Significance
The diversification of host-associated microbial communities depends on barriers to gene flow, imposed by confinement to different hosts or by niche partitioning within single hosts. However, most gut microbiomes are too complex to disentangle the diversification processes. Taking advantage of the simple gut microbiomes of social bees, we demonstrate that bee gut bacteria have diversified both between host species and within single host species through the acquisition of different ecological niches within the same gut. Our study further shows that gut microbiomes differ in spatial distributions within the same host, possibility due to adaptation to specified nutritional niches, such as urea utilization.
Keywords: gene flow, homologous recombination, engineered bacteria, spatial distribution, urea utilization
Abstract
Host-associated microbiomes, particularly gut microbiomes, often harbor related but distinct microbial lineages, but how this diversity arises and is maintained is not well understood. A prerequisite for lineage diversification is reproductive isolation imposed by barriers to gene flow. In host-associated microbes, genetic recombination can be disrupted by confinement to different hosts, for example following host speciation, or by niche partitioning within the same host. Taking advantage of the simple gut microbiome of social bees, we explore the diversification of two groups of gut-associated bacteria, Gilliamella and Snodgrassella, which have evolved for 80 million y with honey bees and bumble bees. Our analyses of sequenced genomes show that these lineages have diversified into discrete populations with limited gene flow. Divergence has occurred between symbionts of different host species and, in some cases, between symbiont lineages within a single host individual. Populations have acquired genes to adapt to specific hosts and ecological niches; for example, Gilliamella lineages differ markedly in abilities to degrade dietary polysaccharides and to use the resulting sugar components. Using engineered fluorescent bacteria in vivo, we show that Gilliamella lineages localize to different hindgut regions, corresponding to differences in their abilities to use spatially concentrated nitrogenous wastes of hosts. Our findings show that bee gut bacteria can diversify due to isolation in different host species and also due to spatial niche partitioning within individual hosts, leading to barriers to gene flow.
Most animal guts contain microbial communities, which often affect health, development, and behavior. These communities are dominated by bacteria and often by bacterial lineages restricted to the guts of one or few host species. Examples of gut-restricted bacteria can be found in humans (1) and other primates (2), ruminants (3), termites (4), and social bees (5), including species of honey bees (genus Apis) and bumble bees (genus Bombus). Studies profiling diversity of gut-restricted bacteria have revealed high levels of fine-scale strain diversity within single host individuals, among individuals within host species, and across related host species; these communities contain clusters of closely related strains, defined on the basis of various sequence divergence cutoffs; the most common cutoff is based on a genome-wide average nucleotide identity (gANI) of 95% (6). Within gut communities, strain clusters may adopt specific spatial (7) or metabolic niches (8) as they compete for resources, engage in interstrain and interspecies warfare (9), and exchange nutrients through cross-feeding (10).
Potentially, the diversification of these strain clusters is linked to different ecological strategies (11). However, differences in ecological roles do not necessarily lead to diversification. In both prokaryotes and eukaryotes, speciation and diversification of lineages depend on reproductive isolation: the lack of homologous recombination among genomes of the diverging lineages (12–14). In communities of microorganisms, the forces leading to divergence of lineages are relatively little studied, and this is specifically true for bacterial communities inhabiting animal guts. One way that a gut bacterial lineage might split into two clusters is through separation or speciation of the host populations (15). Host-restricted gut bacteria are typically exchanged among hosts of the same species through close contact, during social interactions or group-living; examples include most mammals (16), termites (17), cockroaches (18), and social bees (19). This exchange can prevent genetic divergence of bacterial lineages within a host species, but restriction to different host species following host speciation could provide barriers to symbiont exchange. Some evidence for such cospeciation has been reported for gut bacteria of hominins (2), termites (17), and social bees (5). A second route to diversification might be adoption of different ecological niches within the gut. Ecological differences can impose barriers to gene flow, but only if they impose spatial or temporal separation that curtails exchange of chromosomal fragments among strains (11). Demonstrations of ecological niches leading to spatial separation in host-associated microbiomes remain few (8). As yet, the extent to which confinement to different hosts versus niche partitioning within hosts affects bacterial diversification remains unclear.
To investigate diversification of gut bacteria at a fine scale, we advocate for the delineation of clusters based on the extent of gene flow. In this paper, we use “population” for sets of recombining strains, and our use corresponds to the usual meaning of “species” under the biological species concept, although most of these entities do not have formal species names. The delineation of populations based on gene flow has been validated in various organisms, including bacteria (20, 21), viruses (22, 23), and eukaryotes (20). Both host speciation and niche differentiation can disrupt gene flow in host-associated microbes, so analyses aimed at detecting gene flow can elucidate gut microbe diversification. In addition, as gene flow-based approaches delineate populations based on the significant decrease of homologous recombination between populations, genome-wide sequence identity cutoffs are not required for different species (21, 24). Therefore, gene flow-based approaches can describe populations across various bacterial groups in a consistent, and biologically meaningful way.
