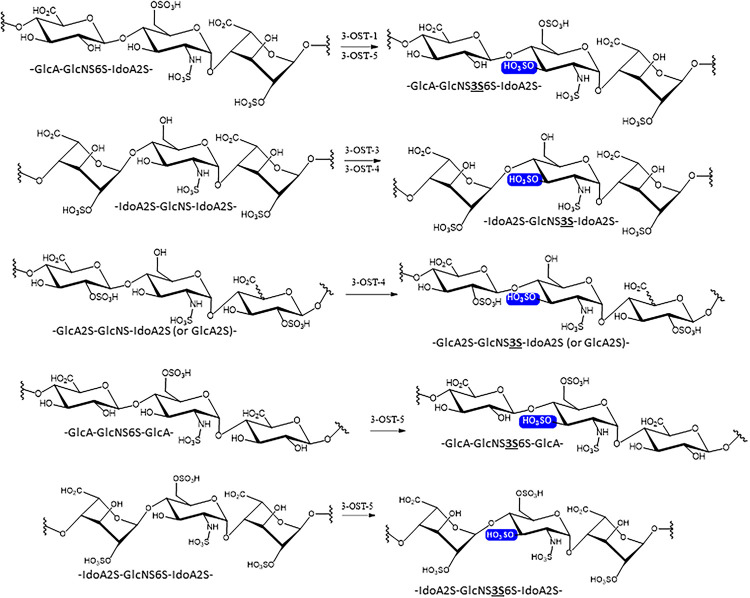
Prior to the advent of chemoenzymatic synthesis, determining the substrate specificity of sulfotransferases relied heavily on disaccharide analysis of product that utilized mostly heterogenous starting material (22). In this regard, limited general concepts of specificity had been developed. Mainly, the 3-OST-1 isoform was capable of generating the anticoagulant pentasacharide sequence GlcNAc6S-GlcA-GlcNS3S6S-IdoA2S-GlcNS6S- while 3-OST-3 (isoforms 3A and 3B) could generate the sequence -IdoA2S-GlcNS3S6S-IdoA2S- that could block HSV infection through disrupting HSV glycoprotein D (gD) protein interactions with cell surface HS (23) (Fig. 1). It was also determined that 3-OST-5 could generate both sequences but whether or not it had preference for another sequence was unknown (24, 25). With the ability to generate a variety of homogenous oligosaccharide substrates, the specificity differences between the isoforms are becoming clearer (23, 25, 26). In addition, a number of these substrates have been cocrystallized with the different 3-OST isoforms allowing the use of site-directed mutagenesis to target and probe-specific interactions between the enzyme and substrate to understand which amino acid residues define the specificity of the individual isoforms (23, 25–29).
Figure 1.
Substrate specificity of 3-OST isoforms. The site of 3-O-sulfation is highlighted with a blue box. The abbreviated trisaccharide sequences are listed underneath of each saccharide structures. The 3-O-sulfation in the abbreviated trisaccharide is emphasized in both bold face and underlined. 3-OST, 3-O-sulfotransferase.
In 2017, Wang et al. (30) published the first detailed analysis of specificity difference between 3-OST-1 and 3-OST-3 with homogeneous substrate examining the effect of 6-O-sulfation. Utilizing oligosaccharide substrates, Wang et al. (30) demonstrated that while 3-OST-1 is completely dependent upon 6-O-sulfation for activity, 3-OST-3 displays greatly reduced activity on 6-O-sulfated oligosaccharides. Surprisingly, kinetic analysis revealed that 6-O-sulfation increased binding affinity to 3-OST-3 with Km values decreasing from 2.7- to 17.6-fold. However, 6-O-sulfation also resulted in decreases in kcat from 7.7- to 20.3-fold. This suggested that 6-O-sulfated substrates bind but may also inhibit 3-OST-3 function. Utilizing identical substrates with and without 6-O-sulfation, Wander et al. (29) demonstrated that a 6-O-sulfated 8-mer could inhibit 3-OST-3 catalyzed product formation on an identical non-6-O-sulfated 8-mer with an IC50 of 0.15 µM. In addition, as the sulfation content increased, the inhibition got stronger. These results support the idea that in vivo, 3-OST-1 likely sulfates post-6-O-sulfation while 3-OST-3 may sulfate pre-6-O-sulfation. How this is regulated in the cell and the role it may play in physiological processes is an area of future interest. Analysis using chemoenzymatically synthesized oligosaccharides also provided new insights into specificity differences between 3-OST-5 versus 3-OST-1 and -3 (28, 29) (Fig. 1). While capable of generating 3-OST-1 and 3-OST-3-like products, it was determined that 3-OST-5 preferentially utilizes 6-O-sulfated oligosaccharides and prefers glucosamine acceptors in the following order: 1) flanked by GlcA residues on both sides, 2) GlcA on nonreducing end side, IdoA2S on reducing side (3-OST1-like), 3) IdoA2S on nonreducing side, GlcA on reducing side, and 4) flanked on both sides with IdoA2S (3-OST3 -like) (Fig. 1).
3-OST isoforms -2, -4, and -6 share the highest sequence conservation with 3-OST-3 so it was unsurprising that when utilizing synthesized 12-mer oligosaccharides, 3-OST-4 showed preference for substates lacking 6-sulfation and containing IdoA2S substrates (31). When presented with an oligosaccharide containing both -GlcA-GlcNS-IdoA2S- and IdoA2S-GlcNS-IdoA2S, 3-OST-4 modified only the GlcNS flanked on both sides by IdoA2S (Fig. 1). Interestingly, 3-OST-4 was also capable of sulfating GlcNS saccharides flanked on either the nonreducing or reducing ends, or both, with the rare 2-O-sulfated GlcA (GlcA2S saccharide). It remains to be seen if 3-OST-3 is also capable of this activity.
