Abstract
Recent events have pushed RNA research into the spotlight. Continued discoveries of RNA with unexpected diverse functions in healthy and diseased cells, such as the role of RNA as both the source and countermeasure to a severe acute respiratory syndrome coronavirus 2 infection, are igniting a new passion for understanding this functionally and structurally versatile molecule. Although RNA structure is key to function, many foundational characteristics of RNA structure are misunderstood, and the default state of RNA is often thought of and depicted as a single floppy strand. The purpose of this perspective is to help adjust mental models, equipping the community to better use the fundamental aspects of RNA structural information in new mechanistic models, enhance experimental design to test these models, and refine data interpretation. We discuss six core observations focused on the inherent nature of RNA structure and how to incorporate these characteristics to better understand RNA structure. We also offer some ideas for future efforts to make validated RNA structural information available and readily used by all researchers.
Keywords: RNA structure, RNA dynamics, RNA folding
RNA research is thriving. Discoveries of new RNAs are frequent, the repertoire of RNA-dependent biological functions is expanding, and ever-more-powerful RNA-centric techniques are being developed. The deluge of data continues to overwhelmingly support the idea that the function of any RNA is always determined to some degree by its structure, and researchers are eager to integrate the role of structure into their understanding of RNA biology. The pandemic caused by the RNA coronavirus severe acute respiratory syndrome coronavirus 2 poignantly reminded the world that RNA is a powerful agent of both disease and therapeutics—RNA is an “A-list molecule.”
The study of RNA and its structure is destined to expand, but unfortunately at times this rapid expansion has been accompanied by a loss of general knowledge about important fundamental properties of RNA structure. Misconceptions and false assumptions take root when RNA’s key properties are overlooked. A solid general understanding of RNA structure is important to develop rigorous mechanistic hypotheses, design experiments to test them, and derive proper interpretations. Now more than ever our community should strive to think and talk about RNA structure in a way that reflects its true behavior. Doing so has practical benefits: For example, Moderna scientists showed the importance of mRNA structure for stability and thus efficacy of the RNA at the heart of their vaccine (1).
Compare, for example, your mental images of messenger RNA (mRNA) and ribosomal RNA (rRNA). To many, the former appears as a squiggly extended line while the latter has a complicated and defined base-paired architecture. The implication is that rRNA is structured and mRNA is not, but is the “limp spaghetti” image of mRNA based on data supporting an absence of structure within a protein-encoding RNA, or is it based on a lack of data, ingrained assumptions, and “lore”? Is it correct to think of RNA as inherently extended, floppy, and “unstructured,” except in special cases?
The common default view of RNA structure is probably somewhat vestigial from when scientists regarded RNA primarily as the transient messenger between DNA and compactly folded proteins. In that paradigm, proteins and DNA had structure-driven jobs to do, while mRNA was mostly a linear template molecule without any needed local or higher-order structure. While some highly specialized RNA molecules (e.g., transfer RNA [tRNA], rRNA, ribozymes, riboswitches, etc.) were known to adopt functional structures, they were likely perceived as exceptions to the norm. The default state of most RNA was probably assumed to be extended, floppy, and “unstructured.” With the discoveries of defined and important structures in diverse RNAs, including mRNAs, from all domains of life and from viruses (2–5), why does this bias often endure? Students have told us that formal training in RNA structure and its direct relationship to function is uncommon. In its absence, human perception is likely molded by what we repeatedly see and hear (6). When the molecules of life are depicted, RNA is often displayed as a wavy line, proteins are drawn by default as compact globular entities, and DNA as a structured double helix. We assert that seeing these standard cartoons repeatedly reinforces assumptions about the default structural states of these molecules.
Likewise, how RNA structure is discussed probably affects thinking. The terms “structured” or “unstructured” RNA are often used to imply a strict binary demarcation, where “structure” is used synonymously with “Watson–Crick pairing,” even though RNA structure requires other types of interactions. While RNA secondary structure prediction algorithms can be useful and powerful, often they are treated as black boxes whose output is assumed to be correct, or as methods to determine which RNAs are “structured” and which are not. The use of more nuanced and descriptive language and a better understanding of the limitations of RNA structure prediction and determination methods could have a powerful positive effect on the field.
Here we review six fundamental properties of RNA structure and dynamics that may not be obvious but whose consideration we hope will promote a richer understanding of RNA. We aim to reemphasize some partially forgotten knowledge, dispel a few myths about the nature of RNA structure, and suggest some best practices for depicting and discussing RNA. We also propose guidelines for making use of automatically generated RNA structure predictions, criteria for guiding and comparing different types of RNA structure investigations, and directions where new tools or shared resources would be broadly beneficial. Note that this perspective is not intended to be a full review of the current or historical literature on these topics but rather a starting point for individual exploration. Our goal is to equip researchers from all disciplines to better integrate RNA structural knowledge into their own work and perhaps spur some fresh thinking about the relationship between RNA structure and function.
Stacking as a Driver for RNA Structure
Reviewing some of RNA’s underlying physical characteristics is worthwhile for understanding RNA structure. Within an RNA strand, the negatively charged ribose–phosphate chain—which makes up two-thirds of the mass of a nucleotide—creates steric and electrostatic constraints on the backbone conformation. Bases, which account for the remaining third of mass, comprise planar aromatic rings decorated with partially charged hydrogen-bond donors and acceptors. Thus, each monomeric nucleotide unit contains charged, polar, and aromatic groups, which can make diverse types of interactions with water, ions, amino acids, small molecules, and other nucleotides. In other words, in their normal aqueous environment RNA nucleotides are highly social—they are made to interact with one another (7, 8).
The realization that every nucleotide in an RNA chain can favorably interact with every other nucleotide is critical for thinking about RNA structure and how it differs from protein structure. In proteins, distinct side-chain characteristics mean some of the 20 amino acids more favorably interact with one another; some are more readily accommodated on the inside of a fold (like phenylalanine due to its aromatic ring), while others are on the solvent interacting surface (like the charged amino acids). Biochemists regularly experience how the type and distribution of amino acids affect the solubility of a given protein. In contrast, although RNA contains a planar aromatic base at every link in its chain, RNA is soluble in salty water. This seeming paradox is resolved because noncovalent interactions drive RNA bases to stack on each other, exposing their charged exocyclic groups to the water molecules and ions in the aqueous environment, leading to RNA “solvation” (9, 10).
Convenient and useful ball-and-stick representations of RNA (and DNA) or “ladders” in popular cartoon depictions generally show “space” between the bases within a helix. In fact, bases stack like coins in a roll with no space between them for solvent (Figs. 1 and 2A). Stacked bases are ∼3.4 Å apart because carbon, nitrogen, and oxygen atoms have all van der Waals radii around 1.7 Å, and these intimate interactions combine with backbone constraints to create the helical conformations we are familiar with (7, 9, 11). Thus, the helical structure of RNA (and DNA) is not induced by Watson–Crick pairing. Rather, within the stacking-induced helical arrangement, base pairs can assemble in A-form helices or other structural elements (12). To understand RNA structure, one must therefore understand base stacking.
