Significance
The current study reveals the functions of FAF1 in protecting cells from ferroptosis, a novel cell death pathway triggered by PUFA peroxidation. In the absence of FAF1, cultured cells and mice are extremely sensitive to ferroptosis when exposed to physiological levels of PUFAs. Mechanistically, FAF1 sequesters PUFAs into the hydrophobic core of a global structure that limits their access to positively charged Fe2+, which catalyzes the peroxidation reaction. These observations suggest that FAF1-mediated protection of PUFA peroxidation plays a critical role in preventing initiation of ferroptosis.
Keywords: ferroptosis, polyunsaturated fatty acids, FAF1
Abstract
Iron-dependent peroxidation of polyunsaturated fatty acids (PUFAs) leads to ferroptosis. While detoxification reactions removing lipid peroxides in phospholipids such as that catalyzed by glutathione peroxidase 4 (GPX4) protect cells from ferroptosis, the mechanism through which cells prevent PUFA peroxidation was not completely understood. We previously identified Fas-associated factor 1 (FAF1) as a protein directly interacting with free PUFAs through its UAS domain. Here we report that this interaction is crucial to protect cells from ferroptosis. In the absence of FAF1, cultured cells became sensitive to ferroptosis upon exposure to physiological levels of PUFAs, and mice developed hepatic injury upon consuming a diet enriched in PUFA. Mechanistically, we demonstrate that FAF1 assembles a globular structure that sequesters free PUFAs into a hydrophobic core, a reaction that prevents PUFA peroxidation by limiting its access to iron. Our study suggests that peroxidation of free PUFAs contributes to ferroptosis, and FAF1 acts upstream of GPX4 to prevents initiation of ferroptosis by limiting peroxidation of free PUFAs.
Both iron and polyunsaturated fatty acids (PUFAs) are essential for survival of mammalian cells. A fundamental biochemical conflict occurs when these two molecules come into close contact, which leads to iron-dependent PUFA peroxidation that causes ferroptosis (1–3). While accumulation of PUFA hydroperoxides in phospholipids is critical to trigger ferroptosis (4, 5), exactly how iron mediates PUFA peroxidation that leads to this accumulation remains unclear. Ferroptosis is inhibited by reactions catalyzed by glutathione peroxidase 4 (GPX4) that reduce PUFA hydroperoxides in phospholipids and those mediated by lipid-soluble antioxidants that remove PUFA radicals (5–8). While these damage control reactions are well recognized to prevent ferroptosis, the mechanism through which cells prevent the initial event that triggers ferroptosis, that is, PUFA peroxidation, remains obscure. It also remains unclear whether peroxidation of free PUFAs contributes to ferroptosis.
Fas-associated factor 1 (FAF1) was originally identified as a tumor suppressor that facilitates proteasomal degradation of β-catenin (9, 10). In the absence of fatty acids (FAs), FAF1 binds to β-catenin, a reaction delivering ubiquitinated β-catenin to proteasomes for degradation (11). Unsaturated but not saturated FAs trigger polymerization of FAF1 through their direct interaction with the UAS domain of the protein (11, 12). Consequently, the interaction between FAF1 and β-catenin is disrupted, leading to stabilization of β-catenin (11). These studies demonstrate that unsaturated FAs inhibit the activity of FAF1 in driving degradation of β-catenin. However, the functions of unsaturated FA-bound FAF1 remain elusive.
In the current study, we demonstrated that even in the absence of a GPX4 inhibitor, cultured cells deficient in FAF1 became sensitive to ferroptosis upon exposure to physiological levels of PUFAs, and mice with a liver-specific knockout of Faf1 developed hepatitis upon consuming a diet enriched in arachidonate (AA), a PUFA. Mechanistically, we revealed that FAF1 inhibited PUFA peroxidation by sequestering free FAs into the hydrophobic core of a globular structure that limits their access to Fe2+. These observations suggest that FAF1 prevents initiation of ferroptosis by limiting peroxidation of free PUFAs.
Results
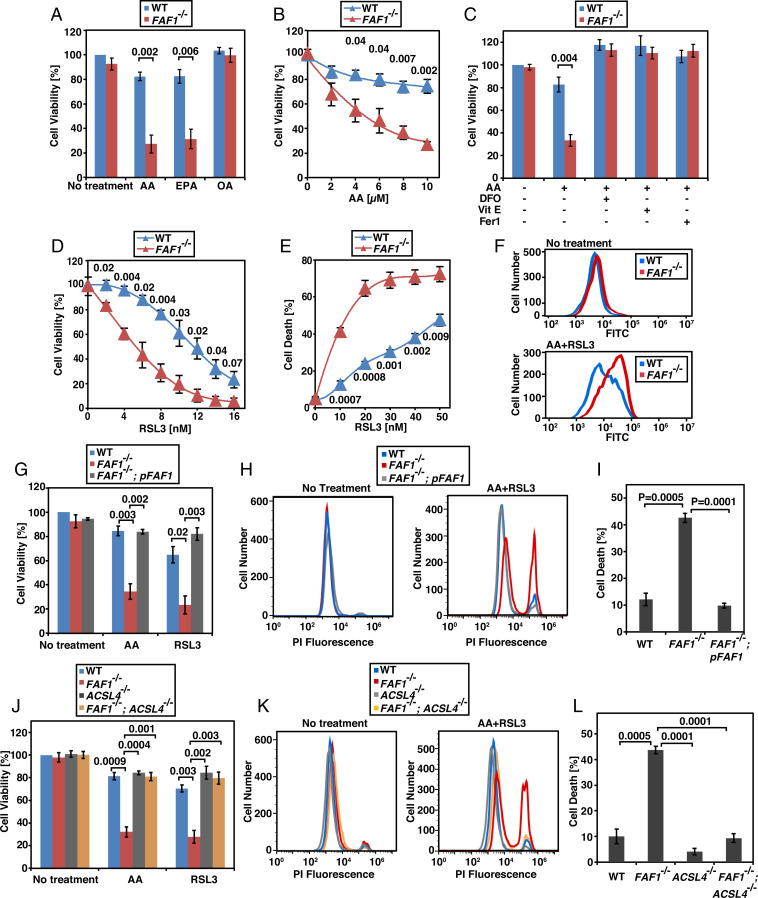
To understand the functions of FAF1 bound with unsaturated FAs, we generated a line of SV589 cells deficient in FAF1 (SI Appendix, Fig. S1A) to analyze their responses to unsaturated FAs. We observed that knockout of FAF1 did not affect survival of cells quantified by intracellular amounts of ATP in the absence of any FA treatment, but this procedure markedly reduced viability of the cells treated with PUFA such as AA or eicosapentaenate (EPA) (Fig. 1A). The AA-induced decrease in viability was concentration dependent (Fig. 1B). Approximately 60 to 70% of FAF1−/− cells died when they were treated with 10 µM AA (Fig. 1 A and B), an amount that is at the lower end of AA concentration in human serum, which is estimated to be 5 to 100 µM (13, 14). In contrast to PUFAs, treatment of FAF1−/− cells with oleate (OA), a monounsaturated FA also interacting with FAF1 (12), did not affect their survival (Fig. 1A). We thus suspected that FAF1−/− cells may be sensitive to ferroptosis, as only PUFAs but not monounsaturated FAs are subject to peroxidation that triggers ferroptosis. Owing to the lack of a molecular marker, ferroptosis is currently defined as a cell death pathway that is inhibited by iron chelators and lipid-soluble antioxidants but sensitized by inhibitors of GPX4 (15). In agreement with this definition, we observed that iron chelator deferoxamine or lipid-soluble antioxidants vitamin E and ferrostatin 1 restored the viability of AA-treated FAF1−/− cells (Fig. 1C), and that FAF1 deficiency decreased the half maximal effective concentration (EC50) value of RSL3, a GPX4 inhibitor (5), on cell viability by ∼50% (Fig. 1D). Cotreatment with AA further sensitized FAF1−/− cells to RSL3, decreasing the EC50 value of the compound by ∼75% (Fig. 1E). FAF1−/− cells were also more vulnerable to death induced by Erastin2, another stimulator of ferroptosis (5) (SI Appendix, Fig. S1B). Consistent with earlier reports that OA was a potent inhibitor of ferroptosis (16, 17), we observed that cotreatment with OA prevented AA-induced death of FAF1−/− cells (SI Appendix, Fig. S1C). Ferroptosis can also be monitored by the fluorescent sensor BODIPY 581/591 C11, the oxidation of which in ferroptotic cells alters the emitted fluorescence from red to green light (18). Flow cytometry quantification of the green fluorescent cells showed that in the absence of AA and RSL3, oxidation of the probe was barely detected in either wild-type (WT) or FAF1−/− cells (Fig. 1 F, Upper). In the presence of AA and RSL3, the probe was oxidized more heavily in FAF1−/− than WT cells (Fig. 1 F, Lower). The same outcome was demonstrated by fluorescent microscopy analysis (SI Appendix, Fig. S1D). Importantly, restoration of FAF1 expression in FAF1−/− cells (FAF1−/−; pFAF1 cells, SI Appendix, Fig. S1A) completely rescued them from AA- and/or RSL3-induced cytotoxicity (Fig. 1 G–I).
