Significance
Polyploidy, which occurs in roughly half of all flowering plants and an even higher percentage of grasses, is thought to be a major driver of adaptation. Higher numbers of copies of each gene in polyploid genomes can increase genetic diversity, which could drive shifts in habitat preference, adaptability, and fitness. To test the effects of increased ploidy, we compared genomic diversity, environmental niche, and fitness responses across climatic gradients between tetraploid and octoploid switchgrass. We found that the octoploids contained novel combinations of the ancestral tetraploid genetic diversity, which was linked to the expansion of switchgrass into unsuitable habitats for tetraploid populations. Our experiments revealed evidence of niche divergence, differential fitness, and a generalist–specialist trade-off between cytotypes.
Keywords: cytotypes, octoploid, Panicum virgatum
Abstract
Polyploidy results from whole-genome duplication and is a unique form of heritable variation with pronounced evolutionary implications. Different ploidy levels, or cytotypes, can exist within a single species, and such systems provide an opportunity to assess how ploidy variation alters phenotypic novelty, adaptability, and fitness, which can, in turn, drive the development of unique ecological niches that promote the coexistence of multiple cytotypes. Switchgrass, Panicum virgatum, is a widespread, perennial C4 grass in North America with multiple naturally occurring cytotypes, primarily tetraploids (4×) and octoploids (8×). Using a combination of genomic, quantitative genetic, landscape, and niche modeling approaches, we detect divergent levels of genetic admixture, evidence of niche differentiation, and differential environmental sensitivity between switchgrass cytotypes. Taken together, these findings support a generalist (8×)–specialist (4×) trade-off. Our results indicate that the 8× represent a unique combination of genetic variation that has allowed the expansion of switchgrass’ ecological niche and thus putatively represents a valuable breeding resource.
Polyploidy, which is the result of whole-genome duplication (WGD) through the doubling of within-species chromosome content (autopolyploidy) or the combination of genomes from distinct taxa that have hybridized (allopolyploidy), can alter the structure and diversity of heritable genetic variance among traits, leading to pronounced evolutionary implications (1–3). Approximately half of all angiosperms are polyploids, and polyploidy is thought to be a major component of adaptation and speciation within this group (1, 2). Specifically, the initial WGD events generate genetic diversity that can be exposed to positive selection eventually associated with shifts in phenotypic novelty, adaptability, and fitness (4). There are a range of potential mechanisms by which increases in ploidy could generate these shifts. For instance, ploidy increases could produce redundant copies of genes, which can mask deleterious mutations, evolve novel or specialized functions (“neo-” or “subfunctionalization”), or afford greater genetic flexibility by either expanding genetic variance or facilitating genomic rearrangement (5). A key question is whether the hypothesized increases in flexibility afforded by higher ploidy translates to a more robust, generalist genotype, as previous empirical results provide no clear answer (e.g., refs. 6–8).
Quantifying how transitions to higher ploidy can generate shifts in fitness and adaptability is difficult when comparing across taxa; however, different ploidy levels, or cytotypes, can exist within a single species, which provides a natural experiment to test how ploidy variation alters genomic diversity (9), adaptive potential (10–12), and ecological niches (3, 13). Despite the ubiquity of ploidy variation in plants and the resulting opportunity to probe the ecological and evolutionary consequences of polyploidy, few studies have had the genomic resources or scope needed to address these dynamics within a mixed-cytotype system. Fortuitously, a widespread, perennial C4 grass in North America, switchgrass (Panicum virgatum), with naturally occurring cytotypes, primarily tetraploids (4×) and octoploids (8×) (14), has been the target of intensive natural sampling and common-garden plantings due to its potential for habitat restoration and biofuel production (15–18). The divergence between gene pools and ecotypes has been closely studied in 4× switchgrass (i.e., ref. 18), but only a few studies have explored the 8× (19–21), and little is known about how this shift to higher-order ploidy has potentially altered fitness, genetic composition, or niche breadth.
Here, we contrast the molecular and quantitative genetic diversity of 4× and 8× switchgrass across naturally occurring genotypes and 10 common gardens to evaluate the basis of ploidy-associated shifts in admixture, adaptive potential, ecological niche, and fitness. Specifically, we discovered 1) recurrent and evolutionary distinct genesis of 8× populations containing novel combinations of genetic diversity; 2) similar morphological/ecotypic divisions within 4× and 8× cytotypes, but divergent cytotype fitness clines, indicating a generalist–specialist trade-off; and 3) niche evolution between 4× and 8× linked to climate adaptation. Combined, our results indicate that mixed-ploidy systems can be used as valuable tools to bolster the resilience of natural and agronomic systems by providing insight on how ploidy variation enables niche divergence, fitness trade-offs, and range expansion.
Results
Recurrent and Evolutionary Distinct Origin of 8× Switchgrass.
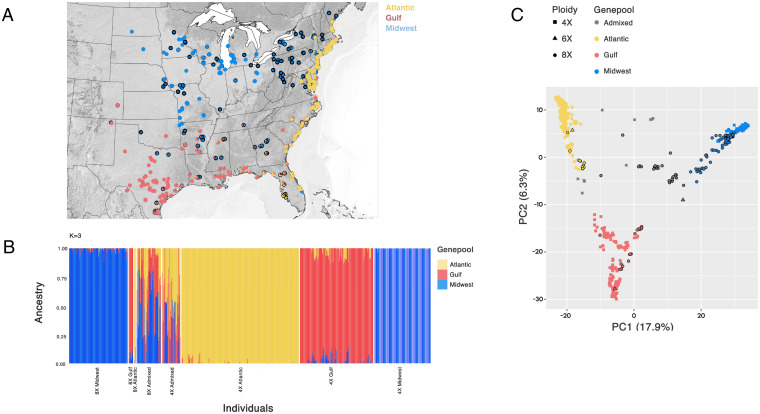
Previous genetic studies focused on switchgrass have mainly utilized the 4× cytotype, which is an allotetraploid with disomic inheritance (2n = 4× = 36) (22, 23), containing two copies each of the N and K subgenomes (4× = KKNN). Little is known about the meiotic behavior of 8× switchgrass (24), but previous analyses suggest that 8× samples are genetically similar to 4× samples (21). The 8× (2n = 8× = 72) switchgrass are generally considered auto-allopolyploids, behaving like an autotetraploid with four copies of each of the two subgenomes present in 4× switchgrass (8× = KKKKNNNN) (20). However, the true inheritance patterns may be heterogeneous across the genome, as seen in other autopolyploids (25, 26). Furthermore, 4× and 8× switchgrass are reproductively isolated due to a postfertilization incompatibility system (27). To date, 8× switchgrass has been assumed to be predominantly composed of a single northern gene pool that is derived from 4× molecular diversity found in Midwestern upland switchgrass, but there has never been a comprehensive survey of the range-wide genetic diversity present in the 8× cytotype. Therefore, to explore the molecular population structure across the natural range of switchgrass, we used a total of 544 deeply resequenced 4× [previously published (18); cultivars and breeding material were omitted for this analysis] and 159 resequenced 8× samples from across the natural range of both cytotypes (Fig. 1A; SI Appendix, Table S1) genotyped at over 58 million single-nucleotide polymorphisms (SNPs).
Fig. 1.
Analyses of population genetic structure. (A) Spatial distributions of the switchgrass subpopulations. Pie charts show the proportion of an individual that was assigned to each subpopulation; 8× samples have a black collar around the pie chart. (B) Bar graphs represent the multilocus genetic assignment of each individual to each ADMIXTURE group. (C) PCA results for both cytotypes, where data points are shaded according to their ADMIXTURE genetic-cluster association.