Fig. 1.
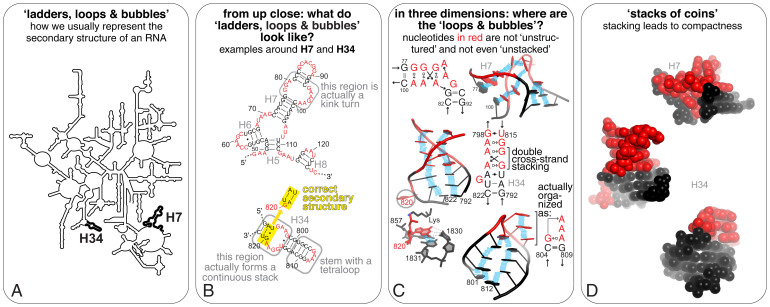
Unintentional bias from cartoon rendering of RNA. (A) A classical cartoon representation of an RNA (this 23S rRNA diagram from ref. 107) reveals mostly global Watson–Crick pairing patterns. (B) Close-ups of the H34 and H7 “helices” highlighted in A to show the sequence of their “internal” and apical loops (red). Boxed regions are displayed in further detail in C. (C) Ball-and-stick cartoon rendering of H7 and H34 to reveal their organization in three dimensions [from PDB ID code 1YHQ (108)]). The “loops and bubbles” unpaired in B—which might therefore be considered “unstructured”—are in fact structured to help orient the emerging stems in three dimensions. Blue bars represent stacking interactions, which occur between all bases. Revised secondary structure diagrams for these regions according to the Leontis–Westhof nomenclature are indicated (76). Note how these similary A/G-rich regions form nonetheless spatially distinct architectures. In three dimensions, G820 is actually bulged out and stacks against U1831 to form a Watson–Crick pair with C1830. (D) H7 and H34 shown as space-filling models using the van der Waals radii for each atom, illustrating the tight packing within these “loops and bubbles,” so that there is no “white space” between base pairs, as cartoons in A–C could lead one to believe.
Fig. 2.
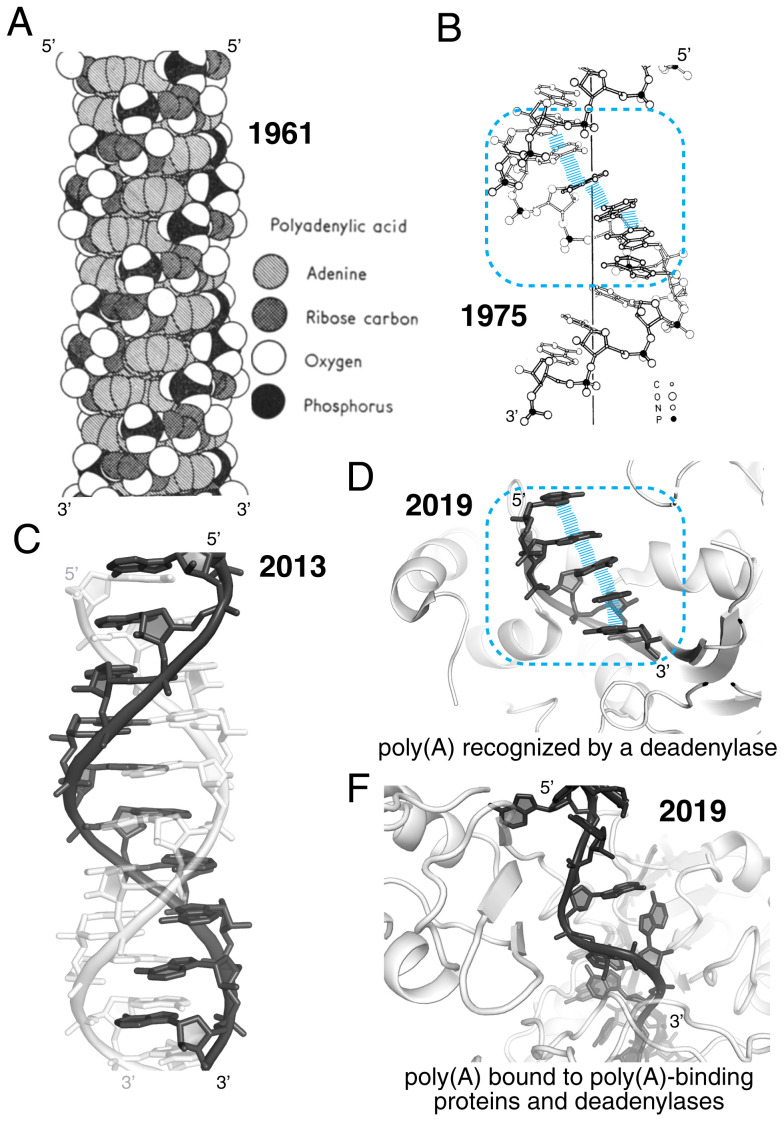
Stacking as a driver of RNA structure: the case of poly(A). (A) The original model by Watson, Crick, Rich, and Davies of the parallel double helix of poly(A) shows extensive stacking of the adenines (15). (B) Structural model for the poly(A) single helix based on the ApApA arrangement in its crystal structure (14). (C) Crystal structure at 1-Å resolution of the complex shown in A (PDB ID code 4JRD) (16). (D) Crystal structure of poly(A) bound to a deadenylase (PDB ID code 6R9J) (19). (E) Section of a 67-nucleotide-long poly(A) bound to a poly(A)-binding protein complex (PDB ID code 6R5K). Intrastrand stacking of the adenines has been replaced by interactions with proteins (20). In B and D, the blue enclosure emphasizes structural similarity supported by stacking interactions (blue lines).
What is the experimental evidence that base stacking drives RNA structure? Starting in the 1950s, biophysical studies of RNA oligonucleotides in solution revealed that unpaired bases stack in helical configurations. Even a single ApA dinucleotide adopts the “beginning of a single-strand helix” (13). This helix was modeled in 1975 on the basis of the crystal structure of ApApA (Fig. 2B) (14). In fact, poly-adenosine [poly(A)] forms a parallel double helix, even though it cannot form Watson–Crick pairs. This “second double helix” was first reported by Watson and Crick in 1961 (Fig. 2A) (15), and its structure was confirmed many decades later (Fig. 2C) (16).
Inherent base stacking leading to helical conformations is not a biophysical fluke but is key to biology (17). For example, the 3′ poly(A) tail that regulates eukaryotic mRNA stability is recognized by deadenylase enzymes primarily based on its stacking characteristics (Fig. 2D) (18, 19), while its structure may be disrupted in other cases (Fig. 2E) (20). Likewise, RNA scientists routinely observe and follow base stacking effects in the laboratory: Thermal denaturation of RNA (or DNA) leads to increased ultraviolet (UV) absorption because as the molecule denatures, base stacking decreases (21).