By applying gene flow-based approaches, we explore the diversification of two bacterial clades, the genera Gilliamella and Snodgrassella, which are exclusively associated with guts of a clade of social bees (Corbiculata), including honey bees (genus Apis), bumble bees (genus Bombus), and stingless bees (tribe Meliponini). Gilliamella and Snodgrassella belong to a consortium of bacterial lineages that have evolved with corbiculate bees for 80 million y (5). While Gilliamella is only found in corbiculate bees, it belongs to the family Orbaceae, which is widely associated with insect guts, having been found in beetles, butterflies, wasps, and flies (19). Based on analyses of complete genome sequences from cultured isolates and of metagenomic sequences, Gilliamella appears to contain considerable strain variation both within and between host species; at least two species, Gilliamella apicola and Gilliamella apis are recognized within single individuals of the western honey bee Apis mellifera (25, 26). Due to ongoing gene acquisition and loss, strains vary in the presence of genes expected to impact host and symbiont ecology, such as those underlying utilization of dietary carbohydrates and interstrain antagonism (27–30). Here we use complete genome sequences of 117 Gilliamella isolates and 57 Snodgrassella isolates from several Apis and Bombus host species, with most intensive sampling from A. mellifera. We define populations on the basis of gene flow at shared loci (21). We find that the diversification of lineages in the bee gut microbiome reflects both host speciation and nutritional niche partitioning within a single gut. We explore one example involving Gilliamella populations. We find differences in capabilities for urea uptake and utilization, and show that these functional differences correspond to differences in spatial distribution within individual honey bee guts.
Results
Core-Gene Sequence-Based Phylogeny and Gene Content Analysis Show Symbiont Diversification between Bee Host Species.
After removing potentially contaminated genomes (SI Appendix, Table S1), we retained complete genome sequences for 117 Gilliamella isolates and 57 Snodgrassella isolates for analyses. For Gilliamella, 63, 43, and 11 of the genomes are from A. mellifera, Bombus species, and other Apis species, respectively; for Snodgrassella, 34, 20, and 3 of the genomes are from A. mellifera, Bombus, and other Apis, respectively (Fig. 1).
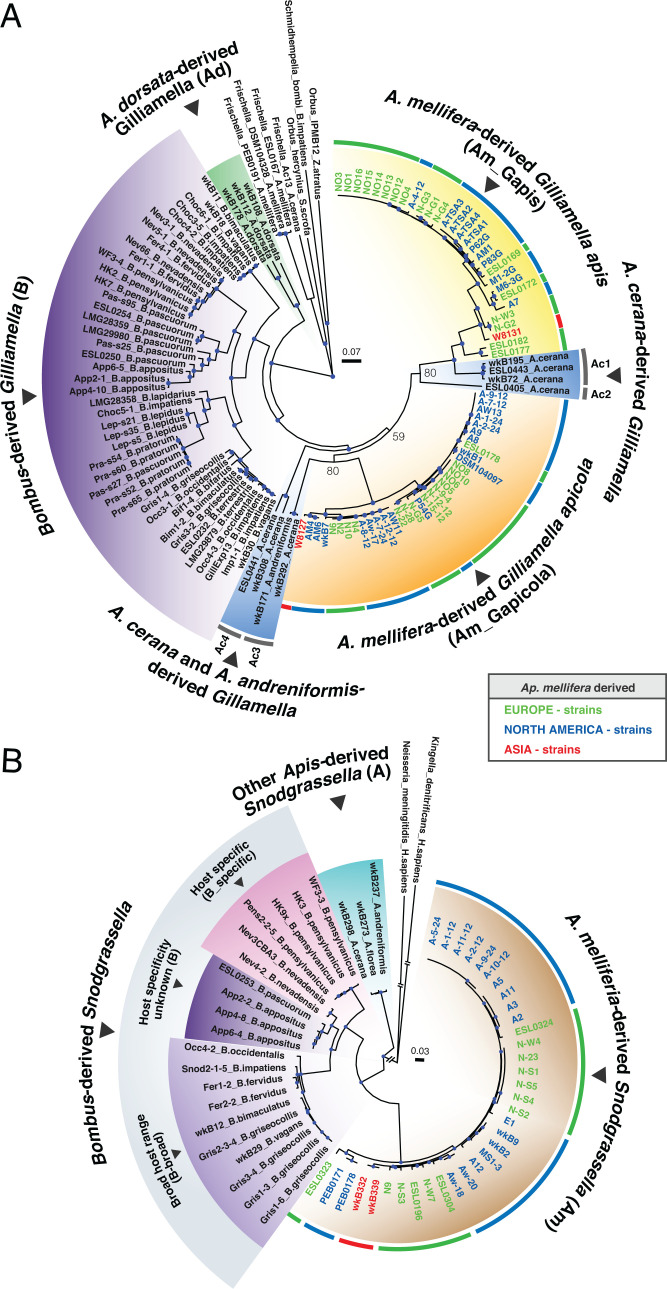
Fig. 1.
