Significance
Despite the important role of human DNA polymerase α (Polα) in genome mutagenesis, there are no structural studies of Polα infidelity. The functional studies are sparse, lack high-resolution approaches, and are performed at a low salt concentration. Here we report the structure of the human Polα catalytic domain in the complex with an incoming deoxycytidine triphosphate (dCTP) and the template:primer containing a T-C mismatch at the growing primer terminus. Pre-steady-state and binding kinetics conducted at a physiological salt concentration revealed that Polα has a remarkably lower affinity to DNA and deoxynucleotide triphosphate (dNTP) than reported previously. Strikingly, we found that the incoming dNTP plays a crucial role in Polα interaction with DNA and in discrimination against a mismatched template:primer. This work is important for understanding the mechanism of Polα infidelity and provides a foundation for future studies.
Keywords: DNA polymerase α, DNA replication, crystal structure, mismatch, kinetic studies
Abstract
Human DNA polymerase α (Polα) does not possess proofreading ability and plays an important role in genome replication and mutagenesis. Polα extends the RNA primers generated by primase and provides a springboard for loading other replication factors. Here we provide the structural and functional analysis of the human Polα interaction with a mismatched template:primer. The structure of the human Polα catalytic domain in the complex with an incoming deoxycytidine triphosphate (dCTP) and the template:primer containing a T-C mismatch at the growing primer terminus was solved at a 2.9 Å resolution. It revealed the absence of significant distortions in the active site and in the conformation of the substrates, except the primer 3′-end. The T-C mismatch acquired a planar geometry where both nucleotides moved toward each other by 0.4 Å and 0.7 Å, respectively, and made one hydrogen bond. The binding studies conducted at a physiological salt concentration revealed that Polα has a low affinity to DNA and is not able to discriminate against a mispaired template:primer in the absence of deoxynucleotide triphosphate (dNTP). Strikingly, in the presence of cognate dNTP, Polα showed a more than 10-fold higher selectivity for a correct duplex versus a mismatched one. According to pre-steady-state kinetic studies, human Polα extends the T-C mismatch with a 249-fold lower efficiency due to reduction of the polymerization rate constant by 38-fold and reduced affinity to the incoming nucleotide by 6.6-fold. Thus, a mismatch at the postinsertion site affects all factors important for primer extension: affinity to both substrates and the rate of DNA polymerization.
Genome replication in eukaryotes relies on three DNA polymerases of the B-family: Polα, Polε, and Polδ (1). Replication of each DNA strand starts from the synthesis of a 9-mer RNA primer by primase, then Polα takes over the primer and extends it with deoxynucleotide triphosphates (dNTPs) (2). Polα works equally well on hybrid and DNA duplexes and generates the DNA primers required for Polε and Polδ (3, 4). Polα and primase make a complex called the primosome (5, 6). Synthesis of the chimeric RNA-DNA primers is regulated by the C-terminal domain of the primase second subunit, which coordinates the operation of two catalytic centers (2). This domain binds tightly to the template and the 5′-end of the primer and controls the access of primase and Polα to the growing primer terminus (7, 8).
It was recently shown that Polα efficiently extends the R-loops generated by replication protein A, which may be employed in vivo for restarting a stalled replication fork (9). The primosome plays a role in innate immunity mediated by the formation of the cytosolic DNA:RNA pool, which regulates the interferon I response (10). The anti-tumor toxin CD437 targets Polα and induces apoptosis in cancer cells and not in normal cells (11).
Human Polα (hPolα) is a heterodimer composed of the catalytic and accessory subunits. The catalytic subunit has a molecular mass of 166 kDa and contains 2 distinct domains: the N-terminal domain (PolαCD, amino acids 338 to 1,250) and the C-terminal domain (1,266 to 1,462). The C terminus tethers the catalytic domain to primase and contains 2 conserved zinc-binding modules (12, 13). PolαCD has the universal “right-hand” DNA polymerase fold (14) with 5 subdomains (SI Appendix, Fig. S1): Catalytic (amino acids 338 to 534 and 761 to 808), exonuclease (535 to 760; inactive), palm (834 to 908 and 968 to 1,076), fingers (909 to 967), and thumb (1,077 to 1,250). The conservative palm plays the important role in substrate binding and catalysis. It has two catalytic aspartates, which coordinate two metal ions required for dNTP binding and for catalysis of the phosphodiester bond formation (4). The thumb is flexible and makes additional contacts with a template:primer, which are not conserved between DNA polymerases of the B-family. The flexible fingers are composed of two antiparallel α-helices and make additional contacts with dNTP. The binding of dNTP by Polα triggers the conformational change in the fingers from the open state to the closed one, which stabilizes dNTP and, therefore, the entire ternary complex.
Polα is less accurate in comparison to Polδ and Polε because it does not possess the exonuclease activity to remove mismatched nucleotides (15). DNA synthesized by error-prone Polα comprises ∼1.5% of the mature genome (16). While Polδ can correct the Polα mistakes (17), the junctions between Okazaki fragments show increased levels of mutations, and it has been proposed that the early loading of nucleosomes and regulatory proteins prevents the efficient removal of mutagenic DNA generated by Polα (16). Thus, Polα may be prearranged for the generation of mutational hotspots at regulatory sites.
Despite the important role of Polα in genome mutagenesis, there are no structural studies of Polα infidelity. The functional studies are sparse and lack high-resolution techniques such as fast kinetics and binding kinetics. Moreover, all previous studies were conducted in the absence of salt or at salt concentrations significantly lower than the physiological salt concentration (4, 17–23).
