Significance
We report a quantitative Raman microscopy method that measures the concentration of protein and lipid in cells at high spatial resolution in living and in fixed samples of tissues, allowing quantitative studies of cell size and organelle regulation both in cell culture and in tissue slices; it can be applied to problems of cell size control, intracellular crowding, and lipid metabolism in the context of cell growth, cell differentiation, cell senescence, and pathology.
Keywords: cell size, quantitative microscopy, single cell mass quantification, lipid imaging, stimulated Raman scattering
Abstract
Cell mass and chemical composition are important aggregate cellular properties that are especially relevant to physiological processes, such as growth control and tissue homeostasis. Despite their importance, it has been difficult to measure these features quantitatively at the individual cell level in intact tissue. Here, we introduce normalized Raman imaging (NoRI), a stimulated Raman scattering (SRS) microscopy method that provides the local concentrations of protein, lipid, and water from live or fixed tissue samples with high spatial resolution. Using NoRI, we demonstrate that protein, lipid, and water concentrations at the single cell are maintained in a tight range in cells under the same physiological conditions and are altered in different physiological states, such as cell cycle stages, attachment to substrates of different stiffness, or by entering senescence. In animal tissues, protein and lipid concentration varies with cell types, yet an unexpected cell-to-cell heterogeneity was found in cerebellar Purkinje cells. The protein and lipid concentration profile provides means to quantitatively compare disease-related pathology, as demonstrated using models of Alzheimer’s disease. This demonstration shows that NoRI is a broadly applicable technique for probing the biological regulation of protein mass, lipid mass, and water mass for studies of cellular and tissue growth, homeostasis, and disease.
Cells must tightly control their volume, mass, and molecular composition to ensure fitness (1) or functionality in a tissue context (2). Cell mass and volume growth are closely coordinated with proliferation (3, 4) and tissue growth (5), and the ratio of mass to volume, that is mass concentration, reflects macromolecular crowding of the intracellular milieu (6) and influences cellular fitness (1). Despite the conceptual simplicity and importance of these physiological features, our understanding how cells regulate and coordinate these aggregate properties has been hampered by the difficulty of accurately measuring them, especially in a tissue context (7). Technological tools to determine single cell mass and volume accurately has been both a driving force but a limited one for addressing these questions. For instance, several methods have been developed for single cell measurement of either cell volume (8–11), cell mass (12–18), or surrogate variables that correlate with cell size (4, 19, 20) (Table 1). Despite the general assumption that cell mass and volume are proportional to each other, cell volume can dramatically deviate from cell mass (5). As the importance of the macromolecular density in cytoplasm has become a focus of interest, a few technologies have been developed to quantitatively characterize the density of the intracellular milieu (21, 22) and subcellular compartments, such as the phase separated compartments (23, 24). However, existing methods lack subcellular resolution or are limited in their applicability in a tissue context. As a result, our current knowledge on the regulation of cell size and cytoplasmic density is drawn from bulk measurements, average behavior of cell populations, suspended or cultured cells in vitro, or from measurement of proxy variables. Furthermore, current single cell mass measurements only provide total biomass and cannot differentiate between protein and lipid mass.
Table 1.
Comparison of NoRI and existing methods for single cell biomass measurement
| Method | Measured parameter | Measurement characteristics | Sample types | References | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mass | Mass density | Absolute quantification* | Subcellular resolution | Chemical composition | Suspended cells | Adherent cell culture | Cells in tissue | Live sample | |||
| NoRI (Normalized Raman Imaging) | Biomass composition and concentration | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | This work |
| SRS (Stimulated Raman Scattering) | Biomass composition and concentration | ✓ | ✓ | ✕ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 25, 29 |
| SMR (Suspended Microchannel Resonator) | Buoyant mass, volume, density | ✓ | ✓ | ✓ | ✕ | ✕ | ✓ | ✕ | ✕ | ✓ | 12–14, 21 |
| QPI (Quantitative Phase Imaging) | Refractive index (dry mass density) | ✓ | ✓ | ✓ | ✓ | ✕ | ✓ | ✓ | ✕ | ✓ | 15–17, 23, 24 |
| Particle tracking, FCS, FRAP | Diffusion | ✕ | ✓ | ✕ | ✓ | ✕ | ✓ | ✓ | ✓ | ✓ | 9, 71 |
| Inertial picobalance | Buoyant mass | ✓ | ✕ | ✓ | ✕ | ✕ | ✕ | ✓ | ✕ | ✓ | 18 |
| Electron microscopy | Number and volume of cellular structures | ✓ | ✓ | ✕ | ✓ | ✕ | ✓ | ✓ | ✓ | ✕ | 8, 9 |
| Light microscopy | Protein or lipid stain, volume of cell or nucleus | ✓ | ✓ | ✕ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 4, 10, 11, 72, 73 |
| Flow cytometry | Light scattering, protein stain, UV absorption | ✓ | ✕ | ✕ | ✕ | ✓ | ✓ | ✕ | ✕ | ✓ | 19, 20 |
*Absolute quantification of mass or mass density in physical units. Volume, area, or intensity in arbitrary units are considered semi-quantitative or relative measurements of biomass.
To develop a method that overcomes the limitations of existing approaches, we turned to stimulated Raman scattering (SRS) microscopy (25), which is ideal for quantitative analysis of cellular materials in tissue. However, existing SRS approaches suffer from heterogeneous signal attenuation due to light scattering in thick samples, limiting their utility for accurate quantification of single cell mass in situ (26). Here we report an alternative approach that enables accurate determination of single cell mass and cytoplasmic mass concentration in live or fixed tissue by computationally removing the effect of light scattering. The key step in normalized Raman imaging (NoRI) is the conversion of the SRS images to absolute concentrations through normalization of the undesirable intensity variation caused by sample light scattering. Hence, we name this method normalized stimulated Raman scattering imaging. Computing the sum of chemical components, including water, is the key element in converting chemical compositions to absolute concentration in a way comparable to those demonstrated in other Raman modalities (27, 28).
NoRI provides two important advantages over existing methods of single cell mass measurement: Protein mass, lipid mass, and water content can be measured separately, and in tissue samples with three-dimensional (3D) resolution (SI Appendix, Fig. S4). Absolute calibration enables cross-comparison across samples and enables further specification of features without internal reference. Using NoRI, we demonstrate that protein and lipid concentrations are reproducible cellular phenotypes, that they are specific to cell types, and that they change with physiological state. NoRI can enable previously inaccessible single cell mass measurements and will allow many open biological questions to be addressed.
Principle of NoRI
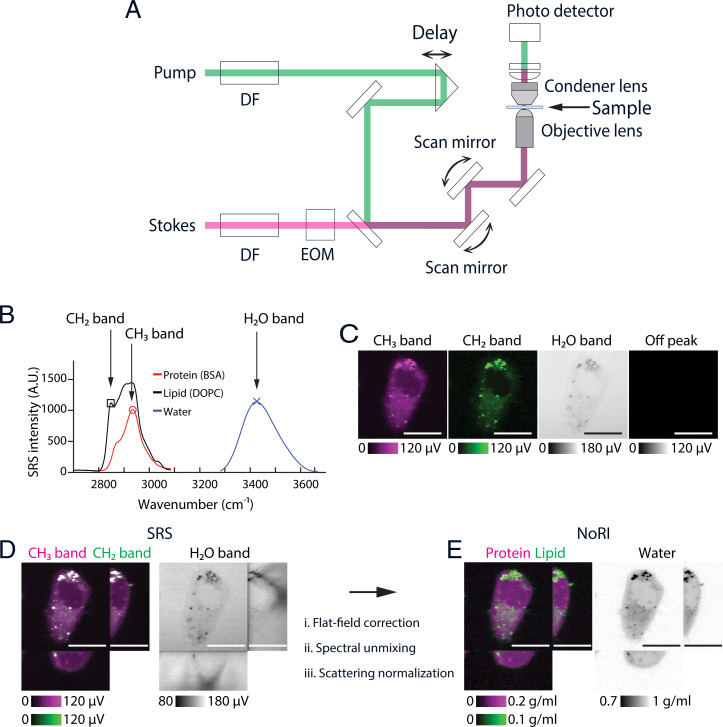
We visualize proteins and lipids under a custom-built SRS microscope (Fig. 1A) by imaging the Raman peaks at 2,935 and 2,853 cm−1 originating from methyl groups in proteins and methylene groups in fatty acids (29). For the normalization of light-scattering, SRS signals from water were measured at the 3,420 cm−1 peak of the oxygen–hydrogen stretching modes (Fig. 1C). These three Raman bands are referred to as the CH3 band, the CH2 band, and the H2O band in the rest of this manuscript (Fig. 1B). The different SRS intensities of proteins, lipids, and water at these bands enable spectral decomposition to protein, lipid, and water (30). The mapping from the CH2, CH3, and H2O SRS intensity to the protein, lipid, and water concentration can be expressed with a 3 × 3 matrix, which we refer to as the “decomposition matrix.” Concentrations computed by conventional methods of spectral decomposition are affected by light scattering that varies within the sample and between different samples and limits quantitative interpretation. To overcome this limitation, we took advantage of the fact that water, proteins, and lipids account for nearly 100% of the chemical composition of most biological samples (34). Hence, we scale the intensity of each component to make the sum of their volumes equal 1 (which is 100% vol/vol) at each voxel. This results in the absolute concentration of protein, lipid, and water in the unit of volume fractions. As an example, Fig. 1D shows the x–y and orthogonal cross-sections of a live A7 cell’s SRS intensity images prenormalization. Especially, the shadows in the orthogonal view of the H2O band highlights the attenuation due to diffraction at the cell edge and intracellular lipid droplets. By contrast, NoRI normalization eliminates these artifacts and provides the absolute concentration of protein, lipid, and water (Fig. 1E and SI Appendix, Fig. S18).