Maximum-likelihood phylogenies of (A) Gilliamella strains and (B) Snodgrassella strains based on nucleotide sequences of core genes. Internal nodes colored in blue denote bootstrap value > 95%. Bootstrap values were added to three internal nodes with low bootstrap support. Major clades have background colors based on their host species. Names and the ring at the outer edge of A. mellifera-derived strains are colored by geographic locations. Abbreviations of major clades are in parentheses. Two clades of Gilliamella from A. mellifera are labeled as G. apis (denoted as Am_Gapis) and G. apicola (denoted as Am_Gapicola), based on the placement of type strains (NO3 and wkB1) in those clades. For Bombus-derived Snodgrassella strains, some are restricted to a single Bombus species (host-specific, denoted as B_specific); some occur in multiple Bombus species (broad-host-range, denoted as B_broad); others do not have host specificity information (denoted as clade B) (25).
After gene annotation and ortholog assignment, we constructed phylogenetic trees for Gilliamella based on 1,141 core orthologous groups and for Snodgrassella based on 1,305 core orthologous groups, defined as single-copy gene families present in at least 80% of the strains. In phylogenies for both Gilliamella and Snodgrassella, strains from the same host species tend to cluster together (Fig. 1). For Gilliamella, the topology among A. mellifera-derived (clades Am_Gapis and Am_Gapicola), Apis cerana-derived (clades Ac1, Ac2, Ac3, and Ac4), and Bombus-derived (clade B) Gilliamella strains matches the host phylogeny, consistent with the long-term codiversification of host and symbiont. However, Apis dorsata-derived Gilliamella strains (clade Ad) branch at the base of the Gilliamella tree, before the split between other Apis strains and Bombus strains. For Snodgrassella, A. mellifera-derived (clade Am) and Bombus-derived strains formed two monophyletic groups and were sister to each other. However, these two groups were sister to Snodgrassella strains from other Apis species (clade A in Fig. 1), which conflicts with the host phylogeny. Thus, the current distribution of symbiont strains across host species is consistent with codiversification occasionally interrupted by switching among host lineages.
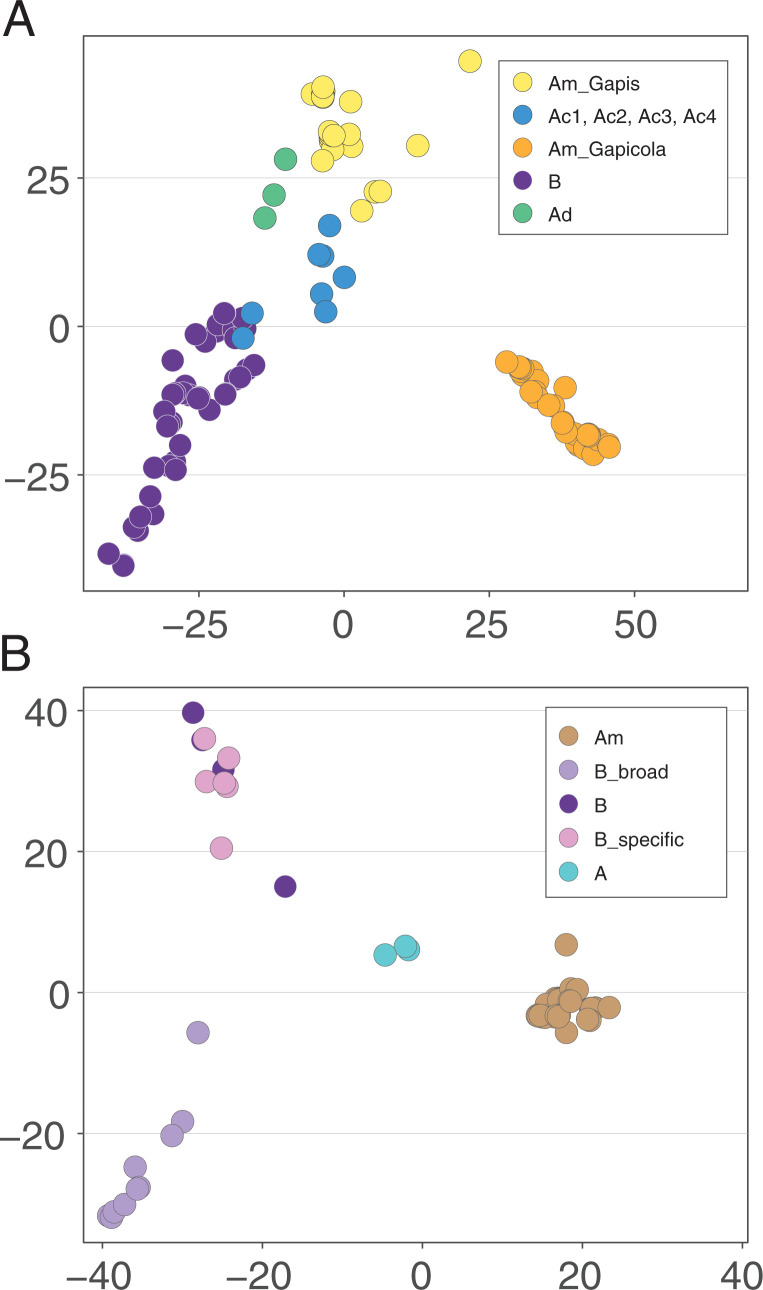