Crystal Structural Analysis of 3-OST Isoforms
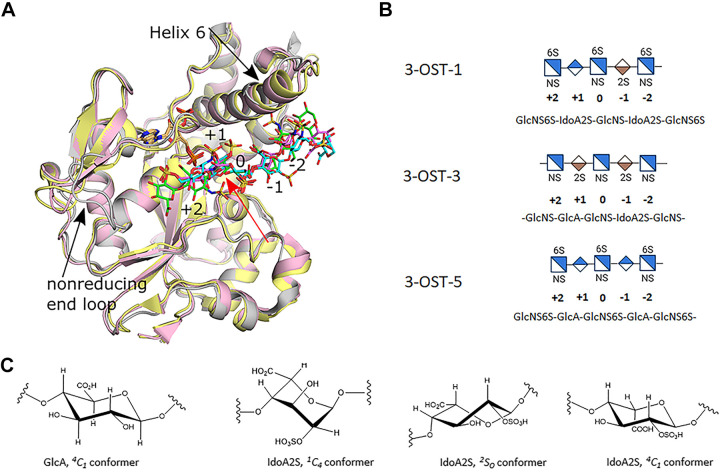
Ternary complex crystal structures of mouse 3-OST-1, as well as human 3-OST-3 and 3-OST-5 have been solved in complex with the inactive cofactor donor 3’-phosphoadenosine 5’-phosphate (PAP) and chemoenzymatically synthesized HS substrates (27–29). Notably, mouse and human 3-OST-1 share 92% sequence identity in the catalytic domain. In particular, the key residues involved in the interaction with saccharide substrates and 3’-phosphoadenosine 5’-phosphosulfate (PAPS) are identical, therefore the structure of human 3-OST-1 should closely resemble the crystal structure of mouse 3-OST-1. Combined with subsequent mutagenesis, this information has allowed for interrogation of the interactions involved in dictating specificity. The catalytic domains of 3-OST-1, -3 (3A and 3B have >99% identity in the catalytic domain), and -5 share high sequence identity (3-OST-1 to 3-OST-5 54%; 3-OST-1 to 3-OST-3 44%; and 3-OST-3 to 3-OST-5 50%) and similar overall structures with RMSDs between 0.88 and 0.95 Å deviation (Fig. 2A). Two main differences exist in the structure of the substrate binding site among these isoforms. In both cases, 3-OST-3 differs from 3-OST-1 and -5 by having a four residue shorter α-helix (known as “helix 6”) that runs across the top of the reducing end of the substrate binding pocket and a two to three residue longer loop on the nonreducing end of the pocket that appears to collapse inward, possibly due to crystal packing influences (Fig. 2A). Although the sequences of the oligosaccharides differ, they all lie in the substrate binding cleft in the same general orientation. Consistent with the biochemical data, 3-OST-1 and 3-OST-3 position the sequences -GlcA-GlcNS6S-IdoA2S and -IdoA2S-GlcNS-IdoA2S- into their active sites, respectively (Fig. 2B). 3-OST-5 was given a substrate where the enzyme could place an IdoA2S in the active site, but instead, -GlcA-GlcNS6S-GlcA- was observed bound consistent with the biochemical results of preference for flanking GlcA saccharides. For all three isoforms, the acceptor glucosamines (position 0) superimpose well with the 3-OH within 0.2 Å of each other and all in a catalytically relevant orientation with respect to the leaving group PAP (Fig. 2A). In addition, residues identified to be important in catalysis are conserved and superimposed well (20). In these structures, differences in substrate sequence and/or conformation begin at the flanking uronic acids on both sides of the acceptor glucosamine (position 0) (Fig. 2A). Glucuronic acids and glucosamine are exclusively found in the 4C1 conformation. The conformation of the IdoA2S saccharides, however, can be influenced by the saccharide sequence context, as well as potentially stabilized by protein interactions, and are typically found in the 1C4 or 2S0 and rarely in the energetically less favorable 4C1 (19, 32) (Fig. 2C). The differences contribute to different trajectories away from the active site.
Figure 2.
Crystal structures of 3-O-sulfotransferase (3-OST) isoforms-1, -3, and -5. A: superpositions of the crystal structures of the catalytic domains for 3-OST-1 (pink, substrate dark pink; PDBcode 3UAN), -3 (gray, substrate green; pdbcode 6XL8), and -5 (light yellow, substrate cyan; pdbcode 7SCE). Positions of saccharides relative to the acceptor glucosamine (0) are labeled (+) for nonreducing end and (−) for the reducing end of the substrate. Black arrows denote largest structural differences between isoforms near the HS binding cleft. The PAP for all three structures is shown in tan and superimpose well. B: the shorthand representations of the central pentasaccharide unit of the oligosaccharide substrates used in the crystal studies are shown. C: conformations of GlcA and IdoA2S found in heparan sulfate/heparin. GlcA, glucuronic acid; HS, heparan sulfate; IdoA2S, 2-O-sulfo iduronic acid; PAP, 3′-phosphoadenosine 5′-phosphate.
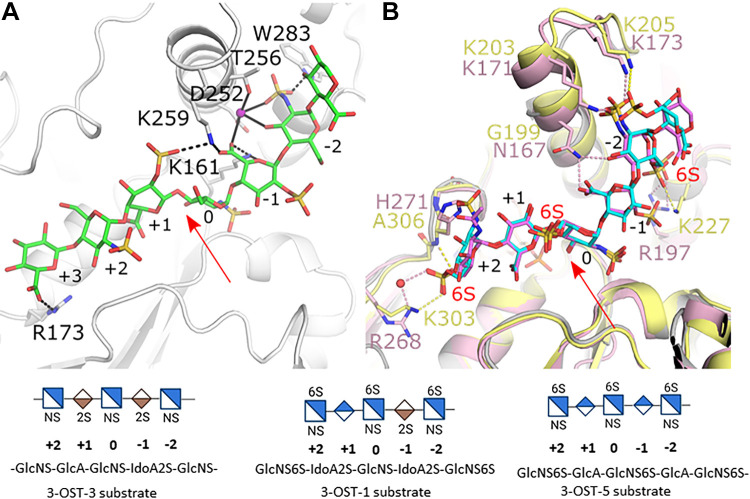
The greatest difference in substrate binding exists on the reducing side of the acceptor glucosamine (Figs. 2A, 3A, and 3B). Both 3-OST-1 and -3 have an IdoA2S at the -1 position, but for 3-OST-1 this saccharide is in the 1C4 conformation while it exists in the 2S0 conformation in 3-OST-3. For 3-OST-5, a GlcA saccharide is present in the 4C1. Contributing to the 3-OST-3 substrate binding is a sodium ion that chelates both the protein via sidechains from residues Asp252 and Thr256 and the substrate using the carboxylate of IdoA2S at the -1 position and the 3-OH and sulfo group from GlcNS at the -2 position (Fig. 3A). While Asp252 is conserved in all 3-OST’s, the equivalent residue to Thr256 is a valine in 3-OST-1 and -5, which would exclude sodium binding. In addition to interacting with the sodium, the sulfo-group of the GlcNS at the -2 position also forms hydrogen bonds with Trp283. The equivalent residue is a Tyr in both 3-OST-1 and -5, which cannot form similar interactions. The T256V and W283A mutations in 3-OST-3 reduce activity by 40% and 92%, respectively (27). This suggests that the Thr and Trp residues contribute to 3-OST-3 specificity and that these interactions may be conserved in 3-OST-2, -4, and -6 substrate binding where these residues are also conserved. The difference in saccharide at the -1 position for the 3-OST-1 and -5 substrates (IdoA2S and GlcA, respectively) generates an offset of the GlcNS6S at the -2 position by ∼1.5 Å, however, the trajectory and orientation of the GlcNS6S at the -2 position is similar (Fig. 3A). In 3-OST-1, Lys171 and Lys173 on the extended helix 6 both interact with the NS group of the GlcNS6S at the -2 position (Fig. 3B). These lysines are conserved in 3-OST-5 (Lys203 and 205, respectively) and while Lys203 is disordered, Lys205 forms a similar interaction with the NS group as Lys173 in 3-OST-1 suggesting how the extended helix may contribute to substrate binding in 3-OST-1 and 3-OST-5.