“Inherently Structured” Does Not Mean “Static”
In the 1970s Barbapapas children’s books, stories revolved around the colorful smiling blob-like characters that adopted all sorts of shapes, in turn a swing, a musical instrument, a boat, a tree, anything to amuse kids. Similarly, although RNA is intrinsically structured, its conformation may change over many time scales, from nanoseconds to milliseconds to longer, depending on the environmental conditions or the presence of ligands (22–24). These built-in dynamics do not make RNA “unstructured”; they are part of its structure and therefore its function. The conformation of an RNA is dictated by a thermodynamic landscape that depends on the sequence of the RNA and the conditions. Depending on the nature of the landscape, a given RNA populates an ensemble of conformations of various compactness, stabilities, and flexibilities. As the length of an RNA increases, the complexity of the landscape and the number of possible nonidentical states increases, but favored states remain. Describing RNA structure (as well as any molecular structure) as consisting of conformational ensembles dictated by statistical thermodynamics is not new but is worth remembering.
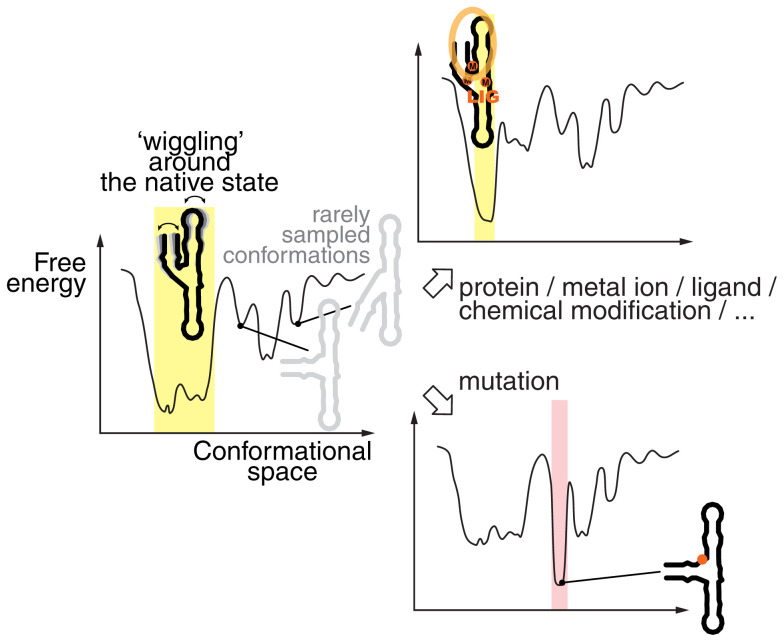
Within a given free-energy landscape, the most thermostable folds populate the valleys; areas above correspond to less favored structures. For some RNA sequences, a single deeper well corresponding to a highly favored conformation prevents frequent sampling of other states (Fig. 3). This is often the native biologically relevant state, and coherent folding to this state is selected by evolution (25–28). However, the bottom of a deep well could have shallow indentations, allowing “wiggling” or “flexing” of the native structure under biological conditions. Examples are mature fully folded tRNAs: Their overall L-shaped fold is maintained but they can flex, bend, or undergo local structural changes as needed to pass through the ribosome or interact with their cognate aminoacyl synthetases (see movie S1 in ref. 29) (30). For other RNAs, the landscape could have a few deep wells that represent more than one favored distinct conformation, which can interconvert. In some cases, these conformations could represent one or more “misfolded” states or stable intermediates (31–33). Some RNA sequences may instead have flatter folding landscapes, or flatter regions within larger landscapes, with several shallow indentations. With these RNAs, incoherent folding leads to multiple readily interconverting transient RNA structures, but not a lack of structure.
Fig. 3.
The conformational landscape of an RNA is rugged and changes according to the environment. The left-most curve shows the free-energy landscape of a hypothetical RNA, highlighting a predominantly populated state, with limited conformational changes within the corresponding conformational space (yellow shading). Other states are energetically less accessible and therefore less populated, as they are separated from the deep “native state” well by high-energy states. However, in a different environment, the binding of proteins, metal ions, metabolites to that RNA, or its chemical modification can further stabilize the native state (Top Right). Conversely, a mutation at a particular position could destabilize the native state and stabilize a nonnative state (pink shading; Bottom Right). In all cases, the RNA is structured but the characteristics of the conformational ensemble can differ. Also, note that a fully extended state is highly energetically unfavorable in all cases. The figure is loosely adapted from refs. 28 and 34.
When RNA is considered as a potential mix of conformations, the inadequacy of the “structured/unstructured RNA” dichotomy becomes clear. “Structured” RNAs that adopt one highly populated state are far from static. Likewise, for an RNA to be truly “unstructured” in the fully extended sense, the landscape would have to be essentially flat with no favored state. This is unrealistic for RNA, as it would require unfavorable breaking of the ubiquitous intramolecular interactions, such as stacking and hydrogen bonding. Even if the possible number of nonidentical states generally increases with RNA length, an overall more favorable collapsed state and likely some specific states will be more populated.
Thinking of RNA as a dynamic ensemble of interconverting conformations has important functional and practical implications (34, 35). In particular, the conformational landscape of a given RNA can change when conditions change, for instance with altered concentrations of ions, shifts in pH, the presence of ligands, interactions with proteins or other molecules, and chemical modifications or mutations of nucleotides (Fig. 3). Single-nucleotide variants can perturb the conformational ensemble, perhaps disfavoring the native biologically relevant structure or favoring misfolded states, leading to disease (36, 37). The kinetics of RNA synthesis may lead to RNA structures that are not necessarily the most stable within the total thermodynamic landscape (“cotranscriptional folding”), and large RNAs may have complex folding pathways with intermediate steps (38–40). Ligand binding to RNA often occurs by capturing a certain conformation from the ensemble, more so than by inducing a new conformation (41, 42). Further illustrating the benefits of considering RNA as a population of states, bioinformaticians showed that chemical probing data are often more reliably explained by an ensemble of various secondary structures than by a single one (34, 43–45).
RNA Is a Compact Molecule
Bases do not “know” what sort of RNA they reside in (mRNA, rRNA, tRNA, small interfering RNA, long noncoding RNA, etc.). Thermodynamically, it is favorable for them in all cases to stack on one another and possibly hydrogen-bond to form structure, even if only transiently. All types of bases can stack and form thermodynamically favorable hydrogen bonds with each other, with the sugar–phosphate backbone, and with the solvent. These combined interactions cause the single strand of RNA to inherently adopt a self-associating state under the aqueous biological conditions, dependent on factors like temperature. As the strand folds back on itself this leads to overall compaction of the molecule, compared to a fully extended state.
Some recent evidence for RNA compactness came from global structure mapping, which showed that 30 to 40% of RNA duplexes in living cells are formed from sequences that are more than 200 nucleotides apart (46). Compactness and long-distance pairing (47), combined with the modularity of RNA (35, 48, 49) and the increase of RNA size over time by accretion (50, 51), result in the 5′ and 3′ ends of any natural RNA being in relatively close spatial proximity (52–55). To create the extended state in which RNA is often depicted would require an input of energy. Interestingly, a similar dichotomy exists between our schematic rendering of proteins with the N and C termini at both ends of a horizontal line, when in fact the ends are in spatial proximity across protein structures (56).