To assess gene content differences among hosts, we applied principal component analysis (PCA) based on all orthologous groups and constructed gene-content trees based on gene presence and absence. Despite the incongruence between the core-gene phylogeny and the host phylogeny, Apis-derived strains tend to group together based on their similar gene content. All Apis-derived Snodgrassella strains clustered together in PCA and formed a monophyletic group in the gene-content tree (Fig. 2 and SI Appendix, Fig. S1). The similar gene contents across Apis-derived Snodgrassella strains, despite evidence for past switching between host species, indicate that gene sets could reflect adaptation to Apis hosts.
Fig. 2.
PCA of all the orthologous groups among (A) Gilliamella and (B) Snodgrassella strains. Abbreviations of major clades and colors of populations are consistent with Fig. 1.
To investigate the importance of shared genes to host adaptation, we assigned Snodgrassella genes as essential genes in culture or as genes beneficial for bee gut colonization, as determined by a previous study using transposon insertion sequencing (31). In total, 26 orthologous groups were found only in Apis-derived Snodgrassella strains. Of these, four were colonization-beneficial genes related to pathways including vitamin B3 biosynthesis and holin. A total of 19 orthologous groups were found only in A. mellifera- and Bombus-derived Snodgrassella strains. Of these, four were colonization-beneficial genes related to lipid transportation and thioesterase. None of the clade-specific orthologous groups corresponded to essential genes.
Population Delineation Based on Gene Flow.
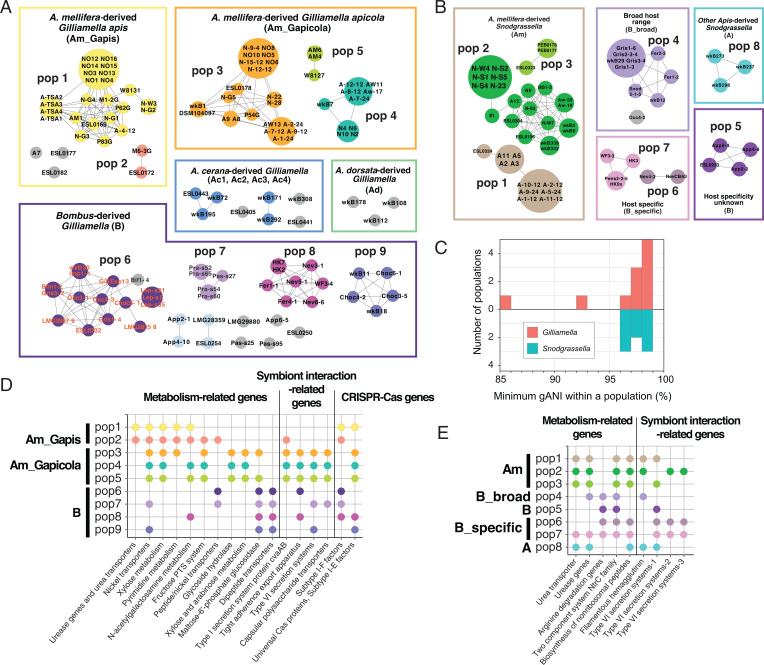
One approach used to delineate bee gut bacterial populations is based on arbitrary sequence similarity cutoffs, such as gANI ≥ 89% (26, 32). Here we investigated Gilliamella and Snodgrassella populations using a gene flow-based approach, PopCOGenT (21), which identifies recombinant regions by measuring the length distributions of identical regions between pairs of genomes. PopCOGenT analysis classified genomes into 28 Gilliamella populations and 9 Snodgrassella populations (Fig. 3). The minimum gANI within Gilliamella populations varied from 86 to 98%, and the minimum gANI values for Snodgrassella populations were ≥96% (Fig. 3 and SI Appendix, Fig. S2). Strains of G. apis and G. apicola belong to distinct populations, verifying their status as separate species within the related honey bee species A. mellifera and A. cerana (25, 26), but G. apis and G. apicola each contained multiple populations, indicating that current nomenclature does not reflect the diverse biological species within Gilliamella.