Fig. 1.
Principles of NoRI. (A) Schematic of the spectral-focusing stimulated Raman scattering microscope. Pump and Stokes femtosecond pulse lasers are chirped by dense flint (DF) glass and intensity modulated by an electrooptical modulator (EOM). Motorized delay is used to fine tune the Raman band. Transmitted pump laser intensity is detected in the transside. (B) Raman spectra of protein, lipid, and water measured by spectral-focusing stimulated Raman scattering microscope. See SI Appendix, Supplementary Methods for the acquisition parameters. Three Raman bands are measured in NoRI: CH3 (red circle, 2,935 cm−1), CH2 (black square, 2,853 cm−1), and H2O (blue X, 3,420 cm−1) bands. (C) Representative SRS images of a live A7 cell at the CH3, CH2, H2O, and off-peak Raman bands. (Scale bars, 20 µm.) (D) Orthogonal view of the SRS images in the CH3, CH2, and H2O Raman bands. (Scale bars, 20 µm.) (E) Orthogonal view of the NoRI measurement of protein, lipid, and water concentrations. (Scale bars, 20 µm.)
To enable this normalization, we devised a calibration and sample imaging procedure that preserves the quantitative relation between the SRS intensity and concentration (in vol/vol) by eliminating other sources of intensity variation. Specifically, the decomposition matrix is measured from the calibration standard samples of known protein, lipid, and water concentrations using the SRS microscope (SI Appendix, Fig. S5). We used bovine serum albumin (BSA) as a protein standard and dioleoylphosphatidylcholine (DOPC) as a lipid standard. Since BSA has similar methyl and methylene group frequency to vertebrate proteomes (SI Appendix, Fig. S16), and DOPC is the most abundant lipid species in lipid membranes, they provide a practical and economical approximation of average proteome and phospholipids. To capture the intrinsic difference in Raman spectra between protein, lipid, and water samples, we measured them under identical optical condition. The calibration images were acquired with the objective lens and the condenser lens at the same positions, and the intensity variation caused by the pointing direction of the laser beams was computationally corrected from the two-dimensional (2D) intensity profile at each tunable-laser wavelength (“flat-field correction mask”) (SI Appendix, Supplementary Methods). Once the decomposition matrix and flat-field correction mask are determined from the calibration samples, biological samples such as live or fixed cells, tissue slices, and small organisms in an unstained or fluorescence-labeled state can be measured. Once the raw SRS intensity images are acquired at the CH3, CH2, and H2O bands (Fig. 1D), the flat-field correction mask generated in the calibration step is applied to the sample images, then the decomposition matrix is applied to each voxel to obtain the unnormalized image of protein, lipid, and water components. Then, normalization is performed at each voxel by dividing by the sum of the prenormalized protein, lipid, and water images (SI Appendix, Fig. S7). The mass concentrations of proteins and lipids is estimated from the volume concentration by multiplying with the mass density of pure protein or lipids (Fig. 1E). We used the densities of BSA and DOPC (1.3643 g/mL and 1.0101 g/mL), respectively, for the mass concentration conversion. We note that lipid droplets, which are composed of neutral lipids and cholesterol esters, have a shifted CH2 band compared to phospholipids, and, therefore, spectral decomposition based on DOPC spectrum is not ideal. On the other hand, lipid droplets occupy a distinct nonaqueous phase and the NoRI output provides an easy means to segment lipid droplets from the rest of the cytoplasm by the thresholding of high lipid concentration (Fig. 2B and SI Appendix, Fig. S10). Therefore, we distinguish lipid droplets from phospholipids by thresholding and converted lipid droplet’s volume to mass using the density of glyceryl trilinoleate (0.925 g/mL).
Fig. 2.
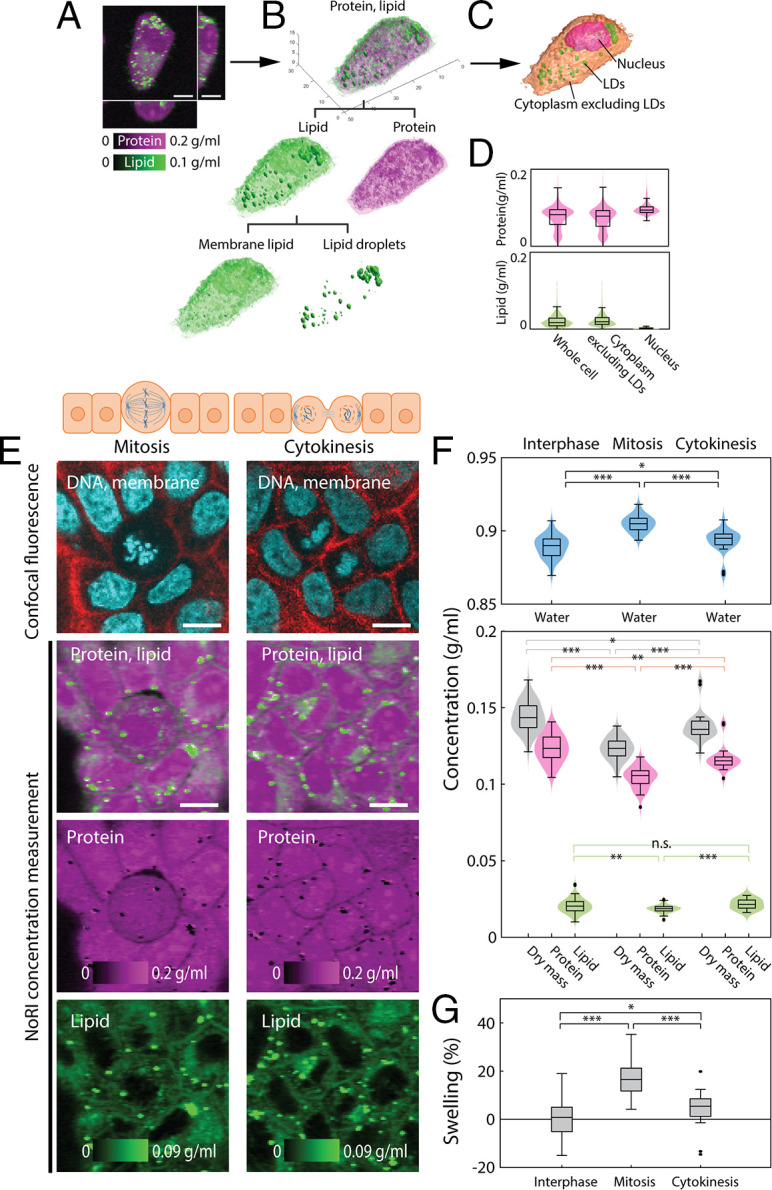
Label-free measurement of protein and lipid shows dilution of cytoplasm in mitotic cells. (A) Volumetric NoRI measurement of a live A7 cell. (Scale bar, 10 µm.) (B) Volumetric visualization of the protein and lipid distribution in the cell. Lipid is further classified as membrane lipids in the aqueous phase of the cytosol and lipid droplets. (C) Label-free volumetric segmentation of a whole cell, nucleus, and lipid droplets from NoRI data. See SI Appendix, Supplementary Methods for the segmentation method. (D) Distribution of protein and lipid concentration within the volume of the whole cell, cytoplasm excluding lipid droplets, and the nucleus in C. (E) Representative live MDCK cells in mitosis and cytokinesis. (Scale bars, 10 µm.) (F) NoRI images of mitotic and interphase cells. Mitotic cells have a more diluted cytoplasm compared to interphase cells, as revealed by lower protein and lipid concentrations and a higher water concentration (water: interphase 0.889 ± 0.008 g/mL, mitosis 0.905 ± 0.006 g/mL, P < 0.0001; dry mass: interphase 0.144 ± 0.009 g/mL, mitosis 0.123 ± 0.007 g/mL, P < 0.0001; protein: interphase 0.123 ± 0.009 g/mL, mitosis 0.105 ± 0.007 g/mL, P < 0.0001; and lipid: interphase 0.021 ± 0.005 g/mL, mitosis 0.019 ± 0.002 g/mL, P = 0.0052). The cytoplasmic concentration recovers toward the interphase value during cytokinesis (water 0.893 ± 0.008 g/mL, dry mass 0.138 ± 0.010 g/mL, protein 0.117 ± 0.008 g/mL, lipid 0.022 ± 0.003 g/mL). Lipid droplets are excluded from the lipid concentration calculation. The number of cells in interphase, metaphase, and cytokinesis are n = 81, n = 57, and n = 24, respectively. (G) Relative difference of cytoplasmic density, here expressed in terms of the relative volume change (“swelling”) based on the degree that a cell of interphase cells’ median cytoplasmic density would have to change to achieve the observed difference in cytoplasmic water concentration. Mean and SD of swelling are 0.5 ± 6.8%, 17.0 ± 6.8%, and 4.2 ± 7.3% for interphase, mitosis, and cytokinesis, respectively (P < 0.001 for interphase and mitosis, P = 0.020 for interphase and cytokinesis). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. n.s. is not significant with P > 0.05.
