Significance
One of the key unresolved questions in the field of molecular chaperones is how they can actively unfold proteins. In this study, we discovered that the Hsp70/Hsp40 chaperone system completely unfolds a native soluble substrate protein, the ligand-binding domain of the glucocorticoid receptor, in a concerted action. Our high-resolution optical tweezers data show in real time how the substrate is attacked by the chaperone machinery. As soon as the hormone has left the binding pocket, up to five Hsp70/Hsp40 complexes bind and unfold the protein in a stepwise manner. This finding constitutes direct evidence that the chaperone machinery can bind to the folded core of the receptor, thus providing a mechanism for Hsp70-induced protein unfolding.
Keywords: single-molecule, protein folding, Hsp70, glucocorticoid receptor, optical tweezers
Abstract
The glucocorticoid receptor (GR) is an important transcription factor and drug target linked to a variety of biological functions and diseases. It is one of the most stringent physiological clients of the Hsp90/Hsp70/Hsp40 chaperone system. In this study, we used single-molecule force spectroscopy by optical tweezers to observe the interaction of the GR’s ligand-binding domain (GR-LBD) with the Hsp70/Hsp40 chaperone system (Hsp70/40). We show in real time that Hsp70/40 can unfold the complete GR-LBD in a stepwise manner. Each unfolding step involves binding of an Hsp70 to the GR-LBD and subsequent adenosine triphosphate (ATP) hydrolysis, stimulated by Hsp40. The kinetics of chaperone-mediated unfolding depend on chaperone concentrations as well as the presence of the nucleotide exchange factor BAG1. We find that Hsp70/40 can stabilize new unfolding intermediates, showing that Hsp70/40 can directly interact with the folded core of the protein when working as an unfoldase. Our results support an unfolding mechanism where Hsp70 can directly bind to folded protein structures and unfold them upon ATP hydrolysis. These results provide important insights into the regulation of GR by Hsp70/40.
The glucocorticoid receptor (GR) is a ligand-dependent transcription factor that is expressed in almost every cell of the human body (1, 2). It consists of a ligand-binding domain (LBD), a DNA-binding domain, and an N-terminal domain (3). Upon hormone activation, the GR translocates from the cytosol to the nucleus, where it binds to specific DNA sequences and thereby regulates the transcription of target genes (4). It is involved in a wide range of critical biological processes including immune response, metabolism, development, and stress response (5, 6). Furthermore, it is connected to a variety of diseases and clinical conditions, such as diabetes (7), asthma (8), depression (9), leukemia (10), osteoporosis (11), inflammatory disorders (12), chronic obstructive pulmonary disease (13), rheumatoid arthritis (14), and allergic rhinitis (15).
Like many members of the superfamily of nuclear receptors, the hormone binding of GR is tightly regulated by the Hsp90/Hsp70/Hsp40 chaperone system (16, 17).
In previous ensemble experiments, it was shown that the Hsp70/Hsp40 chaperone system (Hsp70/40) inhibited ligand binding of ligand-unbound (apo) GR-LBD (18). This finding led to the hypothesis that Hsp70/40 may partially unfold the GR-LBD, thereby dismantling the conformation necessary for ligand binding. Partial unfolding of GR-LBD by Hsp70/40 adds to the increasing evidence that this class of chaperones not only prevents protein aggregation but also acts as an unfoldase and actively unfolds properly folded (19) as well as misfolded and aggregated structures (20). In cases where Hsp70 unfolds proteins by pulling them through pores or disengaging individual polypeptide chains from aggregates, a mechanism called entropic pulling has been proposed (21). However, how such a mechanism could work with small globular proteins such as GR-LBD is not clear.
In a previous study, we described in detail the mechanical unfolding pathway of a single GR-LBD molecule under load and how it is coupled to hormone binding (22). In the present study, we used single-molecule mechanical experiments to investigate the unfolding of GR-LBD driven by Hsp70/40. We show that GR-LBD can be actively unfolded by the consecutive action of up to five Hsp70/40 complexes.
Results
Hsp70/40 Actively Unfolds GR-LBD in a Stepwise Manner.
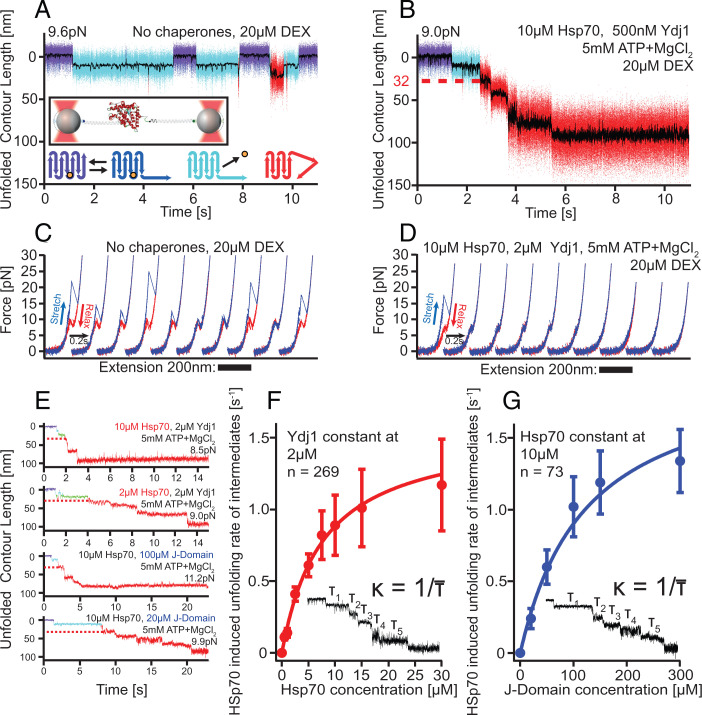
For the single-molecule measurements, we linked the termini of a single GR-LBD molecule to two DNA handles that bind specifically to silica spheres suspended in an optical trap (illustrated in the inset of Fig. 1A; for details see Materials and Methods). To study the action of Hsp70/40 on the structure of GR-LBD, we performed passive-mode experiments where the two optical traps are held at a fixed constant distance. In these experiments, the molecule is subjected to a predefined force bias, set by the distance of the two traps, and the force exerted on the tethered molecule drops upon unfolding-induced lengthening of the molecule. The force bias in the natively folded conformation is noted in all passive-mode graphs. It is typically chosen between 7.5 pN and 11 pN. With progressing unfolding of GR-LBD, the force continuously drops by ∼4 pN.
Fig. 1.