At first, the view of ubiquitous RNA self-association and compactness may seem to contradict studies that showed a low degree of structure in the coding regions of most mRNAs in living cells (57, 58). However, these observations are in fact consistent with the idea that ribosomes, helicases, or other RNA remodeling proteins partially and transiently unwind inherently structured regions (57, 59, 60). Indeed, when translation is inhibited in living cells, mRNAs undergo a decrease in their overall end-to-end length (61, 62). When RNA needs to be accessed, proteins may need to “work at opening” intrinsic RNA structure.
Watson–Crick Pairing Is Important but a Bit Overrated
Watson–Crick base pairing is important, often perceived as THE hallmark of RNA structure and thus an RNA with many potential Watson–Crick base pairs is considered “highly structured.” Should Watson–Crick pairing is used synonymously with “RNA structure”? In fact, in most cases of folded RNA three-dimensional (3D) structures, non-Watson–Crick pairs are critical for creating the tertiary interactions that stabilize the functional conformation (Fig. 4). This information gets lost with the typical rendering of RNA as stems separated by bubbles and loops of “unpaired” bases, which often comes from an RNA structure prediction algorithm (Figs. 1 and 4 and Automatic Predictions of an RNA Secondary Structure).
Fig. 4.
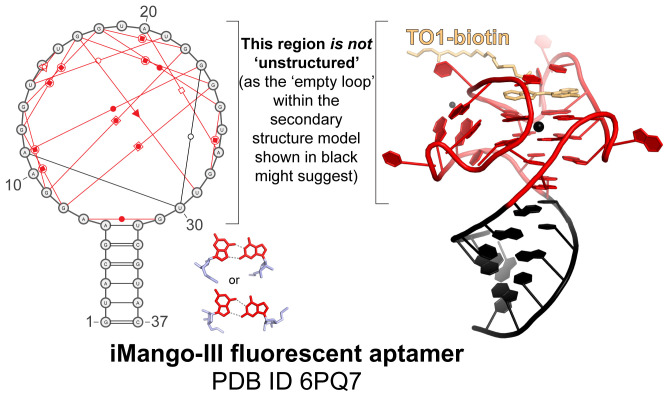
RNA secondary structure diagrams can imply that a structured RNA is “unstructured.” Classical representation of the secondary structure of the iMango-III fluorescent aptamer, as in Fig. 1, next to a ball-and-stick rendering of its crystal structure bound to TO1-biotin [PDB ID code 6PQ7 (109)]. The non-Watson–Crick base pairs [red symbols (76)] are shown as output by RNAPDBee (101). The highlighted examples of G.G pairs (red, base; blue, sugar) involved in this G quadruplex structure occur a total of nine times. Note that the classic secondary structure diagram in black, without the red annotations, could lead one to believe that the large loop is “unstructured” while the helix is “highly structured.” In fact, the entire molecule is structured, and it is the structure of the loop that provides function.
The overemphasis on Watson–Crick pairs in RNA structure possibly has several origins. Most of us learned about DNA before learning about RNA. DNA rarely exists without a complementary strand and consequently forms double-stranded helices with Watson–Crick base pairs. Deviations from these “canonical” pairs are often called “mismatches” that create instability, which is a misnomer for RNA structures where non-Watson–Crick pairs are key for folding and binding to proteins or other ligands (63–65). Likewise, with DNA it is useful to consider the GC content for assessing the overall stability of double helices, as is routinely done when designing hybridizing primers in the laboratory. However, the same thinking is not as meaningful for an uncharacterized RNA, unless one knows a priori that most of the nucleotides are involved in Watson–Crick pairs. In other words, GC content is not a reliable predictor of “highly structured” RNAs.
Overall, the tendency to focus on Watson–Crick pairs may stem from the fact that they are the basis of nucleic acid hybridization and that they are easier to identify, draw, and rationally mutate. Aligning RNA sequence homologs is more straightforward when only Watson–Crick pairs are considered, but in fact it is the deviations from complementarity that are the most useful to confidently determine and align secondary structures. Similarly, computing the most thermostable predicted secondary structure of a given sequence relies on free energy calculations based largely on maximizing the number of Watson–Crick pairs in accordance with measured “nearest-neighbor” parameters (66, 67). However, the state with the most predicted Watson–Crick pairs may not reflect the true biologically relevant secondary structure (see Automatic Predictions of an RNA Secondary Structure).
Long-range tertiary Watson–Crick pairing is often presented as the driver for bringing distal parts of an RNA structure together, but in general base stacking is de facto the primary means to spatially organize the global fold of an RNA. Base stacking and Watson–Crick pairing create stacks of continuous and noncontinuous helices, which define that RNA secondary structure. However, non-Watson–Crick pairing and stacking patterns in helical junctions and internal loops preform a 3D architecture that dictates the angles of emerging helices (compare the angles of H7 and H34 in Fig. 1) (35, 68–70). As a result, specific parts of the RNA are spatially positioned to readily establish interactions often involving nucleotides that are far apart in sequence, but not in three dimensions. Whether these interactions comprise Watson–Crick base pairs (e.g., in pseudoknots, which occur upon base pairing of a single-stranded region in a loop and a complementary sequence elsewhere in the same RNA) or other favorable interactions, the partner regions within a single RNA are only likely to interact if the structure in between allows it.
Experimental evidence of how the global fold precedes the pairing of bases far apart in sequence is obtained when increasing the concentrations of cations (such as K+ and Mg2+) rescues the function of an RNA in which pseudoknot base pairing is disrupted by mutations (71, 72). This occurs because the decrease in conformational stability resulting from the lost base pairs is compensated for by a metal ion-driven increase in the stability of the global architecture, which is dictated by specific stacking and H-bond patterns in loops, junctions, and bulges (73, 74). Looking at it from this angle, pseudoknot formation does not create the global fold; rather, a pseudoknot is stabilized by folding of other elements that bring two complementary sequences closer in space.
Non-Watson–Crick Pairing Is Very Much Underrated
Watson–Crick paired regions form the basis of RNA transcription, translation, and replication. When they occur between different molecules, they provide a code conveniently deciphered by enzymes. Watson–Crick pairing is important for the RNA’s role as a carrier of analog information, but its importance changes within 3D structures of folded RNA molecules. Watson–Crick paired helical regions generally act as spacers with regular geometrical properties essential to position structural elements stabilized by non-Watson–Crick pairs. Within a folded RNA, the “cool” parts are the kinks, turns, and other structural motifs that support the global fold or bind a protein or other ligand (75). These elements rely on non-Watson–Crick pairs (Figs. 1 and 4), which outnumber Watson–Crick pairs in diversity by 11:1 (76). The many functions of RNA beyond a linear information carrier are enabled and dependent on non-Watson–Crick pairing; RNA is what it is because of these interactions.