Here we present the structure of the human PolαCD in the complex with a mismatched template:primer and an incoming dNTP. Pre-steady-state kinetics and binding kinetics were employed to estimate the discrimination of Polα against different mismatches at the postinsertion site. The functional studies were conducted at the physiological salt concentration, which revealed a dramatic salt effect on human Polα interaction with substrates and a relatively low affinity to DNA and dNTP. The analyzed mismatches have a significant effect on catalysis and hPolα interaction with DNA and dNTP. It was revealed that only in the presence of incoming dNTP does Polα become selective for a correct template:primer. Moreover, the discrimination factor against the mismatched duplex is higher at the physiological salt concentration in comparison to conditions with reduced salt.
Results
The T-C Mismatch at the Postinsertion Site Does Not Affect the Structure of hPolα and Its Interaction with Substrates.
The structure of the complex of human PolαCD with deoxycytidine triphosphate (dCTP) and a mismatched DNA template:RNA primer duplex (PolαCD(T-C)) was obtained by cocrystallization and determined at 2.9 Å (SI Appendix, Table S1). The T-C mismatch at the primer 3′-end reflects the misinsertion of deoxycytosine monophosphate (dCMP) opposite the templating Thy during the preceding DNA synthesis step. Thus, the obtained structure catches human Polα in the act of mismatched primer extension with a correct dNTP.
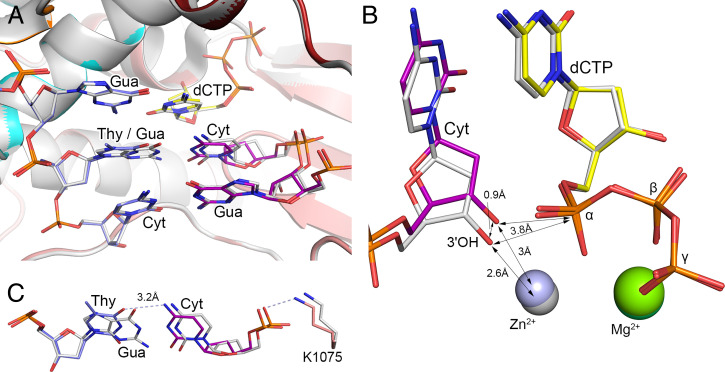
The crystallized hPolαCD (T-C) ternary complex shows very good superposition with a similar complex containing the correct DNA:RNA duplex, with an rmsd of 0.075 Å for 850 Cα atoms (SI Appendix, Fig. S1). The results of structural alignment indicate that the T-C mismatch at the growing primer terminus does not disturb the structures of the template:primer and the Polα catalytic domain. Small changes are localized only near the mismatched base pair (Fig. 1). The T-C mispair has a planar geometry, where Thy and Cyt move toward each other by 0.4 Å and 0.7 Å, respectively, measured by the N1 atoms of Cyt and Thy shift relative to the correct base pair in the aligned ternary complexes. Such convergence allows the formation of one hydrogen bond in the mispair, between N4 atom of Cyt and O4 atom of Thy (Fig. 1C). The position of the Thy phosphate is almost unchanged, while the phosphate of Cyt is shifted by 0.6 Å toward the template. It is interesting that position of the Lys1075 side chain is adjusted to maintain a contact with the Cyt phosphate. Thus, all interactions between hPolα and the template:primer are unaffected by the T-C mismatch at the postinsertion site.
Fig. 1.
Close-up view of the human Polα active site in the aligned ternary complexes containing a matched and mismatched template:primer. (A) Close-up view of the postinsertion and adjacent sites in the aligned ternary complexes. (B) The 3′-OH of the mismatched primer is shifted by 0.9 Å toward the template. The primer 3′-OH was modeled using the Builder tool in the PyMOL Molecular Graphics System. (C) Aligned G-C base pair and T-C mispair at the postinsertion site of PolαCD. The hydrogen bonds in the complex with a T-C mismatch are indicated by light-blue dashed lines. In the complex with a mismatched template:primer, the carbons of dCTP, DNA template, and RNA primer are colored yellow, marine, and purple, respectively. The subdomains of PolαCD—N-terminal, fingers, and palm—are colored orange, cyan, and salmon, respectively. In the complex containing a correct template:primer (PDB code 4qcl), all molecules are colored gray with 10% transparency. Magnesium and zinc ions in the structure with a matched duplex are colored gray and dark green, respectively. The complexes are aligned with an rmsd of 0.075 Å for 850 Cα atoms.
DNA polymerases have 2 canonical metal-binding sites, A and B (24). The metal at site A attracts the electrons from the 3′-OH and facilitates a nucleophilic attack on the α-phosphate of dNTP. The metal at site B is important for dNTP binding because it interacts with all 3 phosphate groups: α, β, and γ. In the hPolα structures with the matched and mismatched template:primer, Mg2+ occupies site B while Zn2+ occupies site A (Fig. 1B). The presence of Zn2+ at site A is probably due to the absence of 3′-OH and the disruption of the octahedral coordination of Mg2+ (4).
The displacement of the mispaired Cyt results in a 0.9 Å shift of the 3′-OH toward the template, which increases the distance to Zn2+ by 0.4 Å (Fig. 1B). The angle between the primer 3′-OH and the α-phosphate of dCTP in the complexes with correct and mismatched duplexes is 165° and 155°, respectively, while the distance between these atoms is not changed. Given the fact that 160° is considered the optimal angle for catalysis, we can conclude that the potential flexibility of the primer 3′-end and an increased distance between the 3′-OH and the catalytic metal are the main factors affecting the efficiency of the T-C mismatch extension.