Fig. 1.
FAF1 deficiency sensitizes cells to ferroptosis. (A–D, G, and J) Viability of SV589-derived cells treated with indicated FAs (10 µM, indicated concentration in B), deferoxamine (DFO, 10 µM), vitamin E (Vit E, 2 µM), ferrostatin 1 (Fer1, 0.1 µM), RSL3 (D, indicated concentration; G and J, 10 nM) for 24 h was measured as described in Materials and Methods, with the value of the untreated WT cells set at 100%. (E) The percentage of dying cells determined through flow cytometry of the cells treated with 10 µM AA and the indicated amount of RSL3 for 12 h followed by propidium iodide (PI) staining. (F) BODIPY 581/591 C11 flow cytometry of the green fluorescence of indicated cells incubated with or without 10 µM AA for 4 h followed by cotreatment with 50 nM RSL3 for 2 h. (H and K) Flow cytometry of cells treated with 10 µM AA and 10 nM RSL3 for 12 h followed by PI staining. (I and L) The percentage of dying cells in experiments shown in H and K, respectively. (A–E, G, I, J, and L) Results are reported as mean ± SEM from three independent experiments. Statistical significance was calculated by unpaired, two-tailed t test.
Another hallmark of ferroptosis is its dependence on long chain acyl-CoA synthetase 4 (ACSL4)-catalyzed activation of PUFAs (4). We thus generated cells deficient in both ACSL4 and FAF1 (SI Appendix, Fig. S1E). Knockout of ACSL4 completely rescued FAF1−/− cells from AA- and/or RSL3-induced ferroptosis (Fig. 1 J–L).
In contrast to treatments inducing ferroptosis, FAF1−/− cells were not more sensitive to apoptosis induced by actinomycin D or etoposide compared to WT cells (SI Appendix, Fig. S1F). AA- or RSL3-induced death of FAF1−/− cells was unaffected by cotreatment with Z-VAD-FMK, an apoptosis inhibitor, necrosulfonamide, a necroptosis inhibitor, or VX-765, a pyroptosis inhibitor (SI Appendix, Fig. S1 G–J). These observations suggest that FAF1 specifically protects cells from ferroptosis. This effect is not restricted to SV589 cells, as knockout of FAF1 in Huh7 cells (SI Appendix, Fig. S1K), a line of human hepatoma cells, also rendered the cells more sensitive to AA- and/or RSL3-induced ferroptosis (SI Appendix, Fig. S1L).
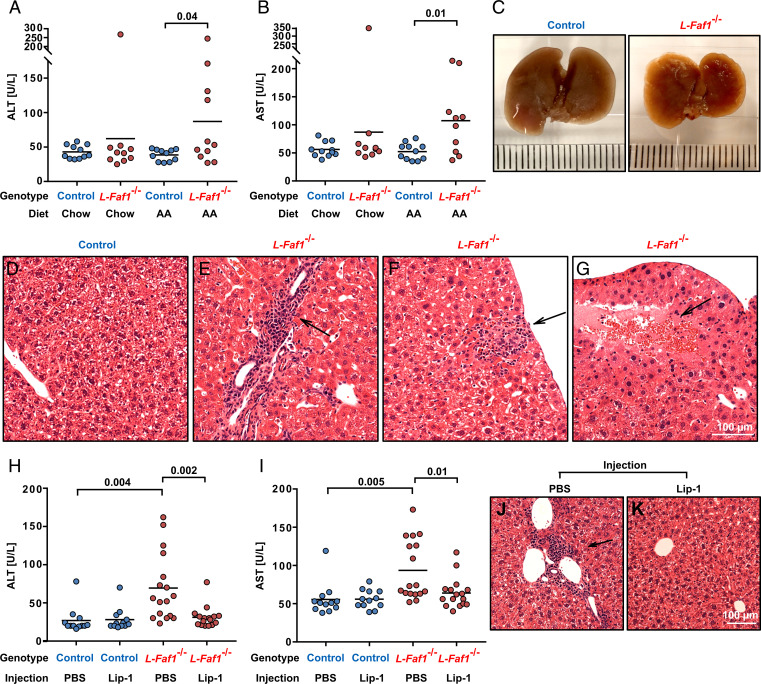
The results shown above demonstrate that FAF1 protects cultured cells from ferroptosis. To determine the physiological relevance of our findings, we generated Faf1f/f mice (SI Appendix, Fig. S2A). Injection of adeno-associated virus (AAV) encoding the Cre-recombinase, but not the control AAV encoding GFP, knocked out Faf1 in livers of these mice (L-Faf1−/−) (SI Appendix, Fig. S2B). Upon feeding with a diet enriched in AA but not control chow diet, ∼50% L-Faf1−/− mice suffered from hepatic damage demonstrated by increased activities of the alanine aminotransferase (ALT) and the aspartate aminotransferase (AST) in serum, the markers indicative of liver injury caused by releasing of these enzymes from dying hepatocytes (Fig. 2 A and B). The texture of their shrunk livers appeared to be uneven (Fig. 2C). Histological analysis revealed that livers from these mice were infiltrated by immune cells (Fig. 2 D and E). Some immune cells migrated to dying hepatocytes indicated by swollen glassy cytoplasm, faint nuclei, and indistinct cell boundary (Fig. 2F). Extensive death of hepatocytes was also observed in many regions of their livers (Fig. 2G). Consistent with a previous mouse model of liver ferroptosis (19), the total and reduced amounts of glutathione were not decreased in L-Faf1−/− mice fed with an AA-enriched diet (SI Appendix, Fig. S2C). This observation suggests that these mice were not subject to general oxidative stresses that deplete glutathione. To further demonstrate that hepatic damage shown in L-Faf1−/− mice fed with AA was caused by ferroptosis, we injected these mice with liproxstatin-1, a potent ferroptosis inhibitor that functions in mice (20). This treatment alleviated liver damage in these mice as demonstrated by reduced levels of serum ALT and AST (Fig. 2 H and I) and by normal histological appearances of the livers without detectable inflammatory foci and death of hepatocytes (Fig. 2 J and K).