Fig. 3.
Gene flow among (A) Gilliamella and (B) Snodgrassella genomes. Near-identical genomes were collapsed by PopCOGenT. Edges connecting two genomes indicate gene flow. Boxes represent the clades in the phylogeny. Major populations from A. mellifera and Bombus are named. Nodes are colored based on population designation. Nodes in gray are strains showed significantly low gene flow with other sampled genomes. Note that one of the Bombus-derived genome Bif1-4 only shows low levels of gene flow with Occ4-3 but not with other genomes; therefore, it is not included in population 6 by PopCOGenT. (C) Minimum nucleotide identity in Gilliamella and Snodgrassella populations. Minimum nucleotide identity was calculated as the gANI between the most divergent members of the same population. Genes related to metabolic pathways, interbacterial antagonistic interactions, and CRISPR-Cas systems are significantly enriched in populations of (D) Gilliamella and (E) Snodgrassella.
As PopCOGenT identifies recombination based on lengths of identical regions flanked by nucleotide polymorphisms, near-identical genomes (with divergence <0.0355%) were collapsed together due to insufficient mutations to detect homologous recombination (21). Many A. mellifera-derived strains from the same geographic location were collapsed; these strains were usually collected from the same hive or nearby hives. Interestingly, several populations defined on the basis of gene flow include A. mellifera-derived strains from different continents (Fig. 1), indicating that the global commercial transportation of A. mellifera hives (33) introduces gene flow among their gut microbiomes.
Functional Genes Enriched in Different Gilliamella Populations.
As bacterial populations adapt to specific ecological niches within hosts, we expect populations to accumulate genes involved in specialized functions. We investigated the enrichment of functional pathways and orthologous groups at different levels, including host species, major symbiont clades, and symbiont populations (SI Appendix, Supplementary Materials and Methods). Since the presence of these ecology-related functional genes varies among genomes, due to gene loss and gain through horizontal transfer, many are distinct from the shared genes used for detecting homologous recombination between pairs of genomes in the PopCOGenT analyses. Genes with inferred functions related to various metabolic pathways, interbacterial antagonistic interactions, and CRISPR-Cas systems are enriched in certain populations (Fig. 3 D and E). Here, we focus on genes related to polysaccharide metabolism and to nitrogen metabolism. This focus is motivated by prior evidence that the bee gut community derives energy from degradation of pollen cell walls and depends on nitrogen waste products for biosynthesis of amino acids (30–32).
Genes related to polysaccharide and sugar metabolism.
Gilliamella is one of several members of the bee gut community that degrade and ferment polysaccharides present in pollen cell walls (30); at least some of these fermentation products are taken up by the host (34). Gilliamella populations possess diverse carbohydrate-active enzymes (CAZymes) for extracellular degradation (designated as GH, for glycosyl hydrolases) and genes related to transport and processing of the products (27, 28, 30).