Hsp70/40 unfolds GR-LBD in a stepwise manner. (A) In the absence of chaperones, holo GR-LBD shows fast opening and closing of the N-terminal “lid” (fast transitions between purple and dark-blue state), ligand dissociation (transition to light-blue state), and ligand rebinding (return to purple/dark-blue flipping) as well as rare partial unfoldings (red) (22). Inset: Experimental scheme for the single-molecule optical tweezers experiment. Bottom: Graphical illustration of states in the upper trace (same color code). The orange sphere symbolizes DEX. (B) Sample trace of Hsp70/40 unfolding apo GR-LBD completely via five intermediates within ∼5 s. Unfolding sets in within ∼1 s after DEX dissociation. The red dashed line marks the 32 nm of unfolded contour length at which the first chaperone-induced unfolding intermediate is located. (C) Stretch-relax cycles of the GR-LBD in the absence of chaperones showing the unfolding and refolding fingerprint of GR-LBD (22). Ten consecutive pulls of the same molecule are presented, including unfolding from ligand-bound states (cycles 1, 4, 8, and 10) and ligand-unbound states (cycles 2, 3, 5, 6, 7, and 9). (D) Ten consecutive traces starting from a natively folded GR-LBD in the presence of Hsp70/40. (E) Sample traces of unfolding at various chaperone concentrations. In all traces, the first 32-nm intermediate is visible (red dashed line). (F) Unfolding rates (inverse of average dwell time per step) versus Hsp70 concentration at constant Ydj1 concentration. Error bars show SEM. (G) Unfolding rates (inverse of average dwell time per step) versus J-domain concentration at constant Hsp70 concentration. Error bars show SEM.
In the absence of chaperones (Fig. 1A), the GR-LBD behaves as previously published (22): At forces of ∼10 pN, the secondary structure element comprising the first 33 N-terminal amino acids (aa) of ligand-bound (holo) GR-LBD opens and closes in rapid equilibrium, resulting in “flipping” transitions between the purple and dark-blue states. These 33 aa act as a “lid” for ligand binding and dissociation. From the lid-open conformation (dark-blue), the ligand dexamethasone (DEX) can dissociate, resulting in the light-blue ligand-unbound conformation. Upon rebinding of ligand, the GR-LBD resumes the flipping transitions indicative of the native ligand-bound state. Occasionally, starting from the light-blue apo GR-LBD state, partial force-induced unfoldings of the GR-LBD can occur (red states). It is important to note that at forces below 10 pN, these partial unfoldings were rare and never led to a complete, let alone irreversible, unfolding of the GR-LBD.
The addition of Hsp70/40 drastically altered the situation, as shown in Fig. 1B. We used the human variant of Hsp70. As Hsp40 protein, we used both the yeast variant Ydj1 as well as its human homolog Hdj2. At chaperone concentrations in the physiological range (23, 24), we saw a stepwise, complete, and irreversible unfolding of apo GR-LBD within ∼5 s. At t = 0 (arbitrary reference frame), the GR-LBD started from the folded holo state again (Fig. 1B, purple/dark-blue flipping transitions). The flipping kinetics were unaffected by the presence of Hsp70/40 (SI Appendix, Fig. 1D). After the first ligand dissociation (Fig. 1B, light-blue state), chaperone unfolding set in after ∼1 s, and the GR-LBD unfolded from the apo state to the entirely unfolded peptide chain via five sequential unfolding intermediates (red phase with steps).
The observed chaperone-induced unfolding only occurred in the presence of all the components Hsp70, Hsp40, MgCl2 and ATP (MgATP). Control experiments showed that omission of any one component led to either incomplete or no unfolding at all (SI Appendix, Fig. 1 B–D).
Note that the force was even lower in Fig. 1B (9.0 pN) than in Fig. 1A (9.6 pN); hence, spontaneous, merely force-induced unfolding can be excluded in Fig. 1B. To rule out potential force calibration errors, we independently confirmed the difference in applied force between Fig. 1 A and B by calculating the force-dependent ratio between the population of the open and closed conformations of the lid (ratio between the times spent in the dark-blue vs. purple states), which is 0.08 in Fig. 1A and 0.06 in Fig. 1B. The higher population of lid-open states confirms that the force in Fig. 1A must have been higher than in Fig. 1B.
Stretch-Relax Cycles Show Inhibition of GR-LBD Refolding by Hsp70/40.
Next, we performed continuous stretch-relax experiments of the GR-LBD in the absence and presence of chaperones (Fig. 1 C and D). In these experiments, one optical trap was held at a fixed position, while the other trap moved back and forth at 500 nm/s, thus continuously unfolding and relaxing GR-LBD. In Fig. 1C, the unfolding and refolding signature fingerprint of the GR-LBD in the absence of chaperones is shown [cf (22)]. Ten consecutive stretch-relax cycles of the same GR-LBD molecule exhibited a mixture of ligand-bound unfoldings (high unfolding force peaks) and ligand-unbound unfoldings (low unfolding force peaks). Note that in every cycle, the GR-LBD folded back completely to the apo lid-open state.
Again, upon addition of Hsp70/40, the situation changed drastically. Now, only the first pull showed the signature unfolding trace of holo GR-LBD. Already in the first relaxation trace, the molecule could not refold to the apo lid-open state, indicating chaperones promptly interfered with folding. After four further stretch-relax cycles, no part of the molecule was able to fold anymore. The stretch-relax cycles now correspond to the wormlike chain traces of the completely unfolded peptide chain of GR-LBD. These experiments show that after the protein was unfolded using mechanical force, the chaperones quickly bound to the unfolded peptide chain, efficiently blocking refolding of GR-LBD.
Concentration-Dependent Kinetics of Chaperone-Induced GR-LBD Unfolding.
In another set of experiments, we tested how chaperone concentrations affect the observed unfolding kinetics. The top two traces in Fig. 1E illustrate the different unfolding rates at high and low Hsp70 concentrations (10 µM in top trace, 2 µM in bottom trace), holding Ydj1 concentration at 2 µM and MgATP at 5 mM constant. Both the entire unfolding time as well as the dwell times of each individual unfolding step depend on Hsp70 concentration. This suggests that each individual unfolding step must involve the binding of Hsp70. Results of experiments where Hsp70 concentration was varied from 500 nM up to 30 µM, while holding Ydj1 constant at 2 µM, are summarized in Fig. 1F. At low concentrations, we observed linear dependence, while toward high concentrations, the curve saturated. Note that over the entire force range from ∼9 pN in the natively folded state down to ∼5 pN in the completely unfolded state, the kinetics did not seem to be force dependent.
Next, we varied Hsp40 concentration while keeping the concentrations of Hsp70 at 10 µM and MgATP at 5 mM constant. We found that high concentrations of full-length Hsp40 (both Ydj1 and Hdj2) interfered destructively with the experiments because of its binding to the substrate (for details see SI Appendix, Fig. 2 B and H). For the Hsp40 variation experiments, we therefore chose a truncated version of Hsp40 consisting only of the J-Domain and the G/F-rich region of Ydj1 (JD). This way, we could eliminate the competition for binding sites between Hsp70 and Hsp40 and exclusively study the effect of the JD on Hsp70’s ATP hydrolysis rate. As shown in the bottom two traces in Fig. 1E, a variation of JD concentration, while holding Hsp70 concentration at 10 µM and MgATP at 5 mM, again modulated the unfolding rate. As before, the dwell time of each unfolding step was affected individually. Note that we needed to add about 50-fold higher concentrations of JD compared to Ydj1 at equal Hsp70 concentrations in order to reach equal unfolding rates of the GR-LBD, in agreement with earlier studies (20, 25).