Fig. 2.
FAF1 protects hepatocytes from ferroptosis in mice fed with AA. (A, B, H, and I) ALT (A and H) and AST (B and I) activities in serum of indicated mice fed with indicated diets (A and B) or AA-enriched diet injected with vehicle control (PBS) or liproxstatin-1 (Lip-1) (H and I) (n = 11 to 17). For each panel, data are combined from two independent experiments with 4 to 9 mice per group. Statistical significance was calculated by unpaired, two-tailed t test. (C) Morphology of the liver from indicated mice fed with AA. (D–G, J, and K) Hematoxylin and eosin-stained images of livers from AA-fed indicated mice (D–G) or that from AA-fed L-Faf1−/− mice subject to indicated injection (J and K). Arrows point to the histological abnormality discussed in the text.
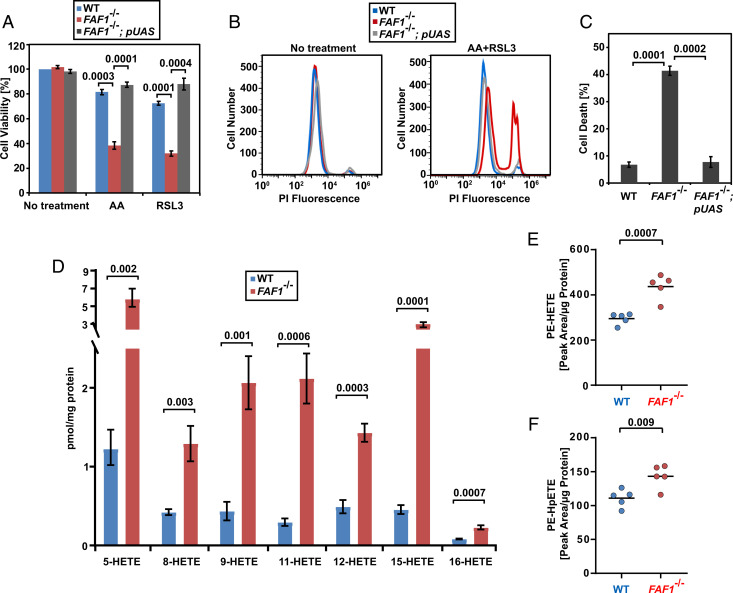
We previously reported that the UAS domain of FAF1 directly interacts with free AA (12). Since expression of the UAS domain of FAF1 alone (FAF1−/−; pUAS cells, SI Appendix, Fig. S3A) was sufficient to rescue FAF1−/− cells from ferroptosis induced by AA and/or RSL3 (Fig. 3 A–C), we suspect that the FAF1–AA interaction may be critical to inhibit ferroptosis in cells through affecting reactions that convert the free FA to other lipids. We first determined whether this interaction affects activation of AA by coenzyme A (CoA), a reaction required for ferroptosis (21). For this purpose, we measured incorporation of AA into phospholipids or neutral lipids using an approach we developed previously (22), as these reactions require activation of AA by CoA. While the amount of free AA was lower in AA-treated FAF1−/− cells than that in WT cells, FAF1 deficiency did not affect AA incorporation into either neutral lipids or phospholipids in cultured cells (SI Appendix, Fig. S3 B–D). Thus, it appears that FAF1 does not affect AA activation by CoA.
Fig. 3.
FAF1 inhibits AA peroxidation in cultured cells. (A–C) Viability of indicated cells was measured as described in Fig. 1 G–I. (D–F) The amount of HETEs (D), PE(18:0_HETE) (E), and PE(18:0_HpETE) (F) normalized against the amount of cellular protein in indicated cells treated with 10 µM AA for 16 h and 50 nM RSL3 for 4 h. (A, C, and D) Results are reported as mean ± SEM from three independent experiments. All statistical significance was determined by unpaired, two-tailed t test.
We then investigated whether the FAF1–AA interaction blocks ferroptosis by inhibiting peroxidation of free AA. Since the immediate AA peroxidation products such as AA hydroperoxyl radicals and hydroperoxyeicosatetraenoic acids (HpETEs) are difficult to measure owing to their lability, we quantified the amount of hydroxyeicosatetraenoic acids (HETEs), the direct reduction products of HpETEs that are stable enough to measure. The amount of HETEs with oxidation of various double bonds in AA was markedly elevated in FAF1−/− cells treated with AA and RSL3 (Fig. 3D). Consistent with increased levels of HETEs, we observed that the amount of malondialdehyde (MDA), an aldehyde generated through auto cleavage of AA peroxides (23), was also elevated in FAF1−/− cells treated with AA (SI Appendix, Fig. S3E). In addition to MDA, AA peroxidation also generates other short chain aldehydes (23). We thus detected carbonylated proteins, which are covalently modified by these short chain aldehydes through protein carbonylation (24), to measure AA peroxidation in cells. This analysis revealed that compared to WT cells, FAF1−/− cells treated with AA accumulated more carbonylated proteins, and this difference was further augmented by cotreatment with RSL3 (SI Appendix, Fig. S3F). Since downstream products of free AA peroxidation such as HETEs, MDA, and carbonylated proteins were all elevated in FAF1−/− cells, AA peroxidation levels were likely increased in these cells. If this is indeed the case, then interaction of free AA with FAF1 may be critical to prevent peroxidation of free AA to produce AA radicals. Since AA- and RSL3-treated FAF1−/− cells died from ferroptosis that is caused by accumulation of phospholipids containing AA peroxides (4, 25), we wondered whether AA radicals, which can enter membranes, may propagate peroxidation reactions to produce these phospholipids (23). Indeed, the amount of phosphatidylethanolamine containing oxidized and peroxidized AA acyl chains (PE-HETE and PE-HpETE, respectively), which were reported to be accumulated in ferroptotic cells (25, 26), was significantly higher in FAF1−/− cells treated with AA and RSL3 (Fig. 3 E and F and SI Appendix, Fig. S3 G and H for mass spectroscopy of PE-HETE and PE-HpETE, respectively). These results suggest that in the absence of FAF1, increased peroxidation of free PUFA may result in peroxidation of PUFA acyl chains in phospholipids that eventually causes ferroptosis.
Consistent with the results obtained in cultured cells, FAF1 deficiency in livers of the mice fed with AA did not change the amount of AA incorporated into neutral lipids or phospholipids (SI Appendix, Fig. S3 I–K) but increased production of MDA (SI Appendix, Fig. S3L).