We next applied NoRI measurement to solution samples of known concentrations to characterize the method’s accuracy in the presence of signal attenuation of diverse origins, including optical aberration from refractive index mismatching and imperfections and temporal instability of the optical system. Objective lenses are typically designed to optimally perform with an immersion media of a specific refractive index, and refractive mismatch of the sample causes optical aberration. For example, SRS intensity of 36% BSA solution sandwiched between a cover glass and a glass slide decreases with imaging depth because the BSA solution has a much higher refractive index than the intended immersion medium of the objective lens (SI Appendix, Fig. S8A). NoRI normalization corrects for this effect and the resulting concentration is homogenous in the entire sample volume (SI Appendix, Fig. S8B). Intensity variation caused by imperfections of the optical system was also removed by the normalization as demonstrated in the water component image of the pure water sample (SI Appendix, Fig. S8 C and D). Environmental instability including ambient temperature drift may cause fluctuations in SRS intensity, thereby hampering time-dependent measurements. As shown in the time trace of SRS signals of a BSA solution at the CH3, CH2, and H2O bands, the intensity fluctuated at about 2% of the mean during 2 h (SI Appendix, Fig. S8 E and F). A conventional approach for normalizing would divide the CH3 and CH2 band signals with that of the H2O band, which indeed reduces the temporal fluctuations (SI Appendix, Fig. S8G). But dividing by the water signal renders the relation between intensity signal and analyte concentration nonlinear (for example, the normalization would diverge when there is little water, as in lipid droplets). NoRI normalization removes the temporal fluctuation while retaining linearity in concentration measurement (SI Appendix, Fig. S8H). We imaged a titration series of BSA solutions and DOPC solutions to demonstrate the linearity of NoRI concentration measurements (SI Appendix, Fig. S9). The NoRI measurements show excellent agreement with the actual concentration of solutions, with sensitivity of ∼15 mg/mL as measured by the SD of the BSA solution sample. Since there is no existing technique to benchmark in situ measurement of the protein and lipid mass of a single cell, we compared the dry mass (sum of protein and lipid concentration) measurement by NoRI with refractive index tomography (Tomocube, HT-2) (35), which is currently the only other method that can measure local mass concentration with subcellular resolution. When we measured fixed HeLa cells by both techniques, we found that dry mass concentration measured by NoRI was in good agreement with the dry mass concentration from refractive index tomography (Pearson correlation coefficient r = 0.736 from SI Appendix, Fig. S12).
Cellular Protein and Lipid Concentration Changes with a Cell’s Physiological State
By providing protein, lipid, and water concentration measurements of a cell or a subcellular compartment, NoRI opens up possibilities in the investigation of single cell physiology. Also, the mass of a cell or nucleus can be computed by integration of the protein and lipid concentration over the volume. Furthermore, nucleus and lipid droplets can be segmented from NoRI images without the use of staining due to their specific lipid concentration profile (Fig. 2A): The nucleus can be recognized from the absence of lipid and lipid droplets from their high lipid concentration (Fig. 2B and SI Appendix, Fig. S13). We demonstrate here several potential applications.
Cells tightly maintain cytoplasmic concentration, as it is directly coupled to cell volume and impacts macromolecular crowding (7, 36), structural integrity of tissue, and is even reported to impact stem cell fate (37). Though the feedback mechanism for maintaining cytoplasmic concentration against external osmotic perturbation is relatively well understood (38), little is known about autonomous change of cytoplasmic concentration under physiological conditions (7). Here we demonstrate that NoRI is ideally suited to probe physiologically controlled cytoplasmic concentration changes. An example of a cell-autonomous change of cytoplasmic concentration occurs during mitosis (39) (Fig. 2E). As a mitotic cell rounds up, a process important for accurate chromosome segregation (40), the cytoplasmic concentration changes due to the forces of osmotic pressure and the contraction of the actomyosin cortex (41, 42). Prior reports of cytoplasmic concentration changes during mitosis required two separate measurements of the single cell’s total dry mass and volume; the cytoplasmic concentration was then calculated by dividing the mass by the volume. This calculation limited the types of samples that could be analyzed to either cell suspensions or sparsely plated monolayers. By contrast, NoRI enables a direct measurement of cytoplasmic concentration and is much more flexible for different sample preparations. By distinguishing proteins from lipids, it provides additional information. To demonstrate this use of NoRI, we measured the cytoplasmic protein, lipid, and dry mass concentration of dividing MDCK (Madin–Darby canine kidney) cells in a confluent culture (Fig. 2E). The coefficient of variation of protein concentration or dry mass concentration was less than 0.07, showing that cytoplasmic concentration is tightly controlled. Also, in agreement with previous reports, the cytoplasmic dry mass concentrations decreased from interphase to mitosis and recovered after cytokinesis (Fig. 2F). The average concentration reduction during mitosis was equivalent to 17% volume swelling relative to the interphase size (Fig. 2G) and was comparable to previous reports (22, 43).
Another factor that could influence the cytoplasmic concentration of proteins and lipids is substrate stiffness. Typically, mammalian cells cultured on a stiff substrate spread to a wider area, take a flatter shape, and have higher cortical tension compared to cells cultured on softer substrates. Despite their larger area, cells on a stiff substrate might have changes in cell volume disproportional to protein content, leading to changes in cytoplasmic concentration (37, 44). Prior studies suggested that the cell volume change due to mechanical effects of the substratum is ion channels and actomyosin cytoskeleton dependent (37), and the role of cortical actomyosin for cell volume regulation is mediated by the YAP/TAZ pathway (44). Using NoRI, we could demonstrate similar phenomena in A7 cells cultured on polyacrylamide gels of different stiffness overnight (Fig. 3A). A7 cells on stiffer substrate had larger area (Fig. 3B) and smaller volume (Fig. 3C), which is consistent with the prior studies. However, counterintuitively, water concentration increased (Fig. 3D) and the cytoplasmic concentrations for total dry mass, protein, and lipid all decreased (Fig. 3 E–G). These results imply a decrease in the cell mass and thereby reveals additional complexity in the effects of mechanical stimuli on cells.
Fig. 3.
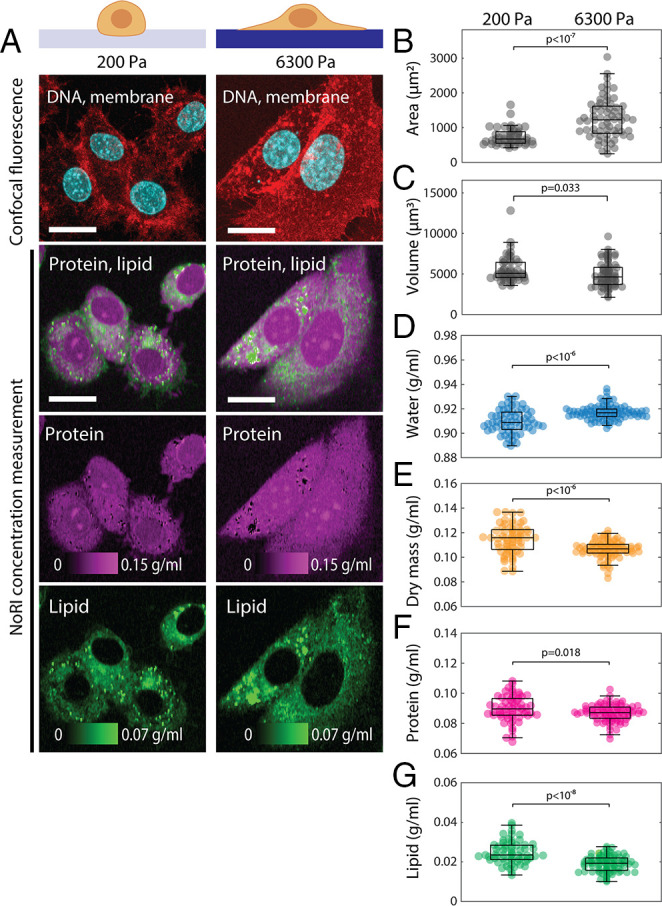
Cytoplasmic density increases in cells cultured on a soft substrate. (A) Representative images of live A7 cells cultured for 24 h on 200 Pa or 6,300 Pa polyacrylamide gels. (Scale bars, 20 µm.) (B) Cell area is greater on the stiff substrate (P < 10−7). Mean and SDs are 740 ± 250 µm2 (n = 46) and 1,264 ± 571 µm2 (n = 67) for 200 Pa and 6,300 Pa, respectively. (C) Cell volume is smaller on the stiff substrate (P = 0.033). Mean and SDs are 5,588 ± 1,672 µm2 (n = 46) and 4,921 ± 1,576 µm2 (n = 67) for 200 Pa and 6,300 Pa, respectively. (D–F) Concentration of water, dry mass (equal to the sum of protein and lipid), protein, and lipid are quantified in the single cell cytoplasm excluding lipid droplets. (D) Cytoplasmic water concentration is higher on the stiff substrate (P < 10−7). Mean and SDs are 0.9098 ± 0.0098 g/mL (n = 61) and 0.9171 ± 0.0058 g/mL (n = 84) for 200 Pa and 6,300 Pa, respectively. (E) Cytoplasmic dry mass concentration is lower on the stiff substrate (P < 10−6). Mean and SDs are 0.1147 ± 0.0118 g/mL (n = 61) and 0.1065 ± 0.0070 g/mL (n = 84) for 200 Pa and 6,300 Pa, respectively. (F) Cytoplasmic protein concentration is lower on the stiff substrate (P = 0.018). Mean and SDs are 0.0899 ± 0.0085 g/mL (n = 61) and 0.0869 ± 0.0055 g/mL (n = 84) for 200 Pa and 6,300 Pa, respectively. (G) Cytoplasmic membrane lipid concentration is lower on the stiff substrate (P < 10−8). Mean and SDs are 0.0248 ± 0.0057 g/mL (n = 61) and 0.0196 ± 0.0040 g/mL (n = 84) for 200 Pa and 6,300 Pa, respectively.