Salt Concentration and dNTP Play a Critical Role in Discrimination of hPolα against Mismatched DNA.
DNA binding studies were conducted on Octet K2, which employs bio-layer interferometry (BLI) technology to monitor molecular interactions in real time. In an advantage to surface plasmon resonance, the BLI technology is fluidics-free and indifferent to the refractive index of the solution. Octet K2 allows one to obtain the rate constants of complex formation (kon), dissociation (koff), and the dissociation constant (KD), which is inversely proportional to complex stability (affinity).
The results of binding kinetics revealed that hPolα binds a DNA duplex with relatively low affinity in the presence of 150 mM NaCl, when dNTP is not provided (KD = 1.4 µM; Table 1). Previous data show a more than 10-fold higher Polα affinity to DNA, which can be explained by the low salt content in the reaction (4, 18). A KD value of 430 nM was obtained for the complex of hPolα and DNA using a fluorescence anisotropy assay (19). The difference with our data may be due to the reduced salt concentration (0.1 M NaCl) and pH (6.5) of the reaction and to the method itself.
Table 1.
Effect of salt, incoming dNTP, and a 3′-terminal mismatch on stability of the complex Polα/template:primer
| 3′-end | dTTP | 100 mM NaCl | 150 mM NaCl | ||||
|---|---|---|---|---|---|---|---|
|
kon mM−1 sec−1 |
koff x10−3 sec−1 |
KD* nM |
kon mM−1 sec−1 |
koff x10−3 sec−1 |
KD* nM |
||
| G-C | − | 342 ± 13 | 14 ± 1.1 | 40.8 ± 1.6 | 242 ± 12 | 340 ± 42 | 1,400 ± 130 |
| + | 291 ± 16 | 8.8 ± 0.12 | 30.2 ± 1.3 | 259 ± 9.9 | 43.5 ± 5.8 | 168 ± 17 | |
| T-C | − | 332 ± 17 | 4.7 ± 0.28 | 14.2 ± 0.13 | 318 ± 21 | 147 ± 18 | 462 ± 43 |
| + | 272 ± 15 | 43.7 ± 6.2 | 161 ± 31 | 314 ± 12 | 597 ± 46 | 1,900 ± 120 | |
| A-C | − | 279 ± 30 | 3.52 ± 0.52 | 12.6 ± 1.3 | 196 ± 13 | 194 ± 16 | 987 ± 18 |
| + | 260 ± 1 | 30.6 ± 0.71 | 118 ± 2.8 | 179 ± 1 | 548 ± 54 | 3,062 ± 287 | |
| C-C | − | 346 ± 51 | 4.05 ± 0.45 | 11.7 ± 0.42 | 290 ± 28 | 188 ± 16 | 646 ± 11 |
| + | 343 ± 34 | 29.2 ± 0.92 | 85.7 ± 9.7 | 227 ± 4 | 578 ± 82 | 2,548 ± 325 | |
Data are presented as mean ± SD.
*KD values are obtained by dividing koff by kon.
To confirm that salt can significantly affect the hPolα/DNA complex, DNA binding was analyzed in the presence of 0.1 M NaCl. Indeed, a reduction of the salt concentration from 0.15 M to 0.1 M increased the affinity to matched and mismatched DNA dramatically, by 34- and 33-fold, respectively (Table 1 and SI Appendix, Fig. S2). The KD value of 40.7 nM obtained here for the correct DNA at 0.1 M NaCl is close to the KD of 58 nM obtained using an electrophoretic mobility gel shift assay (4). In the presence of deoxythymidine triphosphate (dTTP), a salt effect on the hPolα/DNA complex is less severe but still impressive: the reduction of the NaCl concentration to 0.1 M stabilized hPolα complexes with cognate and T-C DNA by 5.6- and 11.8-fold, respectively (Table 1). The elevation of the salt concentration from 150 mM to 200 mM consistently reduced the affinity 7.2-fold for the cognate duplex in the presence of dTTP (KD of 168 nM and 1.2 µM, respectively).
Structural data have shown that the T-C mismatch has a minor effect on the template:primer binding interface of hPolα (SI Appendix, Fig. S1), so it was expected that the hPolα affinity to DNA was not affected. Surprisingly, interaction with a T-C duplex (KD = 462 nM; Table 1) is significantly stronger versus the correct duplex (KD = 1400 nM) at the same conditions (0.15 M salt). The threefold difference in affinity between matched and mismatched template:primers is mainly due to the 2.3-fold lower koff value in the case of a T-C mismatch. Accordingly, the same effect is observed in 0.1 M salt where KD values for matched and mismatched DNA are 40.7 nM and 14.1 nM, respectively. Reasons for the reduced dissociation rate may be the higher flexibility of the primer 3′-end and the shorter distance between the phosphates of the T-C mispair. All eukaryotic replicative DNA polymerases have a rigid DNA binding cleft for the first four base pairs (4). In the case of a T-C mismatch, the relaxed 3′-end of the primer can reduce a steric hindrance and, therefore, stabilize the hPolα/DNA complex.