We hypothesized that 8× switchgrass is derived from ancestral 4× genetic diversity. Analysis of the resequencing data supported the presence of three geographically distinct genetic subpopulations across both 4× and 8× genotypes (referred to as Midwest, Gulf, and Atlantic). These subpopulations are consistent with previous findings that only utilized 4× samples (18), supporting our hypothesis that 8× genotypes are putatively derived from 4× molecular diversity. Due to the complexities of working with a mixed-cytotype system, we verified these patterns using a reference-free k-mer method, which accurately replicated the SNP-based results (Mantel test r = 0.97), indicating that polyploidy did not substantially bias the SNP-based population genetic estimation in this study. Compared to 4× switchgrass, 8× exhibited higher levels of admixture among the three subpopulations, notably between the Midwest and the other two subpopulations, Gulf and Atlantic (4×: 86% have >97% ancestry from a single gene pool, while 4% have >20% ancestry from two gene pools; 8×: 53% have >97% ancestry from a single gene pool, while 29% have >20% ancestry from two gene pools; Fig. 1 B and C). The high levels of genetic admixture detected in the 8×, noticeably absent in the 4×, support the hypothesis that higher ploidy is associated with among-subpopulation mating (23). These results challenge a prevailing assertion that 8× is a secondary taxonomic division within the genetic lineages that compose the cold-adapted upland ecotype of switchgrass (e.g., ref. 28).
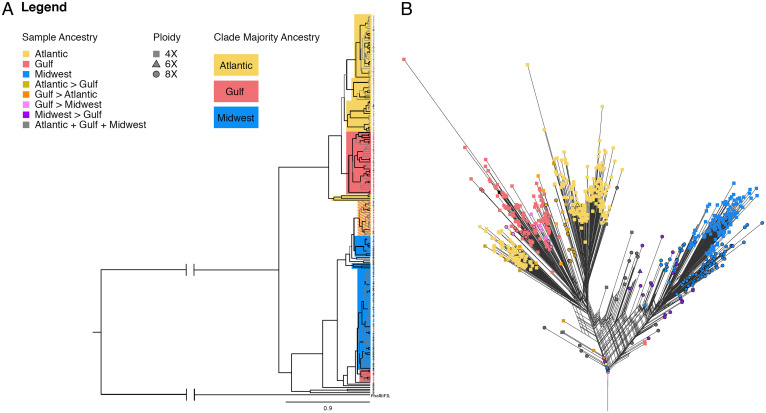
To further explore the process of 8× formation, we compared phylogenetic patterns found using nuclear markers to results based on molecular variation in chloroplast genomes, which generally show higher conservation compared to nuclear genomes and provide valuable tools for understanding the phylogenetic and evolutionary relationships between closely related taxa (29). Specifically, discordance between nuclear and chloroplast phylogenies within species or genera can provide evidence of chloroplast capture resulting from hybridization, though it can also arise from incomplete lineage sorting (30). For a subset of switchgrass genotypes of both cytotypes, we built phylogenetic trees from both nuclear variation and individual chloroplast assemblies to test for congruence between maternal and nuclear genomes between cytotypes (Fig. 2A; SI Appendix, Fig. S1). Among nuclear trees, 8× occur within the distinct clusters of all three gene pools, supporting repeated establishment of 8× lineages and indicating that 4× switchgrass are ancestral to the 8× (21). However, we observed significant discordance between the nuclear and chloroplast trees, whereby numerous individuals that occur within the Gulf and Atlantic clusters of the nuclear tree appear tightly grouped, with the chloroplast haplotypes dominated by individuals with a strong Midwest genetic background (Fig. 2A). Moreover, we estimated the divergence between the five main clades suggested by the tree built from chloroplast haplotypes using BEAST version 2.6.6 (31), with each clade containing both 4× and 8× samples. The estimated dates of the nodes for each of these clades range from 65 thousand years ago (kya) to 86 kya (SI Appendix, Table S1 and Fig. S2). The estimated age of divergence between switchgrass gene pools is estimated at >600 kya (18), indicating that the transition from 4× to 8× happened separately within each gene pool, potentially multiple times, well after the divergence of the gene pools. In fact, identical and near-identical chloroplast haplotypes are shared by 4× and 8× individuals, suggesting a very recent shift in ploidy in some populations (SI Appendix, Table S2). Combined, these observed patterns of population genetic structure, ancestry, and phylogenetic relationships indicate that 8× have emerged multiple times and most likely as the product of disparate parents, including individuals of divergent ancestry. Repeated establishment of 8× populations supports the idea that repeated emergence of 8× switchgrass might promote coexistence (32), which would increase 8× frequency in the overall switchgrass population, providing more chances to avoid minority cytotype exclusion (33).
Fig. 2.
Phylogenetic relationships. (A) Chloroplast tree for a subset of switchgrass samples. Gray branches indicate less than 0.95 posterior probability support, and black branches indicate greater than 0.95 posterior probability support. Colors indicate the nuclear ancestry of the majority of samples in the assigned clade. (B) NeighborNet tree showing nuclear ancestry of all 4× and 8× samples.
Morphological and Fitness Differentiation within Cytotypes of Switchgrass.
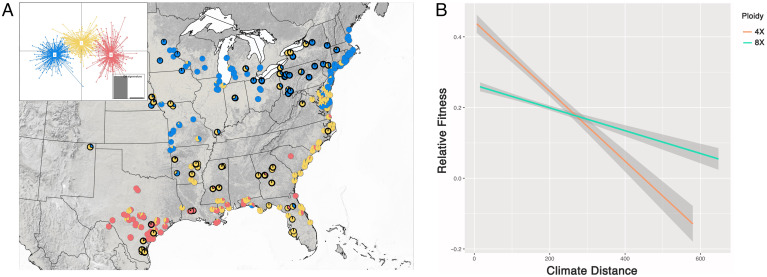
Within 4× lines of switchgrass, documented climate-associated adaptation underscores phenotypic divergence between distinct switchgrass ecotypes (e.g., ref. 18), but the diversity in 8× switchgrass has been assumed to be far less dynamic and has primarily been treated as an exclusively ecotypically upland cytotype (23, 28). However, our results indicate that cytotype evolution is far more complex, where some 8× genotypes are highly admixed composites of upland ecotype and lowland ecotype 4× genetic variation, while others are closely related to either upland or lowland 4× genotypes. Given these disparate patterns of genetic relatedness, it is also possible that 8× phenotypes are far more diverse than previously expected. To explore overall morphological divisions and composition of both cytotypes, we performed in silico classification using a suite of informative morphological data for 408 4× and 102 8× switchgrass samples with sufficient common-garden phenotypic data. Traits of target samples, originally collected from across the entire US geographic range of switchgrass, were measured at both northern and southern common-garden plantings (Austin, TX, and Kellogg Biological Research Station, MI). Assignment was conducted by using a Discriminant Analysis of Principal Components [DAPC; Fig. 3A (34, 35)], which placed the 4× into three broad groupings consistent with both the long-standing division of upland (n = 232) and lowland (n = 96) ecotypes (36–39), as well as emerging evidence of a coastal form (n = 190) (18). Inconsistent with expectations from published literature, our results suggest different subsets of the 8× group, with upland (n = 31), lowland (n = 9), and coastal (n = 62) 4× previously classified through neural networks incorporating expert knowledge (18), suggesting a previously undocumented high degree of phenotypic variability across the octoploid cytotype.
Fig. 3.
Analyses of phenotypic variation at common-garden plantings. (A) De novo morphological assignments for both cytotypes (red, lowland; blue, upland; and gold, coastal); 8× samples are indicated with a black collar, with Inset demonstrating the clustering of the three ecotypes along the first and second discriminant axes. (B) The predicted linear relationship between climate distance (how different planting environment is from sample origin climate) and relative fitness (estimated by the relative biomass measured over two years) for each cytotype. The linear relationships and 95% confidence intervals are depicted for both cytotypes over the same climate distance. Raw data for these predictions are shown in SI Appendix, Fig. S3.