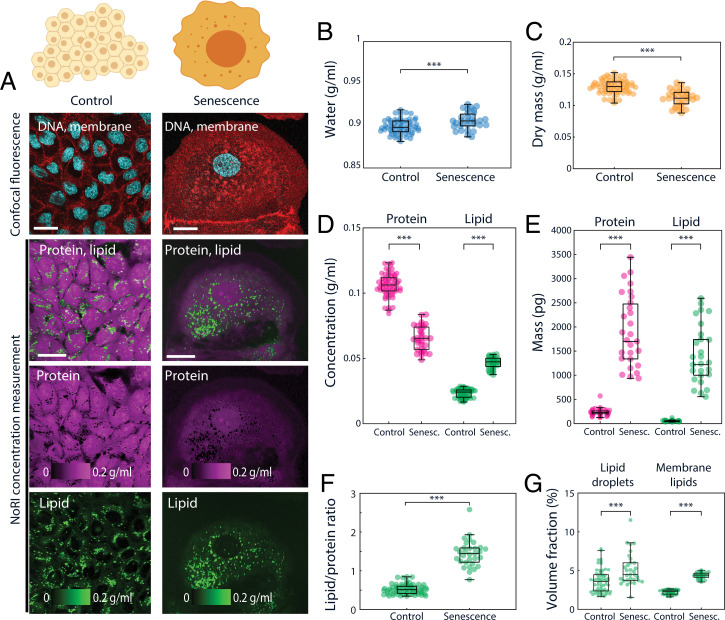
We also measured cytoplasmic concentration changes during cellular senescence. Cellular senescence is accompanied by a wide array of changes, including hypertrophy (45), significant cytoplasm dilution (46, 47), and accumulation of lipids (48). Dilution of cytoplasm is a poorly understood phenomena, and might play a crucial role in cellular aging (46). Cytoplasmic dilution was previously studied using SMR (suspended microchannel resonator), diffusion rate imaging, and ultracentrifugation, but these methods are either constrained to suspended cells or nonphysiological preparation conditions and lack the resolution to discern distinct processes such as lipid accumulation. NoRI provides a flexible way to observe cytoplasmic and lipid density changes in senescence. To explore senescence, we performed NoRI imaging of live MDCK cells and A7 cells induced to undergo senescence by 48 h doxorubicin treatment (100 ng/mL) (Fig. 4 and SI Appendix, Fig. S19). Both MDCK cells and A7 cells showed reduced cytoplasmic dry mass and protein concentrations in senescence compared to proliferating interphase cells (Fig. 4 C and D and SI Appendix, Fig. S19 C and E) in agreement with the prior report. In addition to a decrease in dry mass concentration, senescent cells achieve more significant net hypertrophy due to an increase in cell volume (Fig. 4E and SI Appendix, Fig. S19F). We found that lipid accumulation contributed a significant portion of the net hypertrophy in MDCK cells: The total lipid concentration increased from 0.023 ± 0.003 g/mL to 0.047 ± 0.004 g/mL (Fig. 4D), the total lipid mass increased from 50.5 ± 14.3 pg to 1,377.3 ± 588.6 pg (Fig. 4E), the proportion of lipid to protein increased from 0.53 ± 0.13 to 1.44 ± 0.34 (Fig. 4F), and both of the respective concentrations of lipid droplets and cytosolic membrane lipid increased from 3.6 ± 1.5% and 2.2 ± 0.3% to 5.2 ± 2.1% and 4.4 ± 0.4% (Fig. 4G) with senescence. However, this disproportionate lipid accumulation was cell-type dependent as senescent A7 cells showed balanced accumulation of protein and lipid (SI Appendix, Fig. S 19F), suggesting that there may be heterogeneity in metabolic reprogramming of cellular senescence.
Fig. 4.
Cytoplasmic dilution and lipid accumulation in senescent cells. (A) Representative images of control and senescent MDCK cells. Senescence is induced by a 48-h doxorubicin treatment. (Scale bars, 30 μm.) (B–D) Concentrations of water, protein, membrane lipid, and dry mass of single MDCK cells. Lipid droplets are excluded from lipid and dry mass concentrations. Dry mass is calculated by adding protein mass and lipid mass. Control n = 63, senescent n = 31. (B) Water concentration increases in senescent MDCK cells (P < 10−3). Mean and SD of control is 0.899 ± 0.009 g/mL, senescent is 0.906 ± 0.010 g/mL. (C) Dry mass concentration decreases in senescent MDCK cells (P < 10−9). Mean and SD of control is 0.130 ± 0.011 g/mL, senescent is 0.112 ± 0.013 g/mL. (D) Protein concentration decreases (P < 10−35) and lipid concentration increases (P < 10−47) in senescent MDCK cells. Mean and SD of control: protein 0.106 ± 0.009 g/mL, lipid 0.023 ± 0.003 g/mL and senescent: protein 0.065 ± 0.009 g/mL, lipid 0.047 ± 0.004 g/mL. (E) Senescent cells undergo dramatic hypertrophy in both protein mass (P < 10−31) and lipid mass (P < 10−31). Single cell protein mass and lipid mass were calculated by integrating the respective concentration over cell volume (protein: control 231.9 ± 62.7 pg and senescent 1,891.0 ± 716.3 pg; lipid: control 50.5 ± 14.3 pg and senescent 1,377.3 ± 588.6 pg). (F) Ratio of lipid to protein increases in senescent MDCK cells (P < 10−32). Total lipid is the sum of lipid droplet and membrane lipids. Lipid droplet mass is calculated from lipid droplet volume assuming density of neutral lipid 0.9 g/mL (control 0.53 ± 0.13 and senescent 1.44 ± 0.34). (G) Senescent MDCK cells contain an increased mass of lipid droplets (P < 10−4) and membrane lipids (P < 10−47) (control: lipid droplets 3.6 ± 1.5% [vol/vol], membrane lipids 2.2 ± 0.3% [vol/vol]; senescent: lipid droplets 5.2 ± 2.1% [vol/vol], membrane lipids 4.4 ± 0.4% [vol/vol]). Lipid droplets are detected by thresholding lipid concentration >0.1 g/mL. Membrane lipid quantity is measured by integration of lipid concentration over the cell volume excluding lipid droplets. ***P ≤ 0.001.
Cellular Protein and Lipid Concentration Are Specific to Cell and Tissue Types In Vivo
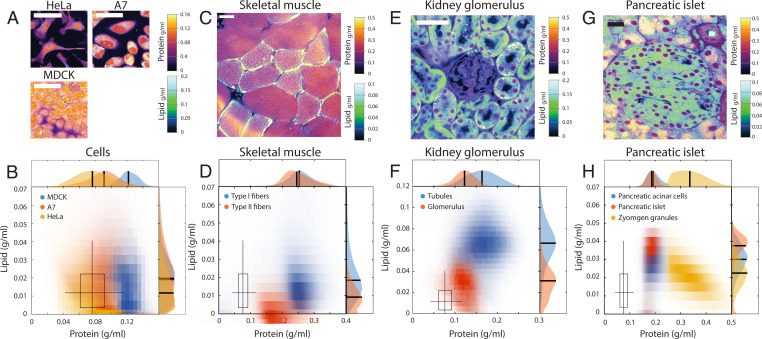
We noted that different cell lines display different protein and lipid concentrations even in identical culture conditions, suggesting cell-type specificity of these properties (Fig. 5 A and B). To extend this observation in animal cell types, we acquired protein and lipid concentration profiles in a diverse array of mouse tissues. Indeed, different cell types displayed distinct protein and lipid concentrations: NoRI correctly identified the differences in lipid concentrations in slow and fast skeletal muscle fibers (49) (Fig. 5 C and D). In the kidney, tubule cells showed higher lipid concentration, presumably due to densely packed mitochondria (50), compared to glomeruli (Fig. 5 E and F). Some intracellular organelles showed distinct protein and lipid concentration profiles compared to the rest of the cytoplasm as in the lipid-free cell nuclei or the protein-dense zymogen granules of pancreatic acinar cells (Fig. 5 G and H). The distinct protein and lipid concentrations of different cell types and organelles enables interpretation of tissue microanatomy in a way that can be more revealing than conventional hematoxylin and eosin–stained histology, specifically with regards to lipid content, which is invisible in conventional histology. Furthermore, the actual protein and lipid concentrations can provide novel information about the physiological or pathological state of the tissue. We demonstrated the potential utility of such quantitative protein and lipid concentration measurements in the following neuroscience applications. First, the quantitative difference in cytoplasmic concentration revealed hidden heterogeneity in an established cell type, the Purkinje neurons of cerebellum. These cells, identified by GAD67 expression (Fig. 6B), showed nearly threefold variation in the cytoplasmic concentration (Fig. 6 C and D), in which lipid and protein concentrations changed proportionately (Fig. 6C). This variability was reproducible in different mouse strains (SI Appendix, Fig. S21B). To rule out the possibility of a fixation artifact, we performed live tissue imaging of acute brain slices and confirmed that this variability is present in live Purkinje neurons (SI Appendix, Fig. S21A) in the same type of preparations used for recording action potentials of these cells. The difference in cytoplasmic concentration could be caused by different levels of protein synthesis or by dilution with water through an osmotic mechanism. Therefore, we sought to determine whether the dense cells have more total dry mass than the light cells or were merely more swollen at the same dry mass. For this purpose, we acquired a volumetric NoRI image of the Purkinje layer (Fig. 6E) and traced the cell body of the Purkinje neurons (Fig. 6F). A total of 12 cells were segmented from Fig. 6E and each cell’s volume, mean cytoplasmic concentration (the sum of protein and lipid concentrations), and total dry mass (the sum of protein mass and lipid mass) were calculated. We found that cytoplasmic concentration decreased with cell volume (Pearson’s R = −0.95), while the total dry mass changed little with cell volume (Pearson’s R = 0.46). This distinction was particularly clear in larger cells with a dry mass greater than 2,500 fL, where the total mass was independent of volume (Pearson’s R = 0.09). This suggests that an osmotic mechanism underlies the variability of cytoplasmic density.