While it is true that isolated non-Watson–Crick pairs are destabilizing within A-form helices, it is equally true that non-Watson–Crick pairs form favorable, specific, and necessary interactions in specific structural contexts (77). A non-Watson–Crick pair in a helix may destabilize that individual helix but enable tertiary interactions that provide overall stabilization to the structure. Given what has been learned about RNA structure over the last four decades, we assert that it is reasonable to refrain from equating the presence of non-Watson–Crick pairs with a lack of structure; implying that an RNA with many non-Watson–Crick pairs is necessarily “unstable” is incorrect without additional information.
Non-Watson–Crick pairings combined with helical stacking give rise to structural motifs that provide the building blocks of many higher-order structures, including ultrastable tetraloops and their receptors, kink-turns, E-loops, etc. While the many possible non-Watson–Crick pairings appear daunting, a well-developed classification, nomenclature, and symbol system has been adopted to describe, talk about, and think about these critical interactions (76). We encourage the RNA community to familiarize themselves with these alternate pairings and the rich structural possibilities they enable and to avoid the oversimplifying “canonical”/”noncanonical” base-pair terminology in favor of one that is more descriptive and precise.
Regarding non-Watson–Crick pairs, using a precise language will help to continuously update mental frameworks. The importance to posttranscriptional chemical modifications is being reemphasized (78, 79), and phenomena such as base tautomerism and protonation remain understudied (80). Because these events can manifest their effects within the context of alternate hydrogen bonding and base pairing schemes, their biological effects are only fully understandable in the context of non-Watson–Crick interactions.
Automatic Predictions of an RNA Secondary Structure: Approach with Caution!
When researchers encounter a novel RNA sequence, they commonly “Mfold” it. Within seconds, a convenient prediction emerges of the lowest predicted free-energy secondary structure of that RNA, computed by a thermodynamics-based algorithm. Unfortunately, often this output from Mfold (or RNAfold, Sfold, and other similar tools (81–83)) is not presented as a prediction, resulting from an algorithm that includes assumptions, but as a representation of the true structure. Alternative pairing possibilities with theoretically higher free energies are generally ignored and the top prediction may even be propagated in the literature without supporting evidence. The danger of only considering the lowest free-energy structure and showcasing it as “The” secondary structure of a particular RNA is that the community ends up taking an untested possibility at face value.
Unfortunately, the outputs from folding programs often substitute for a thorough experimental secondary structure determination. RNA secondary structure prediction software built on rigorous measurements of base-pair stability in different nearest neighbor contexts (84, 85) are valuable tools for rapidly assessing potential Watson–Crick pairing and generating new ideas about RNA structure. However, they consider all nucleotides as equally likely to be involved in secondary structure elements, which often leads to erroneous assumptions about base pairs (86). Furthermore, because these algorithms tend to maximize the predicted number of Watson–Crick pairs (equated with the lowest predicted free energy), they are most accurate when an RNA has many such pairs. The problem is, not all RNAs do.
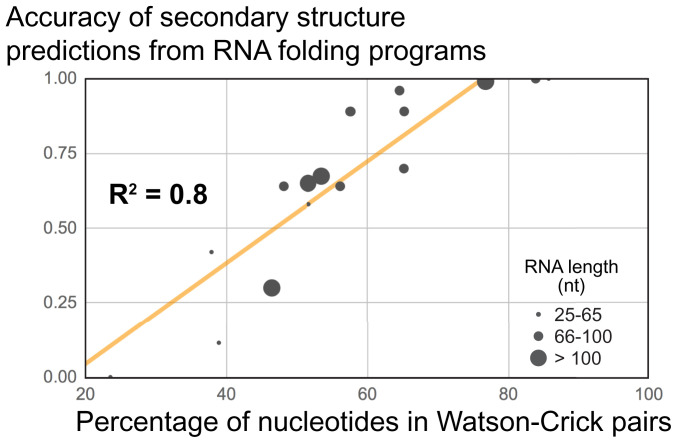
To illustrate this, we used several secondary structure algorithms with various input RNAs for which the correct secondary structure was known, each RNA containing different numbers of Watson–Crick pairs. We observed a strong positive correlation between the accuracy of output models with the number of Watson–Crick pairs within the known structures (Fig. 5; linear regression R2 = 0.8 using 17 recent RNA structures from the Protein Data Bank [PDB]). This holds true regardless of size, as R2 ∼ 0 if the same data were plotted against RNA length. This seemingly trivial exercise highlights the fact that because algorithms are designed to find Watson–Crick pairs, the accuracy of the output depends on whether that RNA actually folds to maximize the number of such pairs. Because one does not know a priori if this is true of a given RNA, the accuracy of the resultant model is difficult to assess without additional data.
Fig. 5.
The higher the percentage of Watson–Crick pairs in an RNA, the better the accuracy of a secondary structure prediction—regardless of RNA length. Accuracy of secondary structure models (0, completely incorrect; 1, 100% correct) as a function of the percentage of nucleotides in Watson–Crick pairs. Each dot represents an RNA >25 nucleotides (excluding duplexes), whose structure was solved by X-ray crystallography and deposited in the PDB between 17 April 2019 and 5 February 2020. The R2 factor from a linear regression analysis is indicated (trendline shown in orange). For details on the determination of the accuracy, see SI Appendix, Supplementary Methods and Table S1.
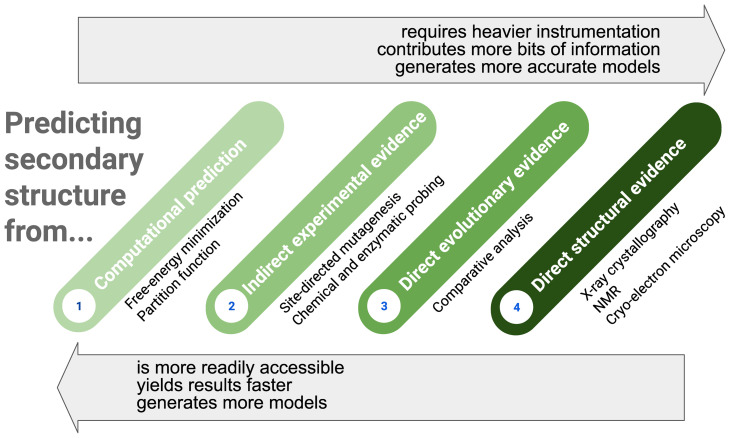
Proper use of secondary structure programs thus requires coupling them with diverse experimental methods in a step-by-step determination of the actual structure (Fig. 6). These may include chemical probing both in vitro and in cells (87–89) and site-directed mutagenesis that provide probabilistic assessments of secondary structures. Multidimensional probing methods combine these two into a rigorous experimental approach that provides information about interactions between specific bases (90), and new methods seek to provide information on the proximity of bases distant in sequence (e.g., refs. 46, 91, and 92). Also, if homologous RNA sequences are available in sufficient numbers and diversity, comparative sequence alignment is a very powerful way to garner evolution-supported evidence for a particular secondary structure prediction (93). Ultimately, a true secondary structure can be verified when the 3D structure is determined. Crystallography, NMR, and now high-resolution cryo-electron microscopy (cryo-EM) represent the crème de la crème, with NMR and cryo-EM also having the ability to potentially detect conformational ensembles and dynamics (94). In general, the risk that a secondary structure model is incorrect decreases with the effort put into determining and testing the model (Fig. 6).