Figure 3.
Differences in HS binding between the 3-O-sulfotransferase (3-OST) isoforms -1, -3, and -5. A: Crystal structure of the catalytic domain of 3-OST-3 binding oligosaccharide substrate (protein gray, HS green; pdbcode 6XL8). B: superposition of crystal structures of 3-OST-1 (pink, substrate dark pink; PDBcode 3UAN) and -5 (light yellow, substrate cyan; pdbcode 7SCE). Residues discussed in text are displayed in stick. Red arrows denote the acceptor 3-OH on glucosamine 0. Pink, black, and yellow dashed lines represent interactions between HS and 3-OST-1, -3, and -5, respectively. Solid black lines represent interactions in 3-OST-3 with the sodium ion. Shorthand representation of the substrates in the active site are shown at the bottom of the figure. HS, heparan sulfate.
On the nonreducing side of the acceptor at saccharide position +1, both 3-OST-1 and 3-OST-5 substrates contain GlcA in the 4C1 conformation and follow similar trajectories across the substrate binding pocket (Fig. 3B). For 3-OST-3, the equivalent saccharide is an IdoA2S observed in the 2SO conformation, resulting in a deviation of the substrate path (Fig. 3, A and B). For 3-OST-3, the 2-O-sulfo group on the +1 IdoA2S is only 3.7 Å from the carboxylate of IdoA2S at the -1 position (Fig. 3A). 3-OST-3 possesses a lysine (residue 259) at the end of its shorter helix 6 that is within hydrogen bonding distance of both the 2-sulfo group and the carboxylate from the two IdoA2Ss that can potentially alleviate the charge repulsion and accommodate flanking IdoA2S residues. This lysine is also conserved in isoforms -2, -4, and -6, but not in 3-OST-1 and -5 (Asn167 and Gly199, respectively) supporting a preference for GlcA at the +1 position for 3-OST-1 and -5. Arg173 of 3-OST-3 forms a hydrogen bond with the carboxylate of GlcA at the +3 position of the pocket. The R173A and R173S mutations retain only 46% and 34% wt activity respectively, suggesting that Arg173 contributes to the specificity 3-OST-3-like isoforms. Arg173 of 3-OST-3 is not conserved in 3-OST-1 or -5 (28). While 3-OST-5 does prefer flanking GlcA to the acceptor glucosamine, it will sulfate the 3-OST-1 substrate. Comparing the structures, 3-OST-5 could likely accommodate the 3-OST-1 substrate within its binding pocket without steric conflict albeit with fewer interactions than when binding to 3-OST-1 (such as Asn167 of 3-OST-1; see Fig. 3B). However, it is likely that the 3-OST-3 substrate would have to bind in a different conformation than found in 3-OST-3 due to potential electrostatic clashes between the IdoA2S saccharides at the +1 and -1 positions and between the substrate and protein.
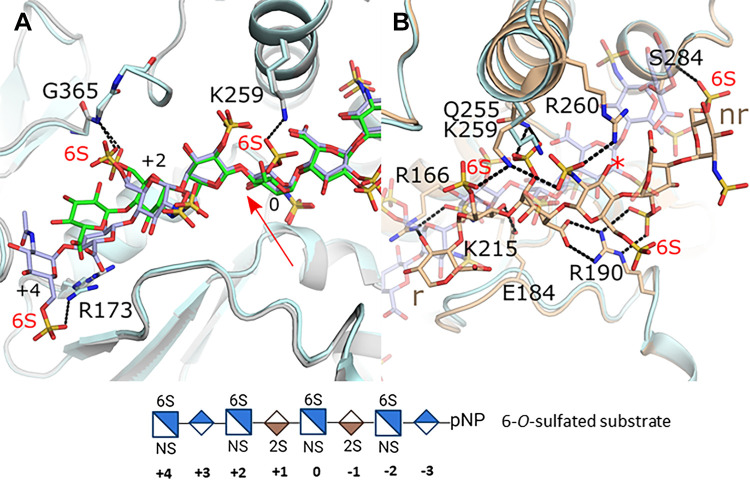
Another major substrate difference between the isoforms is the preferences for a 6-O-sulfated substate by 3-OST-1 and -5. Within the active site of the crystal structures for 3-OST-1 and -5 are three 6-O-sulfated glucosamines. In both structures, the 6-O-sulfo group on the acceptor glucosamine at position 0 does not interact with the protein (Fig. 3B). This is consistent with the in vivo study by Karlsson et al. (33), which demonstrated that when all 3-OSTs were over expressed, the largest increase in 3-O-sulfation appeared on non-6-O-sulfated glucosamines. To better understand how 6-O-sulfation enhanced binding to 3-OST-3 but decreased activity, the structure was solved with a 6-O-sulfated substrate (20). The crystal structure contained two ternary complexes, each binding the substrate in a different orientation (Fig. 4). In one of the 3-OST-3 molecules, the saccharide binds in a productive binding mode similar to the non-6-O-sulfated saccharide (Fig. 4A). In this case the 6-O-sulfo group at the -2 position did not form interactions with the protein while the 6-O-sulfo group of the acceptor glucosamine (position 0) was within hydrogen bonding distance of Lys259. The 6-O-sulfo group on the glucosamine at the +2 position occupied a similar location as seen for 3-OST-1 and -5 but displayed two conformations, both still interacting with the equivalent backbone amide (Gly365). The addition of the 6-O-sulfo groups results in a shift of the +2 glucosamine and subsequent saccharides including the ordering of the +4 glucosamine that forms an interaction between its 6-sulfo group and the sidechain of Arg173 that has shifted to accommodate this substrate. Thus, 6-sulfation generates additional interactions with the protein that may account for the tighter binding. Interestingly, in the second 3-OST-3 molecule in the crystal the substrate bound in a nonproductive mode with reverse polarity across the binding pocket that positioned the acceptor glucosamine far from the active site (Fig. 4B). In this binding mode, in addition to the many other unique protein-saccharide interactions, two of the three observed 6-sulfo groups form interactions with the protein. The first-ordered glucosamine at the nonreducing end forms a novel interaction with the backbone of Ser284 while the 6-O-sulfo group from the reducing end forms a hydrogen bond with the shifted Lys259. The X-ray crystal structures suggest that 6-O-sulfo groups may not