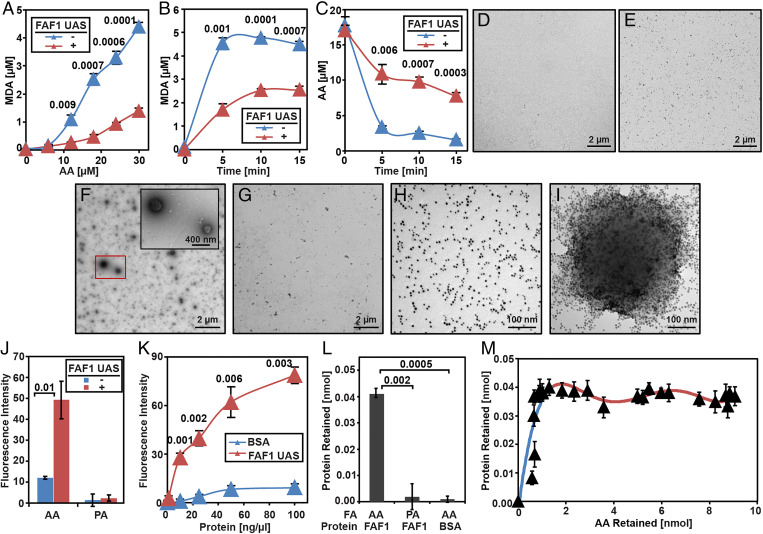
To understand how FAF1 inhibits peroxidation of AA, we set up an in vitro assay to measure peroxidation of AA in the absence or presence of purified FAF1. Incubation with FAF1 inhibited Fe2+-catalyzed AA peroxidation, as illustrated by the reduced production of MDA, one of the AA peroxidation products (SI Appendix, Fig. S4A). Similar to the full-length protein, the UAS domain of FAF1, which directly interacts with AA, also inhibited AA peroxidation, as illustrated by the decreased production of MDA (Fig. 4 A and B) and the decelerated disappearance of AA (Fig. 4C). Since interaction with PUFA stimulates polymerization of the UAS domain of FAF1 (12), we suspected that AA may form a large complex with the UAS domain, and this complex may play an important role in inhibiting AA peroxidation. We thus visualized the complex through negative staining electron microscopy (EM). While AA alone or the purified UAS domain of FAF1 barely generated particles with appreciable size (Fig. 4 D and E), incubation of AA with the UAS domain of FAF1 produced particles of various sizes with radiuses ranging from 50 to 200 nm (Fig. 4F). In contrast, incubation of AA with bovine serum albumin (BSA), a well-characterized FA-binding protein, did not produce these particles (Fig. 4G). Incubation of the UAS domain with palmitate (PA), a saturated FA that does not interact with the UAS domain, failed to produce the particles as well (SI Appendix, Fig. S4B). Inspection of the UAS domain labeled by Ni-NTA-Nanogold revealed that the UAS domain was evenly distributed in the absence of AA (Fig. 4H). Incubation with AA caused the protein to cluster around the surface of the particles observed above (Fig. 4I). In contrast to AA, incubation with PA did not change the distribution of the protein (SI Appendix, Fig. S4C). These observations suggest that AA was assembled into particles coated by the UAS domain at their surface. To determine whether the interior of the particles contained hydrophobic acyl chains of AA, we measured the hydrophobicity of the particles by Nile Red, a compound that is only fluorescent in hydrophobic environments but not aqueous solutions (27). While Nile Red was barely fluorescent in solutions containing AA, addition of the UAS domain markedly increased the fluorescence signal (Fig. 4J). This increase was not observed when the UAS domain was incubated with PA (Fig. 4J). Similar to the UAS domain, incubation of full-length FAF1 with AA but not PA also increased the Nile Red fluorescence signal (SI Appendix, Fig. S4D). In contrast to FAF1, incubation of AA with BSA did not increase the Nile Red fluorescence signals (Fig. 4K).
Fig. 4.
Purified FAF1 sequesters AA from iron-catalyzed peroxidation through the UAS domain. (A–C) Amount of MDA produced (A and B) or AA remained (C) in reactions containing AA (A, indicated amount; B, 30 µM), 50 µM H2O2 and 100 µM FeSO4 with or without 0.3 mg/mL purified UAS domain of FAF1 for 5 min (A) or indicated time (B and C). (D–I) Negative staining EM images of AA (30 µM) (D), UAS domain of FAF1 (0.1 mg/mL) (E), AA mixed with the UAS domain (F), or BSA (0.1 mg/mL) (G), and gold-labeled UAS domain in the absence (H) or presence (I) of AA. (J and K) Nile Red fluorescence of solutions containing 30 µM AA (J and K) or PA (J) and the protein (0.1 mg/mL, indicated concentration in K). (L) Amount of the protein retained by filtration of solutions containing indicated proteins (50 µg/mL) and FAs (50 µM). (M) Amount of the UAS domain plotted against that of AA retained by the filtration of solutions containing 50 µg/mL UAS domain and 0 to 400 µM AA. (A–C and J–M) Results are reported as mean ± SEM from three independent experiments. Statistical significance was determined by unpaired, two-tailed t test.
The observations shown above suggest that sequestering AA into a hydrophobic core of the complex assembled by FAF1 may be critical to inhibit AA peroxidation, presumably by limiting access of the FA to positively charged Fe2+. If this is the case, then other conditions generating a similar structure may also inhibit AA peroxidation. It was reported that in the absence of any protein, unsaturated FAs such as OA under acidic pH formed particles with a hydrophobic core and a size similar to that of the UAS domain–AA complex observed above (28). However, it remains unclear whether AA exhibits similar behavior under acidic pH. EM analysis indicated that AA itself in the absence of any protein formed a similar structure under acidic pH (SI Appendix, Fig. S4 E and F). The interior of the particles also appeared to be hydrophobic, as reducing pH of the solution containing AA alone raised the fluorescence intensity of Nile Red (SI Appendix, Fig. S4G). Consistent with the hypothesis that the sequestration of AA molecules into hydrophobic environments inhibits their peroxidation, reducing pH decreased the Fe2+-catalyzed production of MDA in the absence of any protein (SI Appendix, Fig. S4H).
The assembly of the FAF1–AA complex raises the question about the stoichiometry between the protein and the FA. To address this question, we isolated the complex by filtration through 100-kDa molecular weight cutoff membranes. This procedure specifically retained proteins in the complex, as only the UAS domain was trapped by the filtration when it was incubated with AA; other conditions that do not support formation of the complex did not result in retention of the proteins (Fig. 4L). To measure the stoichiometry, we mixed AA with the radioactive FA tracer so that the amount of the FA retained following the same filtration could be determined through measurement of radioactivity. Plotting the amount of the protein retained versus total AA added into the reaction revealed a saturation curve, indicating that the maximal amount of the protein was incorporated into the complex when the total AA concentration exceeded ∼50 µM (SI Appendix, Fig. S4I). In contrast, plotting the amount of AA retained versus total AA added into the reaction showed that much higher amounts of total AA, ∼360 µM, were required to reach saturation (SI Appendix, Fig. S4J). These observations suggest a two-phase stoichiometry during formation of the complex, which is better illustrated when the amount of the protein retained was plotted against the amount of the FA trapped during the filtration (Fig. 4M). During the first phase when the protein was in excess (Fig. 4M, blue line), one molecule of the UAS domain can sequester ∼25 molecules of AA into the complex. During the second phase when the FA was in excess, the same amount of protein can sequester increasing amounts of AA into the complex up to a stoichiometric ratio of ∼1:200 (Fig. 4M, red line). This high protein-to-AA stoichiometric ratio suggests that FAF1 may be extremely efficient in protecting AA from peroxidation. In comparison, one molecule of BSA, which also inhibits AA peroxidation (29), can only bind five molecules of AA (30).