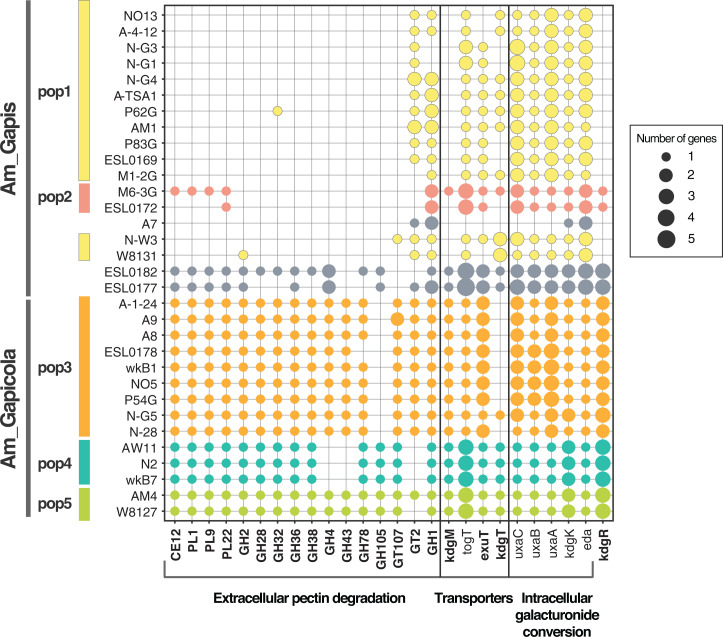
Functional enrichment analysis suggests that Gilliamella populations differ in use of substrates derived from pollen cell walls. The Am_Gapicola clade possesses all key genes for extracellular degradation of pectin (PL1, PL9, PL22, and CE12), whereas Gilliamella Am_Gapis population 1 has lost genes for secreted enzymes but retains most of the inner membrane transporters (togT, exuT, kdgT) and galacturonide conversion genes (Fig. 4). Am_Gapis population 1 has also lost the transporter on the outer membrane, oligogalacturonate-specific porin family protein (kdgM), as well as its transcriptional regulator kdgR. Differences in CAZyme repertoires were also found among the Gilliamella Am_Gapicola clades: for example, population 3 has lost the GH105 gene; population 4 has lost GH4, GH43, and GT2 (Fig. 4). Thus, these populations are predicted to differ in abilities to degrade different polysaccharides.
Fig. 4.
Distribution of genes related to pectin degradation in Gilliamella strains. Near-identical genomes identified by PopCOGenT are combined and represented by one genome. Strains are ordered based on their positions in the phylogeny. Strains are colored based on population designation. Strains in gray do not have gene flow with other sampled genomes. Genes significantly enriched in Am_Gapis or Am_Gapicola are in bold. Note that only the extracellular pectin degradation genes that significantly differ among populations are included in the figure.
The degradation of polysaccharides produces a mixture of monosaccharides that can be used by some Gilliamella strains as carbon substrates (35). Gilliamella populations differ in abilities to use specific monosaccharides. Am_Gapicola populations possess the genes for using xylose, arabinose, mannose, rhamnose, and galactose, whereas Am_Gapis populations have lost the transporters for xylose, arabinose, and key enzymes for galactose metabolism (SI Appendix, Fig. S3). Am_Gapis population 2 but not population 1 possesses the mannose-6-phosphate isomerase gene, manA, suggesting differences in ability to use mannose (35).
Genes related to nitrogenous waste utilization.
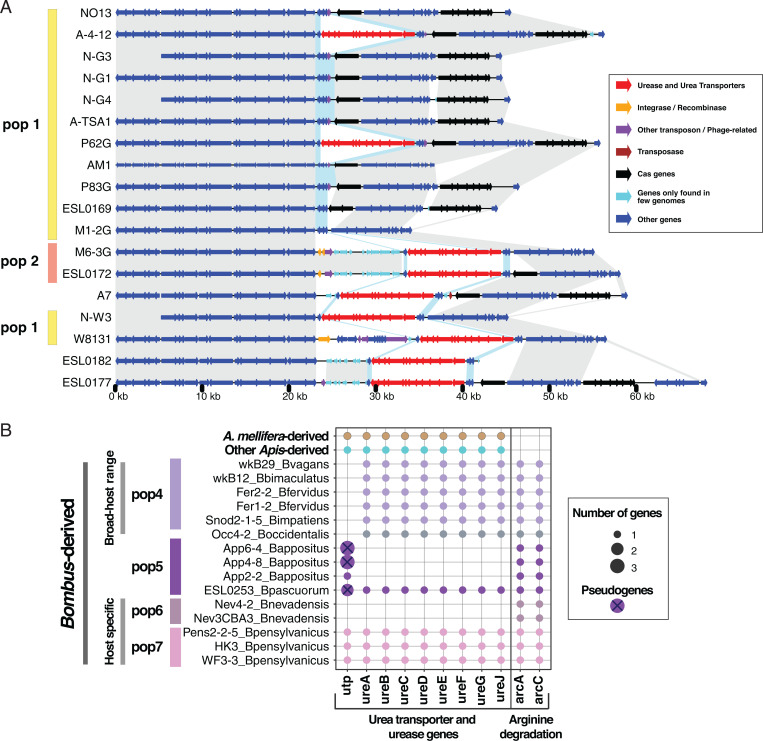
The Malpighian tubules empty nitrogenous wastes as uric acid and urea into the gut lumen at the pylorus sphincter forming the midgut–hindgut junction, providing an abundant source of nitrogen potentially usable by bacteria. A striking difference among Gilliamella populations involves genes related to urea utilization (Fig. 3D). All strains in Am_Gapis population 2, two strains (A-4-12 and P62G) in population 1, as well as strains in the Am_Gapis clade not assigned to any population (Fig. 5), possess an 11-kb genomic island that encodes the urea ABC-type transporters urtABCDE and urease genes ureABCDEFG, which degrade urea to ammonia (Fig. 5 and SI Appendix, Fig. S4). To investigate the origin of the urea transporters and urease genes found in Gilliamella, we searched the amino acid sequences against the National Center for Biotechnology Information (NCBI) nonredundant protein database and constructed phylogenetic trees for each urea-related gene. Although other bee gut bacterial species, Snodgrassella and Bartonella, possess homologs of these genes, the Gilliamella copies are mostly closely related to genes from Sodalis and other Enterobacteriaceae (SI Appendix, Fig. S5), suggesting that G. apis gained these genes through horizontal gene transfer from a member of the Enterobacteriaceae.