To get closer to in vivo conditions, we have analyzed how dTTP at near physiological concentrations affects hPolα interaction with cognate and mismatched duplexes. DNA polymerases require two divalent metals for dNTP binding, so dTTP was supplied together with Mg2+. Upon the addition of 50 µM dTTP and 5 mM MgCl2 into reaction, affinity to the correct and T-C template:primers changes dramatically and in opposite directions (Table 1 and SI Appendix, Fig. S2). KD values for the correct and T-C DNA decrease 8.3-fold and increase 4.1-fold, respectively. In both cases, the rate of dissociation determines the change in affinity. Thus, in the presence of the correct dNTP and 0.15 M NaCl, hPolα binds the correct DNA with an 11.3-fold higher affinity versus the mismatched one.
A different effect of dTTP on hPolα interaction with the cognate and T-C template:primers was observed in 0.1 M NaCl, with affinity being increased by only 35% and decreased 11.4-fold, respectively (Table 1). Thus, in 0.1 M salt, an incoming dNTP has an 8.5-fold stronger effect on T-C DNA versus the correct DNA. In contrast, in 0.15 M NaCl, dNTP has a twofold weaker effect on mismatched DNA versus the correct one. The relatively low koff value of 0.014 s−1 for the hPolα/DNA complex in 0.1 M salt could explain the small effect of dTTP on the affinity to the correct template:primer. Of note, in the presence of the correct dNTP and 0.1 M NaCl, hPolα has only a 5.3-fold higher affinity to the correct DNA versus the mismatched DNA, which results in a 2.1-fold lower discrimination against the T-C mismatch when compared to the conditions with 0.15 M salt (Table 1).
The effect of two other 3′ terminal mismatches, C-C and A-C, on the hPolα interaction with DNA was analyzed (Table 1 and SI Appendix, Fig. S2). In the absence of dNTP, these mismatches lead to higher stability of the Polα/DNA complex, as shown above for the T-C mismatch. Upon dTTP addition, affinity to DNA with a C-C mismatch reduces 7.3- and 3.9-fold in 0.1 M and 0.15 M salt, respectively. In the case of an A-C mismatch, the corresponding values are 9.4- and 3.1-fold. In the presence of dNTP, the C-C mismatch destabilizes the hPolα/DNA complex in 0.1 M and 0.15 M NaCl by a factor of 2.8 and 15.2, respectively. For the A-C DNA, the corresponding values are 3.9 and 18.2. Therefore, the discrimination against the C-C and A-C mismatches is, respectively, 5.4- and 4.7-fold stronger in 0.15 M salt than in 0.1 M salt. These results indicate that salt concentration has a dramatic effect on the hPolα interaction with DNA, on selectivity for the cognate template:primer, and on the role of dNTP in the stability of the Polα/DNA complex.
The Mismatch at the Growing Primer End Affects Catalysis and Affinity to Incoming dNTP.
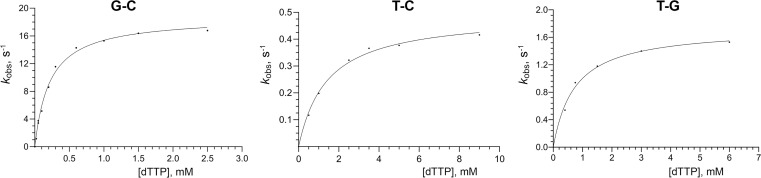
Pre-steady-state kinetic studies were undertaken at the physiological salt concentration to estimate the effect of the T-C mismatch at the postinsertion site on catalysis and affinity to dTTP. Single-nucleotide incorporation experiments were done under single-turnover conditions by providing an excess of Polα over DNA. This assay allowed an estimation of the maximal polymerization rate (kpol) and the apparent dissociation constant (Kd) for the incoming nucleotide. hPolαCD (5 µM) was incubated with Cy3-labeled DNA (0.5 µM) and quickly mixed with varying concentrations of dTTP under rapid chemical quench conditions. For each dTTP concentration, the fraction of the extended primer was plotted against time (SI Appendix, Fig. S3), and data were fit to a single-exponential equation. The obtained rate constant values were plotted against dTTP concentration (Fig. 2) to calculate the kpol and KD (Table 2).
Fig. 2.
Single-turnover kinetics of matched and mismatched primer extension by human Polα. The 3′-terminal base pair or mispair is indicated above the graphs. The primer extension rates are plotted against the dTTP concentration, and data fit to a hyperbolic equation to obtain KD and kpol values.
Table 2.
Effect of T-C and T-G mismatches at the postinsertion site on efficiency of DNA primer extension by human Polα
| 3′-end |
kpol s−1 |
KD
(dTTP) mM |
Efficiency* mM−1 s−1 |
|---|---|---|---|
| G-C | 18.7 ± 0.41 | 0.218 ± 0.017 | 85.8 |
| T-C | 0.492 ± 0.015 (38)† | 1.43 ± 0.14 (6.56) | 0.344 (249) |
| T-G | 1.74 ± 0.062 (10.8) | 0.732 ± 0.088 (3.36) | 2.38 (36.1) |
Data are presented as mean ± SD.
*Efficiency values are calculated by dividing kpol by KD.
†Discrimination factor is indicated in parentheses.