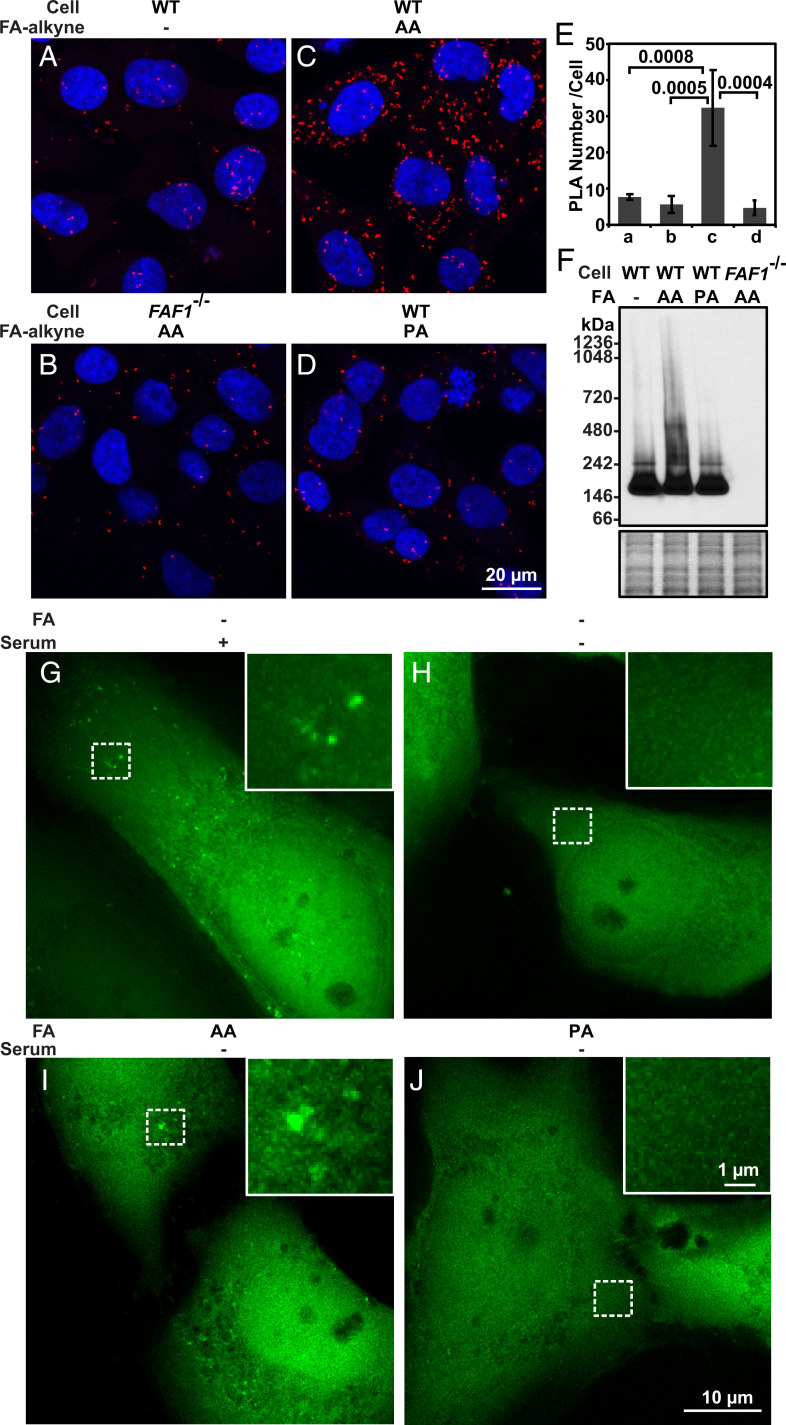
To determine the physiological relevance of the in vitro findings shown above, we used the approach of click chemistry coupled with the proximity ligation assay (PLA) to confirm the interaction between FAF1 and free PUFA inside cells. For this purpose, cells treated with triacsin C, an acyl-CoA synthetase inhibitor (31), were incubated with ω-alkynyl AA, a clickable analog of AA. We then covalently attached Oregon Green 488 to the AA analog through the click reaction, stained the cells with antibodies against Oregon Green 488 and FAF1, and performed a PLA (32). The fluorescent puncta, which signaled for their interaction, were barely detected in WT cells treated with normal AA or FAF1−/− cells treated with the clickable AA analog (Fig. 5 A and B). In contrast, a PLA signal was readily visible in WT cells treated with the clickable AA analog (Fig. 5C). The specificity of this interaction was demonstrated by the lack of the interaction signal generated between FAF1 and a clickable analog of PA, a saturated FA that does not interact with FAF1 in vitro (12) (Fig. 5D). Quantification of the fluorescent puncta per cell in these images demonstrates the specific interaction between FAF1 and free AA in cultured cells (Fig. 5E). If FAF1 assembles AA into a protein–FA complex showed in Fig. 4, AA should also cause polymerization of FAF1 in cultured cells. To test this hypothesis, we treated cells with various FAs and determined the polymerization status of FAF1 through Blue Native polyacrylamide gel electrophoresis (BN-PAGE) followed by immunoblot analysis. FAF1 displayed a high-molecular weight smear when the cells were treated with AA but not PA, and this signal was FAF1 specific, as no signal was detected in FAF1−/− cells (Fig. 5F).
Fig. 5.
Assembly of FAF1–AA complex in cultured cells. (A–D) PLA interaction signal (red puncta) of SV589-derived cells subjected to indicated treatments. (E) PLA signal per cell reported as mean ± SD from 63 to 95 cells in experiments shown in A–D. (F) Immunoblot analysis of FAF1 following BN-PAGE in the indicated cells treated with the indicated FAs as described in Materials and Methods (Upper gel). InstantBlue-stained SDS-PAGE of the same samples serves as a loading control (Lower gel). (G–J) Live cell imaging analysis of FAF1−/−; pEGFP-FAF1 cells before (G) and after serum withdrawal with supplementation of the indicated FAs (H–J) as described in Materials and Methods.
We also visualized FAF1 distribution in cells to detect the FAF1–AA complex. Owing to the sensitivity of the complex to detergents, microscopic approaches that require cell permeabilization by detergents are not suitable for this purpose. We thus performed a live cell imaging analysis to address this question. To this end, we stably transfected FAF1−/− cells with a plasmid encoding GFP-tagged FAF1 and selected a clone of the cells in which expression of the transgene was similar to that of the endogenous protein (FAF1−/−; pEGFP-FAF1 cells, SI Appendix, Fig. S5A). In addition to the diffused cytosolic staining pattern, some FAF1 appeared to form puncta in cells cultured in fetal calf serum (FCS) even without treatment with exogenous AA (Fig. 5G). These FAF1 puncta disappeared upon withdrawal of serum (Fig. 5H). Thus, it appears that AA supplied in serum was sufficient to drive the formation of the protein–FA complex. Consistent with this scenario, these puncta specifically maintained in cells cultured in serum-free condition supplemented with AA but not PA (Fig. 5 I and J).