simply exclude oligosaccharides from binding in a catalytically relevant orientation, but rather allow for additional interactions that potentially enhance binding as well as allow for binding in nonproductive modes (29). The presence of 3-O-sulfo groups in addition to the 6-O-sulfo groups appeared to increase inhibition suggesting that product inhibition may enhance the inhibition observed with 6-O-sulfated oligosaccharides (29). In addition to a nonproductive binding mode, inhibition with both substrate and product could occur through dead-end complexes whereby binding of the oligosaccharide stabilizes the binding of PAP (3'-phosphoadenosine 5'-phosphate), inhibiting its exchange or blocking access of PAPS to its binding pocket. Product inhibition has also been observed for 3-OST-5 (28).
Figure 4.
Binding by a 6-O-sulfated oligosaccharide substrate to 3-OST-3. A: superposition of 3-OST-3 binding HS without 6-O-sulfo groups (protein gray, HS green; pdbcode 6XL8) with 3-OST-3 binding 6-O-sulfo containing HS in a productive binding mode consistent with catalysis (protein light blue, HS light purple; PDB 6XKG molecule B). Red arrow denotes acceptor 3OH position. B: superposition of the productive binding mode of HS with 6-O-sulfo group (protein light blue, HS light purple/transparent; PDB 6XKG 3-OST-3 molecule B) with 3-OST-3 binding the same 8-mer oligosaccharide in a nonproductive manner (protein and HS in tan; PDB 6XKG 3-OST-3 molecule A). The red asterisk is positioned at the 3OH that would typically be found bound at the active site near the PAP as in A. The structure of the octasaccharide substrate with 6-O-sulfation used in the crystal study is shown at the bottom of the figure. HS, heparan sulfate; PAP, 3′-phosphoadenosine 5′-phosphate; 3-OST, 3-O-sulfotransferase.
These studies have helped explain the substrate preferences for 3-OST-1 versus 3-OST-3 as well as identify a novel preferred substrate for 3-OST-5. We have also garnered insight into the molecular requirements for 6-sulfation for 3-OST-1 and 3-OST-5 and how 6-O-sulfation may contribute to 3-OST-3 inactivity via strong productive and nonproductive binding. This information is critical in understanding how to utilize the various enzymes in sequential modification during chemoenzymatic synthesis. These studies suggest that the temporal regulation of 3-OST and 6-OST isoforms may be utilized by the cell to generate HS with different 3-O-sulfation patterns with respect to the saccharide content.
Analysis of 3-O-Sulfated HS from Biological Sources
There is considerable interest in the HS-research field to analyze 3-O-sulfated HS from biological sources with a high degree of accuracy. In addition to its low abundance, the 3-O-sulfated glucosamines are present in different saccharide sequences. For example, Chen et al. (34) isolated five different tetrasaccharides from pharmaceutical heparin after digestion by heparin lyases. Others also reportedly isolated different hexasaccharides, octasaccharides, and octadecasaccharides carrying 3-O-sulfation from pharmaceutical heparin (35, 36). The low abundance in 3-O-sulfated HS and the structural variability that exists necessitates the need for a highly sensitive approach to carry out the analysis. A commonly used approach among biologists to measure the level of 3-O-sulfated HS is to use fluorescently labeled antithrombin (37) or antibody (38), which is compatible with typical histochemistry studies. However, the results merely offer qualitative measurement for the level of 3-O-sulfated HS and the results lack the information on specific saccharides around the 3-O-sulfation sites, which is critical for determining the responsible 3-OST isoform. In fact, antithrombin can bind to multiple saccharide sequences containing 3-O-sulfated glucosamine residue. NMR analysis has been the gold standard approach to detect the saccharide sequences among 3-O-sulfated HS variants (39), but this method requires very large quantity of sample and highly trained individuals. NMR analysis is currently too difficult to become a standard method for the analysis of 3-O-sulfation from biological sources particularly in a high-throughput fashion.
Another approach to analyze HS is to reduce HS polysaccharide into smaller structural units followed by compositional analysis of the individual structural units. Disaccharide compositional analysis is a common technique for analyzing HS. The process involves digestion with nitrous acid to degrade the polysaccharides into disaccharides. This allows for assessment of the degree of 3-O-sulfated HS in the sample based on the amount of 3-O-sulfated disaccharides isolated (40). This approach, however, involves the use of radioactive isotope labelling for the detection (41). The method is applied in analyzing pharmaceutical heparin, which the amount of sample is unlimited, or the HS from cell lines, from which the HS is metabolically labeled with radioactive isotopes. Despite the success, whether nitrous acid degradation leads to desulfation of the disaccharide products has not been fully addressed, raising the question over the degree of accuracy.