An extension of a matched primer by hPolα shows a kpol of 18.7 s−1 and an apparent KD of 218 µM (Table 2). The T-C mismatch decreases the kpol 38-fold and the affinity to incoming dTTP 6.6-fold, which results in a 249-fold lower efficiency of the mismatched DNA duplex extension versus the correct DNA duplex extension. The obtained constants allow the calculation of the polymerization rate for matched and mismatched DNA at 50 µM dTTP, resulting in 3.5 s−1 and 0.017 s−1, respectively. Thus, at a near physiological dNTP concentration, the rate of T-C mismatch extension is 206-fold lower versus that of a G-C base pair. It is worth noting that the value of 0.017 s−1 is 35-fold lower than the koff for T-C DNA in the presence of 50 μM dTTP (0.597 s−1; Table 1). This fact indicates that the majority of Polα complexes with mismatched DNA are nonproductive regarding catalysis and that they dissociate without primer extension.
In addition, the extension of the T-G mismatch was analyzed at the same conditions as for the T-C mismatch. The T-G mismatch reduces kpol 10.8-fold and the affinity to incoming dTTP 3.4-fold, which results in a 36-fold lower efficiency of mismatched primer extension versus the correct primer extension (Table 2). Therefore, in comparison to the T-C mismatch, the T-G mispair has a 3.5- and twofold weaker effect on catalysis and affinity to dNTP, respectively. Thus, human Polα extends the T-G mismatch with a 6.9-fold higher efficiency in comparison to the T-C mispair. This result is consistent with structural studies showing two hydrogen bonds in the T-G base pair (25) and only one hydrogen bond in the T-C mispair with potentially flexible Cyt (Fig. 1).
A steady-state kinetic assay showed that human Polα discriminates against the T-G mismatch at the growing primer terminus by a factor of 483 (18), which is 13-fold higher than the one obtained in this study (Table 2). It is interesting that a reduction in the affinity to dNTP made the major contribution to this factor, and the rate of T-G mismatch extension was reduced only by 21%. Experiments with yeast Polα showed similar discrimination against the T-G mismatch at the postinsertion site and no effect on catalysis (17). Such a small effect on catalysis versus our data is likely due to the type of kinetic approach and to the absence of salt in reaction. Notably, in both studies mentioned above, the KM values for incoming dNTP in the case of cognate DNA were less than 1 µM, which is more than 2 orders of magnitude lower versus the KD value obtained in the current study in the presence of 0.15 M salt (Table 2). Thus, salt concentration has a remarkable effect on Polα interaction with both substrates (DNA and dNTP), probably due to the hydrophilic interaction interface with approximately 30 hydrogen bonds (4).
Discussion
In this work, we conducted structural and functional studies to investigate how the mismatch at the postinsertion site affects primer extension by human Polα. As an example, we used the T-C mismatch, which is considered relatively easy to generate and to bypass because it does not make a steric hindrance in the DNA duplex and in the active site. By utilizing the gap-filling assay, it was shown that human Polα generates the T-C mismatch with relatively high probability (20). These data are consistent with the structural analysis of the ternary complex of RB69 DNA polymerase with a nascent T-dCTP mispair, where the position of the templating Thy and the incoming dCTP are not changed in comparison to a correct base pair, and there is a gap between them (26). Two water molecules occupy this gap and mediate interaction between the bases of these noncognate nucleotides.
Primer extension by DNA polymerases is based on phosphodiester bond formation after formation of the ternary complex, where the 3′-OH of the primer makes a nucleophilic attack on the α-phosphate of dNTP. The divalent metal coordinated at the metal binding site A works as a catalyst by attracting electrons from the 3′-OH of a primer. For efficient catalysis of this reaction, the correct positioning of the primer 3′-OH is important. This defines the optimal distances from 3′-OH to the divalent metal and to the α-phosphate and the optimal angle between the 3′-OH and the α-phosphate.
According to the structural analysis of the ternary complex, the T-C mismatch at the growing primer terminus does not affect hPolα interaction with a template:primer. The mispaired Cyt of the primer moves toward the template by 0.9 Å, which increases the distance between the 3′-OH and the catalytic metal by 0.4 Å (Fig. 1). Similar displacement of the primer Cyt was observed in the structure of human Polβ in the binary complex with DNA containing a T-C mismatch at the growing primer end (25). Surprisingly, the ternary complex of Polβ with the same mismatch showed the frayed distorted conformation of Cyt, which precludes primer extension (25). Most likely, that structure is an artifact of crystal packing and/or is due to soaking the crystal of a binary complex in a dNTP solution. In support of this suggestion, Polβ extends the T-C mismatch fairly well, with only a twofold lower efficiency in comparison to the T-G mismatch (25).
The obtained structure likely reflects only one of the T-C mismatch conformations at the postinsertion site, which is stabilized by the noncanonical hydrogen bond between Thy and Cyt. An additional factor contributing to the current conformation is the missing contact between the removed 3′-OH of a primer and the metal at site A. Thus, structural data predict the potential wobbling of Cyt between positions found in structures with a correct base pair and a mispair (Fig. 1). Analysis of these structures shows the absence of obstacles, which can impede such wobbling. Moreover, just one weak hydrogen bond cannot stabilize the T-C mispair in the conformation observed in the structure (Fig. 1C). The proposed instability of this mispair and the primer 3′-end would certainly affect the catalysis of the phosphodiester bond formation and stacking with a nascent base pair, resulting in destabilization of the ternary complex and, therefore, in reduced affinity of hPolα to both substrates. In addition, the mismatch at the primer 3′-end can increase the probability of duplex melting.