To determine the physiological relevance of the protein–FA stoichiometry, we measured the amount of FAF1 protein in cells by immunoblot analysis calibrated by known amounts of purified FAF1 (SI Appendix, Fig. S5B), and that of free AA in cells through mass spectroscopy analysis. This analysis revealed that in cells cultured in FCS the absence of additional AA treatment, the molar ratio of FAF1 to free AA in cells was ∼1:300 (Table 1). Considering that the amount of protein-to-FA ratio can reach 1:200 in the complex formed in vitro, up to two-thirds of free AA in cells under this condition may be sequestered by FAF1 thereby protected from peroxidation. WT cells expressing FAF1 were thus less dependent on GPX4-catalyzed detoxification reaction for their survival; consequently, they were more resistant to RSL3 compared to cells deficient in FAF1 (Fig. 1D). Treating cells with additional AA increased the amount of free AA in cells by approximately five-fold (Table 1), making FAF1 more limiting in protecting cells from ferroptosis. Under this condition, the cells relied more on GPX4 for their survival, and therefore were more sensitive to RSL3 compared to cells that were not treated with AA (SI Appendix, Fig. S5C). In the absence of FAF1, levels of Fe2+-catalyzed free AA peroxidation under this condition should be even higher. As a result, amounts of phospholipids containing AA peroxides may overwhelm the GPX4-catalyzed detoxification reaction so that FAF1−/− cells succumbed to death even in the absence of RSL3 (Fig. 1B).
Table 1.
Quantification of FAF1 and free AA in cells
| AA Treatment | FAF1 (nmol/g total protein) | Free AA (nmol/g total protein) |
|---|---|---|
| — | 2.20 ± 0.06 | 705.52 ± 138.22 |
| 10 µM | 1.80 ± 0.21 | 3,452.91 ± 291.63 |
Amounts of FAF1 protein and free AA in cells treated with or without AA for 12 h. Results are expressed as mean ± SD from four independent experiments.
Discussion
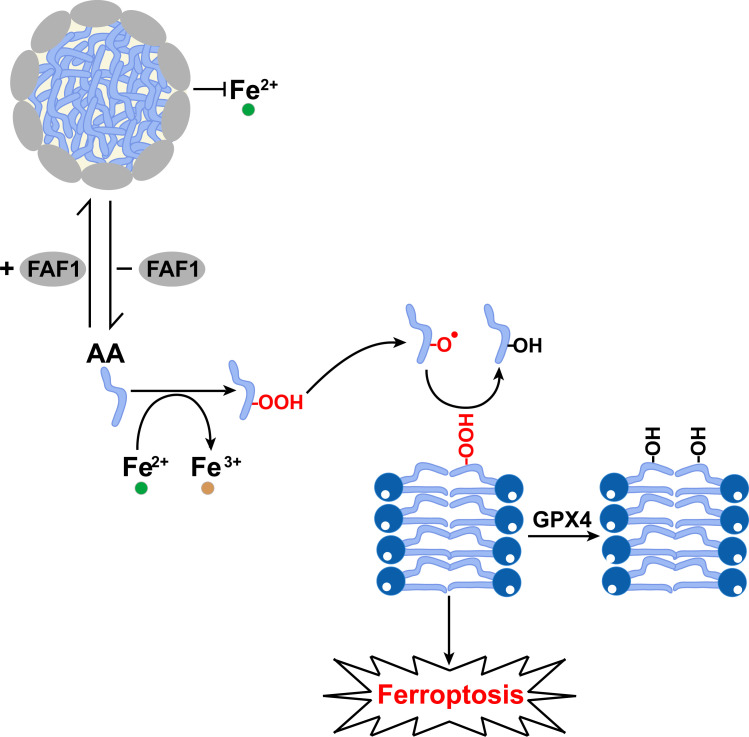
The major implications of our findings are summarized in Fig. 6. Both FAF1 and free PUFAs are present in cytosol; in the presence of FAF1, the protein sequesters PUFAs into the hydrophobic core of the FAF1–PUFA complex so that PUFAs are not accessible to peroxidation catalyzed by Fe2+ present in cells. In the absence of FAF1-mediated protection, PUFAs are more readily accessible to Fe2+-catalyzed peroxidation. While we were unable to measure the direct products of PUFA peroxidation, such as PUFA radicals and HpETE, we observed increased generation of downstream products, such as HETEs, MDA, and carbonylated proteins in cells. If these indirect measurements indeed reflect the extent of PUFA peroxidation, the PUFA peroxide radicals should be increased in cells deficient in FAF1. These radicals, which can enter membranes, may propagate peroxidation reactions of PUFA-containing phospholipids (23), the formation of which depends on ACSL4-catalyzed activation of PUFAs. Accordingly, we observed increased amounts of PE-HpETE and its reduced product PE-HETE in FAF1−/− cells. Ferroptosis may be executed when the amount of peroxidized PUFA-containing phospholipids exceeds the detoxification capacity of GPX4 and FSP1 (6, 7, 33). Thus, our study demonstrates that peroxidation of free PUFAs contributes to ferroptosis, presumably by producing lipid-soluble radicals that can propagate peroxidation reaction in membranes. According to this model, FAF1 acts upstream of GPX4 to protect cells from ferroptosis by preventing peroxidation of free PUFAs. This scenario is consistent with the findings that knockout of FAF1 stimulated ferroptosis in AA-treated cells and AA-fed mouse livers even in the absence of a GPX4 inhibitor. While this model explains how FAF1 protects cells from ferroptosis through the UAS domain, it remains unclear whether FAF1 may inhibit ferroptosis through other mechanisms mediated by other domains of the protein.
Fig. 6.
Graphic illustration of the function of FAF1 in protecting cells from ferroptosis. In the presence of FAF1, free AA is sequestered into a lipid–protein complex that prevents their access to Fe2+. In the absence of FAF1, free AA is more easily accessible to Fe2+, resulting in increased peroxidation of the fatty acids. The PUFA peroxides produced may be converted to PUFA radicals, which can enter phospholipid bilayers to catalyze peroxidation of PUFA-containing phospholipids, the formation of which depends on ACSL4-catalyzed activation of PUFAs. Ferroptosis may be triggered when the amount of peroxidized phospholipids produced overwhelms the scavenging activity of GPX4.
The current study demonstrates that FAF1 assembles PUFAs into a complex that prevents their peroxidation. This conclusion is directly supported in vitro by the observations that sequestration of AA by the purified FAF1 inhibited Fe2+-catalyzed peroxidation. This observation is physiologically relevant, as the direct interaction of FAF1 with AA in cells resulted in polymerization of the protein, and this interaction was critical to prevent ferroptosis in cultured cells treated with AA and hepatocytes of mice fed with a diet enriched in AA. Our live cell imaging analysis showing cytosolic FAF1 puncta formed in the presence of AA provides evidence to support the existence of the FAF1–AA complex in cells. However, owing to the sensitivity of the complex to detergents, we are unable to directly visualize the complex in cells using EM, which requires cell permeabilization by detergents.