The simplest model to account for both these asymptotic dependencies in Fig. 1 F and G assumes that each transition from one unfolding intermediate to the next requires two events: 1) the binding of one Hsp70 and 2) the interaction of Ydj1/JD with Hsp70, catalyzing ATP hydrolysis. In the limit of high concentrations of both Hsp70 and Hsp40, saturation will be reached through ATP hydrolysis approaching its maximal Hsp40-catalyzed rate. This model can be mathematically formulated as follows:
| [1] |
with : unfolding rate of GR-LBD from one unfolding intermediate to the next; : on-rate of Hsp70; : Hsp70 concentration; on-rate of Hsp40; : Hsp40 concentration; and : fastest possible hydrolysis rate.
Fits in Fig. 1 F and G are global fits to both datasets using the fitting parameters summarized in SI Appendix, Table 1.
Chaperone Concentration and Nucleotide Exchange Modulate the GR-LBD Folding/Unfolding Kinetics.
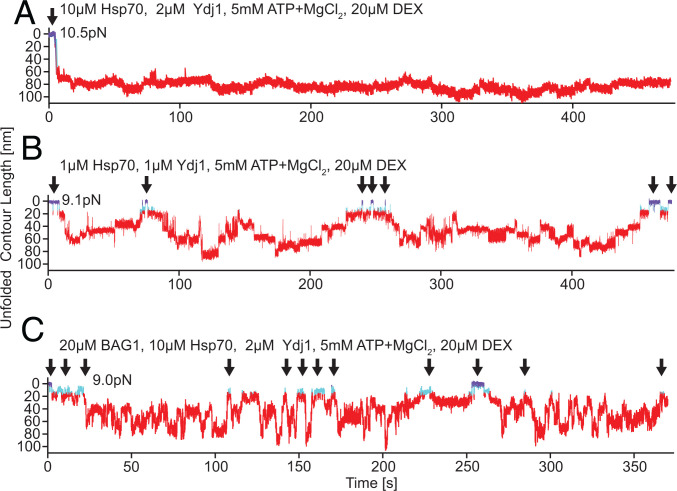
As discussed in the previous section,, Hsp70/40-induced unfolding of GR-LBD is irreversible at high, albeit still physiological, chaperone concentrations (23, 24), (, ). Even long observations in passive mode (>400 s in Fig. 2A) did not show any significant refolding of GR-LBD apart from small fluctuations around the fully unfolded contour length. This explains the persistence of the previously reported inability of GR-LBD to bind ligand in the presence of chaperones (18). Since refolding of the GR-LBD is not only transiently but also permanently blocked, ligand can never rebind.
Fig. 2.
Reversibility of chaperone-induced unfolding depends on chaperone concentrations and the presence of nucleotide exchange factor BAG1. (A) Sample trace of Hsp70/40-induced unfolding of GR-LBD at high chaperone concentration. Unfolding from the fully folded DEX bound state (arrow) proceeds rapidly within 5 s, and GR-LBD stays permanently unfolded (>400 s) with only small fluctuations around the fully unfolded length. The color code of states is the same as in Fig. 1A. (B) Sample trace of Hsp70/40-induced unfolding of GR-LBD at low chaperone concentrations where the GR-LBD undergoes repeated transitions between fully folded DEX bound states (arrows) and fully unfolded states. (C) Sample trace at high chaperone concentrations (same as in A) in the presence of the nucleotide exchange factor BAG1. GR-LBD undergoes rapid transitions between unfolded and refolded states (arrows). At 250 s, GR-LBD even populates the fully folded holo state marked in purple.
The reason for this inhibition of refolding must be multiple Hsp70s bound to the peptide chain of the GR-LBD. After hydrolysis, the Hsp70s bound to the GR-LBD are in the adenosine diphosphate (ADP)–bound state. ADP dissociation times are on the order of minutes (26). When an Hsp70 that is bound to the GR-LBD loses its ADP and binds ATP, this opens Hsp70’s substrate binding domain and allows the GR-LBD to escape. However, at high enough chaperone concentrations, whenever one of the multiple Hsp70s dissociates from GR-LBD, there are enough Hsp70s around to take its place immediately and bind to this now unoccupied binding site.
At low enough chaperone concentrations on the other hand (Fig. 2B), the GR-LBD can recover from chaperone-induced unfolding (i.e., can refold completely and bind ligand). In Fig. 2B, instead of a strongly directed and irreversible unfolding, we observed drift-like sequences of alternating steps down and up in contour length. Over the time course of 450 s, the molecule reached the fully unfolded state twice (around 120 s and around 180 s) and several times the fully folded state (arrows).
The addition of the nucleotide exchange factor BAG1 to Hsp70/40 showed a striking effect (Fig. 2C). BAG1 removed ADP from Hsp70, thereby allowing a new ATP to bind faster. Upon binding of ATP, Hsp70 opens its substrate-binding domain, hence releasing substrate. Note that the only difference between Fig. 2 A and C is the addition of BAG1. While in Fig. 2A, GR-LBD remained unfolded indefinitely, in Fig. 2C it refolded several times to the light-blue ligand unbound state (arrows), and at ∼250 s, it rebound ligand.
Chaperone Unfolding Also Works at Zero Force.
The complete chaperone-induced unfolding we observed in our optical trap experiments always occurred under mechanical load, raising the question of whether Hsp70/40 can drive such complete unfolding also in the absence of load. First, as a control experiment, we tested whether chaperones can unfold holo GR-LBD. To this end, we incubated holo GR-LBD with high chaperone concentrations (10 µM Hsp70, 2 µM Ydj1, 5 mM MgATP) in a buffer containing 50 µM DEX for 3 h. After incubation, we investigated this sample in optical trapping experiments. All GR-LBD molecules assayed (n = 15) were natively folded and ligand bound in the first stretching attempt (SI Appendix, Fig. 3A). The first stretching attempts of four different molecules are shown. This is evidence that Hsp70/40 could not unfold holo GR-LBD in our experiments.
Next, we prepared apo GR-LBD. Through multiple buffer exchanges, we reduced the concentration of DEX in the buffer to less than 0.1 nM. We incubated the sample after buffer exchanges for >12 h in the absence of chaperones. In optical trapping experiments with this sample in the absence of chaperones, all GR-LBDs (n = 10) were ligand unbound in the first stretching cycle. The first stretching cycles of four different molecules are shown in SI Appendix, Fig. 3B. All molecules lacked the flipping transition indicative of holo GR-LBD [cf encircled areas SI Appendix, Fig. 3A and (22)].
In optical trapping experiments of the same apo GR-LBD sample with additional 40 min of incubation at high chaperone concentrations (10 µM Hsp70, 2 µM Ydj1), all GR-LBDs (n = 10) were completely unfolded already before the first stretching cycle. The first stretching cycles of four different molecules are shown in SI Appendix, Fig. 3C. They lack any sign of a folded structure, but merely follow the extension characteristic of an unfolded GR-LBD polypeptide chain. It should be noted that in the absence of chaperones, we never found a GR-LBD that was unfolded already before the first stretching cycle.
Comparison of Unfolding Intermediates in Force-Induced and Chaperone-Induced Unfoldings of the GR-LBD.