Fig. 6.
Hierarchy in the methods commonly used to predict the secondary structure of an RNA. The confidence level in an RNA secondary structure model depends on the number of pieces of computational and experimental information used to predict that model (roughly increasing from left to right), which relates to the time to obtain that model (also increasing from left to right), which in turn is linked to accessibility to advanced instrumentation and expertise in structure determination methods.
Going Forward
Upgrading the Discourse about RNA.
As more colleagues enter the fray of RNA research, our responsibility as an RNA community is to ensure that RNA is discussed in ways that reflect its true nature. RNA is far from being “like DNA, with some extra things…,” and generalizations and oversimplifications only cloud understanding. For instance, as a community we could refrain from referring to a particular RNA as being “highly structured,” “structured,” or “unstructured” and choose instead specific language that describes the existing observations regarding its structure and dynamics. Similarly, describing non-Watson–Crick pairs as “mismatches” and “noncanonical pairs” only propagates the misconception that such pairs are less important than Watson–Crick pairs. The word “mismatch” also denies the evolutionary pathway that led to the conservation of a non-Watson–Crick pair at a particular position. The G.G and A.G pairs shown in Figs. 1 and 4, for example, are in fact perfect matches as the RNAs evolved to maintain such pairs. Simply but accurately talking about RNA will help the RNA research field advance in full force.
Developing User-Friendly Applications for Depicting RNA.
Unexpectedly, software for easily drawing RNA secondary structures is uncommon. Existing programs also tend to solely consider Watson–Crick pairs (traditionally the sole hallmark of “secondary” structure) and/or target only certain RNA families [see, for example, XRNA (http://rna.ucsc.edu/rnacenter/xrna/xrna_faq.html), Forna (95), or RiboVision (96)]. Tools have been devised that output non-Watson–Crick pairs from a 3D structure (97, 98), but drawing software does not generally include ways to easily depict these. Generally speaking, no drawing tool has really established itself beyond the laboratory it originated from, except perhaps Varna (99) and R2R (100). For full adoption by the research community, the challenge remains to design a tool that would be performant but easy to use. Efforts in that direction have been made most recently for example with RNAPDBee (101), RNA2Drawer (102), R2DT (103), and RNArtist (https://github.com/fjossinet/RNArtist).
Classifying Secondary Structure Predictions by Confidence Level.
Rigorous RNA secondary structure determination necessarily requires integrating data from multiple methods and constant refinement as new data are available. Hence, we suggest classifying secondary structure models based on four (or more) confidence levels: Level 1 for an ensemble of unvalidated computational predictions, Level 2 for models further supported by indirect experimental evidence of structure (such as site-directed mutagenesis and chemical or enzymatic probing), Level 3 for a structure resulting from covariation analysis, and Level 4 for the model derived from a solved high-resolution 3D structure, etc. (Fig. 6). Some of the existing depositories like Rfam (104) or the NDB (105), among others, could be involved in validating, standardizing, archiving, and disseminating secondary-structure models, as is the norm for sequences, RNA sequence alignments (which by definition are based on secondary structure), and tertiary structures. Establishing a secondary-structure database would harmonize the RNA community around core values for an equitable usage and portrayal of structural information.
Integrating Disseminated Knowledge for Reliable Predictions.
Cross-comparing and combining all available data about a particular RNA and related molecules remains the go-to approach for deriving reliable 2D and possibly 3D structure predictions. Such a “meta-analysis” is generally done by experienced researchers using multiple tools and information hubs à la RNACentral (106). Integrating existing tools would help make available information more widely available to a broader group of nonexperts. This could perhaps eliminate the propagation of incorrect or outdated secondary-structure information, link secondary-structure models to other data, and stimulate the development of harmonized ways to portray and validate secondary-structure predictions. The available data are deep and rich but need to be more accessible and better-integrated; this will disseminate the ability to use RNA structure to inform further experiments and boost confidence in RNA research from the general public.
Concluding Remarks
The time is ripe to shake up some assumptions about RNA. Representing RNA as a wavy line is convenient, but when convenience betrays biology, flaws emerge. Every RNA is capable of self-association, and structured RNAs are not rare, even though the structures may be fleeting, dynamic, or altered by interacting proteins. Simply but accurately rendering and talking about RNA will help the RNA research field advance in full force. This becomes a necessity as RNA scientists study RNAs of increasing size and complexity, with diverse regulatory or therapeutic roles. RNA has gone mainstream, so let’s make sure RNA structure properties return to the front seat.
Supplementary Material
Acknowledgments
We thank current and past members of the J.S.K. laboratory (Ben Akiyama, Steve Bonilla, David Costantino, Daniel Eiler, Erik Hartwick, Rachel Jones, Zoe O’Donoghue, Maddie Sherlock, and Matt Szucs) for stimulating discussions and in particular Andrea MacFadden, Anna-Lena Steckelberg, and Brian Wimberly for coming up with a chart that inspired Fig. 6. We also thank Drs. Jennifer Doudna, Olivia Rissland, Neel Mukherjee, Tom Blumenthal, and Eric Westhof for insightful comments on the manuscript. RNA structure-centric research by the authors is supported by NIH grants R35GM118070 (J.S.K.), R01AI133348 (J.S.K.), and R21AI157244 (Q.V. and J.S.K.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112677119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Mauger D. M., et al. , mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. U.S.A. 116, 24075–24083 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaafar Z. A., Kieft J. S., Viral RNA structure-based strategies to manipulate translation. Nat. Rev. Microbiol. 17, 110–123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luttermann C., Meyers G., A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 282, 7056–7065 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Mercer T. R., Mattick J. S., Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Ding Y., et al. , In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Dechêne A., Stahl C., Hansen J., Wänke M., The truth about the truth: A meta-analytic review of the truth effect. Pers. Soc. Psychol. Rev. 14, 238–257 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Martel P., Base crystallization and base stacking in water. Eur. J. Biochem. 96, 213–219 (1979). [DOI] [PubMed] [Google Scholar]
- 8.Ts’o P. O. P., Melvin I. S., Olson A. C., Interaction and association of bases and nucleosides in aqueous solutions. J. Am. Chem. Soc. 85, 1289–1296 (1963). [DOI] [PubMed] [Google Scholar]
- 9.Saenger W., Principles of Nucleic Acid Structure (Springer, New York, 1984). [Google Scholar]
- 10.Westhof E., Ed., Water and Biological Macromolecules (The Macmillan Press LTD, 1993). [Google Scholar]
- 11.Sundaralingam M., Structure and conformation of nucleosides and nucleotides and their analogs as determined by x-ray diffraction. Ann. N. Y. Acad. Sci. 255, 3–42 (1975). [DOI] [PubMed] [Google Scholar]
- 12.Westhof E., Fritsch V., RNA folding: Beyond Watson-Crick pairs. Structure 8, R55–R65 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Holcomb D. N., Tinoco I., Conformation of polyriboadenylic acid: pH and temperature dependence. Biopolymers 3, 121–133 (1965). [Google Scholar]