Exactly how AA is packaged into the complex remains unclear. We observed that the complex contained different protein-to-FA ratio dependent on different concentrations of AA. When the FA was limiting, the complex contained a FA-to-protein ratio of 25. When the FA was in excess, more AA was incorporated into the complex so that this ratio can reach 200. These observations suggest that the FA and protein may assemble into a basic unit when the protein is in excess, and more AA might be dragged into these basic units when the FA is in excess. Measurement of free AA and FAF1 protein in cells suggests that the latter case might be more physiologically relevant. According to the stoichiometric ratio determined in vitro, ∼70% of free AA in cultured cells could be protected from peroxidation by sequestration into the FAF1–AA complex. In the absence of FAF1, the majority of AA might be prone to peroxidation, causing FAF1−/− cells to rely more on GPX4-catalyzed detoxification reactions for their survival. These cells were thus more sensitive to the GPX4 inhibitor, RSL3, particularly when the cells were treated with AA.
Considerable evidence demonstrates that ferroptosis is triggered by phospholipids containing peroxidized PUFAs (4, 25). In cells expressing arachidonate 15-lipoxygenase B (ALOX15B), the enzyme-catalyzed peroxidation of PUFA acyl chains in phospholipids may be primarily responsible for producing these lipids (26, 34, 35). However, ALOX15B is only expressed in limited tissues. In cells that do not express ALOX15B, which include hepatocytes and the SV589 fibroblasts used in the current study, Fe2+-catalyzed peroxidation of free PUFAs appears more likely to be the initiation event of ferroptosis, as the cation is difficult to reach the hydrophobic interior of membranes to catalyze peroxidation of PUFA acyl chains attached to phospholipids. In contrast to lipoxygenase-catalyzed peroxidation, Fe2+-catalyzed peroxidation of PUFAs is not position specific. Our observation that FAF1 deficiency led to increased production of HETEs without position specificity is consistent with the idea that they were produced by Fe2+-catalyzed peroxidation of AA. Once generated, free PUFA peroxide radicals may then catalyze peroxidation of PUFA acyl chains at phospholipid bilayers. Thus, FAF1-mediated inhibition of iron-catalyzed peroxidation of free PUFAs should limit production of phospholipids containing the peroxidized FAs.
Together with our previous report (11), this study establishes FAF1 as a FA sensor that allows cell proliferation and survival when there is an abundant supply of PUFAs. Assembly of the FAF1–PUFA complex not only enables cell proliferation by stabilization of β-catenin (11) but also protects cells from ferroptosis by limiting exposure of PUFAs to iron. Because of these critical functions, deficiency in FAF1 might make cells vulnerable upon exposure to high levels of PUFAs. In this regard, it is interesting to note that FAF1 is genomically deleted in ∼6% of glioblastoma, presumably because of its growth advantage through stabilization of β-catenin (36). It will be interesting to determine whether these tumors are uniquely sensitive to treatments that raise levels of PUFAs in the tumor cells.
Materials and Methods
Animal Studies and Diets.
All animal experiments described in this work were approved and conducted under the oversight of the University of Texas (UT) Southwestern Institutional Animal Care and Use Committee. All mice were housed in colony cages in a room with a 12-h light/12-h dark cycle. Faf1f/f mice (C57BL/6) were generated by inserting LoxP sites into introns 2 and 3 of the Faf1 gene through the CRISPR-Cas9 approach by UT Southwestern Transgenic Core. Genotyping was performed by PCR with the primers A (5′-GATTGGGGGCCAGGAGTTG-3′) and B (5′-AGGGAGTGGGTATTGCAAAC-3′), which produced a 1,730-bp or 1,650-bp fragment for the alleles with or without the two insertions of LoxP, respectively (SI Appendix, Fig. S2A). To generate liver-specific Faf1−/− mice, male littermates of 5- to 8-wk-old Faf1f/f mice originated from two independent lines were injected with AAV/DJ-CAG-iCRE (Vector Biolabs) at a dose of 4 × 1011 viral particles/mouse via the tail vein to delete exon 3 of Faf1 in livers that resulted in disruption of Faf1 ORF. AAV/DJ-CAG-GFP (Vector Biolabs) was injected as the control. Feeding experiments were started 2 wk after the injection. For this purpose, mice were fed a standard chow diet (Teklad Global Rodent Diet 2016) supplemented with 400 mg/kg ferrous sulfate in the absence or presence of 2% arachidonic ethyl for 14 d. Some of the mice fed with AA were administered liproxstatin-1 (10 mg/kg body weight) or the vehicle control (1% Dimethylsulfoxide in phosphate-buffered saline [PBS]) once a day by intraperitoneal injection during the feeding period. Following euthanization with isoflurane, blood was collected and the activity of ALT and AST in serum was assayed by the University of Texas Southwestern Metabolic Phenotyping Core. Livers were harvested and fixed in 4% paraformaldehyde overnight, and the histology analysis was performed by the University of Texas Southwestern Histology Core.
Cell Viability Assay.
On day 0, cells were seeded at 1,000 cells per well in 96-well plates. On day 1, cells were treated as described in the figure legends. Cell viability was assessed by CellTiter-Glo luminescent cell viability assay according to the manufacturer’s recommendations.
Cell Death Assay.
On day 0, cells were seeded at 200,000 cells per 10-cm plate. On day 1, cells were treated as described in the figure legends. Propidium iodide ready flow reagent (Thermo Fisher Scientific) was added to the cells suspended in 200 μL PBS containing 0.1% BSA according to the instructions of the manufacturer. Flow cytometry was performed with a BD Accuri C6 plus instrument, and the data were analyzed by FlowJo V10 software.
Lipid Peroxidation.
For cell culture experiments, indicated cells were set up at 500,000 cells per dish on day 0. On day 1, MDA produced in each dish of the cells was measured using a lipid peroxidation assay kit according to the manufacturer’s instructions. Lipid analysis measuring HETEs was performed at the University of California, San Diego Lipidomics Core (37), with more detailed protocols provided in SI Appendix, SI Materials and Methods. For mouse experiments, MDA was assayed with the same approach from livers of the mice (20 mg) shown in Fig. 2A. For in vitro assay, the amount of MDA was measured according to conditions described in the figure legends in buffer A (100 mM Tris⋅HCl, 150 mM NaCl, pH = 7.5) except for SI Appendix, Fig. S4H in which buffer B (50 mM Tris-maleate, 150 mM NaCl) with the indicated pH was used. The full-length FAF1 and the UAS domain of FAF1 was purified exactly as previously described (12).
Negative Staining EM.
Indicated FAs and/or proteins incubated in buffer A for 10 min at room temperature were loaded onto carbon-coated, glow-discharged 400-mesh copper grids (Electron Microscopy Sciences) for 5 min, washed twice, stained by two drops of 0.75% uranyl formate (Electron Microscopy Sciences) for 20 s, and air dried. Electron micrographs were collected on a JEOL 1400+ transmission electron microscope. For immune-gold negative staining EM, purified hexa-histidine–tagged UAS domain was attached to 5 nm Ni-NTA-Nanogold (Nanoprobes) according to the manufacturer’s recommendations. Electron micrographs were collected on a JEOL 1200-EX transmission electron microscope.
Nile Red Assay.