Fig. 5.
Different cell and tissue types have distinct protein and lipid concentrations. (A and B) Protein and lipid concentration distribution in fixed HeLa cells, live A7 cells, and live interphase MDCK cells. (Scale bars, 50 μm.) The boxplot marks the median, 25% and 75% quartiles of fixed HeLa cell concentrations with whiskers marking 5% and 95% ranges. The boxplot is reproduced in D, F, and H for comparison. (C and D) Protein and lipid concentration of lipid-rich type I myofibers (n = 56) and lipid-lean type II myofibers (n = 28) in the traverse section of fixed murine skeletal muscle. (Scale bar, 20 μm.) Type I fibers have higher lipid concentrations than type II fibers. Protein concentrations are similar in the two types. (E and F) Fixed mouse kidney tissue. Glomerulus and tubules can be distinguished by distinct protein and lipid concentrations. (Scale bar, 100 μm.) (G and H) A pancreatic islet from fixed mouse pancreas tissue. (Scale bar, 100 μm.) Acinar cells contained large number of protein-dense vesicles, which are most likely zymogen granules for storing and secreting digestive enzymes. Cell nuclei are distinguishable by the absence of lipid. Protein-dense membrane surrounds blood vessels and ducts.
Fig. 6.
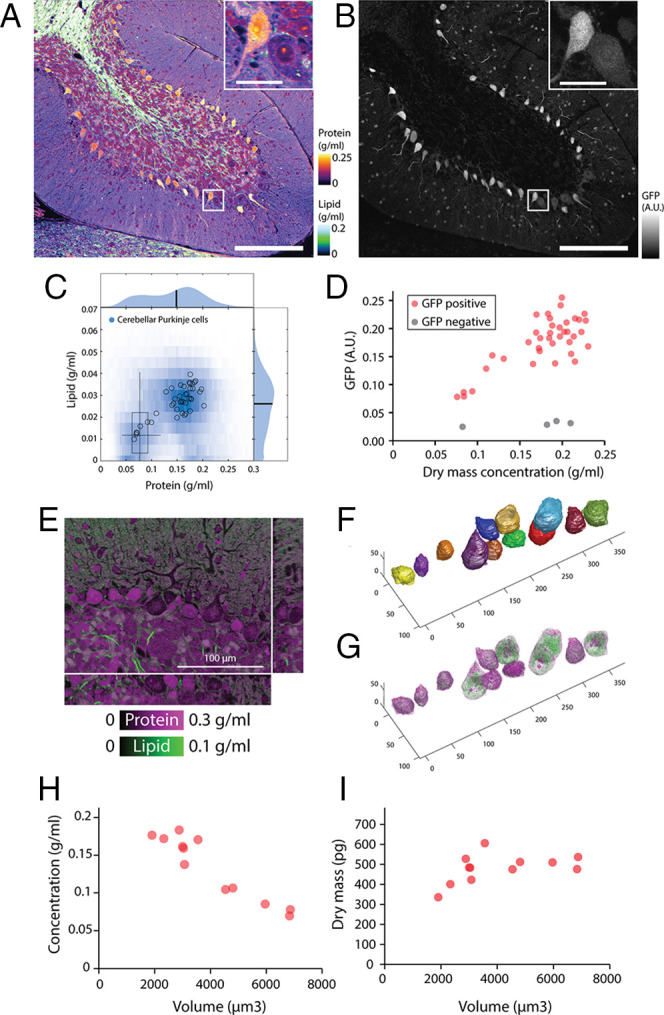
Bimodal cytoplasmic densities of Purkinje cells in the cerebellum. (A) NoRI image of fixed mouse cerebellum lobe 10. (Scale bar, 200 μm.) (Inset) Detailed view of boxed area. (Inset Scale bar, 30 μm.) (B) Confocal fluorescence of GAD67-GFP in the same area as A. Purkinje cells are identified by expression of GAD67-GFP. (C) Protein and lipid concentration distribution of all Purkinje cells (blue density plot) and individual Purkinje cells (open circles). The boxplot displays the concentration distribution of fixed HeLa cells in Fig. 5B for comparison. (D) GAD67-GFP expression level correlates with dry mass concentration in Purkinje cells (n = 38). GAD67-GFP negative cells from the Purkinje layer are also shown (n = 4). (E) Volumetric NoRI measurement of fixed mouse cerebellum. (Scale bar, 100 µm.) (F) Cell body of Purkinje neurons are manually segmented from the label-free NoRI image in E. Cell boundaries are determined from the distinct protein and lipid concentration of cells compared to the surrounding tissue. (G) Volumetric visualization of the protein and lipid distribution in the cells. (H) Cerebellar Purkinje cells decrease in cytoplasmic dry mass density with increasing cell body volume (n = 12). (I) Dry mass of the cell body of the cells shown in D.
Tissue-Type–Specific Protein and Lipid Concentration Changes with Disease
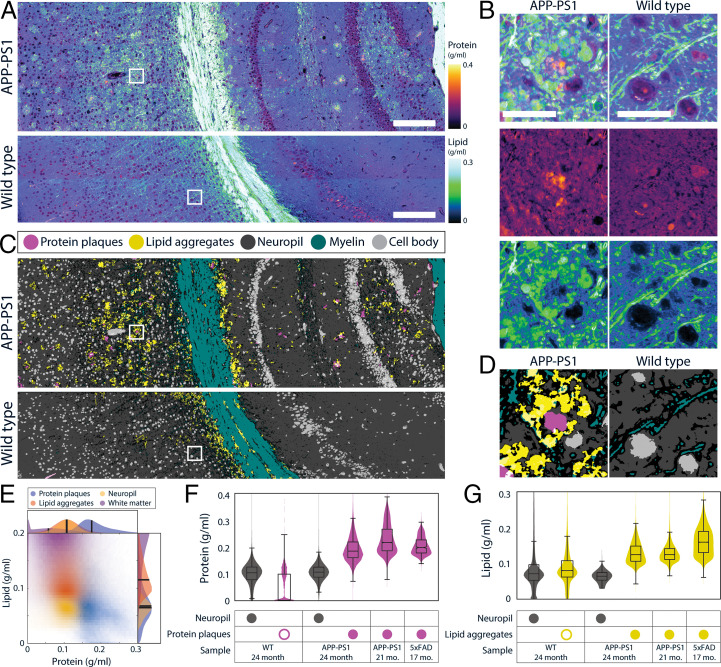
We explored the utility of protein and lipid measurement as a novel quantitative biomarker of disease pathology using murine models of Alzheimer’s disease (51) (Fig. 7). SRS microscopy had been previously applied to this model and showed the capability to detect amyloid plaques by the CH3 Raman band (52, 53). Furthermore, it was shown that the CH2 Raman band can detect lipid aggregation (52, 53) in the corona of senile plaques (the area surrounding amyloid plaques) (54). Consistent with these prior reports, we observed amyloid plaques surrounded by lipid aggregates in the APP-PS1 brain but not in the wild-type brain (Fig. 7 A and B). Since NoRI provides protein and lipid concentrations, we asked whether automated pathology analysis can be performed utilizing the absolute nature of NoRI data. In this regard, we found that the protein and lipid concentrations of cell bodies, white matter, and neuropil (dense entanglement of dendrites, unmyelinated axons, and synapses that surround cell bodies in gray matter) are similar between normal brain tissue and the normal appearing areas of APP-PS1 brain tissue. The consistency of concentration profiles enabled us to detect amyloid plaques and lipid aggregates by using a simple unsophisticated segmentation method (SI Appendix, Supplementary Methods, section 6.2). Briefly, cell bodies and white matter were detected by thresholding lipid images at <0.030 g/mL and at >0.147 g/mL, respectively. Trainable WEKA segmentation, an ImageJ machine learning plug-in (55), was used to segment amyloid plaques in protein images and neuropil in lipid images. The remaining areas of intermediate lipid concentration were segmented to lipid aggregates and myelin using their morphological differences (56). In total, we segmented the tissue into five classes—protein plaques, lipid aggregates, neuropil, myelin, and cell body (Fig. 7 C and D), utilizing their distinct absolute concentration profiles and morphologies. The lipid aggregates were different from lipid droplets based on their much lower lipid concentration (Fig. 7E), which is consistent with the model of neurite dystrophy origin (54). A biological replicate of APP-PS1 showed a similar concentration profile in the lesions, whereas an alternative Alzheimer mouse model 5xFAD showed higher lipid concentration than those in APP-PS1 (Fig. 7 F and G and SI Appendix, Fig. S22). Whether the quantitative difference in the concentrations consistently signifies differences of pathophysiology in neurite dystrophy will be the subject of future work.