- 14.Saenger W., Riecke J., Suck D., A structural model for the polyadenylic acid single helix. J. Mol. Biol. 93, 529–534 (1975). [DOI] [PubMed] [Google Scholar]
- 15.Rich A., Davies D. R., Crick F. H., Watson J. D., The molecular structure of polyadenylic acid. J. Mol. Biol. 3, 71–86 (1961). [DOI] [PubMed] [Google Scholar]
- 16.Safaee N., et al. , Structure of the parallel duplex of poly(A) RNA: Evaluation of a 50 year-old prediction. Angew. Chem. Int. Ed. Engl. 52, 10370–10373 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Howard F. B., Frazier J., Singer M. F., Miles H. T., Helix formation between polyribonucleotides and purines, purine nucleosides and nucleotides. II. J. Mol. Biol. 16, 415–439 (1966). [DOI] [PubMed] [Google Scholar]
- 18.Tang T. T. L., Passmore L. A., Recognition of poly(A) RNA through its intrinsic helical structure. Cold Spring Harb. Symp. Quant. Biol. 84, 21–30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang T. T. L., Stowell J. A. W., Hill C. H., Passmore L. A., The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases. Nat. Struct. Mol. Biol. 26, 433–442 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schäfer I. B., et al. , Molecular basis for poly(A) RNP architecture and recognition by the Pan2-Pan3 deadenylase. Cell 177, 1619–1631.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devoe H., Tinoco I. Jr., The hypochromism of helical polynucleotides. J. Mol. Biol. 4, 518–527 (1962). [DOI] [PubMed] [Google Scholar]
- 22.Al-Hashimi H. M., Walter N. G., RNA dynamics: It is about time. Curr. Opin. Struct. Biol. 18, 321–329 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q., Al-Hashimi H. M., Domain-elongation NMR spectroscopy yields new insights into RNA dynamics and adaptive recognition. RNA 15, 1941–1948 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi X., Mollova E. T., Pljevaljcić G., Millar D. P., Herschlag D., Probing the dynamics of the P1 helix within the Tetrahymena group I intron. J. Am. Chem. Soc. 131, 9571–9578 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan T., Sosnick T. R., Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat. Struct. Biol. 4, 931–938 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Pan J., Thirumalai D., Woodson S. A., Folding of RNA involves parallel pathways. J. Mol. Biol. 273, 7–13 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Thirumalai D., Lee N., Woodson S. A., Klimov D., Early events in RNA folding. Annu. Rev. Phys. Chem. 52, 751–762 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Thirumalai D., Woodson S. A., Kinetics of folding of proteins and RNA. Acc. Chem. Res. 29, 433–439 (1996). [Google Scholar]
- 29.Dunkle J. A., et al. , Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332, 981–984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantara W. A., et al. , Modifications modulate anticodon loop dynamics and codon recognition of E. coli tRNA(Arg1,2). J. Mol. Biol. 416, 579–597 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Hsu C.-F., et al. , Formation of frameshift-stimulating RNA pseudoknots is facilitated by remodeling of their folding intermediates. Nucleic Acids Res. 49, 6941–6957 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.St-Pierre P., et al. , A structural intermediate pre-organizes the add adenine riboswitch for ligand recognition. Nucleic Acids Res. 49, 5891–5904 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell R., et al. , The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J. Mol. Biol. 363, 531–544 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Ganser L. R., Kelly M. L., Herschlag D., Al-Hashimi H. M., The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 20, 474–489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cruz J. A., Westhof E., The dynamic landscapes of RNA architecture. Cell 136, 604–609 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Halvorsen M., Martin J. S., Broadaway S., Laederach A., Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 6, e1001074 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan Y., et al. , Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505, 706–709 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell R., Herschlag D., New pathways in folding of the Tetrahymena group I RNA enzyme. J. Mol. Biol. 291, 1155–1167 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Pyle A. M., Fedorova O., Waldsich C., Folding of group II introns: A model system for large, multidomain RNAs? Trends Biochem. Sci. 32, 138–145 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Mitchell D. III, Russell R., Folding pathways of the Tetrahymena ribozyme. J. Mol. Biol. 426, 2300–2312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberman J. A., Wedekind J. E., Riboswitch structure in the ligand-free state. Wiley Interdiscip. Rev. RNA 3, 369–384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vicens Q., Mondragón E., Batey R. T., Molecular sensing by the aptamer domain of the FMN riboswitch: A general model for ligand binding by conformational selection. Nucleic Acids Res. 39, 8586–8598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spasic A., Assmann S. M., Bevilacqua P. C., Mathews D. H., Modeling RNA secondary structure folding ensembles using SHAPE mapping data. Nucleic Acids Res. 46, 314–323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morandi E., et al. , Genome-scale deconvolution of RNA structure ensembles. Nat. Methods 18, 249–252 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Tomezsko P. J., et al. , Determination of RNA structural diversity and its role in HIV-1 RNA splicing. Nature 582, 438–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Z., et al. , RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultes E. A., Spasic A., Mohanty U., Bartel D. P., Compact and ordered collapse of randomly generated RNA sequences. Nat. Struct. Mol. Biol. 12, 1130–1136 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Gracia B., et al. , RNA structural modules control the rate and pathway of RNA folding and assembly. J. Mol. Biol. 428, 3972–3985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leontis N. B., Lescoute A., Westhof E., The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 16, 279–287 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrov A. S., et al. , History of the ribosome and the origin of translation. Proc. Natl. Acad. Sci. U.S.A. 112, 15396–15401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Francklyn C., Schimmel P., Aminoacylation of RNA minihelices with alanine. Nature 337, 478–481 (1989). [DOI] [PubMed] [Google Scholar]
- 52.Lai W.-J. C., et al. , mRNAs and lncRNAs intrinsically form secondary structures with short end-to-end distances. Nat. Commun. 9, 4328 (2018). [DOI] [PMC free article] [PubMed]
- 53.Vicens Q., Kieft J. S., Rissland O. S., Revisiting the closed-loop model and the nature of mRNA 5′-3′ communication. Mol. Cell 72, 805–812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoffe A. M., Prinsen P., Gelbart W. M., Ben-Shaul A., The ends of a large RNA molecule are necessarily close. Nucleic Acids Res. 39, 292–299 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leija-Martínez N., et al. , The separation between the 5′-3′ ends in long RNA molecules is short and nearly constant. Nucleic Acids Res. 42, 13963–13968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishna M. M. G., Englander S. W., The N-terminal to C-terminal motif in protein folding and function. Proc. Natl. Acad. Sci. U.S.A. 102, 1053–1058 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J. S., Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwok C. K., Tang Y., Assmann S. M., Bevilacqua P. C., The RNA structurome: Transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 40, 221–232 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Bhaskaran H., Russell R., Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature 449, 1014–1018 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Q., Fairman M. E., Jankowsky E., DEAD-box-protein-assisted RNA structure conversion towards and against thermodynamic equilibrium values. J. Mol. Biol. 368, 1087–1100 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adivarahan S., et al. , Spatial organization of single mRNPs at different stages of the gene expression pathway. Mol. Cell 72, 727–738.