Given our observations of extensive molecular and phenotypic diversity in the 8× cytotype, a pressing question is whether naturally occurring switchgrass cytotypes exhibit divergent climate adaptation and are associated with variable fitness responses across a range of environments. Specifically, shifts in ecological tolerance (i.e., shifts in the range of suitable climate conditions) might have occurred from: 1) alterations in genetic composition associated with higher ploidy levels and hybridization; or 2) divergent adaptation in reproductively isolated cytotypes (27, 40). Within switchgrass, biomass is considered a proxy for plant fitness (17), capturing both aspects of vegetative and reproductive vigor and output. This relationship is supported by the results from two field experiments conducted by Palik et al. (41), which revealed a high correlation between biomass and number of seeds per plant (R2 = 0.83). To control for the effects of underlying genetic structure and ensure sufficient sample sizes, we compared the relative fitness, estimated by biomass, of 4× and 8× in the Midwest subpopulation (ancestry coefficient > 0.9) at 10 common-garden sites against the climate distance from the origin climate of each collection location (Fig. 3B; SI Appendix, Fig. S3). Our results showed that, in common gardens with climates most similar to the climate of origin (the climate of the original collection sites), 4× genotypes have higher relative fitness than 8× genotypes, but fitness declines more slowly for 8× than 4×, with 8× eventually having higher relative fitness than 4× in climates more different from the climate of origin (ploidy and climate distance interaction, F = 44.68, P = 3.23e-11).
Distinct Niches Linked to Genetic Variability Supports Cytotype Coexistence.
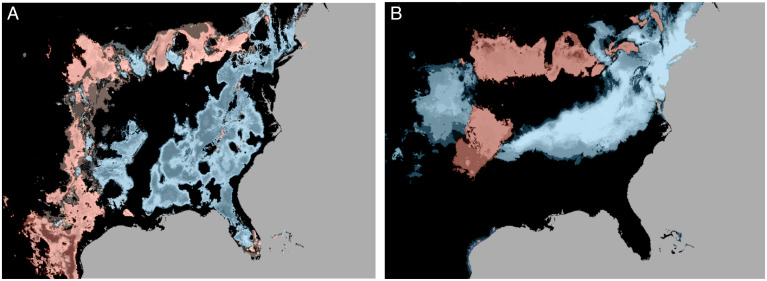
Ploidy increases may expand the diversity of suitable habitats potentially facilitating range expansion (42). To test this hypothesis, we used environmental niche modeling (ENM) based on occurrence data to explore potential niche differences between 8× and 4× overall and within the Midwest subpopulation, where within-cytotype replication was high enough to permit robust inference within subpopulations (43, 44). Although there are 8× within the Gulf and Atlantic subpopulations, low representation of these samples in our dataset (n < 15 for both) preclude a similar ENM approach for these target groups. Using the identity test to compare niche similarity (45), we observed statistically different niches in both the overall set (D = 0.799, P = 0.002) and within the Midwest subpopulation (D = 0.685, P = 0.002), suggesting different environmental distributions of each switchgrass cytotype (Fig. 4). Higher habitat suitability for 8× in pronounced northeastern/southeastern 4× range gaps suggests that increased ploidy allowed colonization of habitat previously unsuitable to switchgrass. ENM results from the Midwest indicate a wider breadth of habitat suitability for the 8× (B2, a metric of niche breadth; see ref. 46; of 8× = 0.676, B2 of 4× = 0.391), but, throughout much of the central Midwest, 4× switchgrass have relatively higher habitat suitability. These findings provide landscape-scale support for the strong specialist vs. generalist trade-off we observed in the common gardens, which may enable cytotype coexistence.
Fig. 4.
ENM. Black indicates similar niche suitability. More intense blue coloration indicates higher 8× habitat suitability, while more intense red coloration indicates higher 4× habitat suitability. (A) Differences in habitat suitability for all 4× and 8×. (B) Differences in habitat suitability for 4× and 8× belonging to the Midwestern subpopulation.
It is possible that the broader environmental, phenotypic, and ecological distributions of the 8× cytotype is, in part, caused by wide crosses and reticulate admixture that may have accompanied ploidy increases. Our population genomic analyses indicated a putative among-subpopulation hybrid origin of some 8×, which potentiates a role for increased genetic variation in facilitating ecological niche evolution between cytotypes (40, 47). In the Midwest subpopulation, for instance, the ancestry coefficients from ADMIXTURE are similar between 8× and 4×, but discordance between the chloroplast and nuclear phylogenetic trees along with higher levels of admixture (Figs. 1 and 2) between the Gulf and Midwest further south suggest that the 8× Midwest subpopulation might harbor alleles from the diverged Southern “Gulf” and “Atlantic” clades. As observed in some 4× genotypes (18), admixture from locally adapted gene pools could have enabled range expansion of 8× genotypes. To evaluate this potential mechanism, we investigated signals of Atlantic and Gulf ancestry across the genomes of Midwest switchgrass using ancestry-informative alleles from SNPs with high Fst (> 0.5) between the three gene pools (i.e., Gulf vs. Midwest and Atlantic vs. Midwest). The mean ancestry signal of both Gulf and Atlantic gene pools was higher in the Midwest 8× than 4× (MW8× Atlantic: 0.147, MW4× Atlantic: 0.0596; Mann–Whitney u test P value for Atlantic < 1.4 e-15; MW8× Gulf: 0.152, MW4× Gulf: 0.0552; Mann–Whitney u test P value for Gulf < 2.2e-16; SI Appendix, Fig. S4). Moreover, there is evidence of geographic variability in the ancestry signal, as Midwest 8× samples in the west demonstrate a higher ratio of Gulf to Atlantic ancestry signal compared to Midwest 8× samples in the east (Mann–Whitney u test P value < 1.7e-10; SI Appendix, Fig. S5).
To explicitly test if retained genomic introgression regions were putatively under selection in 8× populations, we used a series of targeted redundancy analyses (RDAs) to relate the presence of Gulf and Atlantic ancestry in 8× individuals with climatic and geographic factors. Gulf and Atlantic ancestry informative SNPs were >2× more strongly associated with climate (percentage of variance explained: Gulf = 42.1% and Atlantic = 41.3%) than geography (Gulf = 16.6% and Atlantic = 16.2%). When partitioning out the effect of geographic distance, for the Gulf and Atlantic, respectively, we identified 1,108 and 1,120 outlier candidate introgression SNPs that were strongly associated with aspects of climate. We compared these sets of SNPs in the Midwest 8× with the most pronounced shifts in allele frequencies away from the Midwest background and toward Atlantic (n = 705) or Gulf (n = 753) frequencies (Allele Frequency Shift Approach [AFS]). Of these, 126 SNPs in the Atlantic and 157 Gulf AFS outliers met the criteria to be included in the RDAs run using all Midwest samples, with 67 (53.2%) and 88 (56.1%) being significantly correlated with climate from the RDA approach. In comparison, only 4.7% and 5.0% of all SNPs in the RDA and within the same minor allele frequency (MAF) range have significant RDA correlations, an 11.3× and 11.2× enrichment in the AFS outliers. Many of the AFS outliers were on chromosomes 07K, 07N, and 08N (65.1% in Atlantic and 57.3% in Gulf), raising the possibility that these chromosomes are driving the enrichment pattern. However, even when these chromosomes are removed, 20 of 44 (45.5%) and 33 of 67 (49.3%) of AFS outliers have significant RDA results, compared to 4.7% and 5.3% of all SNPs in the RDA across the same chromosome set and similar MAF range, a 9.9× and 9.3× enrichment in the AFS outliers. This suggests that in the Midwest 8×, genomic regions containing SNPs with alleles at frequencies similar to those found in Atlantic and/or Gulf switchgrass play an outsized role in climate adaptation compared to the genome on the whole.