Fig. 5.
Gilliamella and Snodgrassella strains possess urea transporters and urease genes. (A) Synteny of urea transporter genes, urease genes, and flanking regions in Gilliamella. Genes are colored based on their functions. Homologous regions are shaded in gray. Homologous genes next to the urea transporters and urease genes are colored in blue shade. (B) Distribution of the urea transporter and urease genes in Snodgrassella. Strains are colored based on their populations. Crossed dots are likely pseudogenes harboring internal stop codons. As A. mellifera-derived and other Apis-derived genomes possess the same number of urea-related genes, they are collapsed into one representative genome for visualization. Near-identical genomes identified by PopCOGenT are collapsed and represented by one genome.
Interestingly, urea transporter (utp) and urease (ureABCDEFGJ) genes are also significantly enriched in some Snodgrassella populations (Fig. 5), suggesting that urea recycling could be important for Snodgrassella host adaptation. Apis-derived strains and B_specific population 7 possess both the utp and the urease genes, while the B_broad populations have lost the transporter, and population 6, the other host-specific population, has lost all urea-related genes. Of the four strains in population 5, three strains have lost all the urease genes while keeping the urea transporter; however, the transporter genes in population 5 appear nonfunctional, as they contain multiple internal stop codons and transposases. While lacking genes for converting urea to ammonia, Bombus-derived strains possess arginine deiminase genes (arcAC) that enable degradation of arginine to ammonia via citrulline (Fig. 5).
Genes related to symbiont interactions.
Gene-enrichment analysis suggests that Gilliamella populations differ in their interactions with the gut community and in their modes of gut colonization. Most Am_Gapis populations lack genes related to capsular polysaccharide transporters, type I (colicin V secretion proteins), type II (Tight adherence [Tad] export apparatus), and type VI secretion systems (SI Appendix, Fig. S6). These genes are important for interbacterial competition and biofilm formation. Am_Gapis population 1 also lacks the cvaA and cvaB genes that underlie the type I secretion system, which enables antibacterial peptide secretion.
CRISPR-Cas spacers largely confined to distinct populations.
Gilliamella strains carry CRISPR-Cas systems to defend against viruses (28). Except for a few Am_Gapis strains (N-W3 and N-G2), most of the Am_Gapis strains carry both subtype I-E and I-F Cas proteins (SI Appendix, Fig. S7) (36). We compared CRISPR spacers across strains. Spacers represent genomic segments of bacteriophage from past infections, so matching spacers reveals sets of Gilliamella strains that have undergone infection by the same or very similar bacteriophage. Strains in the same population often share similar CRISPR spacers. For example, Am_Gapicola populations 3, 4, and 5 share extensive spacers within each population, but no similar spacers were found between populations.
Spatial Segregation of Gilliamella Populations in the Bee Gut.