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khong A., Parker R., mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J. Cell Biol. 217, 4124–4140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hermann T., Westhof E., Non-Watson-Crick base pairs in RNA-protein recognition. Chem. Biol. 6, R335–R343 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Hermann T., Patel D. J., Adaptive recognition by nucleic acid aptamers. Science 287, 820–825 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Leontis N. B., Westhof E., Analysis of RNA motifs. Curr. Opin. Struct. Biol. 13, 300–308 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Rivas E., The four ingredients of single-sequence RNA secondary structure prediction. A unifying perspective. RNA Biol. 10, 1185–1196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rivas E., Evolutionary conservation of RNA sequence and structure. Wiley Interdiscip. Rev. RNA 12, e1649 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lescoute A., Westhof E., The interaction networks of structured RNAs. Nucleic Acids Res. 34, 6587–6604 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lescoute A., Westhof E., Topology of three-way junctions in folded RNAs. RNA 12, 83–93 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laing C., Schlick T., Analysis of four-way junctions in RNA structures. J. Mol. Biol. 390, 547–559 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fica S. M., Mefford M. A., Piccirilli J. A., Staley J. P., Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat. Struct. Mol. Biol. 21, 464–471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roth A., Nahvi A., Lee M., Jona I., Breaker R. R., Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA 12, 607–619 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Draper D. E., Folding of RNA tertiary structure: Linkages between backbone phosphates, ions, and water. Biopolymers 99, 1105–1113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Auffinger P., Grover N., Westhof E., Metal ion binding to RNA. Met. Ions Life Sci. 9, 1–35 (2011). [PubMed] [Google Scholar]
- 75.Butcher S. E., Pyle A. M., The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 44, 1302–1311 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Leontis N. B., Westhof E., Geometric nomenclature and classification of RNA base pairs. RNA 7, 499–512 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turner D. H., Thermodynamics of base pairing. Curr. Opin. Struct. Biol. 6, 299–304 (1996). [DOI] [PubMed] [Google Scholar]
- 78.Seelam P. P., Sharma P., Mitra A., Structural landscape of base pairs containing post-transcriptional modifications in RNA. RNA 23, 847–859 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frye M., Harada B. T., Behm M., He C., RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Westhof E., Yusupov M., Yusupova G., Recognition of Watson-Crick base pairs: Constraints and limits due to geometric selection and tautomerism. F1000Prime Rep. 6, 19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chan C. Y., Lawrence C. E., Ding Y., Structure clustering features on the Sfold Web server. Bioinformatics 21, 3926–3928 (2005). [DOI] [PubMed] [Google Scholar]
- 82.Zuker M., Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gruber A. R., Lorenz R., Bernhart S. H., Neuböck R., Hofacker I. L., The Vienna RNA websuite. Nucleic Acids Res. 36, W70–W74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DiChiacchio L., Mathews D. H., Predicting RNA-RNA interactions using RNAstructure. Methods Mol. Biol. 1490, 51–62 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Turner D. H., Mathews D. H., NNDB: The nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 38, D280–D282 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doshi K. J., Cannone J. J., Cobaugh C. W., Gutell R. R., Evaluation of the suitability of free-energy minimization using nearest-neighbor energy parameters for RNA secondary structure prediction. BMC Bioinformatics 5, 105 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mitchell D. III, Assmann S. M., Bevilacqua P. C., Probing RNA structure in vivo. Curr. Opin. Struct. Biol. 59, 151–158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kubota M., Tran C., Spitale R. C., Progress and challenges for chemical probing of RNA structure inside living cells. Nat. Chem. Biol. 11, 933–941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mailler E., Paillart J.-C., Marquet R., Smyth R. P., Vivet-Boudou V., The evolution of RNA structural probing methods: From gels to next-generation sequencing. Wiley Interdiscip. Rev. RNA 10, e1518 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Tian S., Das R., RNA structure through multidimensional chemical mapping. Q. Rev. Biophys. 49, e7 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Mustoe A. M., Lama N. N., Irving P. S., Olson S. W., Weeks K. M., RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc. Natl. Acad. Sci. U.S.A. 116, 24574–24582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krokhotin A., Mustoe A. M., Weeks K. M., Dokholyan N. V., Direct identification of base-paired RNA nucleotides by correlated chemical probing. RNA 23, 6–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rivas E., Clements J., Eddy S. R., A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods 14, 45–48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kaledhonkar S., et al. , Late steps in bacterial translation initiation visualized using time-resolved cryo-EM. Nature 570, 400–404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lorenz R., et al. , ViennaRNA package 2.0. Algorithms Mol. Biol. 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bernier C. R., et al. , RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss. 169, 195–207 (2014). [DOI] [PubMed] [Google Scholar]
- 97.Jossinet F., Westhof E., Sequence to structure (S2S): Display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics 21, 3320–3321 (2005). [DOI] [PubMed] [Google Scholar]
- 98.Jossinet F., Ludwig T. E., Westhof E., Assemble: An interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics 26, 2057–2059 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Darty K., Denise A., Ponty Y., VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weinberg Z., Breaker R. R., R2R--software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12, 3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zok T., et al. , RNApdbee 2.0: Multifunctional tool for RNA structure annotation. Nucleic Acids Res. 46 (W1), W30–W35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Johnson P. Z., Kasprzak W. K., Shapiro B. A., Simon A. E., RNA2Drawer: Geometrically strict drawing of nucleic acid structures with graphical structure editing and highlighting of complementary subsequences. RNA Biol. 16, 1667–1671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sweeney B. A., et al. , R2DT is a framework for predicting and visualising RNA secondary structure using templates. Nat. Commun. 12, 3494 (2021). [DOI] [PMC free article] [PubMed]
- 104.Kalvari I., et al. , Rfam 14: Expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 49 (D1), D192–D200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Coimbatore Narayanan B., et al. , The nucleic acid database: New features and capabilities. Nucleic Acids Res. 42, D114–D122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Petrov A. I., et al. ; The RNAcentral Consortium, RNAcentral: A comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 45 (D1), D128–D134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Petrov A. S., et al. , Secondary structures of rRNAs from all three domains of life. PLoS One 9, e88222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tu D., Blaha G., Moore P. B., Steitz T. A., Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121, 257–270 (2005). [DOI] [PubMed] [Google Scholar]
- 109.Trachman R. J. III, et al. , Co-crystal structure of the iMango-III fluorescent RNA aptamer using an X-ray free-electron laser. Acta Crystallogr. F Struct. Biol. Commun. 75, 547–551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.