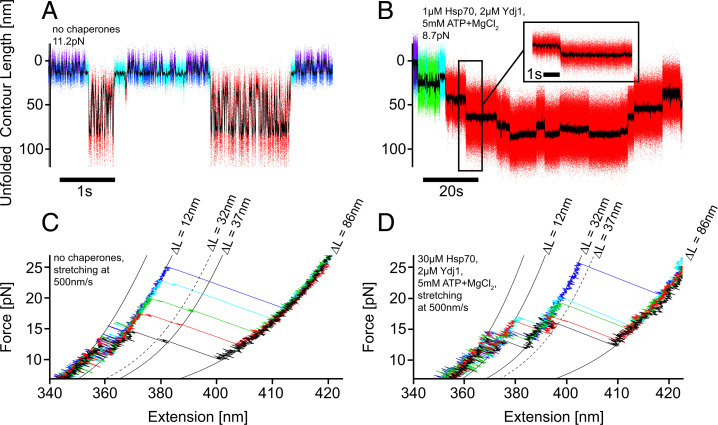
To obtain insight into the mechanism used by Hsp70/40 to unfold GR-LBD, we compared unfolding traces in which the protein was unfolded purely by mechanical forces with traces where unfolding was chaperone induced. Fig. 3 A and B illustrate the striking differences. Under the influence of a high force of 11.2 pN and in the absence of chaperones (Fig. 3A), the GR-LBD underwent very rapid equilibrium transitions between the folded holo state and the completely unfolded state, populating many short-lived intermediates. These intermediates and the corresponding force-dependent transition rates were analyzed in our previous studies (22). Note that refolding from the completely unfolded peptide chain to the natively folded apo state (light-blue) can occur within as little as 0.01 s, even against the forces applied. In contrast, in the presence of chaperones (Fig. 3B), the individual steps were slow (seconds) and no additional fast transitions between unfolding intermediates occurred. This indicates the unfolding as well as the folding steps are under complete control of the chaperones.
Fig. 3.
Comparison of force-induced and chaperone-induced unfoldings of GR-LBD in passive mode and stretch-relax experiments. (A) Multiple cycles of complete unfolding and refolding of GR-LBD in equilibrium at a high force of 11.2 pN. The molecule transitions between folded and unfolded states populating several short-lived intermediates [dwell times on the order of ∼0.01 s, cf (22)]. The color code of states is the same as in Fig. 1A. For comparison, in (B) chaperone-induced unfolding and refolding of GR-LBD are shown. The fast kinetics is completely absent, and only long-lived intermediates are populated (dwell times on the order of 5 s). Again, the color code of states is the same as in Fig. 1A, with the addition of a green Ydj1-bound state. (C) Superposition of five force-induced stretching traces (out of n > 1,000) of holo GR-LBD in the absence of chaperones. Only a barely visible, very short-lived intermediate at 37-nm unfolded contour length is populated. The traces are colored for better visual clarity. (D) Superposition of five selected force-induced stretching traces of holo GR-LBD at very high chaperone concentrations (30 µM Hsp70, 2 µM Ydj1, 5 mM MgATP). The unfolding traces exhibit the population of a very stable and long-lived intermediate at 32-nm unfolded contour length. The high stability of this chaperone-induced unfolding intermediate suggests direct binding of the chaperone to the folded core of the protein. The traces are colored for better visual clarity.
It is important to note that we found great variation in the shape of the unfolding traces we observed when GR-LBD was unfolded by Hsp70/40. This variability is likely related to Hsp70 binding to different sites at N terminus and C terminus as well as unfolding steps driven by mechanical force and backward steps through successful refolding attempts of GR-LBD. However, in all traces where we could clearly identify the five unfolding levels (selected traces in SI Appendix, Fig. 4), we consistently observed a first unfolding intermediate at ∼32-nm unfolded polypeptide length. Comparison with a DnaK (bacterial Hsp70) binding site prediction algorithm (27) showed that the binding site with the strongest Hsp70 affinity lies precisely at the 32-nm position of unfolded polypeptide.
Comparing stretch-relax experiments performed with GR-LBD at 500 nm/s in the absence of chaperones and at very high chaperone concentration, we found additional evidence that chaperones actively control the unfolding process. A zoom into a set of five pulls in the absence of chaperones (Fig. 3C) showed that the complete unfolding process from the folded to the unfolded state occurred without any long-lived population of intermediates. This is consistent with the very short intermediate dwell times we found under these conditions in passive mode experiments (Fig. 3A). Beyond an occasionally visible, very short–lived intermediate at a contour length of 37 nm, no further intermediates could be observed. In contrast, in ca. 8% of pulling traces (5 out of 61) obtained at very high chaperone concentrations (30 µM Hsp70, 2 µM Hsp40, 5 mM MgATP), we observed strikingly long-lived intermediate states at a contour length of ∼32 nm, matching our experimental results in passive mode. We could never observe the 32-nm intermediate in stretch-relax cycles in the absence of chaperones (n > 1,000). When one of the components (Hsp70, Hsp40, MgATP) was left out, such intermediates were never observed either (0 out of 31). This finding provides evidence that Hsp70 had attacked the folded GR-LBD structure and unfolded it to an intermediate state that was stabilized by direct interaction with Hsp70.
Discussion
Hsp70/40 Shows Holdase as well as Unfoldase Activity.
The mechanism by which Hsp70/40 performs its chaperoning tasks on substrates has been a matter of debate for decades (28). A large body of literature provides evidence that Hsp70 has holdase activity (i.e., can strongly bind to unfolded chains, thus blocking aggregation and refolding) (29–32). Consistent with this, our results show that after mechanical unfolding, the protein chain remains permanently unfolded if chaperones are added to the solution, indicating that the chain becomes decorated with Hsp70 molecules (Figs. 1D and 2A). The fact that the nucleotide exchange factor BAG1 abolishes the chaperone-induced inhibition of GR-LBD refolding further corroborates this observation (Fig. 2C).
Hsp70 has also been implicated in the unfolding of natively and nonnatively structured proteins, thus exhibiting unfoldase activity (18–20, 29, 31, 33, 34). Various models have been suggested for such a mechanism, including entropic pulling to induce the breakdown of clathrin cages (34) and amyloid aggregates (20), a ratchet mechanism in the context of translocation of proteins through membrane pores (35), or a similar mechanism inducing the inactivation of (36). Probably, the mechanisms are plentiful and vary from substrate to substrate.
Our results provide direct evidence that Hsp70/40 is able to unfold the GR-LBD completely in a sequence of multiple Hsp70/40-binding events and Hsp40-stimulated hydrolysis. A concerted action of many complexes (up to five in our experiments) leading to a stepwise unfolding may serve as a more general mechanism for how the energy of multiple ATP hydrolyses can be used to unfold stable proteins.
It is important to note that this Hsp70/40-induced complete unfolding also happens in the absence of force (SI Appendix, Fig. 3C) and is therefore not just a consequence of the force exerted by the optical traps.
Whether or not the complete GR-LBD is unfolded will depend on the concentrations of chaperones as well as cofactors such as BAG1, which are able to shift the equilibrium between folded and unfolded states by stimulating ADP release (Fig. 2C). Kirschke et al. (18) proposed that Hsp70/40 unfolds GR-LBD only partially, but their hydrogen deuterium exchange mass spectrometry experiments were conducted at a molar ratio of GR-LBD and Hsp70 close to 1:1, thus precluding multiple Hsp70 bindings to one GR-LBD molecule (37). While the physiological GR-LBD:Hsp70/40 ratio may vary in the cell, our scenario is relevant given that cellular Hsp70 concentrations generally exceed GR concentration (23, 24, 38).