Nitrous acid degradation method has been gradually replaced with enzymatic degradation, using mixtures of three heparin lyases (42). A complete enzyme digestion to disaccharide can be readily achieved under mild conditions and the resulting HS disaccharides are well separated by liquid chromatography (LC). Typically, eight different disaccharides are liberated from HS after heparin lyase digestion, and the disaccharide standards are commercially available. Coupling a mass spectrometer to LC dramatically increases the sensitivity for the disaccharide compositional analysis (43). Currently, liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) is being widely used, offering very high sensitivity for the analysis. To add the quantitative capability for the HS disaccharide compositional analysis, a series of 13C-labeled HS disaccharide calibrants are included. The LC-MS/MS method has been demonstrated to offer sufficient sensitivity to quantify HS from plasma and tissues (44, 45). Unfortunately, the standard disaccharide analysis generally is not well suited for the analysis of 3-O-sulfated HS for two reasons. First, the majority of structural motifs containing the 3-O-sulfation are resistant to the digestion by heparin lyases, forming tetrasaccharides not disaccharides (46, 47). In addition, 3-O-sulfated tetrasaccharides and disaccharide undergo spontaneous degradation, further complicating the efforts to quantify the level of 3-O-sulfated HS (46, 48).
With improved access to synthetic 3-O-sulfated oligosaccharides, two methods have been recently published to analyze 3-O-sulfated HS (33, 49). In one study, Karlsson et al. (33) report using a standard disaccharide analysis. The authors claimed that 3-O-sulfated heparin or HS was degraded to 3-O-sulfated disaccharides after using excessive amounts of heparin lyases. The disaccharides are conjugated with 2-aminoacridone (AMAC), which coupled a fluorophore to the disaccharides. Authors then demonstrated separation of both 3-O-sulfated disaccharides and non-3-O-sulfated disaccharides using a LC method coupled with a fluorescence detector. The method was primarily designed for determining the levels of 3-O-sulfated saccharide motifs from the bioengineered HS produced by Chinese hamster ovary (CHO) cell culture. The method was also used for the analysis of different brands of low-molecular weight heparin. Examples for using this method for the analysis of HS directly isolated from animal tissues or biological fluids to demonstrate its sensitivity were not presented.
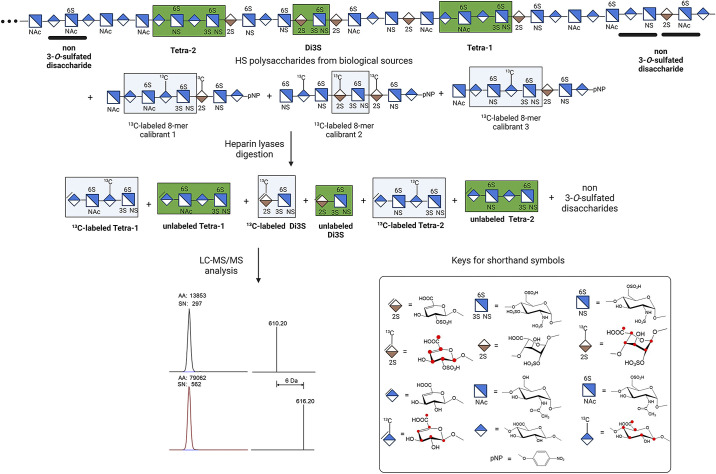
Another study by Wang et al. (49) reported the use of 13C-labeled oligosaccharides to analyze 3-O-sulfated HS from biological sources (Fig. 5). In this method, authors synthesized three 13C-labeled 8-mers carrying GlcNS3S6S in different saccharide contexts. The calibrants are mixed with HS from biological sources followed by the digestion with heparin lyases to yield both 13C-labeled and unlabeled 3-O-sulfated tetrasaccharides (Tetra-1 and Tetra-2), one 3-O-sulfated disaccharide (Di3S) and other non-3-O-sulfated disaccharides (Fig. 5). The samples were then subjected to LC-MS/MS analysis. Both 13C-labeled and unlabeled tetrasaccharides were detected. The molecular weight of 13C-labeled tetrasaccharides are 6 Da higher than their unlabeled tetrasaccharide counterparts. The ratio of the peak areas from 13C-labeled tetrasaccharides and unlabeled tetrasaccharide counterparts allows for quantitation of unlabeled 3-O-sulfated tetrasccharides (Fig. 5). In addition to analyzing the 3-O-sulfation context and quantity in pharmaceutical heparin, the method can determine the relatively low level of 3-O-sulfated HS from mouse plasma after the animals received subcutaneous injection of low-molecular weight heparin. In the same study, this method was also used to detect the level of 3-O-sulfated HS from brain tissue of patients with Alzheimer’s disease (49). An elevation of specific 3-O-sulfated HS with a sequence of ΔUA-GlcNAc6S-GlcA-GlcNS3S6S was detected in Alzheimer’s disease patients. Although authors did not specify the detection limits for the method, they report, for the first time, the detection of 3-O-sulfated HS in human brain, corroborating the findings that the levels of 3-OST-2, -3, and -4 are increased in hippocampus from patients with Alzheimer’s disease (50).
Figure 5.
Analysis of 3-O-sulfated HS using a LC-MS/MS method. The HS polysaccharides from biological sources are mixed with known amounts of 13C-labeled 8-mer calibrants. The mixture is subjected to the digestion with heparin lyases. The 3-O-sulfated oligosaccharides, presented as Tetra-1, Tetra-2, and Di3S are liberated along with non-3-O-sulfated disaccharides. The products are then analyzed by LC-MS/MS. Comparing the ratio of peak area 13C-labeled Tetra-1 and unlabeled Tetra, one can determine the amount of Tetra-1 from the HS polysaccharides. Red dots denote the location of the 13C-atoms. HS, heparan sulfate; LC-MS/MS, liquid chromatography coupled with tandem mass spectrometry.
The use of 13C-labeled oligosaccharides plays critical roles in enabling the analysis of 3-O-sulfated HS from biological samples. The 13C-labeled oligosaccharides are chemically stable and the yield of oligosaccharides from one batch synthesis is sufficient for the analysis of hundreds of thousands of samples with semihigh-throughput capability. In addition, tetrasaccharide analysis allows for a better understanding of the surrounding saccharide/sulfation pattern than disaccharide analysis, providing information as to which isoform of 3-OST may have been involved in generating the 3-O-sulfation. One limitation for the analysis of 3-O-sulfated HS is that the actual number of 3-O-sulfated structures in nature is unknown. Further expansion for the synthesis of diversified 13C-labeled 3-O-sulfated oligosaccharide calibrants will improve the coverage of the accuracy for the 3-O-sulfated structural analysis. The ability to identify the relevant isoform helps to assign biological context for the individual isoforms.