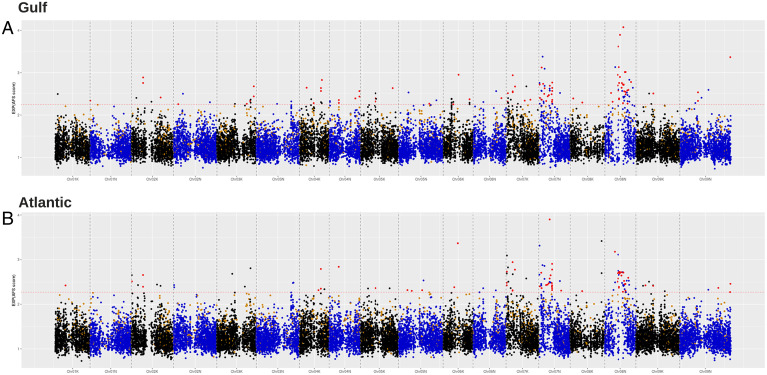
The SNPs in Midwest 8× with significant climate correlations from the RDA and allele frequency shifts toward Atlantic (n = 67) or Gulf (n = 88) demonstrated a strong relationship with primarily two climate variables: temperature seasonality (Bioclimatic variable 4) and precipitation in the driest quarter (Bioclimatic variable 17). These SNPs are distributed throughout the genome, but two regions, the left arm of Chr07N and the pericentromere of Chr08N, are enriched for these overlapping candidate SNPs (Fig. 5). Specifically, on Chr08N, there is a broad signal along a 16 Mb region of the pericentromere associated with precipitation in the driest quarter. We detected elevated linkage disequilibrium (LD) in the Midwest subpopulation in this 16 Mb region, where SNPs separated by 10 to 100 kb have LD values 3.7× greater than SNPs outside the pericentromeric region, compared to 2.5× and 2.6× in the Atlantic and Gulf subpopulations, respectively (SI Appendix, Fig. S6). Elevated LD in this region extends even farther, with SNPs separated by 1 to 10 Mb still having LD values 5.3× higher than background levels, compared to 2.8× and 1.7× for the Atlantic and Gulf subpopulations. The broad pattern on Chr08N is consistent with patterns expected from an inversion in Midwest 8× with origins from Southern switchgrass (i.e., Gulf and Atlantic).
Fig. 5.
Shifts in allele frequency from Midwest ancestry toward Gulf (A) and Atlantic (B) ancestry. The red SNPs are the overlapping outliers identified by both an environmental approach (RDA outliers) and the strongest shifts in allele frequency.
Discussion
Examining ploidy variation within species provides a rare opportunity to examine patterns of ploidy establishment, coexistence of mixed cytotypes, and subsequent divergence (3) without the confounding effects of speciation. Using a combination of genomic, phenotypic, and niche modeling approaches, we discovered that the switchgrass 8× are not, as originally assumed, a uniform upland clade (e.g., refs. 23 and 28), but a genetically and phenotypically diverse polyphyletic group that arises within all known switchgrass ecotypes and gene pools (18). Moreover, compared to the 4×, 8× display higher performance stability over increasing climate distance and distinct ecological niches with suitable habitat in noticeable 4× range gaps, indicating weaker signals of local adaptation and broader environmental tolerances. These results indicate that increases in switchgrass ploidy are correlated with stronger “general-purpose genotypes” (i.e., ref. 48), allowing them to persist in a wider range of environments: a classic generalist–specialist trade-off. When comparing diploids with higher ploidy levels, there has been a long-standing debate about whether shifts in tolerance are attributable to general-purpose genotypes (e.g., refs. 48–51) or if higher ploidy genotypes exist as a series of variable specialists (52, 53). Our results provide strong support that increases in ploidy in similar genetic backgrounds result in increased ecological tolerance that allows range expansion (54), but possibly at the cost of fitness or competitiveness under optimal conditions. Recent transcriptome analyses with another mixed-cytotype system (common reed) support such a trade-off, indicating that tetraploids completed their life cycle faster, but octoploids developed more stress tolerance (55).
Our results indicate higher levels of admixture of diverged gene pools in the 8× than 4×, suggesting that the occurrence of wide crosses might have promoted a shift to higher ploidy. For example, in the Midwest genetic subpopulation, 8× exhibit higher levels of Southern (Atlantic and Gulf subpopulations) switchgrass ancestry than the 4×, with many of the genomic regions containing Southern ancestry putatively under selection by the environment. This indicates that the elevated Southern switchgrass variation contained in the 8×, which is likely related to their evolutionary origins, served as adaptive introgressions that allowed the expansion of the upland switchgrass range into habitat unsuitable for the 4× (56, 57). Specifically, when considering Chr08N, the LD patterns, high frequency of Southern switchgrass alleles, and correlation with climate variables raises the intriguing possibility that this region contains an inversion introduced by gene flow and acting as an “adaptive cassette” (58) and driven to near fixation by positive selection. Overall, the genome-wide results indicate that elevated levels of Southern switchgrass variation in 8× Midwest switchgrass putatively promoted the evolution of the switchgrass range into previously unsuitable habitat for 4× switchgrass, emphasizing the beneficial effects of increased genetic variation resulting from higher ploidy levels (10, 47, 59). The signals detected here offer a mechanistic basis for colonization and subsequent coexistence of mixed-cytotype systems in their native ranges.
While the majority of biofuel switchgrass cultivars are 4× (60), our results challenge a traditional assumption that 4× span a considerably broader geographical range. Contrasting patterns of relative fitness and adaptation between the cytotypes across broad environmental gradients within the same gene pool suggest that increases in ploidy level have resulted in useful combinations of genetic diversity not present in the 4×. This is notable, as much of the genomic diversity associated with environmental selection in the switchgrass 4× was unique to each subpopulation (18), and, if some of the unique gene variants were to be combined, the resulting progeny would have adaptation traits allowing expansion to new ecological niches (61). Our results indicate that the 8× already represent such a unique combination of genetic variation that has allowed the expansion of switchgrass’ ecological niche and thus represent a valuable breeding resource. With the acceleration of anthropogenic climate change, our results suggest that careful examination of the genomic divergence present in systems with intraspecific ploidy variation could hold answers for utilizing natural diversity to ameliorate the effects of ongoing environmental shifts.
Materials and Methods
Flow Cytometry.
To determine the ploidy level of target switchgrass samples, we utilized a LSRFortessa SORP Flow Cytometer (BD Biosciences). A total of 200 to 300 mg of young leaf tissue was macerated by using a razor blade and then treated with 1 mL of CyStain PI Absolute P nuclei extraction buffer (Sysmex Flow Cytometry) mixed with 1 μL of 2-mercaptoethanol for 15 min. Free nuclei were isolated by using a CellTrics 30-μm filter (Sysmex) followed by 20-min treatment with 2 mL of CyStain PI Absolute P staining buffer (Sysmex), 12 μL of propidium iodide, and 6 μL of RNase A for 20 min on wet ice. Subsequently, prepped samples were run on the flow cytometer to assess nuclei size, with a minimum of 10,000 nuclei analyzed per individual. Flow-cytometer results were analyzed by using FlowJo software (BD Biosciences), and average units of fluorescence per nuclei were used to bin samples into three categories. Using binning parameters established with flow-cytometry data from switchgrass samples of verified ploidy, ploidy level of the samples was assigned as 4× if the cell population had 40,000 to 80,000 units of fluorescence, 6× for 80,000 to 100,000 units, and 8× for 100,000 to 140,000 units.
Determining Ploidy with nQuire.
We use the program nQuire (62) to determine the ploidy of samples in our dataset. We tested multiple coverage thresholds and found that requiring 20+ coverage in at least 500,000 variable positions across the genome provided ploidy calls that agree with flow-cytometry results and expectations based on previous population-structure results. Samples with fewer than 500,000 variable positions with 20+ coverage were removed from the analysis. Based on the nQuire results, the final dataset includes 544 4×, 3 6×, and 159 8× samples.
Ploidy Considerations in Mixed Dataset.
A challenge of comparing between individuals of differing ploidy is generating genotypes that are comparable across cytotypes. Simulations show that simply using diploid genotypes for all cytotypes can generate biases when examining population structure (63). As such, we generated dosage genotypes for all samples reflecting the portion of the genotype that is the reference allele. The dosage genotypes ranged from 0 to 2, with 4× switchgrass containing dosage genotypes of 0, 1, or 2 reference alleles and 8× switchgrass containing dosage genotypes of 0, 0.5, 1, 1.5, and 2, reflecting 0 through 4 copies of the reference allele. We also generated diploid genotypes (three potential genotypes) for all samples to use with ADMIXTURE, which does not accept dosage genotypes.
SNP Calling.