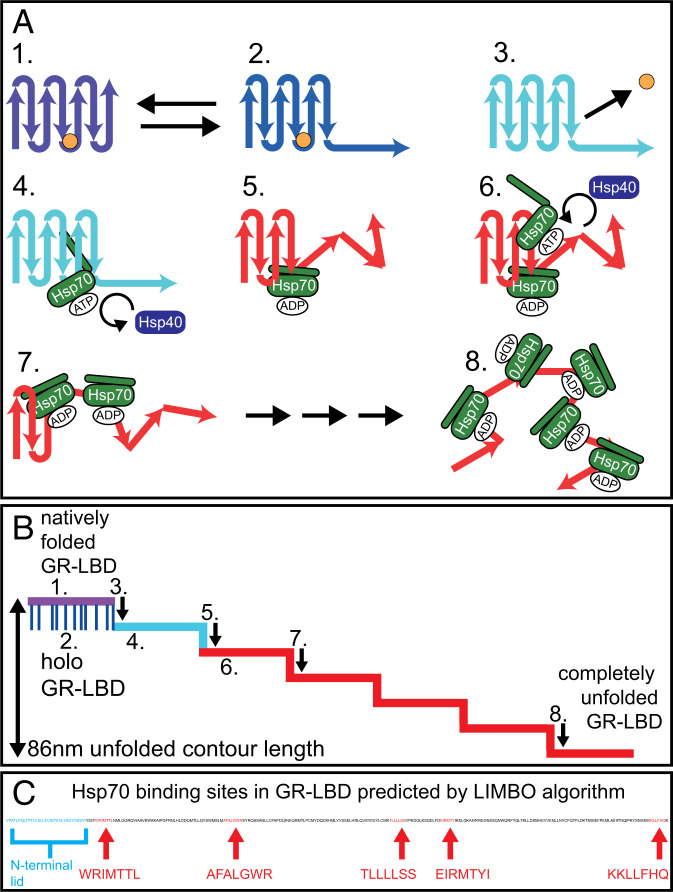
To orchestrate complete substrate unfolding, Hsp70s must bind to motifs that gradually become exposed as unfolding progresses. The LIMBO algorithm for DnaK-binding motif prediction (27) detects five evenly distributed DnaK-binding sites in the 224-aa sequence of the folded part of apo GR-LBD (Fig. 4C). Sterically, the length of the unfolded GR-LBD polypeptide allows the binding of five Hsp70 molecules. In the case of the similar-sized protein rhodanese, binding of up to seven DnaKs could be modeled onto its unfolded chain (33).
Fig. 4.
Model of Hsp70/40-induced unfolding of GR-LBD. The same color code as in all previous passive-mode traces was used to identify the states of GR-LBD: Purple: natively folded, DEX bound, lid closed; dark-blue: DEX bound, lid open; light-blue: DEX unbound, apo structure still folded; red: unfolded by chaperones. (A, 1) GR-LBD is in the natively folded holo state with its N-terminal lid closed (purple state). The hormone DEX is depicted as a small orange circle. (A, 2) Holo GR-LBD undergoes rapid flipping transitions between the lid-closed (purple) and the lid-open (dark-blue) states. (A, 3) After DEX dissociation, the lid (i.e., the first 33 aa) remains unfolded permanently (light-blue state). (A, 4) Chaperones can now attack apo GR-LBD. The first Hsp70 (in its open ATP bound state) binds to the 32-nm binding motif, which is located in the folded remainder of apo GR-LBD. 5.) Upon Hsp40-stimulated ATP hydrolysis, Hsp70 unfolds (“chews open”) the N-terminal upstream part of apo GR-LBD until the 32-nm binding motif. The associated step in contour length occurs almost simultaneously with ATP hydrolysis. Hsp70 then remains bound to the unfolded peptide chain in the ADP-bound state until ADP dissociation and rebinding of a new ATP occurs (minutes). (A, 6–8) Consecutive binding of up to four more Hsp70 molecules and J-protein–catalyzed ATP hydrolysis induced further unfoldings. The mechanism may also involve a combination of ratchet and/or entropic pulling mechanisms. At the end of the unfolding process, GR-LBD is a completely unfolded peptide chain decorated with up to five Hsp70s bound in the ADP state. The unfolding most likely starts at the N terminus, but does not necessarily have to proceed sequentially as depicted. (B) Schematic illustration of a typical Hsp70/40-induced unfolding of GR-LBD. Steps 1–8 are mapped according to the model in A. (C) Position and sequence of the Hsp70-binding sites in the sequence of GR-LBD as predicted by the LIMBO algorithm (27).
GR-LBD Unfolding Involves Multiple Steps, Each Consisting of Hsp70/40 Binding and Subsequent ATP Hydrolysis.
Our simple kinetic model (Eq. 1) assumes that each of the multiple unfolding steps involves the binding of one Hsp70 and subsequent hydrolysis of one ATP, stimulated by Hsp40. ATPases associated with diverse cellular activities (AAA) such as ClpX, by contrast, use many more ATPs to complete full unfolding (39–41).
As for the energetics of the unfolding process, five molecules of ATP [∼100 kBT (42, 43)], carry enough energy to unfold a DEX-unbound GR-LBD [225 aa, 41 kBT (22)] completely.
According to our model, the dwell times on each unfolding level are then the sum of binding times of one Hsp70 and Hsp40 as well as the time the very hydrolysis step takes. The model thereby fully explains the dependence of the unfolding intermediate dwell times on chaperone concentrations (Fig. 1E). It also accounts for the saturation behavior of the graphs in Fig. 1 F and G: At very high concentrations of either one chaperone, the binding rate of the other chaperone as well as become limiting. The highest unfolding rate per step achieved in our experiments was 1.34/s 0.22/s, in agreement with previously measured hydrolysis rates (26, 44). In our model, a potential delay between hydrolysis and unfolding is lumped into .
The chronological sequence of events we propose in our model is as follows: At high chaperone concentrations, Hsp70 binds within a short period of time (e.g., at 10 µM Hsp70, on average within , cf SI Appendix, Table 1) after a new unfolding intermediate has been populated and a new dwell period has started. ATP hydrolysis is stimulated by Hsp40 and ends the dwell time by inducing the transition to the next intermediate at a longer contour length (e.g., at 2 µM Ydj1, the dwell time on the intermediate is , cf SI Appendix, Table 1). Reducing the concentration of either one of the chaperones will delay either Hsp70 binding or ATP hydrolysis and thereby elongate the dwell time spent on the unfolding intermediate.
The opposite case of a refolding step (e.g., the clearly visible upward steps in Figs. 2B and 3B) then corresponds to the dissociation of Hsp70 followed by the refolding of a part of the GR-LBD’s peptide chain that had previously been occupied by Hsp70. Note that the folding process kinetically seems to be under full control of the Hsp70 detachment kinetics, and no misfolded states (22) can be observed. Refolding steps can be accelerated through the addition of the nucleotide exchange factor BAG1 (cf Fig. 2 A and C).