Fig. 7.
Altered protein and lipid concentrations in brain tissue from Alzheimer’s disease genetic models. (A) Protein and lipid concentration images of APP-PS1 Alzheimer model mouse and wild-type (WT) mouse brains. (Scale bars, 100 μm.) (B) Detailed view of A. APP-PS1 brain shows senile plaques with protein-dense core and lipid-rich corona. In the WT brain, high concentration lipid is localized to myelin fibers. (Scale bar, 30 μm.) (C) Tissue segmentation by using a pixel classifier based on the protein and lipid concentrations and morphological features. Both wild-type and APP-PS1 images are processed by the same parameters. Each pixel is classified as protein plaques (pink), lipid aggregates (yellow), neuropil (dark gray), myelin (green), or cell body (light gray). Ambiguous pixels at the border of two classes were left unclassified (black). Protein plaques and lipid aggregates are scattered throughout the gray matter of APP-PS1 mouse brain tissue. (D) Detailed view of C. (E) Measurement of protein and lipid concentrations of protein plaques, lipid aggregates, neuropil, and white matter classes in the APP-PS1 image in A. (F) Quantitative analysis of protein concentration in WT (n = 1) and APP-PS1 (n = 2) and 5xFAD (n = 1), two murine Alzheimer’s disease models. Protein concentration is shown for neuropil and protein plaque classes detected by the pixel classifier described above. A small number of pixels in the WT were misclassified as protein plaques (open pink circle), but their protein concentration was significantly lower than that of true protein plaques (filled pink circles). The presence of protein plaques can be quantitatively characterized by the elevated protein concentration of the protein plaque class. Control neuropil protein 0.099 ± 0.042 g/mL, protein plaques 0.044 ± 0.056 g/mL 24 mo. APP-PS1 neuropil protein 0.106 ± 0.031 g/mL, protein plaques 0.193 ± 0.047 g/mL 21 mo. APP-PS1 protein plaques 0.229 ± 0.055 g/mL 17 mo. 5xFAD protein plaques 0.205 ± 0.031 g/mL. (G) Quantitative analysis of lipid concentration in the same samples as K. Lipid concentration is shown for neuropil and lipid aggregate classes identified by the pixel classification, as described above. The presence of lipid aggregates can be detected by the high lipid concentration of lipid plaque class (filled yellow circle), which contrasts with the low lipid concentrations (open yellow circle) seen in the WT, which has a lipid concentration in neuropil 0.089 ± 0.071 g/mL. Some pixels in WT are misclassified as lipid aggregates but they can be discounted as revealed by their low lipid concentration 0.090 ± 0.053 g/mL 24-mo-old. APP-PS1 mouse tissue shows lipid concentration in lipid aggregates of 0.129 ± 0.030 g/mL, which is significantly higher than that of neuropil, 0.064 ± 0.015 g/mL. The concentrations of lipid aggregates in the 21-mo-old APP-PS1 samples and in the 16-mo-old 5xFAD mouse are similarly high at 0.129 ± 0.028 g/mL and 0.162 ± 0.040 g/mL, respectively.
Discussion
We report here on a method, NoRI, which enables quantification of protein and lipid biomass concentrations at high spatial resolution in three-dimensional samples. Compared to existing methods, NoRI’s advantage is its in situ measurement capability to distinguish protein biomass and lipid biomass. Its working principle is based on the realization that water, protein, and lipids occupy nearly 100% (vol/vol) of wet biological samples. Although unaccounted components such as polysaccharides could in principle limit the accuracy of NoRI measurement, the size of this error is small in typical mammalian cells where the three most abundant components (water, protein, and lipid) account for ∼93% (wt/vol) of the wet mass (34). Moreover, the absolute size of the error due to unaccounted components scales with the absolute concentrations of the respective components. For example, when a sample has 75% (wt/vol) water, 18% (wt/vol) protein, and 7% (wt/vol) polysaccharides, the error in normalization is equal to or less than 7% of the respective absolute concentrations, i.e., <0.0525 g/mL for water and <0.0126 g/mL for proteins. We note that the absence of water in lipid droplets does not impede the normalization even though we utilize the H2O band for normalization. This is because the reference is the sum of all components, which is, in this case, dominated by lipid signals. Owing to its simplicity, this approach can be combined with other formats of SRS microscopy, spectral separation methods, and for the analysis of chemical components in addition to protein, lipids, and water. For example, the algorithm can be combined with an epi-detection SRS microscope (57) to achieve in vivo measurement in live animals. Since nucleic acid displays a distinct peak in the high-wavenumber Raman band, which permits spectral separation from proteins and lipids (58), the NoRI normalization method can be used to measure the absolute concentration of nucleic acids along with protein, lipids, and water by measuring four Raman peaks of protein, lipids, water, and nucleic acids (SI Appendix, Fig. S15). To implement the NoRI algorithm, quantitative reproducibility is required in the SRS instrumentation (SI Appendix, Fig. S8). To this end we built an SRS microscope with a chirped femtosecond pulse laser based on the spectral-focusing principle (59), with particular specifications to meet such reproducibility requirements (SI Appendix, Supplementary Methods), and obtained the sensitivity of ∼0.015 g/mL. The sensitivity can be further improved by averaging (SI Appendix, Fig. S14B). The spatial resolution of NoRI is determined from the resolution of the SRS microscope, which is similar to that of a two-photon microscope (25). The theoretical resolution at 770 to 805 nm wavelength was 0.57 µm and 1.58 µm in lateral and axial dimensions. Actual resolution inside tissue is usually lower due to the distortion of the laser focus by the sample and extended in z dimension owing to the local refractive index. The spatial resolution in tissue, as measured from the intensity profile of a small lipid droplet embedded at the 44-µm depth of a 100-µm-thick tissue, was <1.18 µm and 1.90 µm in lateral and axial dimensions, respectively (SI Appendix, Fig. S4). The acquisition rate of NoRI imaging is limited by the time required for taking three frames of SRS imaging. Each SRS image takes 1 s per frame. It takes an additional 10 s each time the laser wavelength is switched to change the Raman band. Hence, the slow switching time of wavelength limits applications to rapid events that happen on the order of tens of seconds. For live cell imaging, 2D NoRI images can be acquired without incubation chambers. But a stage-top incubation chamber was essential for 3D live cell volumetric imaging to avoid rapid cell shape change from stress. When scanning a large area or volume of stable samples (e.g., fixed tissue), acquisition time can be minimized by acquiring all images in one Raman band at once. Pigmented samples such as red blood cells or melanocytes induce two-photon absorption (TPA), which interferes with faithful SRS intensity measurement (60). TPA should either be measured from the off-Raman band and computationally subtracted from the Raman band signals (61) or eliminated by bleaching the pigments (62) (SI Appendix, Fig. S17). As we use light sources in 770 to 805 nm and 1,045 nm, which fall in the near-infrared window of biological tissues, dispersion is not significant within the imaging depth (SI Appendix, Supplementary Methods, section 9), yet some biological samples exhibit optical anisotropy and display different SRS intensities in a polarization sensitive way (63). The polarization dependence may provide additional information and is left for future work.
Both actomyosin cortex contraction and water influx are required for mitotic rounding (39, 41). Yet the mechanism of water regulation during mitosis is relatively less well understood compared to the cytoskeletal components. A few proteins, including membrane proteins and ion channels, had been previously identified to reduce mitotic cell swelling in an RNAi screen where cortical stiffening and volume change was measured using atomic force microscopy (AFM) and confocal microscopy (64). Phenotypic screening using AFM is primarily aimed at mechanical properties such as stiffness and, for characterizing water flux; intracellular pressure is calculated by combining the force measurement with a biophysical model (Laplace law applied to the 3D shape of the cell) (39). Cytoplasmic concentration measurements by NoRI provide a powerful alternative for probing components of intracellular water regulation. In this report, we observed dilution of cytoplasm in mitotic cells, which was consistent with previous reports by other groups (22, 43) (Fig. 2). However, there are also contradicting reports in which concentration of cytoplasm before or during mitosis was observed (17, 65, 66). We note that the cells that swell during mitosis have an interphase cytoplasm concentration between 0.095 and 0.190 g/mL and reduced cytoplasm concentration of 0.085 to 0.167 g/mL during metaphase [HT29 cells (43), L1210 cells (22), and MDCK cells in this study]. By contrast, the cells in the latter reports, where cytoplasm was concentrated during mitosis, started at an extremely low interphase cytoplasm concentration of 0.025 g/mL and increased to a metaphase cytoplasm concentration of 0.06 g/mL (mouse embryonic stem cells) (17). The discrepancy may be explained if the absolute cytoplasmic density rather than relative volume change is regulated during mitosis. Whether an optimal cytoplasmic density exists for mitosis is an open question which NoRI is well suited to address.