The samples were sequenced by using Illumina HiSeq ×10 and Illumina NovaSeq 6000 paired-end sequencing (2 × 150 bp) at HudsonAlpha Institute for Biotechnology, Huntsville, AL, and the Department of Energy Joint Genome Institute, Berkeley, CA. To account for variable library sizes, reads were pruned to a max coverage of 50×. Reads were mapped to the P. virgatum v5 assembly (18) by using bwa-mem (64). Duplicate reads were filtered by using Picard (http://broadinstitute.github.io/picard) and realigned around indels by using GATK 3.0 (65). Multisample SNP calling was done by using SAMtools mpileup (66) and Varscan V2.4.0 (67) with a minimum coverage of eight and a minimum alternate allele count of four. Alleles were determined by using a binomial test (hypothesized probability of success = 0.5). However, for the tetrasomic calls, a binomial test was performed for three hypothesized probabilities (0.25, 0.5, and 0.75) using the binom.test function in R based on a maximum P value and the contribution of reads that match reference: Five tetrasomic genotypes were determined, equivalent to AAAA, AAAB, AABB, AAAB, and BBBB (or 0/4; 1/3; 2/2; 3/1; and 4/0).
Sample Filtering.
The unfiltered sample set includes the samples from Lovell et al. (18) and additional, primarily 8× resequenced samples to generate a sample set of 1,059 samples. We focused our analyses on natural populations, so any samples that originated from breeding populations or cultivars were removed. Any samples that were assigned using principal component analysis (PCA) or ADMIXTURE to a different gene pool from the rest of the samples from the same sampling source were removed. We calculated distance matrices and neighbor-joining (NJ) trees in R (68, 69), and samples assigned to a different clade in an NJ tree than the rest of the samples from the same sampling source were removed. Samples that looked to be from the same or closely related sampling source, but originated from geographically distant locations, were filtered based on the geographic patterns in that clade of the NJ tree. Any samples that did not fulfill the sequencing-coverage cutoffs for nQuire were also removed.
Population Genetic Approaches.
We use the adegenet R package to run PCA on our genotypes (34). We used dosage genotypes to minimize biases in the PCA results based on ploidy differences (63), though the results for our analyses were consistent regardless of the use of disomic, tetrasomic, or polyploid genotypes (SI Appendix, Fig. S7).
We also used ADMIXTURE (70) using diploid genotypes to estimate the number of populations and the proportion of ancestry of the samples in the dataset. We ran PCA and ADMIXTURE on the entire dataset and separately for the inferred Midwest gene pools.
Neighbor Net Tree.
Ancestral genotypes were estimated following ref. 18. Briefly, coding sequences (CDSs) were identified that share common ancestry with Panicum hallii and Sorghum bicolor. For each orthology network, two multiple sequence alignments were generated in mafft (71), one with and one without the switchgrass sequence, followed by extraction of marginal character states in Phangorn (72) using the maximum-likelihood algorithm. This process generated ancestral state genotypes at 10,874,224 SNPs in CDS regions, of which 100,000 were randomly selected, and a variant call format (VCF) file for the entire sample set and the ancestral genotype was generated for these SNPs. The VCF was converted to the NEXUS format by using vcf2phylip (73). We used SplitsTree (74) to generate a NeighborNet tree and the phangorn package in R (72) for visualization.
Chloroplast Analyses.
Chloroplast assembly for each of the samples was performed by using NOVOPlasty (75). The seed sequence used for assembly was identified by using BLAT (76) with Kanlow chloroplast sequence (GenBank accession no. HQ731441.1). The assemblies were aligned by mafft v7.471 (71).
We used the chloroplast alignment and generated unweighted pair group method with arithmetic mean (UPGMA) and neighbor-joining trees and calculated bootstrap values using the ape package in R (77). For improved interpretation and visualization, we used these trees to select a subset of 218 samples (SI Appendix, Table S3), removing some closely related samples, particularly those with identical or near-identical haplotypes, but making sure to retain samples that clustered closely in the chloroplast trees, but had different ploidy or nuclear ancestry patterns. We then estimated the chloroplast tree using BEAST v2.6.6 (31), using the same parameters as detailed for “model 1” used below for estimating divergence date.
Using the UPGMA and neighbor-joining trees of all samples, we also selected paired 4× and 8× samples that belong to the same clade (bootstrap values > 0.9) that contain only samples from one gene pool or only samples from Southern switchgrass, according to population structure analysis of the nuclear genome. This process generated a set of 13 4× and 13 8× switchgrass samples along with P. hallii var. fil. We estimated the divergence dates between chloroplast haplotypes in this sample subset using BEAST v2.6.6 (31), using 8.35 million years ago (mya) as the prior for the divergence date between switchgrass and P. hallii (18).
We ran BEAST using three sets of parameters to test the robustness of the time estimates. For all tests, we used Site Model:Substitution Rate:estimate; Site Model:Substitution Rate:Gamma Category Count:4; Site Model:Shape:estimate; Site Model:Subst Model:HKY:Frequencies=empirical; Click Model:strict clock:clock.rate=1.0; MCMC:Chain Length:1,000,000; MCMC:tracelog:500; MCMC:treelog:500. For the switchgrass–P. hallii divergence prior, we generated a new prior, selecting P. hallii and one switchgrass library, with “Normal” distribution, not monophyletic, mean=8.35 (mya). For model 1, we used Priors:Tree.t:Coalescent Constant Population; Priors:clockRate:Gamma:Alpha=0.001,Beta=1,000. For model 2, we used Priors:Tree.t:Coalescent Constant Population; Priors:clockRate:Gamma using default values(Alpha=2,Beta=2). For model 3, we used Priors:Tree.t:Coalescent Exponential Population; Priors:clockRate:Gamma:Alpha=0.001,Beta=1,000. Model 1 generated the smallest 95% intervals for the estimated parameters, but all three models generated highly similar results. The final tree contains five main clades with >99% posterior probability, each clade containing both 4× and 8× samples. “Southern” consists of individuals from the Gulf gene pool, though the overall clade also contains 4× samples with Atlantic ancestry. “Midwest A” and “Midwest B” contain samples from Midwest gene-pool clades. “Gulf” and “Atlantic” contain samples from clades with Gulf and Atlantic ancestry, respectively (SI Appendix, Table S4).
K-Mer Approach.
To observe the effects of potential biases in calling 8× variation from a 4× reference, we employed a reference-free k-mer (sequences of DNA of length “k”)-based approach to analyze the population structure and compare the overall patterns to the results obtained using SNPs. Reference-free and alignment-free methods of assessing population genetic structure have been shown to reduce some of the inherent biases involved in polyploid genetics (78). Here, we used the k-mer hashing method employed by the Mash program to confirm population genetic patterns derived from traditional SNP-based analyses (79). In this analysis, raw sequence data from whole-genome sequencing are decomposed into k-mers, and then unique k-mers are tabulated for each individual in the dataset. The resulting tables are hashed by using the MinHash algorithm to facilitate computational comparison (80). The Jaccard distance between individual hashes then is a reliable estimate of the genetic distance between samples.
Our analysis followed the method used in VanWallendael and Alvarez (78) on 40 individuals randomly selected from the study. Briefly, we first trimmed fastq sequence files randomly to equal size to reduce library-size biases using fastq-tools (https://github.com/dcjones/fastq-tools). We then performed k-mer decomposition, sampling 100,000 random k-mers (–s 100,000) of length 21 bp (–k 21) and removing k-mers detected only once to avoid sequencing errors (–m 2). We used the inferred genetic distance matrix to perform Principal Coordinate Analysis and compared the results to SNP-based analyses using Mantel tests.
Morphological/Ecotype Classifications.