Cell-Based Synthesis of 3-O-Sulfated HS
A cell-based approach has recently been utilized to study the products of 3-O-sulfotransferases isoforms (33). Using a CHO cell line deficient in chondroitin/dermatan sulfate and overexpressing individual 3OST’s, Karlsson et al. (33) used disaccharide analysis to study the changes in HS produced by the cell. While 3-OST-6 did not appear to show changes in 3-O-sulfation, all other cell lines expressing 3-OSTs showed the largest increase in 3-sulfation on acceptor glucosamines with no 6-sulfation, including 3-OST-1. 3-OST-1 expressing cells only showed an increase in 3-sulfation on glucosamines with an unsulfated uronic acid on the nonreducing side as expected. 3-OST-2 and -5 both showed the largest increase in 3-O-sulfation on glucosamine linked to uronic acids with a 2-O-sulfo group on the nonreducing side while 3-OST-3A, -3B, and -4 all showed similar increases with both 2-O-sulfo and no 2-O-sulfo on the uronic acid on the reducing side. This methodology is currently unable to distinguish the identity of the uronic acid on either side of the acceptor or the presence of 6-O-sulfation on proximal glucosamines. However, this study revealed that HS produced by the modified CHO cells overexpressing 3-OST isoforms -3A, -3B, and -4 could all generate anticoagulant HS. The HS produced with 3-OST-4 demonstrated reduced binding of platelet factor 4 (PF4), which theoretically would reduce likelihood of inducing heparin-induced thrombocytopenia (HIT). This is consistent with the work performed by Li et al. (31), which demonstrated that 3-OST-4 could be used to generate anti-FXa HS. In addition, Wang et al. (30) demonstrated that 3-OST-3 could produce a homogenous anticoagulant octasaccharide containing a glucosamine flanked on both sides by IdoA2S saccharides with a similar binding affinity to AT and IC50 for inhibiting the activity of FXa as that of the pentasaccharide anticoagulant but with faster clearance times in a mouse model. The accumulation of this information suggests the possibility to utilize different 3-OST isoforms to produce protamine reversible anticoagulant oligosaccharides, with reduced HIT, and faster clearance rates providing for tighter control of clotting during surgical procedures or for other applications. Whether the substrate specificities of 3-OST isoforms expressed in modified cell-based assay accurately represent in vivo synthesis in animals and humans remain unknown. The newly emerged LC-MS/MS-based method for the analysis of 3-O-sulfated HS from animal and human tissues should provide the analytical tool to answer the question.
Conclusions
HS isolated from biological sources is a structurally complex mixture. 3-O-sulfated HS is a unique subpopulation because of its low abundance and close relationship with biological functions of HS. The lack of the ability to study the 3-O-sulfated HS has been a hurdle in HS research. New tools have been developed for investigating the biological roles of 3-O-sulfated HS. The progress is largely attributed to the availability of structurally homogeneous oligosaccharides. It should be noted that a 3-O-sulfated HS oligosaccharide microarray array is now available, offering a new approach to studying the binding of proteins and 3-O-sulfated oligosaccharides in a high-throughput manner (16). The improved knowledge of the substrate specificity of 3-OST isoforms will further enhance the ability to synthesize the oligosaccharides and the LC-MS/MS-based method for the analysis of 3-O-sulfated HS from biological sources. Because 3-OST enzymes and saccharide substrates involve both specific and nonspecific interactions, the crystal structural information offers a model to understand how proteins select HS binding partners to exhibit the biological functions.
GRANTS
This work was supported in part by the Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health Grant 1ZIA ES102645 (to L.C.P.) and by NIH Grants HL094463 and HL144970 (to J.L.).
DISCLOSURES
J.L. is a founder of Glycan Therapeutics and has equity. J.L. laboratory at University of North Carolina has received a gift from Glycan Therapeutics to support research in glycosciences. L.C.P. declares no competing interest.
This article is part of the special collection “Deciphering the Role of Proteoglycans and Glycosaminoglycans in Health and Disease.” Liliana Schaefer, MD, served as Guest Editor of this collection.
AUTHOR CONTRIBUTIONS
J.L. and L.C.P. conceived and designed research; L.C.P. analyzed data; L.C.P. interpreted results of experiments; J.L. and L.C.P. prepared figures; J.L. and L.C.P. drafted manuscript; J.L. and L.C.P. edited and revised manuscript; J.L. and L.C.P. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank A.M. Kaminski and Lalith Perera for thoughtful discussion and critical reading of the manuscript.
REFERENCES
- 1.Linhardt RJ, Liu J. Synthetic heparin. Curr Opin Pharmacol 12: 217–219, 2012. doi: 10.1016/j.coph.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold K, Liao Y-E, Liu J. Potential use of anti-inflammatory synthetic heparan sulfate to attenuate liver damage. Biomedicines 8: 503, 2020. doi: 10.3390/biomedicines8110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer 5: 526–542, 2005. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 4.Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183: 1043–1057, 2020. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thacker B, Xu D, Lawrence R, Esko J. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol 35: 60–72, 2014. doi: 10.1016/j.matbio.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atha DH, Lormeau J-C, Petitou M, Rosenberg RD, Choay J. Contribution of monosaccharide residues in heparin binding to antithrombin III. Biochemistry 24: 6723–6729, 1985. doi: 10.1021/bi00344a063. [DOI] [PubMed] [Google Scholar]