Cell shape can dramatically change depending on the stiffness of the substrate and this can be accompanied by cell volume change (44). Cells on soft substrates usually appear more round, while cells on stiff substrates spread to a larger area. But the spreading cells could actually have smaller volumes, if they had reduced height (67, 68). Such cell volume changes may have functional implications such as cell fate and mediated by changes in the intracellular macromolecular crowding (37) or membrane tension (44). We note that A7 cells on a higher stiffness gel had less dense cytoplasm as well as smaller cell volume in comparison to the same cells cultured on a softer gel (Fig. 3). This confirms that cytoplasm density cannot be assumed to be inversely proportional to the cell volume, which depends on the untested assumption of constant cellular dry mass. We believe that NoRI will be a useful tool for investigating mechanical regulation of cell volume by providing a direct measurement of the cytoplasm concentration and cell volume, which is also not limited to flat substrates, as are existing methods (44).
Neurohr and Amon (7) observed that senescent yeast cells undergo a large dilution in cytoplasm (50%) measured by SMR. However, in mammalian cells, the fragility of detached senescent cells prevented direct measurement using SMR and genetically encoded multimeric nanoparticles (GEMs) were instead used to assess the change in intracellular crowding from diffusion rate measurement. Several mechanisms of how cytoplasm density may impact cellular function have been proposed, including dilution of unstable proteins, change of diffusion rates and reaction kinetics, and change in DNA-to-cytoplasm ratio, but all of these conclusions are at present speculation. By providing direct measurement of cytoplasmic concentration and the respective concentration of protein and lipids, NoRI is poised to aid further studies on the role of cytoplasmic dilution in cellular senescence. Using NoRI, we have confirmed that senescent mammalian cells undergo cytoplasmic dilution, but by a much smaller magnitude than found in yeast (Fig. 5). Furthermore, along with hypertrophy, senescent cells are known to up-regulate lipid metabolism pathways and accumulate lipid droplets (45, 48, 69). NoRI enables quantitative characterization of senescent cell hypertrophy and the respective contributions of protein and lipids. Using this capability, we have already demonstrated the heterogeneity in senescence-related lipid accumulation in different cell types. When subjected to an identical treatment and culture condition, lipid accumulation was accelerated more than protein accumulation in senescent MDCK cells (Fig. 5F), but protein and lipid accumulated proportionately in senescent A7 cells (SI Appendix, Fig. S19G). We believe that NoRI should help resolve issues surrounding quantitative analysis of cell growth not only in cellular senescence but in a broad range of contexts, including cell cycle and differentiation.
The difference of cytoplasmic density between cell types or cellular states has been known for a long time and was the basis for cell sorting by density gradient centrifugation (70, 71). Also, some forms of active cell volume change such as shrinking of apoptotic cells had been described (38). However, it was only in recent years that the development of new technologies enabled measurement of single cell cytoplasmic concentration and brought to focus the biological regulation of cytoplasmic concentration (7, 36). Changes in cytoplasmic concentration can influence protein complex formation and biochemical reaction rates via global tuning of mass action and diffusion rates (72). Such global concentration tuning could have a critical effect on the activity of signaling molecules whose concentrations are close to their activation threshold (7). Osmoregulatory components (36), macromolecular synthesis (9), and mechanotransduction signals (37) are among potential factors that can modulate cytoplasmic concentration. The functional significance of cytoplasmic concentration changes may be context dependent and vary with cell types: NoRI imaging of animal tissues reveals the distinct concentration profiles of different cell types (Fig. 5), heterogeneity (Fig. 6), and disease-related changes (Fig. 7). Further characterization of cytoplasmic protein and lipid concentration of various cell types in different health and disease states could be a fascinating subject for future studies. Also, our initial observation in Alzheimer’s disease models in mice suggests that protein and lipid concentrations could be a good quantitative disease biomarker. Finally, the use of NoRI has reminded us of the limitations of conventional histological observations of tissue. The color intensity of histological images is usually normalized to a curve to facilitate computer-based analysis, especially to remove the color and intensity variation due to different staining protocols (70). By contrast, absolute protein and lipid concentrations, as measured by NoRI, are inherently physiologically meaningful and could provide new information. We expect NoRI to make important contributions in understanding the regulatory mechanism and functional significance of cytoplasmic concentration, as well as to open up new questions regarding the density of subcellular compartments, tissue-specific problems, and questions on the coordination of lipid and protein metabolism in cell size control.
Supplementary Material
Acknowledgments
We thank George Q. Daley and Ralph Weissleder for supporting the development of NoRI. S.O. and M.W.K. were supported by the National Institute of General Medical Sciences (NIGMS) of the NIH under Award R01GM026875. C.L. and C.J.T. were supported by NIH HD03443. W. Yang, A.L., and X.S.X. were supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the NIH under Grant R01EB017254. W. Yang was supported by the Office of the Provost, Faculty of Arts and Sciences, and Center for Advanced Imaging at Harvard University. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard University. D.F. was supported by a Beckman Young Investigator Award. C.R. and W. Yin were supported by NIH R01AG055413. M.B. was supported by NIGMS under Award R35GM137895. We thank Claudio Vinegoni, Wade Regehr, Stephani Rudoph, Jui-Hsia Weng, Ahmed Rattani, Tony Tsai, Anastasia Shindyapina, José Pedro Castro, and Vadim Gladyshev for the generous donation of mouse tissue samples and zebrafish embryos; Fake "Frank" Lu, Yuanzhen Suo, Xili Liu, Doaa Megahed, Scott Gruver, Scott Rata, and Victor Luria for helpful discussions; and Emily Cronin-Furman, Robert Rendano, and Jian Liu for technical assistance. We thank YongKeun Park and Tomocube Inc. for generously providing a demo HT-2 microscope.
Footnotes
Reviewers: J.L.-S., Janelia Farm Research Campus; M.P., Institut Curie; H.R., Institut Fresnel Marseille; and L.W., California Institute of Technology.
Competing interest statement: S.O., C.L., D.F., and M.W.K. filed a patent application for NoRI. X.S.X. has a financial interest in Invenio Imaging, Inc.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117938119/-/DCSupplemental.
Data Availability
The raw data of figures are available from the corresponding author upon request. The Matlab code used in this study is available at GitHub, https://github.com/kirschnerlab/NoRI. All other study data are included in the article and/or SI Appendix.