To assign in silico morphological classifications to infer similarity to known switchgrass ecotypes, we utilized an assumption-free approach that reduces possible assignment biases by avoiding user-defined training groups. Specifically, a DAPC (34, 35) approach was run in adegenet (34) on the same set of 16 informative plant traits (leaf: length, width, length/width ratio, area, lamina thickness and lamina/midrib thickness ratio; whole plant: number of tillers, tiller height, product of tiller height × number, tiller height/count ratio, panicle height, panicle height/count ratio, leaf canopy height and tiller/leaf height ratio; phenology: date of green-up and date of panicle emergence) used to call ecotypes in Lovell et al. (18), which were collected at a northern (KBSM in Hickory Corners, MI) and southern (PKLE in Austin, TX) common garden in 2019. Prior groups were determined by first transforming the phenotypic data using PCA, and then the first 10 principal components (PCs) were used in a k-means algorithm to classify individuals into three possible groupings to match the documented ecotypic variation present in the 4× (18), aiming to maximize the variation between groups. Next, DAPC was implemented on the 10 retained PCs to provide an efficient description of the morphological clusters using two synthetic variables, which are linear combinations of the original phenotypic variables that have the largest between-group variance and the smallest within-group variance (i.e., the discriminant functions). Since this approach is based on Discriminant Analysis, we also obtained posterior membership probabilities for each individual, thus allowing us to assess variation present in both cytotypes.
Relative Fitness over Climate Distance.
For this analysis, a plant present at 1 of the 10 common-garden sites with a Midwestern genetic background was considered a replicate for either the 8× (n = 912) or 4× (n = 638) cytotype. At four of the common-garden sites (KBSM, FRMI, CLMB, and PKLE), some of the same 8× genotypes were planted multiple times, but each of these were considered independent replicates, given that they each provide separate estimates of 8× Midwest fitness. Relative fitness for each plant was estimated by summing collected biomass values over 2 y (2019 and 2020) and dividing it by the maximum summed biomass value for each cytotype, respectively. Climate distance from collection origin to planting location was calculated for each replicate. Specifically, to assess climate distance, we calculated the outer distance between matrices of six environmental predictors from WorldClim [BIO1 = annual mean temperature, BIO2 = mean diurnal range, BIO4 = temperature seasonality, BIO5 = maximum temperature of warmest month, BIO16 = precipitation of wettest quarter, and BIO17 = precipitation of driest quarter (81)] that were previously determined to be important in determining the distribution of switchgrass (18) for the common gardens and collection locations for each plant. For each cytotype, we plotted the linear relationship between relative fitness and climate distance, subsequently comparing and testing if the slopes of the two cytotypes differed by using ANOVA to analyze the interaction between ploidy and climate distance.
ENM and Associated Hypotheses Testing.
Using the ENMtools R package, we built four ENMs to simulate modern-day potential ranges and explicitly test potential niche differences between 8× and 4× overall and within the Midwest subpopulation (43, 44). The final datasets, based on the filtered samples, used to build the SDMs comprised 236 (4×), 88 (8×), 79 (8× Midwest), and 53 (4× Midwest) occurrence records. Six environmental predictors were used in our final ENM modeling (BIO1 = annual mean temperature, BIO2 = mean diurnal range, BIO4 = temperature seasonality, BIO5 = maximum temperature of warmest month, BIO16 = precipitation of wettest quarter, and BIO17 = precipitation of driest quarter). ENMs were then generated within the ENM tools package by using a generalized linear model algorithm. For each model, the occurrence data were coupled with 1,000 pseudoabsence data generated randomly within the modeled study area. Models were trained with 80% of the coupled occurrences and pseudoabsence data and tested with the remaining 20%. Each of the four ENMs was replicated 500 times, which were evaluated via the area under the receiver operating characteristic curve [AUC (82)]. Models with AUC values above 0.75 are considered potentially useful (83), and, in this case, all models had AUC values above this threshold (4× = 0.867, 8× = 0.800, 4× Midwest = 0.876, and 8× Midwest = 0.803). Estimates of niche breadth (B2 based on ref. 46) were obtained for each ENM. Lastly, we conducted niche identity tests (n = 500 replications) for both the overall cytotype and Midwest models to determine, in each instance, if the two groups’ occurrences in the environment are random draws from a shared underlying distribution (45). Departure from the null hypothesis in this test could potentially indicate the occurrence of niche evolution among other possibilities (84). These analyses focused on the Midwestern subpopulation due to low sample size of 8× Gulf and Atlantic.
Southern Switchgrass Ancestry Analyses.
We chose three training sets, one from each gene pool, composed of 40 4× individuals that were at the most extreme ends of the PCA distributions, had high ancestry to one of the subpopulations in the k = 3 ADMIXTURE analysis, and then filtered to include as many populations and as much geographic variation as possible. We then used vcfTools (85) to identify SNPs with Fst > 0.5 between pairwise comparisons of gene pools, followed by slight LD pruning with plink (86) (LD < 0.99 in 1,000-bp windows). We filtered the Midwest-vs.-Gulf and Midwest-vs.-Atlantic high-Fst SNPs by missing data in all the Midwest samples (<20% missing data), resulting in 58,829 high-Fst SNPs for Midwest-vs.-Gulf and 59,119 high-Fst SNPs for Midwest-vs.-Atlantic. For each high-Fst SNP, one or both alleles can be informative about ancestry, and we called an allele as informative about ancestry if it was absent from one training set or its frequency in one training set was above 0.5 and 2.5× greater than the frequency in the other training set. An allele that is not ancestry-informative is present in both training sets at high enough frequency, so that it cannot be used to infer from which population it originated. Using these criteria, the Midwest-vs.-Gulf comparison has 20,273 (34.5%) SNPs, where both alleles are ancestry-informative, and 17,933 SNPs (30.5%) and 20,612 (35.0%) SNPs, where one allele is Midwest-informative or Gulf-informative, respectively, and the other allele is not ancestry-informative. For the Midwest-vs.-Atlantic comparison, 20,525 SNPs (34.7%) have both alleles as informative about ancestry, and 21,669 SNPs (36.7%) and 16,918 SNPs (28.6%) have Midwest-informative and Gulf-informative, respectively, for one allele, and the other allele is not ancestry-informative. We used dosage of Atlantic-informative and Gulf-informative alleles at each SNP for RDA and to estimate the amount of Southern switchgrass ancestry in each sample.
We estimated the shift in allele frequency (AFS) in a Midwest test population from the Midwest training set toward either the Gulf or Atlantic training sets by calculating the following: “train_range” = allele frequency difference between the Atlantic or Gulf and the Midwest training sets; “test_range” = allele frequency difference between test population and MW training set. Multiply any SNPs with a negative value in “train_range” by −1, so that a positive “test_range” value means a shift toward the allele frequency in the Atlantic/Gulf training set. “AFS score” = test_range/train_range and represents the percentage of the allele-frequency difference between the training sets that the population has shifted toward the Atlantic/Gulf frequency. AFS score = 1 means allele frequency is the same as Atlantic/Gulf training set; AFS score = 0 means allele frequency is the same as the Midwest training set. We used a cutoff of three SDs from the mean to identify candidate SNPs based on elevated AFS score.
RDAs.
We implemented RDA in the R package vegan (87) to: 1) calculate the proportion of variation in the lowland ancestry SNPs that can be uniquely explained by climate, geography, and their joint effect (88); and 2) identify specific SNPs strongly associated with the environment (89, 90). To partition explainable variance attributable to climate and geography, we ran three models: 1) one full model with climate (BIO1 = annual mean temperature, BIO2 = mean diurnal range, BIO4 = temperature seasonality, BIO5 = maximum temperature of warmest month, BIO16 = precipitation of wettest quarter, and BIO17 = precipitation of driest quarter) and geography (latitude and longitude of each sampled individual); 2) one model with climate and a conditioned matrix consisting of geographic information; and 3) a final model with geographic information conditioned using a matrix of the climate variables. The inertia (i.e., variance) values from the constrained matrix of each model were compared to determine the relative importance of climate, geography, and their joint effect. For each model, multivariate linear regression was used on the lowland ancestry calls of each candidate SNP and the extracted climate variables to produce a matrix of fitted values. PCA was then performed on this matrix to generate canonical axes that were linear combinations of the predictors (91). Each significant axis was approximately normally distributed (n = 3 for both Gulf and Atlantic analyses). SNPs loading at the tails more likely indicate selection related to the predictors (i.e., climate), so we identified all markers that fell within 2.5 SDs (two-tail P = 0.012) from the center as putative lowland-ancestry SNPs under selection (90, 92).