It is likely that the structure reflects the most stable conformation of the complex, which may be due to obtaining the crystals at a salt concentration below the physiological level (see Materials and Methods). The catalytic domain of Polα is very flexible and can acquire different conformations. For example, the thumb domain can partially detach from the DNA duplex, and the fingers can acquire an open conformation even in the presence of an incoming dNTP (4). We cannot exclude the possibility that in the structure reported here, the conformation of the mismatch-containing complex is not the same as one that dominates in vivo in the presence of 0.15 M salt and where Polα may bind the substrates in a more discriminative mode. Despite this possibility, we think that the obtained structural and functional data are generally consistent, taking into account the critical role of the interaction between the two substrates in the stabilization of the entire Polα/template:primer/dNTP complex.
The functional studies conducted at the physiological salt concentration revealed that the generation of the T-C mismatch at the primer 3′-end significantly affects all parameters of primer extension by hPolα: the interaction with both substrates and catalysis. This mismatch reduces the polymerization rate 38-fold and the affinity to a template:primer and dNTP 11.3- and 6.6-fold, respectively. Regarding activity, the efficiency of the T-C mismatch extension is 249-fold lower versus the matched primer. An impact of the T-C mismatch on catalysis is likely due to two factors: an increased distance between the 3′-OH and the catalytic metal and potential wobbling of the primer 3′-end. The last factor should also compromise stacking between the mispair and the nascent base pair, resulting in a reduced affinity of Polα to both substrates.
hPolα extends the T-G mispair with a 6.9-fold higher efficiency in comparison to the T-C mispair, with catalysis and affinity to dNTP being reduced only by 10.8- and 3.4-fold, respectively, in comparison with a matched duplex (Table 2). We suggest that the higher efficiency of the T-G mismatch extension versus the T-C mismatch extension is mediated by the increased stability of this mispair, due to the formation of two hydrogen bonds between Thy and Gua (27). This in turn results in more stable stacking with a nascent base pair and, therefore, in a twofold higher affinity of Polα to incoming dNTP. A 3.5-fold higher kpol value for the T-G mismatch extension versus the T-C mismatch extension may be due to the higher conformational stability of Gua and its 3′-OH.
In the absence of salt, the T-G mismatch has a 13-fold stronger effect on the efficiency of the primer extension (18). Of note, the main discrimination factor is an affinity to incoming dTTP, which was reduced 368-fold (from 0.57 µM to 210 µM) by this mismatch. Strikingly, at 0.15 M salt the T-G mismatch has a two orders of magnitude weaker effect on the affinity to dNTP (Table 2) than in the absence of salt. We suggest that the value obtained in this study at the physiological salt concentration is more realistic because the T-G mispair is one of the most stable and easiest to generate and extend (20).
The binding studies conducted in the presence of 0.15 M NaCl revealed that the incoming dNTP, which binds at the insertion site and forms a nascent base pair with a templating nucleotide, does make hPolα selective for a matched template:primer. The proposed reason for this effect is stacking between the base pairs at the insertion and postinsertion sites. Therefore, the interaction between two substrates stabilizes the Polα/DNA/dNTP ternany complex, which results in the increased affinity of hPolα to DNA. Mispairs at the postinsertion site likely impair stacking with the nascent base pair and with an adjacent base pair in the DNA duplex. Some mispairs like T-C and T-G have a planar geometry and can establish one or two hydrogen bonds, but they are less stable than Watson–Crick base pairs, which affects stacking with neighboring base pairs. It is expected that the weakened interaction between a nascent base pair and a mismatched template:primer reduces the stability of the entire complex and the Polα affinity to each substrate.
Higher selectivity of Polα for a cognate template:primer in the presence of dNTP and at the physiological salt concentration may be explained by the proper solvation of Polα and substrates, which can affect their conformation and flexibility. The important role of dNTP in Polα interaction with a template:primer indicates that DNA binding studies for DNA polymerases would be more relevant to in vivo conditions when conducted in the presence of dNTPs and at the physiological salt concentration. It is expected that salt concentration has a dramatic effect on the stability of the ternary complexes of other DNA polymerases, at least of the B-family. The relatively low discrimination of human Polα against a mismatched template:primer explains the efficient extension of different mismatched DNA duplexes by cognate and noncognate dNTPs reported recently by our group (4).
Materials and Methods
Oligonucleotides and Reagents.
Oligonucleotides were manufactured by IDT Inc. Deoxyribonucleotides used for crystallization and primer extension were obtained from Fisher Scientific. Reagents for crystallization were obtained from Hampton Research.
Protein Expression and Purification.
Cloning, expression, and purification to the homogeneity of PolαCD have been described elsewhere (28). Peak fractions obtained from the Heparin HP HiTrap column (GE Healthcare) were combined and dialyzed to the buffer specific for each application.
Crystallization, Data Collection, and Structure Determination.
The DNA duplex was obtained at a 0.2 mM concentration by annealing at 43 °C for 30 min (after heating at 80 °C for 1 min) in buffer containing 10 mM Tris⋅HCl, pH 7.9, and 70 mM KCl. The sequences for the DNA template and RNA primer were 5′-ATAGTCGCTCCAGGC (the region complementary to a primer is underlined) and 5′-rGrCrCrUrGrGrArGrCrG/ddC/, respectively (/ddC/is a dideoxycytidine used to prevent primer extension; SI Appendix, Table S2). PolαCD was dialyzed to 10 mM Tris⋅HCl, pH 7.7, 0.1 M KCl, 1% glycerol, 1 mM DTT, and 1.2 mM MgCl2. After the addition of 4 mM dCTP and mismatched DNA at 10% excess, the sample was concentrated to 16 mg/mL and flash-frozen in aliquots.