- 7.Shukla D, Rowe CL, Dong Y, Racaniello VR, Spear PG. The murine homolog (MpH) of human herpesvirus entry protein B (HveB) mediates entry of pseudorabies virus but not herpes simplex virus types 1 and 2. J Virol 73: 4493–4497, 1999. doi: 10.1128/JVI.73.5.4493-4497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thacker BE, Seamen E, Lawrence R, Parker MW, Xu Y, Liu J, Vander KCW, Esko JD. Expanding the 3-O-sulfate proteome–enhanced binding of neuropilin-1 to 3-O-sulfated heparan sulfate modulates its activity. ACS Chem Biol 11: 971–980, 2016. doi: 10.1021/acschembio.5b00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel VN, Lombaert IMA, Cowherd SN, Shworak N, Xu Y, Liu J, Hoffman MP. Hs3st3-modified heparan sulfate controls KIT+ progenitor expansion by regulating 3-O-sulfotransferases. Dev Cell 29: 662–673, 2014. doi: 10.1016/j.devcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smits NC, Kobayashi T, Srivastava PK, Skopelja S, Ivy JA, Elwood DJ, Stan RV, Tsongalis GJ, Sellke FW, Gross PL, Cole MD, DeVries JT, Kaplan AV, Robb JF, Williams SM, Shworak NW. HS3ST1 genotype regulates antithrombin’s inflammomodulatory tone and associates with atherosclerosis. Matrix Biol 63: 69–90, 2017. doi: 10.1016/j.matbio.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Zhu Y, Song X, Xiao Y, Su G, Liu X, Wang Z, Xu Y, Liu J, Eliezer D, Ramlall TE, Lippens G, Gibson J, Zhang F, Linhardt RJ, Wang L, Wang C. 3-O-sulfation of heparan sulfate enhances tau interaction and cellular uptake. Angew Chem Int Ed Engl 59: 1818–1827, 2020. doi: 10.1002/anie.201913029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamimura K, Rhodes JM, Ueda R, McNeely M, Shukla D, Kimata K, Spear PG, Shworak NW, Nakato H. Regulation of notch signaling by Drosophila heparan sulfate 3-O sulfotransferase. J Cell Biol 166: 1069–1079, 2004. doi: 10.1083/jcb.200403077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Díaz-Balzac CA, Lázaro-Peña MI, Tecle E, Gomez N, Bülow HE. Complex cooperative functions of heparan sulfate proteoglycans shape nervous system development in Caenorhabditis elegans. G3 (Bethesda) 4: 1859–1870, 2014. doi: 10.1534/g3.114.012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsau C, Ito M, Gromova A, Hoffman MP, Meech R, Makarenkova HP. Barx2 and Fgf10 regulate ocular glands branching morphogenesis by controlling extracellular matrix remodeling. Development 138: 3307–3317, 2011. doi: 10.1242/dev.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Pedersen LC. Anticoagulant heparan sulfate: structural specificity and biosynthesis. Appl Microbiol Biotechnol 74: 263–272, 2007. doi: 10.1007/s00253-006-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chopra P, Jposhi A, Wu J, Lu W, Yadavalli T, Wolfert MA, Shukla D, Zaia J, Boons G-J. The 3-O-sulfation of heparan sulfate modulates protein binding and lyase degradation. Proc Natl Acad Sci USA 118: e2012935118, 2021. doi: 10.1073/pnas.2012935118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Cai C, Chandarajoti K, Hsieh P, Lin Y, Pham TQ, Sparkenbaugh EM, Sheng J, Key NS, Pawlinski RL, Harris EN, Linhardt RJ, Liu J. Homogeneous and reversible low-molecular weight heparins with reversible anticoagulant activity. Nat Chem Biol 10: 248–250, 2014. doi: 10.1038/nchembio.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Pagadala V, Jester HM, Lim AM, Pham TQ, Goulas AMP, Liu J, Linhardt RJ. Chemoenzymatic synthesis of heparan sulfate and heparin oligosaccharides and NMR analysis: Paving the way to a diverse library for glycobiologists. Chem Sci 8: 7932–7940, 2017.doi: 10.1039/c7sc03541a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Linhardt RJ. Chemoenzymatic synthesis of heparan sulfate and heparin. Nat Prod Rep 31: 1676–1685, 2014. doi: 10.1039/c4np00076e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edavettal SC, Lee KA, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J Biol Chem 279: 25789–25797, 2004. doi: 10.1074/jbc.M401089200. [DOI] [PubMed] [Google Scholar]
- 21.Edavettal SC, Carrick K, Shah RR, Pedersen LC, Tropsha A, Pope RM, Liu J. A conformational change in heparan sulfate 3-O-sulfotransferase-1 is induced by binding to heparan sulfate. Biochemistry 43: 4680–4688, 2004. doi: 10.1021/bi0499112. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Shworak NW, Sinaÿ P, Schwartz JJ, Zhang L, Fritze LMS, Rosenberg RD. Expression of heparan sulfate d-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. J Biol Chem 274: 5185–5192, 1999. doi: 10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Moon AF, Sheng J, Pedersen LC. Understanding the substrate specificity of the heparan sulfate sulfotransferases by an integrated synthetic crystallographic approach. Curr Opin Struct Biol 22: 550–557, 2012. doi: 10.1016/j.sbi.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia G, Chen J, Tiwari V, Ju W, Li J-P, Malmström A, Shukla D, Liu J. Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J Biol Chem 277: 37912–37919, 2002. doi: 10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- 25.Xu D, Moon A, Song D, Pedersen LC, Liu J. Engineering sulfotransferases to modify heparan sulfate. Nat Chem Biol 4: 200–202, 2008. doi: 10.1038/nchembio.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon A, Edavettal SC, Krahn JX, Munoz EM, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Structural analysis of the sulfotransferase (3-O-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor of herpes simplex virus 1. J Biol Chem 279: 45185–45193, 2004. doi: 10.1074/jbc.M405013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon AF, Xu Y, Woody S, Krahn JM, Linhardt RJ, Liu J, Pedersen LC. Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin. Proc Natl Acad Sci USA 109: 5265–5270, 2012. doi: 10.1073/pnas.1117923109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wander R, Kaminski AM, Wang Z, Stancanelli E, Xu Y, Pagadala V, Li J, Krahn JM, Pham TQ, Liu J, Pedersen LC. Structural and substrate specificity analysis of 3-O-sulfotransferase isoform 5 to synthesize heparan sulfate. ACS Catal 11: 14956–14966, 2021. doi: 10.1021/acscatal.1c04520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wander R, Kaminski AM, Xu Y, Pagadala V, Krahn JM, Pham TQ, Liu J, Pedersen LC. Deciphering the substrate recognition mechanisms of the heparan sulfate 3-O-sulfotransferase-3. RSC Chem Biol 2: 1239–1248, 2021. doi: 10.1039/d1cb00079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Hsieh P-H, Xu Y, Thieker D, Chai EJE, Xie S, Cooley B, Woods RJ, Chi L, Liu J. Synthesis of 3-O-sulfated oligosaccharides to understand the relationship between structures and functions of heparan sulfate. J Am Chem Soc 139: 5249–5256, 2017. doi: 10.1021/jacs.7b01923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Su G, Xu Y, Arnold K, Pagadala V, Wang C, Liu J. Synthesis of 3-O-sulfated heparan sulfate oligosaccharides using 3-O-sulfotransferase isoform 4. ACS Chem Biol 16: 2026–2035, 2021. doi: 10.1021/acschembio.1021c00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsieh P-H, Thieker DF, Guerrini M, Woods RJ, Liu J. Uncovering the relationship between sulphation patterns and conformation of iduronic acid in heparan sulphate. Sci Rep 6: 29602, 2016. doi: 10.1038/srep29602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson R, Chopra P, Joshi A, Yang Z, Vakhrushev SY, Clausen TM, Painter CD, Szekeres GP, Chen Y-H, Sandoval DR, Hansen L, Esko JD, Pagel K, Dyer DP, Turnbull JE, Clausen H, Boons G-J, Miller RL. Dissecting structure-function of 3-O-sulfated heparin and engineered heparan sulfates. Sci Adv 7: eabl6026, 2021. doi: 10.1126/sciadv.abl6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Lin L, Agyekum I, Zhang X, St. Ange K, Yu Y, Zhang F, Liu J, Amster IJ, Linhardt RJ. Structural analysis of heparin-derived 3-O-sulfated tetrasaccharides: antithrombin binding site variants. J Pharm Sci 106: 973–981, 2017. doi: 10.1016/j.xphs.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guerrini M, Elli S, Mourier P, Rudd TR, Gaudesi D, Casu B, Boudier C, Torri G, Viskov C. An unusual antithrombin-binding heparin octasaccharide with an additional 3-O-sulfated glucosamine in the active pentasaccharide sequence. Biochem J 449: 343–351, 2013. doi: 10.1042/BJ20121309. [DOI] [PubMed] [Google Scholar]
- 36.Mourier PAJ, Guichard OY, Herman F, Viskov C. Isolation of a pure octadecasaccharide with antithrombin activity from an ultra-low-molecular-weight heparin. Anal Biochem 453: 7–15, 2014. doi: 10.1016/j.ab.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Kalita M, Chua JS, Boothello RS, Joice A, Antelope O, Roy A, Anandh Babu PV, Saijoh Y, Desai UR, Kuberan B. Visualizing antithrombin-binding 3-O-sulfated heparan sulfate motifs on cell surfaces. Chem Commun (Camb) 56: 14423–14426, 2020. doi: 10.1039/d0cc05893a. [DOI] [PubMed] [Google Scholar]
- 38.Ten Dam GB, Kurup S, van de Westerlo EM, Versteeg EM, Lindahl U, Spillmann D, van Kuppevelt TH. 3-O-sulfated oligosaccharide structures are recognized by anti-heparan sulfate antibody HS4C3. J Biol Chem 281: 4654–4662, 2006. doi: 10.1074/jbc.M506357200. [DOI] [PubMed] [Google Scholar]
- 39.de Agostini AI, Dong J-C, de Vantéry Arrighi C, Ramus M-A, Dentand-Quadri I, Thalmann S, Ventura P, Ibecheole V, Monge F, Fischer A-M, HajMohammadi S, Shworak NW, Zhang L, Zhang Z, Linhardt RJ. Human follicular fluid heparan sulfate contains abundant 3-O-sulfated chains with anticoagulant activity. J Biol Chem 283: 28115–28124, 2008. doi: 10.1074/jbc.M805338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Shworak NW, Fritze LMS, Edelberg JM, Rosenberg RD. Purification of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. J Biol Chem . 271: 27072–27082, 1996. doi: 10.1074/jbc.271.43.27072. [DOI] [PubMed] [Google Scholar]
- 41.Lindahl U, Kusche-Gullberg M, Kjellén L. Regulated diversity of heparan sulfate. J Biol Chem 273: 24979–24982, 1998. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 42.Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Sequencing complex polysaccharides. Science 286: 537–542, 1999. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- 43.Li G, Li L, Tian F, Zhang L, Xue C, Linhardt RJ. Glycosaminoglycanomics of cultured cells using a rapid and sensitive LC-MS/MS approach. ACS Chem Biol 10: 1303–1310, 2015. doi: 10.1021/acschembio.5b00011. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Arnold K, Xu Y, Pagadala V, Su G, Myatt H, Linhardt RJ, Liu J. Quantitative analysis of heparan sulfate using isotopically labeled calibrants. Commun Biol 3: 425, 2020. doi: 10.1038/s42003-020-01150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Dhurandhare VD, Mahung CA, Arnold K, Li J, Su G, Xu D, Maile R, Liu J. Improving the sensitivity for quantifying heparan sulfate from biological samples. Anal Chem 93: 11191–11199, 2021. doi: 10.1021/acs.analchem.1c01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhurandhare VM, Pagadala V, Ferreira A, Muynck LD, Liu J. Synthesis of 3-O-sulfated disaccharide and tetrasaccharide standards for compositional analysis of heparan sulfate. Biochemistry 59: 3186–3192, 2020. doi: 10.1021/acs.biochem.9b00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada S, Yoshida K, Sugiura M, Sugahara K, Khoo KH, Morris HR, Dell A. Structural studies on the bacterial lyase-resistant tetrasaccharides derived from the antithrombin III-binding site of porcine intestinal heparin. J Biol Chem 268: 4780–4787, 1993. doi: 10.1016/S0021-9258(18)53465-7. [DOI] [PubMed] [Google Scholar]
- 48.Huang Y, Mao Y, Zong C, Lin C, Boons G-J, Zaia J. Discovery of a heparan sulfate 3-O-sulfation specific pealing reaction. Anal Chem 87: 592–600, 2015. doi: 10.1021/ac503248k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Arnold K, Dhurandahare VM, Xu Y, Pagadala V, Labra E, Jeske W, Fareed J, Gearing M, Liu J. Analysis of 3-O-sulfated heparan sulfate using isotopically labeled oligosaccharide calibrants. Anal Chem 94: 2950–2957, 2022. [Erratum in Anal Chem 94: 4134, 2022]. doi: 10.1021/acs.analchem.1c04965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sepulveda-Diaz JE, Alavi Naini SM, Huynh MB, Ouidja MO, Yanicostas C, Chantepie S, Villares J, Lamari F, Jospin E, van Kuppevelt TH, Mensah-Nyagan AG, Raisman-Vozari R, Soussi-Yanicostas N, Papy-Garcia D. HS3ST2 expression is critical for the abnormal phosphorylation of tau in Alzheimer’s disease-related tau pathology. Brain 138: 1339–1354, 2015. doi: 10.1093/brain/awv056. [DOI] [PMC free article] [PubMed] [Google Scholar]