LD Analysis.
For LD analysis, r2 was calculated in PLINK v1.9 (93) and VCFtools (85) between hiFst SNPs across all samples and within each of the three gene pools using diploid-only genotypes. LD heatmaps were visualized by using the LDheatmap R package (94).
Supplementary Material
Acknowledgments
We thank J. Randall (North Carolina Botanical Garden), M. Casler (US Department of Agriculture–Agricultural Research Service [USDA-ARS]), A. Stottlemeyer (The Ohio State University Newark), and M. Harrison (Germplasm Resources Information Network) for generously sharing seed collections of switchgrass. We thank the HudsonAlpha Genomic Services Lab for loading Illumina X10 sequencing runs and the Joint Genome Institute (JGI) production group for sequencing library ASHOA. We are grateful for JGI sequencing capacity contributed to the community sequencing effort by the Bioenergy Research Centers through the Bioenergy Science Center led by the Oak Ridge National Laboratory. We thank Lisa Vormwald, Matthew Smith, Perla Dubeney, John Sanley, Nick Ryan, Todd Bortnem, Tim Vugteveen, Scott Hoffman, Matt Donahue, and Justin Shih for propagating, planting, and managing switchgrass field plantings. This research was supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Program Grants DE-SC0014156 and DE-SC0021126 (to T.E.J.) and DE-SC0017883 (to D.B.L.). The work (Proposal 10.46936/10.25585/60001049) conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231. This material is based upon work supported in part by the Great Lakes Bioenergy Research Center, US Department of Energy, Office of Science, Office of Biological and Environmental Research under Award DE-SC0018409. The work conducted by Argonne National Laboratory is supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Program under Contract DE-AC02-06CH11357. The field research was supported by the USDA-ARS, an equal opportunity employer.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118879119/-/DCSupplemental.
Data Availability
Raw sequencing data for switchgrass included in our analyses are available at the NIH NCBI Sequence Read Archive (accession nos. all included in the supplemental information). Useful SNP sets and generalized versions of the code used in our analyses are publicly available at GitHub (https://github.com/jdnapier/A-generalist-specialist-tradeoff-between-switchgrass-cytotypes-impacts-climate-adaptation-and-geogra) (95).
References
- 1.Ramsey J., Schemske D. W., Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501 (1998). [Google Scholar]
- 2.Soltis P. S., Marchant D. B., Van de Peer Y., Soltis D. E., Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 35, 119–125 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Kolář F., Čertner M., Suda J., Schönswetter P., Husband B. C., Mixed-ploidy species: Progress and opportunities in polyploid research. Trends Plant Sci. 22, 1041–1055 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Alix K., Gérard P. R., Schwarzacher T., Heslop-Harrison J. S. P., Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Ann. Bot. 120, 183–194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otto S. P., The evolutionary consequences of polyploidy. Cell 131, 452–462 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Lowry E., Lester S. E., The biogeography of plant reproduction: Potential determinants of species’ range sizes. J. Biogeogr. 33, 1975–1982 (2006). [Google Scholar]
- 7.Hijmans R. J., et al. , Geographical and environmental range expansion through polyploidy in wild potatoes (Solanum section Petota). Glob. Ecol. Biogeogr. 16, 485–495 (2007). [Google Scholar]
- 8.Johnson A. L., Govindarajulu R., Ashman T. L., Bioclimatic evaluation of geographical range in Fragaria (Rosaceae): Consequences of variation in breeding system, ploidy and species age. Bot. J. Linn. Soc. 176, 99–114 (2014). [Google Scholar]
- 9.te Beest M., et al. , The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 109, 19–45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parisod C., Holderegger R., Brochmann C., Evolutionary consequences of autopolyploidy. New Phytol. 186, 5–17 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Manzaneda A. J., et al. , Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae). New Phytol. 193, 797–805 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin S. L., Husband B. C., Adaptation of diploid and tetraploid chamerion angustifolium to elevation but not local environment. Evolution 67, 1780–1791 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Baniaga A. E., Marx H. E., Arrigo N., Barker M. S., Polyploid plants have faster rates of multivariate niche differentiation than their diploid relatives. Ecol. Lett. 23, 68–78 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Costich D. E., Friebe B., Sheehan M. J., Casler M. D., Buckler E. S., Genome‐size variation in switchgrass (Panicum virgatum): Flow cytometry and cytology reveal rampant aneuploidy. Plant Genome 3, 130–141 (2010). [Google Scholar]
- 15.Sanderson M. A., et al. , Switchgrass as a sustainable bioenergy crop. Bioresour. Technol. 56, 83–93 (1996). [Google Scholar]
- 16.Wright L., “Historical Perspective on how and why switchgrass was selected as a ‘Model’ high-potential energy crop” (Tech. Rep. ORNL/TM-2007/109, Oak Ridge National Laboratory, Oak Ridge, TN, 2007).
- 17.Lowry D. B., et al. , QTL × environment interactions underlie adaptive divergence in switchgrass across a large latitudinal gradient. Proc. Natl. Acad. Sci. U.S.A. 116, 12933–12941 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovell J. T., et al. , Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass. Nature 590, 438–444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMillan C., Weiler J., Cytogeography of Panicum virgatum in central North America. Am. J. Bot. 46, 590–593 (1959). [Google Scholar]
- 20.Lu F., et al. , Switchgrass genomic diversity, ploidy, and evolution: Novel insights from a network-based SNP discovery protocol. PLoS Genet. 9, e1003215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabowski P. P., Morris G. P., Casler M. D., Borevitz J. O., Population genomic variation reveals roles of history, adaptation and ploidy in switchgrass. Mol. Ecol. 23, 4059–4073 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada M., et al. , Complete switchgrass genetic maps reveal subgenome collinearity, preferential pairing and multilocus interactions. Genetics 185, 745–760 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Triplett J. K., Wang Y., Zhong J., Kellogg E. A., Five nuclear loci resolve the polyploid history of switchgrass (Panicum virgatum L.) and relatives. PLoS One 7, e38702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley R. D., Vogel K. P., Chromosome numbers of released cultivars of switchgrass, indiangrass, big bluestem, and sand bluestem. Crop Sci. 22, 1082–1083 (1982). [Google Scholar]
- 25.Ramsey J., Schemske D. W., Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33, 589–639 (2002). [Google Scholar]
- 26.Mason A. S., Wendel J. F., Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front. Genet. 11, 1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez-Reyna J. M., Vogel K. P., Incompatibility systems in switchgrass. Crop Sci. 42, 1800–1805 (2002). [Google Scholar]
- 28.Zhang Y., et al. , Natural hybrids and gene flow between upland and lowland switchgrass. Crop Sci. 51, 2626–2641 (2011). [Google Scholar]
- 29.Bi Y., et al. , Chloroplast genomic resources for phylogeny and DNA barcoding: A case study on Fritillaria. Sci. Rep. 8, 1184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soltis D. E., Kuzoff R. K., Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxifragaceae). Evolution 49, 727–742 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Bouckaert R., et al. , BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 15, e1006650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felber F., et al. , Establishment of a tetraploid cytotype in a diploid population: Effect of relative fitness of the cytotypes. J. Evol. Biol. 4, 195–207 (1991). [Google Scholar]
- 33.Levin D. A., Minority cytotype exclusion in local plant populations. Taxon 24, 35–43 (1975). [Google Scholar]
- 34.Jombart T., adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Jombart T., Collins C., “A tutorial for discriminant analysis of principal components (DAPC) using adegenet 2.0.0” (Tech. Rep. MRC Centre for Outbreak Analysis and Modelling, Imperial College London, London, 2015).