The screening of crystallization conditions was performed with the sitting-drop vapor diffusion method at 295 K by mixing 1 µL ternary complex solution with 1 µL reservoir solution. Initial screen solutions producing tiny crystals were optimized to produce cube-shaped crystals at 295 K with reservoir solution containing 0.8 mM zinc sulfate, 8.8% vol/vol PEG MME 550, 50 mM MES-NaOH, and pH 6.5. The crystals were soaked in cryoprotectant solution for a few seconds, scooped in a nylon-fiber loop, and flash-cooled in a dry nitrogen stream at 100 K. The best cryoprotectant solution contained 0.8 mM zinc sulfate, 12% vol/vol PEG MME 550, 50 mM MES-NaOH, pH 6.5, and 20% ethylene glycol. The crystals were grown in 50 to 100 mM salt, considering all potassium, sodium, and chloride ions present in the protein and reservoir solutions, and due to shrinking of the droplet during crystallization. In the crystals used in diffraction data collection, the salt concentration could be lower than 50 mM due to the brief immersion of a crystal into the cryoprotectant.
All preliminary diffraction data were obtained on a Rigaku R-AXIS IV imaging plate using Osmic VariMaxTM HR mirror-focused CuKα radiation from a Rigaku FR-E rotating anode operated at 45 kV and 45 mA. Complete diffraction data sets were collected using synchrotron X-rays on the Argonne National Laboratory Advanced Photon source beamline 24-ID-E. All intensity data were indexed, integrated, and scaled with the HKL-2000 program package (29).
The coordinates of the protein from the PolαCD/DNA:RNA/dCTP complex (PDB accession code) (4) were used as a starting model for the refinement. The 2Fo – Fc Fourier map revealed positions of an RNA primer-DNA template duplex with the clearly visible T-C mismatch (SI Appendix, Fig. S4), dCTP, metal ions, polyethylene glycol, and some solvent molecules. The positions of magnesium and zinc ions were determined using an anomalous difference Fourier map. All these ligands were added to the model and refined with CNS version 1.1 (30). Model inspection and adjustments were performed using Turbo-Frodo. The figures containing molecular structures were prepared using PyMoL (31). The crystal parameters, data processing, and refinement statistics are summarized in SI Appendix, Table S1.
Binding Studies.
The analysis of the PolαCD/DNA binding kinetics was done at 23 °C on Octet K2 (Sartorius AG). This device uses BLI technology to monitor molecular interactions in real time. Purified PolαCD was dialyzed to 30 mM Tris-Hepes, pH 7.8, 200 mM NaCl, 1% glycerol, and 2 mM TCEP; concentrated to 100 µM; and flash-frozen in aliquots. The quality of the Polα sample was checked by dynamic light scattering using DynaPro NanoStar (Wyatt Technology, Santa Barbara) to confirm that >99% of protein was in monomeric form. The template with a biotin-TEG at the 5′-overhang (SI Appendix, Table S2) was annealed to the primer and immobilized on the streptavidin-coated biosensor (SAX, Sartorius AG). The primer contains a dideoxy cytosine at the 3′-end and was added at twofold molar excess with regard to the template. SAX sensors were loaded with DNA-biotin at 50 nM concentration for 7 min at 500 rpm. Then sensors were blocked by incubating for 2 min in 10 µg/mL biocytin. In the first row of a 96-well microplate (Greiner Bio-One), the first six wells contained the buffer, consisting of 30 mM Tris-Hepes, pH 7.8, 150 mM NaCl, 2 mM TCEP, and 0.002% Tween 20. The next six wells contained the twofold dilutions of PolαCD in the same buffer. All wells in the next row contained only the buffer for the reference. The average value and the error are calculated from at least three independent experiments.
Pre-Steady-State Kinetic Studies.
Pre-steady-state kinetic studies were performed on the QFM-4000 rapid chemical quench apparatus (BioLogic, France) at 23 °C. The Polα sample was prepared in the same way as for the binding studies. Reactions contained 2.5 µM PolαCD, 250 nM DNA, varying concentrations of dTTP, 30 mM Tris-Hepes, pH 7.8, 0.15 M NaCl, 8 mM MgCl2, 2 mM TCEP, and 0.2 mg/mL BSA. In reactions with 9 mM dTTP, the concentration of MgCl2 was increased to 11 mM to provide some excess over dNTP. Polα was incubated with a Cy3-labeled 15-mer primer annealed to a 25-mer DNA template (SI Appendix, Table S2), to allow for the formation of the binary complex, and was rapidly mixed with dTTP and MgCl2 followed by quenching with 0.3 M EDTA. Products were collected in a tube and separated by denaturing urea PAGE. The Cy3-labeled products were visualized by Typhoon FLA 9500 (GE Healthcare) and quantified by ImageJ, version 1.5.3 (NIH). The fraction of the extended primer was calculated by dividing the amount of the extended primer by the amount of primer added to the reaction. For each dTTP concentration, the percentage of the extended primer was plotted against time and the data were fit to a single exponential equation:
| [1] |
where A is the amplitude, kobs is the observed rate for dNTP incorporation, and t is the time. The kobs was plotted against dTTP concentration and the data were fit to the following equation:
| [2] |
using the GraphPad Prizm software to obtain kpol, the maximum rate of nucleotide incorporation, and KD, the apparent dissociation constant for the incoming nucleotide.