The Role of Hsp40.
The exact role of the Hsp40 cochaperone in Hsp70/40 interaction with substrate has been an important matter of debate (45, 46). It is a well-known fact that Hsp40 stimulates Hsp70’s ATP hydrolysis via its J-Domain (47, 48). Furthermore, Hsp40 can interact with substrate and seems to target Hsp70 to intended binding sites, supporting a “tagging” model (49–51).
Most recent studies of the Hsp70/40 and also Hsp90 interaction with GR-LBD used Ydj1 as the Hsp40 cochaperone (18, 37). We started with the same approach using Ydj1, but later also conducted control experiments with Hdj2 and JD.
For all three different variants of Hsp40, when added in combination with Hsp70 and MgATP, we were able to show complete and stepwise unfolding of the GR-LBD (SI Appendix, Fig. 2 E–G). Also, we directly demonstrated the stimulation of Hsp70’s ATP hydrolysis rate by Hsp40 through variation of JD concentration (Fig. 1 E and G). Nonetheless, there were remarkable differences in the effect of these four Hsp40 variants on GR-LBD, both with and without Hsp70/MgATP.
A striking difference between full-length Ydj1 and the truncated JD construct was the fact that we needed ∼50-fold higher concentrations of JD compared to Ydj1 to achieve similar unfolding rates at the same Hsp70 concentration. For instance, ∼100 µM of JD versus 2 µM of Ydj1 was necessary in combination with 10 µM Hsp70 for an unfolding rate of ∼0.8/s (see graphs in Fig. 1 F and G). This effect has been reported before (20, 25). The 50-fold higher efficiency of Ydj1 in promoting Hsp70/40 unfolding of GR-LBD compared to JD can only be due to a region truncated in the JD mutant. This finding is consistent with those regions creating transient interactions with the substrate, thus recruiting Hsp70 more efficiently to these sites (49–51).
As a control experiment and to further investigate the possibility of a tagging mechanism of Hsp40, we also measured GR-LBD in combination with all Hsp40 variants in the absence of Hsp70 (SI Appendix, Fig. 2 A–C). We identified a state where Ydj1 binds to apo GR-LBD (green state in SI Appendix, Fig. 2A), but never to holo GR-LBD. This Ydj1-bound state reproducibly occurred at an unfolded contour length of ∼20 nm. Ydj1 binding was competitive with DEX binding. However, this state was not populated by Hdj2 (SI Appendix, Fig. 2B). Also, at high concentrations of Ydj1 (>2 µM), this state in fact seemed to inhibit or delay the unfoldase activity of Hsp70 (SI Appendix, Fig. 2H). Finally, Hsp70/40 unfoldings did not require this particular Ydj1-bound state, since this state was not observed for Hdj2, even though unfolding was induced with similar efficiency (SI Appendix, Fig. 2 B and F). Moreover, unfoldings with Hsp70/Ydj1 often did not show this Ydj1-bound state on the way (e.g., Fig. 1B shows no green state on the way). We therefore conclude that this particular Ydj1-bound state is not required for efficient GR-LBD unfolding.
Apart from the Ydj1-bound state at ∼20 nm, we did not detect any further binding of Ydj1 to the GR-LBD. In force-induced complete unfoldings of the GR-LBD in the presence of Ydj1, GR-LBD did not exhibit any long-lived intermediates, but in fact showed identical kinetics as in force-induced unfoldings in the absence of chaperones (SI Appendix, Fig. 2D). If Ydj1 binds to GR-LBD to target Hsp70 to specific binding sites in the sense of a “tagging mechanism,” this binding must be weak and transient, as we could not observe it in our experiments.
In the case of Hdj2, we observed irreversible binding to unfolded GR-LBD at high concentrations exceeding 2 µM (SI Appendix, Fig. 2B). This finding is consistent with earlier reports (33) where the Escherichia coli homolog of Hsp40 DnaJ bound irreversibly to denatured rhodanese. Since much lower concentrations of Hsp40 can already stimulate ATP hydrolysis by Hsp70, this strong and irreversible binding is likely not the physiologically relevant mode of action.
JD on its own had no effect on the GR-LBD even at very high concentrations of 100 µM (SI Appendix, Fig. 2C).
The Mechanism of Hsp70/40 Unfolding of GR-LBD.
In general, unfoldase mechanisms can be divided into the two categories of passive “ratchet mechanisms” and “active mechanisms” (52), similar to the paradigm juxtaposition of “conformational selection” versus “induced fit” (53). In the context of Hsp70/40 unfolding of the GR-LBD, a pure ratchet mechanism would require frequent spontaneous conformational fluctuations of the GR-LBD. Hsp70/40 could then bind to more unfolded conformations (conformational selection) and thereby inhibit their refolding. This would rectify the fluctuations into the direction of further unfolded contour lengths and eventually lead to an unfolding of the GR-LBD. An active unfolding mechanism would have to involve the conversion of the hydrolysis-driven closing of Hsp70’s substrate-binding domain around the GR-LBD’s peptide chain into a force that unfolds the GR-LBD. Afterward, refolding of the actively unfolded parts of GR-LBD would have to be inhibited.
In the following, we discuss a ratchet as well as two active unfolding mechanisms in the light of our experimental results.
Ratchet.
As stated in the previous paragraph, a ratchet mechanism would have to rely on the presence of spontaneous conformational fluctuations of the GR-LBD toward further unfolded states, which Hsp70 could then rectify. In our experiments, we did not observe such fluctuations. Looking at the Hsp70/40 unfolding in Fig. 3B, for example, the unfolding and refolding steps in the presence of chaperones exhibited long dwell times on the order of ∼5 s without significant fluctuations present, neither short- nor long-lived. A pure ratchet mechanism is hence unlikely.
One might object that the fluctuations could be too fast for our sampling rate (30 kHz) to be recorded or that the smoothing of our data conceals them. However, even if this were the case, the fluctuations would still only sum up to extremely small amounts of cumulative dwell time in more unfolded conformations, rendering it impossible for Hsp70/40 to bind and conduct ATP hydrolysis within these short time intervals. Analysis of chaperone-induced unfolding traces using a hidden Markov model (HMM) rendered an upper limit for the population of such potential (and hence, in our assay, invisible) transiently unfolded states of <0.5% at 10 µM Hsp70 and 2 µM Ydj1. Our experiments involving the variation of chaperone concentration (Fig. 1 F and G) rendered an on-rate of ATP-bound Hsp70 of , in agreement with previous studies that reported a comparable on-rate of ATP-bound Hsp70 to substrate of (54–56). By contrast, binding of Hsp70 to conformational fluctuations that are only populated <0.5% of the time would require a much higher actual on-rate of ATP-bound Hsp70 to substrate of .
We cannot exclude transient unfoldings that are invisible in our assay because they do not lead to changes in contour length, but the above arguments also apply to these. In addition, we know from the previous discussion that both Hsp70 binding and ATP hydrolysis are obligatory for each unfolding step. Several attempts of binding may be necessary until Hsp70 commits to hydrolysis, rendering a ratchet mechanism even less likely. This leads to the conclusion that Hsp70/40 most probably unfolds GR-LBD through an active unfolding mechanism, not a ratchet.