- 36.C. L. Porter, Jr, An analysis of variation between upland and lowland switchgrass, Panicum virgatum L., in central Oklahoma. Ecology 47, 980–992 (1966). [Google Scholar]
- 37.Casler M. D., Vogel K. P., Taliaferro C. M., Wynia R. L., Latitudinal adaptation of switchgrass populations. Crop Sci. 44, 293–303 (2004). [Google Scholar]
- 38.Casler M. D., Stendal C. A., Kapich L., Vogel K. P., Genetic diversity, plant adaptation regions, and gene pools for switchgrass. Crop Sci. 47, 2261–2273 (2007). [Google Scholar]
- 39.Lowry D. B., et al. , Adaptations between ecotypes and along environmental gradients in Panicum virgatum. Am. Nat. 183, 682–692 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Ramsey J., Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. U.S.A. 108, 7096–7101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palik D. J., Snow A. A., Stottlemyer A. L., Miriti M. N., Heaton E. A., Relative performance of non-local cultivars and local, wild populations of switchgrass (Panicum virgatum) in competition experiments. PLoS One 11, e0154444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leitch A. R., Leitch I. J., Genomic plasticity and the diversity of polyploid plants. Science 320, 481–483 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Warren D. L., Beaumont L. J., Dinnage R., Baumgartner J. B., New methods for measuring ENM breadth and overlap in environmental space. Ecography 42, 444–446 (2019). [Google Scholar]
- 44.Warren D. L., et al. , ENMTools 1.0: An R package for comparative ecological biogeography. Ecography 44, 504–511 (2021). [Google Scholar]
- 45.Warren D. L., Glor R. E., Turelli M., Environmental niche equivalency versus conservatism: Quantitative approaches to niche evolution. Evolution 62, 2868–2883 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Levins R., Evolution in Changing Environments (Princeton University Press, Princeton, NJ, 1968). [Google Scholar]
- 47.Coughlan J. M., Han S., Stefanović S., Dickinson T. A., Widespread generalist clones are associated with range and niche expansion in allopolyploids of Pacific Northwest Hawthorns (Crataegus L.). Mol. Ecol. 26, 5484–5499 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Baker H. G., “Characteristics and modes of origin of weeds” in The Genetics of Colonizing Species, Stebbins G. L., Bake H. G., Eds. (Academic Press, New York, 1965), pp. 147–168. [Google Scholar]
- 49.Bardil A., de Almeida J. D., Combes M. C., Lashermes P., Bertrand B., Genomic expression dominance in the natural allopolyploid Coffea arabica is massively affected by growth temperature. New Phytol. 192, 760–774 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Madlung A., Polyploidy and its effect on evolutionary success: Old questions revisited with new tools. Heredity 110, 99–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madlung A., Wendel J. F., Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet. Genome Res. 140, 270–285 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Van Valen L., Morphological variation and width of ecological niche. Am. Nat. 99, 377–390 (1965). [Google Scholar]
- 53.Brittingham H. A., Koski M. H., Ashman T. L., Higher ploidy is associated with reduced range breadth in the Potentilleae tribe. Am. J. Bot. 105, 700–710 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Stebbins G. L., Polyploidy, hybridization, and the invasion of new habitats. Ann. Mo. Bot. Gard. 72, 824–832 (1985). [Google Scholar]
- 55.Wang C., et al. , Transcriptome analysis of tetraploid and octoploid common reed (Phragmites australis). Front Plant Sci 12, 653183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bossdorf O., Lipowsky A., Prati D., Selection of preadapted populations allowed Senecio inaequidens to invade Central Europe. Divers. Distrib. 14, 676–685 (2008). [Google Scholar]
- 57.Treier U. A., et al. , Shift in cytotype frequency and niche space in the invasive plant Centaurea maculosa. Ecology 90, 1366–1377 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Kirkpatrick M., Barrett B., Chromosome inversions, adaptive cassettes and the evolution of species’ ranges. Mol. Ecol. 24, 2046–2055 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Soltis P. S., Soltis D. E., The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. U.S.A. 97, 7051–7057 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Casler M. D., Vogel K. P., Harrison M., Switchgrass germplasm resources. Crop Sci. 55, 2463–2478 (2015). [Google Scholar]
- 61.Sacks E. J., Multiple genomes give switchgrass an advantage. Nature 590, 394–395 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Weiß C. L., Pais M., Cano L. M., Kamoun S., Burbano H. A., nQuire: A statistical framework for ploidy estimation using next generation sequencing. BMC Bioinformatics 19, 122 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meirmans P. G., Liu S., van Tienderen P. H., The analysis of polyploid genetic data. J. Hered. 109, 283–296 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKenna A., et al. , The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koboldt D. C., et al. , VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Yu G., et al. , Ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
- 70.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schliep K. P., phangorn: Phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ortiz E. M., (2019) vcf2phylip v.2.0: Convert a VCF matrix into several matrix formats for phylogenetic analysis. Zenodo. https://doi.org/ 10.5281/zenodo.2540861. Accessed 20 February 2021. [DOI]
- 74.Huson D. H., Bryant D., Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Dierckxsens N., Mardulyn P., Smits G., NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18–e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kent W. J., BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paradis E., Schliep K., ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 78.VanWallendael A., Alvarez M., Alignment-free methods for polyploid genomes: Quick and reliable genetic distance estimation. Mol. Ecol. Resour. 22, 612–622 (2022). [DOI] [PubMed] [Google Scholar]
- 79.Ondov B. D., et al. , Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Broder A. Z., “On the resemblance and containment of documents” in Proceedings. Compression and Complexity of SEQUENCES 1997 (Cat. No. 97TB100171) (IEEE, Piscataway, NJ, 1997), pp. 21–29.
- 81.Fick S. E., Hijmans R. J., WorldClim 2: New 1‐km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017). [Google Scholar]
- 82.Fawcett T., An introduction to ROC analysis. Pattern Recognit. Lett. 27, 861–874 (2006). [Google Scholar]
- 83.Elith J., “Quantitative methods for modeling species habitat: Comparative performance and an application to Australian plants” in Quantitative Methods for Conservation Biology, Ferson S., Burgman M., Eds. (Springer, New York, 2000), pp. 39–58. [Google Scholar]
- 84.Warren D. L., Cardillo M., Rosauer D. F., Bolnick D. I., Mistaking geography for biology: Inferring processes from species distributions. Trends Ecol. Evol. 29, 572–580 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Danecek P., et al. ; 1000 Genomes Project Analysis Group, The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Purcell S., et al. , PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oksanen J., et al. , Vegan: Community Ecology Package. R Package (Version 2.3-3, 2016). https://cran.r-project.org/web/packages/vegan/index.html. Accessed 6 August 2021.
- 88.Gugger P. F., Ikegami M., Sork V. L., Influence of late Quaternary climate change on present patterns of genetic variation in valley oak, Quercus lobata Née. Mol. Ecol. 22, 3598–3612 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Rellstab C., Gugerli F., Eckert A. J., Hancock A. M., Holderegger R., A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 24, 4348–4370 (2015). [DOI] [PubMed] [Google Scholar]
- 90.Forester B. R., Lasky J. R., Wagner H. H., Urban D. L., Comparing methods for detecting multilocus adaptation with multivariate genotype-environment associations. Mol. Ecol. 27, 2215–2233 (2018). [DOI] [PubMed] [Google Scholar]
- 91.Legendre P., Legendre L., Numerical Ecology (Elsevier, Amsterdam, 2012). [Google Scholar]
- 92.Forester B. R., Jones M. R., Joost S., Landguth E. L., Lasky J. R., Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes. Mol. Ecol. 25, 104–120 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Chang C. C., et al. , Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, s13742–s13015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shin J., Blay S., Graham J., McNeney B., LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 16, 1–10 (2006). [Google Scholar]
- 95.J. D. Napier, P. P. Grabowski, A-generalist-specialist-tradeoff-between-switchgrass-cytotypes-impacts-climate-adaptation-and-geogra. GitHub. https://github.com/jdnapier/A-generalist-specialist-tradeoff-between-switchgrass-cytotypes-impacts-climate-adaptation-and-geogra. Deposited 9 February 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data for switchgrass included in our analyses are available at the NIH NCBI Sequence Read Archive (accession nos. all included in the supplemental information). Useful SNP sets and generalized versions of the code used in our analyses are publicly available at GitHub (https://github.com/jdnapier/A-generalist-specialist-tradeoff-between-switchgrass-cytotypes-impacts-climate-adaptation-and-geogra) (95).