Supplementary Material
Acknowledgments
We thank K. Jordan for editing this manuscript. This work was supported by the National Institute of General Medical Sciences (NIGMS, of the NIH) grant R35 GM127085 to T.H.T. The University of Nebraska Medical Center Genomics Core receives partial support from the NIGMS INBRE-P20GM103427 grant as well as the Fred & Pamela Buffett Cancer Center support grant P30 CA036727. The Eppley Institute′s X-ray Crystallography Core Facility is supported by Cancer Center support grant P30 CA036727. This work is also based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the NIGMS (P30 GM124165). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
Footnotes
Competing interest statement: The authors declare no competing interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111744119/-/DCSupplemental.
Data Availability
The crystal structure coordinates of PolαCD (T-C) have been deposited in PDB under accession number 7n2m. The coordinates of the protein from the PolαCD/DNA:RNA/dCTP complex were retrieved from PDB under accession code 4qcl (4).
References
- 1.Burgers P. M. J., Kunkel T. A., Eukaryotic DNA replication fork. Annu. Rev. Biochem. 86, 417–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranovskiy A. G., et al. , Mechanism of concerted RNA-DNA primer synthesis by the human primosome. J. Biol. Chem. 291, 10006–10020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perera R. L., et al. , Mechanism for priming DNA synthesis by yeast DNA polymerase α. eLife 2, e00482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baranovskiy A. G., et al. , Activity and fidelity of human DNA polymerase α depend on primer structure. J. Biol. Chem. 293, 6824–6843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baranovskiy A. G., Tahirov T. H., Elaborated action of the human primosome. Genes (Basel) 8, 62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Núñez-Ramírez R., et al. , Flexible tethering of primase and DNA Pol α in the eukaryotic primosome. Nucleic Acids Res. 39, 8187–8199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baranovskiy A. G., et al. , Insight into the human DNA primase interaction with template-primer. J. Biol. Chem. 291, 4793–4802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zerbe L. K., Kuchta R. D., The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry 41, 4891–4900 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Mazina O. M., et al. , Replication protein A binds RNA and promotes R-loop formation. J. Biol. Chem. 295, 14203–14213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Starokadomskyy P., et al. , DNA polymerase-α regulates the activation of type I interferons through cytosolic RNA:DNA synthesis. Nat. Immunol. 17, 495–504 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han T., et al. , The antitumor toxin CD437 is a direct inhibitor of DNA polymerase α. Nat. Chem. Biol. 12, 511–515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klinge S., Núñez-Ramírez R., Llorca O., Pellegrini L., 3D architecture of DNA Pol α reveals the functional core of multi-subunit replicative polymerases. EMBO J. 28, 1978–1987 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suwa Y., et al. , Crystal structure of the human pol α B subunit in complex with the C-terminal domain of the catalytic subunit. J. Biol. Chem. 290, 14328–14337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J., et al. , Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell 89, 1087–1099 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Pavlov Y. I., Shcherbakova P. V., Rogozin I. B., Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int. Rev. Cytol. 255, 41–132 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Reijns M. A. M., et al. , Lagging-strand replication shapes the mutational landscape of the genome. Nature 518, 502–506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavlov Y. I., et al. , Evidence that errors made by DNA polymerase α are corrected by DNA polymerase delta. Curr. Biol. 16, 202–207 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Copeland W. C., Lam N. K., Wang T. S., Fidelity studies of the human DNA polymerase α. The most conserved region among α-like DNA polymerases is responsible for metal-induced infidelity in DNA synthesis. J. Biol. Chem. 268, 11041–11049 (1993). [PubMed] [Google Scholar]
- 19.Coloma J., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K., Human DNA polymerase α in binary complex with a DNA:DNA template-primer. Sci. Rep. 6, 23784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka S., et al. , Functions of base selection step in human DNA polymerase α. DNA Repair (Amst.) 9, 534–541 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Lund T. J., et al. , B family DNA polymerases asymmetrically recognize pyrimidines and purines. Biochemistry 50, 7243–7250 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong Q., Wang T. S., Mutational studies of human DNA polymerase α. Lysine 950 in the third most conserved region of α-like DNA polymerases is involved in binding the deoxynucleoside triphosphate. J. Biol. Chem. 270, 21563–21570 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Thomas D. C., et al. , Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry 30, 11751–11759 (1991). [DOI] [PubMed] [Google Scholar]
- 24.Yang W., Lee J. Y., Nowotny M., Making and breaking nucleic acids: Two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Batra V. K., Beard W. A., Pedersen L. C., Wilson S. H., Structures of DNA polymerase mispaired DNA termini transitioning to pre-catalytic complexes support an induced-fit fidelity mechanism. Structure 24, 1863–1875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia S., Wang J., Konigsberg W. H., DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase. J. Am. Chem. Soc. 135, 193–202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia S., Konigsberg W. H., Mispairs with Watson-Crick base-pair geometry observed in ternary complexes of an RB69 DNA polymerase variant. Protein Sci. 23, 508–513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baranovskiy A. G., et al. , Structural basis for inhibition of DNA replication by aphidicolin. Nucleic Acids Res. 42, 14013–14021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otwinowski Z., Minor W., Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Brünger A. T., et al. , Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- 31.DeLano W. L., The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA, 2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structure coordinates of PolαCD (T-C) have been deposited in PDB under accession number 7n2m. The coordinates of the protein from the PolαCD/DNA:RNA/dCTP complex were retrieved from PDB under accession code 4qcl (4).