References
- 1.Klumpp S., Scott M., Pedersen S., Hwa T., Molecular crowding limits translation and cell growth. Proc. Natl. Acad. Sci. U.S.A. 110, 16754–16759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginzberg M. B., Kafri R., Kirschner M., On being the right (cell) size. Science 348, 1245075 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzur A., Kafri R., LeBleu V. S., Lahav G., Kirschner M. W., Cell growth and size homeostasis in proliferating animal cells. Science 325, 167–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kafri R. et al., Dynamics extracted from fixed cells reveal feedback linking cell growth to cell cycle. Nature 494, 480–483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper K. L. et al., Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions. Nature 495, 375–378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Berg J., Boersma A. J., Poolman B., Microorganisms maintain crowding homeostasis. Nat. Rev. Microbiol. 15, 309–318 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Neurohr G. E., Amon A., Relevance and regulation of cell density. Trends Cell Biol. 30, 213–225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunziker E. B., Kapfinger E., Saager C., Hypertrophy of growth plate chondrocytes in vivo is accompanied by modulations in the activity state and surface area of their cytoplasmic organelles. Histochem. Cell Biol. 112, 115–123 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Delarue M. et al., mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174, 338–349.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie S., Skotheim J. M. A., A G1 sizer coordinates growth and division in the mouse epidermis. Curr. Biol. 30, 916–924.e2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottier C. et al., Dynamic measurement of the height and volume of migrating cells by a novel fluorescence microscopy technique. Lab Chip 11, 3855–3863 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Godin M. et al., Using buoyant mass to measure the growth of single cells. Nat. Methods 7, 387–390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Son S. et al., Direct observation of mammalian cell growth and size regulation. Nat. Methods 9, 910–912 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryan A. K. et al., Measuring single cell mass, volume, and density with dual suspended microchannel resonators. Lab Chip 14, 569–576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barer R., Interference microscopy and mass determination. Nature 169, 366–367 (1952). [DOI] [PubMed] [Google Scholar]
- 16.Popescu G. et al., Optical imaging of cell mass and growth dynamics. Am. J. Physiol. Cell Physiol. 295, C538–C544 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandoz P. A., Tremblay C., van der Goot F. G., Frechin M., Image-based analysis of living mammalian cells using label-free 3D refractive index maps reveals new organelle dynamics and dry mass flux. PLoS Biol. 17, e3000553 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Martín D. et al., Inertial picobalance reveals fast mass fluctuations in mammalian cells. Nature 550, 500–505 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Tzur A., Moore J. K., Jorgensen P., Shapiro H. M., Kirschner M. W., Optimizing optical flow cytometry for cell volume-based sorting and analysis. PLoS One 6, e16053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro H. M., Practical Flow Cytometry (Wiley-Liss, New York, ed. 4, 2003). [Google Scholar]
- 21.Grover W. H. et al., Measuring single-cell density. Proc. Natl. Acad. Sci. U.S.A. 108, 10992–10996 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Son S. et al., Resonant microchannel volume and mass measurements show that suspended cells swell during mitosis. J. Cell Biol. 211, 757–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi W. et al., Tomographic phase microscopy. Nat. Methods 4, 717–719 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Shin S., Kim D., Kim K., Park Y., Super-resolution three-dimensional fluorescence and optical diffraction tomography of live cells using structured illumination generated by a digital micromirror device. Sci. Rep. 8, 9183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freudiger C. W. et al., Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322, 1857–1861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill A. H., Manifold B., Fu D., Tissue imaging depth limit of stimulated Raman scattering microscopy. Biomed. Opt. Express 11, 762–774 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huizinga A., Bot A. C. C., de Mul F. F. M., Vrensen G. F. J. M., Greve J., Local variation in absolute water content of human and rabbit eye lenses measured by Raman microspectroscopy. Exp. Eye Res. 48, 487–496 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Karuna A. et al., Label-free volumetric quantitative imaging of the human somatic cell division by hyperspectral coherent anti-stokes Raman scattering. Anal. Chem. 91, 2813–2821 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Cheng J.-X., Xie X. S., Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 350, aaa8870 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Fu D. et al., Quantitative chemical imaging with multiplex stimulated Raman scattering microscopy. J. Am. Chem. Soc. 134, 3623–3626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedman S. L., Roll F. J., Isolation and culture of hepatic lipocytes, Kupffer cells, and sinusoidal endothelial cells by density gradient centrifugation with Stractan. Anal. Biochem. 161, 207–218 (1987). [DOI] [PubMed] [Google Scholar]
- 32.Xu F., Zhang Q., Wang H., Establishing a density-based method to separate proliferating and senescent cells from bone marrow stromal cells. Aging (Albany NY) 12, 15050–15057 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dill K. A., Ghosh K., Schmit J. D., Physical limits of cells and proteomes. Proc. Natl. Acad. Sci. U.S.A. 108, 17876–17882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alberts B. et al., “The chemical components of a cell” in Molecular Biology of the Cell, M. Anderson, S. Granum, Eds. (Garland Science, ed. 5, 2008), p. 63. [Google Scholar]
- 35.Shin S. et al., “Optical diffraction tomography using a digital micromirror device for stable measurements of 4D refractive index tomography of cells” in Quantitative Phase Imaging II, G. Popescu, Y. Park, Eds. (2016), vol. 9718, p. 971814. [Google Scholar]
- 36.Cadart C., Venkova L., Recho P., Lagomarsino M. C., Piel M., The physics of cell-size regulation across timescales. Nat. Phys. 15, 993–1004 (2019). [Google Scholar]
- 37.Guo M. et al., Cell volume change through water efflux impacts cell stiffness and stem cell fate. Proc. Natl. Acad. Sci. U.S.A. 114, E8618–E8627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann E. K., Lambert I. H., Pedersen S. F., Physiology of cell volume regulation in vertebrates. Physiol. Rev. 89, 193–277 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Taubenberger A. V., Baum B., Matthews H. K., The mechanics of mitotic cell rounding. Front. Cell Dev. Biol. 8, 687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cadart C., Zlotek-Zlotkiewicz E., Le Berre M., Piel M., Matthews H. K., Exploring the function of cell shape and size during mitosis. Dev. Cell 29, 159–169 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Stewart M. P. et al., Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature 469, 226–230 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Fischer-Friedrich E., Hyman A. A., Jülicher F., Müller D. J., Helenius J., Quantification of surface tension and internal pressure generated by single mitotic cells. Sci. Rep. 4, 6213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zlotek-Zlotkiewicz E., Monnier S., Cappello G., Le Berre M., Piel M., Optical volume and mass measurements show that mammalian cells swell during mitosis. J. Cell Biol. 211, 765–774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez Gonzalez N. et al., Cell tension and mechanical regulation of cell volume. Mol. Biol. Cell 29, 2591–2600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernandez-Segura A., Nehme J., Demaria M., Hallmarks of cellular senescence. Trends Cell Biol. 28, 436–453 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Neurohr G. E. et al., Excessive cell growth causes cytoplasm dilution and contributes to senescence. Cell 176, 1083–1097.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okumura N. et al., Density-gradient centrifugation enables the purification of cultured corneal endothelial cells for cell therapy by eliminating senescent cells. Sci. Rep. 5, 15005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flor A. C., Wolfgeher D., Wu D., Kron S. J., A signature of enhanced lipid metabolism, lipid peroxidation and aldehyde stress in therapy-induced senescence. Cell Death Discov. 3, 17075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fiehn W., Peter J. B., Lipid composition of muscles of nearly homogeneous fiber type. Exp. Neurol. 39, 372–380 (1973). [DOI] [PubMed] [Google Scholar]
- 50.Larsson L., The ultrastructure of the developing proximal tubule in the rat kidney. J. Ultrastruct. Res. 51, 119–139 (1975). [DOI] [PubMed] [Google Scholar]
- 51.Radde R. et al., Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 7, 940–946 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji M. et al., Label-free imaging of amyloid plaques in Alzheimer’s disease with stimulated Raman scattering microscopy. Sci. Adv. 4, eaat7715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schweikhard V., Baral A., Krishnamachari V., Hay W. C., Fuhrmann M., Label-free characterization of Amyloid-β-plaques and associated lipids in brain tissues using stimulated Raman scattering microscopy. bioRxiv [Preprint] (2019). 10.1101/789248 (Accessed 21 March 2022). [DOI]
- 54.Duyckaerts C., Delatour B., Potier M. C., Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 118, 5–36 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Arganda-Carreras I. et al., Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Püspöki Z., Storath M., Sage D., Unser M., Transforms and operators for directional bioimage analysis: A survey. Adv. Anat. Embryol. Cell Biol. 219, 69–93 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Slipchenko M. N. et al., Vibrational imaging of tablets by epi-detected stimulated Raman scattering microscopy. Analyst (Lond.) 135, 2613–2619 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Lu F.-K. et al., Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. Proc. Natl. Acad. Sci. U.S.A. 112, 11624–11629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu D., Holtom G., Freudiger C., Zhang X., Xie X. S., Hyperspectral imaging with stimulated Raman scattering by chirped femtosecond lasers. J. Phys. Chem. B 117, 4634–4640 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berto P., Andresen E. R., Rigneault H., Background-free stimulated Raman spectroscopy and microscopy. Phys. Rev. Lett. 112, 053905 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Shi L. et al., Optical imaging of metabolic dynamics in animals. Nat. Commun. 9, 2995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chung J. Y. et al., A melanin-bleaching methodology for molecular and histopathological analysis of formalin-fixed paraffin-embedded tissue. Lab. Invest. 96, 1116–1127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z., Zheng W., Hsu C. Y. S., Huang Z., Polarization-resolved hyperspectral stimulated Raman scattering microscopy for label-free biomolecular imaging of the tooth. Appl. Phys. Lett. 108, 033701 (2016). [Google Scholar]
- 64.Toyoda Y. et al., Genome-scale single-cell mechanical phenotyping reveals disease-related genes involved in mitotic rounding. Nat. Commun. 8, 1266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boucrot E., Kirchhausen T., Mammalian cells change volume during mitosis. PLoS One 3, e1477 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Habela C. W., Sontheimer H., Cytoplasmic volume condensation is an integral part of mitosis. Cell Cycle 6, 1613–1620 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie K., Yang Y., Jiang H., Controlling cellular volume via mechanical and physical properties of substrate. Biophys. J. 114, 675–687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adar R. M., Safran S. A., Active volume regulation in adhered cells. Proc. Natl. Acad. Sci. U.S.A. 117, 5604–5609 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Millner A., Atilla-Gokcumen G. E., Lipid players of cellular senescence. Metabolites 10, 1–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Eycke Y. R., Allard J., Salmon I., Debeir O., Decaestecker C., Image processing in digital pathology: An opportunity to solve inter-batch variability of immunohistochemical staining. Sci. Rep. 7, 42964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Joyner R. P. et al., A glucose-starvation response regulates the diffusion of macromolecules. eLife 5, e09376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berenson D. F., Zatulovskiy E., Xie S., Skotheim J. M., Constitutive expression of a fluorescent protein reports the size of live human cells. Mol. Biol. Cell 30, 2985–2995 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Varinli H., Osmond-McLeod M. J., Molloy P. L., Vallotton P., LipiD-QuanT: A novel method to quantify lipid accumulation in live cells. J. Lipid Res. 56, 2206–2216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of figures are available from the corresponding author upon request. The Matlab code used in this study is available at GitHub, https://github.com/kirschnerlab/NoRI. All other study data are included in the article and/or SI Appendix.