Indicated FAs and proteins incubated in buffer A containing 1.2 µg/mL Nile Red or buffer B with various pH containing 1.6 µg/mL Nile Red were assayed for fluorescence with excitation and emission wave length set at 526 and 586 nm, respectively, using a Biotek Synergy Neo2 for SI Appendix, Fig. S4D or a Biotek Synergy 4 for the rest of the experiments.
In Vitro Stoichiometric Assay.
Purified UAS domain mixed with AA containing 0.0005% of 3H-arachidonic acid tracer (mol/mol) incubated in buffer A (80 µL) was filtered through 100-kDa molecular weight cutoff membranes (Pall Corporation). The amount of protein and AA in flow-through was measured by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) and scintillation counting using Tri-Carb 2800TR (PerkinElmer), respectively. The amount of protein retained was calculated by subtracting the values in the flow-through in the presence of AA from that in the absence of FA. The amount of AA retained was calculated by subtracting the values in the flow-through in the presence of the protein from that in the absence of the protein for each AA concentration.
Proximity Ligation Assay.
On day 0, cells were seeded at 60,000 cells per well containing a 12-mm round coverslip in 24-well plates. On day 1, cells were switched to medium supplemented with 5% delipidated FCS and 1 µM A939572 (SCD1 inhibitor) for 24 h. On day 2, cells treated with or without 50 µM ω-alkynyl AA and 150 µM AA for 4 h were subjected to click reaction to covalently attach Oregon Green 488 (Thermo Fisher Scientific) to the clickable analog of AA followed by proximity ligation assay as previously described (32), using 10 µg/mL rabbit anti-FAF1 and goat anti-fluorescein/Oregon Green (Thermo Fisher Scientific) as the primary antibody and Duolink in situ PLA probe anti-rabbit MINUS and Duolink in situ PLA probe anti-goat PLUS (Sigma-Aldrich) as the oligonucleotide-conjugated secondary antibody. Fluorescent images were acquired with excitation and emission wave lengths set at 531 and 592 nm, respectively. ImageJ software was used for quantification of the interaction signal.
Materials and other methods can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Michael Brown and Joseph Goldstein for their constant supports, Lisa Beatty and Camille Harry for cell culture, Chelsea Burroughs and Nancy Heard for graphic illustration, and Sarah Martin and Katy Lowe for mass spectroscopy analysis. We would also like to acknowledge University of Texas Southwestern Medical Center EM Core, Live Cell Imaging Core, Histology Core, and Metabolic Phenotyping Core for their technical support. Lipid analysis measuring various HETEs was performed at the University of California, San Diego Lipidomics Core. This work was supported by the NIH (GM-116106, GM-140851, and HL-20948) and the Welch Foundation (I-1832).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. B.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107189119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Conrad M., et al. , Regulation of lipid peroxidation and ferroptosis in diverse species. Genes Dev. 32, 602–619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng H., Stockwell B. R., Unsolved mysteries: How does lipid peroxidation cause ferroptosis? PLoS Biol. 16, e2006203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W. S., Stockwell B. R., Ferroptosis: Death by lipid peroxidation. Trends Cell Biol. 26, 165–176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doll S., et al. , ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 13, 91–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W. S., et al. , Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bersuker K., et al. , The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doll S., et al. , FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Dixon S. J., et al. , Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L., et al. , Fas-associated factor 1 antagonizes Wnt signaling by promoting β-catenin degradation. Mol. Biol. Cell 22, 1617–1624 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L., et al. , Fas-associated factor 1 is a scaffold protein that promotes β-transducin repeat-containing protein (β-TrCP)-mediated β-catenin ubiquitination and degradation. J. Biol. Chem. 287, 30701–30710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H., et al. , Unsaturated fatty acids stimulate tumor growth through stabilization of β-catenin. Cell Rep. 13, 495–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H., et al. , UAS domain of Ubxd8 and FAF1 polymerizes upon interaction with long-chain unsaturated fatty acids. J. Lipid Res. 54, 2144–2152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Blanco G., et al. , Serum levels of arachidonic acid metabolites change during prostate cancer progression. Prostate 74, 618–627 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Pompeia C., Lima T., Curi R., Arachidonic acid cytotoxicity: Can arachidonic acid be a physiological mediator of cell death? Cell Biochem. Funct. 21, 97–104 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Galluzzi L., et al. , Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W. S., et al. , Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 113, E4966–E4975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magtanong L., et al. , Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem. Biol. 26, 420–432.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacDonald M. L., Murray I. V. J., Axelsen P. H., Mass spectrometric analysis demonstrates that BODIPY 581/591 C11 overestimates and inhibits oxidative lipid damage. Free Radic. Biol. Med. 42, 1392–1397 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Carlson B. A., et al. , Glutathione peroxidase 4 and vitamin E cooperatively prevent hepatocellular degeneration. Redox Biol. 9, 22–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedmann Angeli J. P., et al. , Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon S. J., et al. , Human haploid cell genetics reveals roles for lipid metabolism genes in nonapoptotic cell death. ACS Chem. Biol. 10, 1604–1609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vale G., et al. , Three-phase liquid extraction: A simple and fast method for lipidomic workflows. J. Lipid Res. 60, 694–706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayala A., Muñoz M. F., Argüelles S., Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 360438 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fritz K. S., Petersen D. R., Exploring the biology of lipid peroxidation-derived protein carbonylation. Chem. Res. Toxicol. 24, 1411–1419 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagan V. E., et al. , Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 13, 81–90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wenzel S. E., et al. , PEBP1 wardens ferroptosis by enabling lipoxygenase generation of lipid death signals. Cell 171, 628–641.e26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenspan P., Fowler S. D., Spectrofluorometric studies of the lipid probe, nile red. J. Lipid Res. 26, 781–789 (1985). [PubMed] [Google Scholar]
- 28.Rendón A., et al. , Model systems of precursor cellular membranes: Long-chain alcohols stabilize spontaneously formed oleic acid vesicles. Biophys. J. 102, 278–286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukuzawa K., et al. , Antioxidant effect of bovine serum albumin on membrane lipid peroxidation induced by iron chelate and superoxide. Biochim. Biophys. Acta 1668, 145–155 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Curry S., Mandelkow H., Brick P., Franks N., Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 5, 827–835 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Tomoda H., Igarashi K., Cyong J. C., Omura S., Evidence for an essential role of long chain acyl-CoA synthetase in animal cell proliferation. Inhibition of long chain acyl-CoA synthetase by triacsins caused inhibition of Raji cell proliferation. J. Biol. Chem. 266, 4214–4219 (1991). [PubMed] [Google Scholar]
- 32.Gao X., Hannoush R. N., Single-cell in situ imaging of palmitoylation in fatty-acylated proteins. Nat. Protoc. 9, 2607–2623 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Dixon S. J., Stockwell B. R., The hallmarks of ferroptosis. Annu. Rev. Cancer Biol. 3, 35–54 (2019). [Google Scholar]
- 34.Bender G., Schexnaydre E. E., Murphy R. C., Uhlson C., Newcomer M. E., Membrane-dependent activities of human 15-LOX-2 and its murine counterpart: Implications for murine models of atherosclerosis. J. Biol. Chem. 291, 19413–19424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mashima R., Okuyama T., The role of lipoxygenases in pathophysiology; new insights and future perspectives. Redox Biol. 6, 297–310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng S., et al. , A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev. 27, 1462–1472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quehenberger O., et al. , Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51, 3299–3305 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.