Entropic Pulling.
Entropic pulling (21) would require Hsp70 to bind to an unfolded part of GR-LBD, but extremely close to the still folded remainder of the structure. It would then be entropically more favorable for Hsp70 to be located farther away from the remaining folded part of GR-LBD, which would lead to repulsive forces between the Hsp70 bound to the unfolded peptide chain and the folded portion of the intermediate. The consecutive action of five Hsp70s binding to evenly distributed binding sites (Fig. 4C) might suffice to unfold the 224-aa folded remainder of apo GR-LBD. This would, however, require that Hsp70-binding sites and the domain boundaries of each unfolding intermediate match precisely within less than 1 nm.
Moreover, given the relative sizes of GR-LBD (30 kDa) and Hsp70 (70 kDa), entropic pulling seems even more unlikely. With the GR-LBD being less than half the size of Hsp70, neither of the molecules provides a large enough excluded volume, comparable to a membrane or a large aggregate, for an entropic pulling mechanism to work.
While we cannot rule out an entropic pulling mechanism entirely, it is unlikely to constitute the major mechanism for Hsp70/40-induced GR-LBD unfolding.
Direct Interaction between Folded GR-LBD and Hsp70.
The contour lengths and number of unfolding intermediates induced by Hsp70 varied substantially in our experiments. However, the first intermediate occurring at 32-nm unfolded contour length is very reproducible in passive-mode traces that exhibit clear unfolding steps (SI Appendix, Fig. 4). The same long-lived intermediate at 32 nm also shows up in force-extension traces at high chaperone concentrations (Fig. 3D). Strikingly, in stretch-relax cycles, this intermediate is not populated in the absence of chaperones, where we instead observed only a short-lived intermediate at 37 nm (Fig. 3C). Neither a ratchet nor an entropic pulling mechanism can explain the population of this intermediate, since for both mechanisms we would expect to find the same intermediates as in force-induced unfolding (i.e., the short-lived 37-nm intermediate). Among the DnaK-binding sites predicted by the LIMBO algorithm (27) (mapped in Fig. 4C), the binding site with the highest score lies exactly at 32-nm contour length from the N terminus. Apparently, Hsp70 can bind to this binding site within the folded core of GR-LBD and, upon ATP hydrolysis, unfold the upstream part of the polypeptide toward the N terminus. It may well be that the aa residues containing the Hsp70-binding motif are already transiently locally unfolded, facilitating Hsp70 binding. Our assay would be blind to such partial unfoldings as long as the overall contour length does not change. Note that the 32-nm intermediate only forms in Hsp70-induced unfoldings. Hsp70 likely still forms a direct contact with the folded GR-LBD even after inducing the first unfolding step. Such a direct interaction could explain the longevity of the 32-nm intermediate observed in pulling traces, where this intermediate exists up to forces >25 pN (Fig. 3D).
Direct binding of Hsp70 to folded structures and associated stabilization of proteins have been reported before (57, 58). While in those studies active unfolding by Hsp70/40 could not be observed, such chaperone-bound intermediates might serve as starting points for the active unfolding we observed. We propose a “chewing” mechanism as a mode for Hsp70 to act as an unfoldase, where Hsp70 binds to a folded structure and upon Hsp40-mediated ATP hydrolysis forces open a part of the protein attacked. While we have a strong indication for such a mechanism for the first 32-nm intermediate, we believe that for later unfolding steps, a combination of all three discussed mechanisms may occur. The sequence of events we postulated for Hsp70-induced GR-LBD unfolding is summarized in Fig. 4 A and B: Upon lid opening and subsequent DEX unbinding, the folded portion of GR-LBD exposes the 32-nm binding site. Hsp70 can attack and, after Hsp40-mediated ATP hydrolysis, unfolds the N-terminal part of GR-LBD up to this binding site. This sequence is repeated until unfolding is completed. As outlined above, the later unfolding steps could be a combination of chewing, ratchet, or entropic pulling.
Conclusion
In summary, we showed that Hsp70/40 acts both as holdase and as unfoldase for GR-LBD. The unfolding mechanism involving direct binding to the folded core may prove important also in the many other molecular processes in which Hsp70/40 works as an unfoldase. While Hsp70/40 can completely unfold GR-LBD under the in vitro conditions used in our study, it will be of interest how the tightly regulated interplay with the Hsp90 system and other cochaperones (Hop, p23) will modify this view. While EM structural studies have already given us insight into these complexes, future single-molecule experiments have the potential to shed light on the kinetics and intermediates of the vital process of GR activation.
Materials and Methods
All protein constructs were prepared using standard recombinant techniques described in SI Appendix. The experiments were carried out using the custom-built dual-beam optical tweezers described in (59). Wormlike-chain fitting was conducted as described in SI Appendix. HMM-based assignment of states was performed as in (60).
Supplementary Material
Acknowledgments
We thank Katarzyna Tych for technical help and discussions. This work was supported by the German Research Foundation, Sonderforschungsbereich 1035, Projektnummer 201302640, Projects A5 (to M.R.) and A3 (to J.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119076119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Weikum E. R., Knuesel M. T., Ortlund E. A., Yamamoto K. R., Glucocorticoid receptor control of transcription: Precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 18, 159–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chrousos G. P., Kino T., Glucocorticoid signaling in the cell. Expanding clinical implications to complex human behavioral and somatic disorders. Ann. N. Y. Acad. Sci. 1179, 153–166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giguère V., Hollenberg S. M., Rosenfeld M. G., Evans R. M., Functional domains of the human glucocorticoid receptor. Cell 46, 645–652 (1986). [DOI] [PubMed] [Google Scholar]
- 4.Tronche F., Kellendonk C., Reichardt H. M., Schütz G., Genetic dissection of glucocorticoid receptor function in mice. Curr. Opin. Genet. Dev. 8, 532–538 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Baschant U., Tuckermann J., The role of the glucocorticoid receptor in inflammation and immunity. J. Steroid Biochem. Mol. Biol. 120, 69–75 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Sapolsky R. M., Romero L. M., Munck A. U., How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev. 21, 55–89 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Jacobson P. B., et al. , Hepatic glucocorticoid receptor antagonism is sufficient to reduce elevated hepatic glucose output and improve glucose control in animal models of type 2 diabetes. J. Pharmacol. Exp. Ther. 314, 191–200 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Sousa A. R., Lane S. J., Cidlowski J. A., Staynov D. Z., Lee T. H., Glucocorticoid resistance in asthma is associated with elevated in vivo expression of the glucocorticoid receptor β-isoform. J. Allergy Clin. Immunol. 105, 943–950 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Chourbaji S., Gass P., Glucocorticoid receptor transgenic mice as models for depression. Brain Res. Rev. 57, 554–560 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Renner K., Ausserlechner M. J., Kofler R., A conceptual view on glucocorticoid-lnduced apoptosis, cell cycle arrest and glucocorticoid resistance in lymphoblastic leukemia. Curr. Mol. Med. 3, 707–717 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Canalis E., Mazziotti G., Giustina A., Bilezikian J. P., Glucocorticoid-induced osteoporosis: Pathophysiology and therapy. Osteoporos. Int. 18, 1319–1328 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Necela B. M., Cidlowski J. A., Mechanisms of glucocorticoid receptor action in noninflammatory and inflammatory cells. Proc. Am. Thorac. Soc. 1, 239–246 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Barnes P. J., Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 131, 636–645 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Laan R. F., Jansen T. L., van Riel P. L., Glucocorticosteroids in the management of rheumatoid arthritis. Rheumatology (Oxford) 38, 6–12 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Barnes P. J., Therapeutic strategies for allergic diseases. Nature 402 (6760 suppl.), B31–B38 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Vandevyver S., Dejager L., Libert C., On the trail of the glucocorticoid receptor: Into the nucleus and back. Traffic 13, 364–374 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Lorenz O. R., et al. , Modulation of the Hsp90 chaperone cycle by a stringent client protein. Mol. Cell 53, 941–953 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Kirschke E., Goswami D., Southworth D., Griffin P. R., Agard D. A., Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157, 1685–1697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahiya V., et al. , Coordinated conformational processing of the tumor suppressor protein p53 by the Hsp70 and Hsp90 chaperone machineries. Mol. Cell 74, 816–830.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Wentink A. S., et al. , Molecular dissection of amyloid disaggregation by human HSP70. Nature 587, 483–488 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Goloubinoff P., De Los Rios P., The mechanism of Hsp70 chaperones: (Entropic) pulling the models together. Trends Biochem. Sci. 32, 372–380 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Suren T., et al. , Single-molecule force spectroscopy reveals folding steps associated with hormone binding and activation of the glucocorticoid receptor. Proc. Natl. Acad. Sci. U.S.A. 115, 11688–11693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nollen E. A., Brunsting J. F., Song J., Kampinga H. H., Morimoto R. I., Bag1 functions in vivo as a negative regulator of Hsp70 chaperone activity. Mol. Cell. Biol. 20, 1083–1088 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diamant S., Goloubinoff P., Temperature-controlled activity of DnaK-DnaJ-GrpE chaperones: Protein-folding arrest and recovery during and after heat shock depends on the substrate protein and the GrpE concentration. Biochemistry 37, 9688–9694 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Liberek K., Wall D., Georgopoulos C., The DnaJ chaperone catalytically activates the DnaK chaperone to preferentially bind the sigma 32 heat shock transcriptional regulator. Proc. Natl. Acad. Sci. U.S.A. 92, 6224–6228 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell R., Wali Karzai A., Mehl A. F., McMacken R., DnaJ dramatically stimulates ATP hydrolysis by DnaK: Insight into targeting of Hsp70 proteins to polypeptide substrates. Biochemistry 38, 4165–4176 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Van Durme J., et al. , Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comput. Biol. 5, e1000475 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clerico E. M., et al. , Hsp70 molecular chaperones: Multifunctional allosteric holding and unfolding machines. Biochem. J. 476, 1653–1677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma S. K., Christen P., Goloubinoff P., Disaggregating chaperones: An unfolding story. Curr. Protein Pept. Sci. 10, 432–446 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Winardhi R. S., Tang Q., You H., Sheetz M., Yan J., The holdase function of Escherichia coli Hsp70 (DnaK) chaperone. bioRxiv [Preprint] (2018). 10.1101/305854 (Accessed 21 March 2022). [DOI]
- 31.Slepenkov S. V., Witt S. N., The unfolding story of the Escherichia coli Hsp70 DnaK: Is DnaK a holdase or an unfoldase? Mol. Microbiol. 45, 1197–1206 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Perales-Calvo J., Giganti D., Stirnemann G., Garcia-Manyes S., The force-dependent mechanism of DnaK-mediated mechanical folding. Sci. Adv. 4, eaaq0243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kellner R., et al. , Single-molecule spectroscopy reveals chaperone-mediated expansion of substrate protein. Proc. Natl. Acad. Sci. U.S.A. 111, 13355–13360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sousa R., et al. , Clathrin-coat disassembly illuminates the mechanisms of Hsp70 force generation. Nat. Struct. Mol. Biol. 23, 821–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neupert W., Herrmann J. M., Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez F., et al. , Molecular basis for regulation of the heat shock transcription factor σ32 by the DnaK and DnaJ chaperones. Mol. Cell 32, 347–358 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Wang R. Y.-R., et al. , Structure of Hsp90–Hsp70–Hop–GR reveals the Hsp90 client-loading mechanism. Nature 601, 460–464 (2022). [DOI] [PMC free article] [PubMed]
- 38.Rossini G. P., Malaguti C., Nanomolar concentrations of untransformed glucocorticoid receptor in nuclei of intact cells. J. Steroid Biochem. Mol. Biol. 51, 291–298 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Maillard R. A., et al. , ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell 145, 459–469 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baker T. A., Sauer R. T., Clp X. P., An ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 1823, 15–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordova J. C., et al. , Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell 158, 647–658 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milo R., Phillips R., Cell Biology by the Numbers (Garland Science, 2015). [Google Scholar]
- 43.Astumian R. D., Bier M., Mechanochemical coupling of the motion of molecular motors to ATP hydrolysis. Biophys. J. 70, 637–653 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laufen T., et al. , Mechanism of regulation of Hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. U.S.A. 96, 5452–5457 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayer M. P., Gierasch L. M., Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 294, 2085–2097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan C.-Y., Lee S., Cyr D. M., Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones 8, 309–316 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Misselwitz B., Staeck O., Rapoport T. A., J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell 2, 593–603 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Kityk R., Kopp J., Mayer M. P., Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Mol. Cell 69, 227–237.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Kampinga H. H., Craig E. A., The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cyr D. M., Langer T., Douglas M. G., DnaJ-like proteins: Molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 19, 176–181 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Cheetham M. E., Caplan A. J., Structure, function and evolution of DnaJ: Conservation and adaptation of chaperone function. Cell Stress Chaperones 3, 28–36 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayer M. P., Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 38, 507–514 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Changeux J.-P., Edelstein S., Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep. 3, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmid D., Baici A., Gehring H., Christen P., Kinetics of molecular chaperone action. Science 263, 971–973 (1994). [DOI] [PubMed] [Google Scholar]
- 55.Gisler S. M., Pierpaoli E. V., Christen P., Catapult mechanism renders the chaperone action of Hsp70 unidirectional. J. Mol. Biol. 279, 833–840 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Nguyen B., Hartich D., Seifert U., Rios P. L., Thermodynamic bounds on the ultra- and infra-affinity of Hsp70 for its substrates. Biophys. J. 113, 362–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mashaghi A., et al. , Alternative modes of client binding enable functional plasticity of Hsp70. Nature 539, 448–451 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Sharma S., et al. , Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell 133, 142–153 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Grison M., Merkel U., Kostan J., Djinović-Carugo K., Rief M., α-Actinin/titin interaction: A dynamic and mechanically stable cluster of bonds in the muscle Z-disk. Proc. Natl. Acad. Sci. U.S.A. 114, 1015–1020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stigler J., Ziegler F., Gieseke A., Gebhardt J. C. M., Rief M., The complex folding network of single calmodulin molecules. Science 334, 512–516 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.