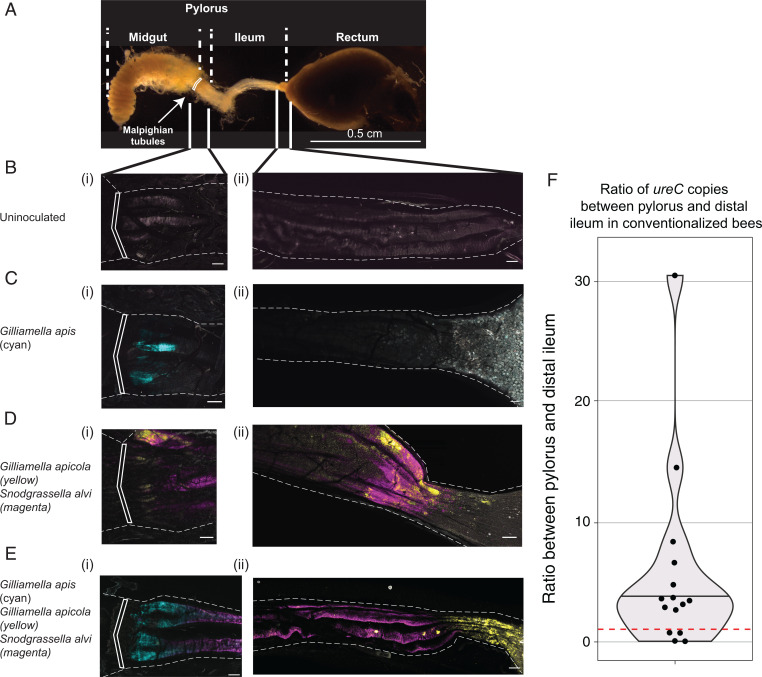
As the Malpighian tubules secrete nitrogenous wastes into the pylorus region (Fig. 6A), we predicted that the differences in prevalence of urea transporters and urease genes among populations reflected different ecological niches corresponding to differences in physical locations of Gilliamella populations along the A. mellifera gut. G. apis populations with the urea metabolism-related genes are expected to localize near the Malpighian tubules at the anterior end of the hindgut. To explore this issue, we constructed strains of A. mellifera-derived G. apis (strain M6-3G from Am_Gapis population 2), G. apicola (strain wkB7 from Am_Gapicola population 4), and Snodgrassella (strain wkB2 from Snodgrassella population 2) that constitutively express distinct fluorescent proteins. We inoculated these fluorescent strains into cohorts of emerged bees, maintained these bees under hive-like conditions, and 7 d later we dissected these bees and imaged their whole guts using fluorescence microscopy (Fig. 6).
Fig. 6.
Spatial partitioning of G. apis and G. apicola in bee guts. Honey bees were colonized with Snodgrassella expressing E2-Crimson (magenta), G. apicola expressing red chromoprotein (yellow), or G. apis expressing green fluorescent protein (cyan) alone or in combination. After 7 d, we dissected bee guts and imaged them using fluorescent confocal microscopy. (A) Dissected gut of an adult honey bee, and schematic of gut regions. We compared the (i) pylorus and (ii) distal ileum-rectum junction for different samples (B–E). White boxes across A–E indicate the pyloric sphincter regions. (Scale bars: 100 µm in B–E.) (B) Uncolonized bees show minimal background fluorescence in pylorus and ileum. (C) Monocolonized G. apis (cyan) colonizes the pylorus, with minimal colonization of ileum (i-ii). (D) Cocolonization of G. apicola (yellow) and Snodgrassella (magenta) show slight pylorus colonization and robust ileum colonization. (E) Bees coinoculated with G. apis (cyan), G. apicola (yellow), and Snodgrassella (magenta). Cocolonized species retain similar localization to monoculture strains, with G. apis outcompeting G. apicola at the pylorus. For each inoculation condition, 6 to 12 bees were inspected during each of three independent colonization trials. Representative images are shown. (F) Ratio of ureC gene copies between pylorus and distal ileum in 15 age-controlled bees with a conventional gut microbiome. ureC gene copy number was normalized based on 16S rRNA gene copy number. Samples with ratio above 1 (red dotted line) have higher abundance of G. apis in pylorus than in ileum (Wilcoxon signed-rank test P = 0.012).
Initially, we investigated how G. apis colonized bees when inoculated alone. In all 18 inspected bees, G. apis robustly colonized the pylorus, immediately after the pyloric sphincter between the midgut and hindgut, the most proximal region of the ileum (Fig. 6C). This site immediately follows the pyloric sphincter at which Malpighian tubules empty nitrogenous wastes including urea into the lumen of the digestive tract (Fig. 6A). In contrast to G. apis, G. apicola repeatedly failed to effectively colonize when administered alone. However, G. apicola did colonize when coinoculated with Snodgrassella in 18 inspected bees (Fig. 6D). As seen previously (19, 37), both Snodgrassella and G. apicola robustly colonized the ileum, with G. apicola apparently enriched near the distal portion of the ileum near the rectum. Both Snodgrassella and G. apicola could partially colonize the region near the pylorus, but this colonization was less robust than that of G. apis and appeared further away from the pyloric sphincter.
Next, we coinoculated these fluorescent strains all together in bees at approximately equal optical density, as a minimal defined community of engineered strains. Surprisingly, in the 18 inspected bees, coinoculation had overall little effect on where individual species colonized the bee gut. While in monoculture both Snodgrassella and G. apicola could minimally colonize the pylorus, when G. apis was present, G. apis reliably outcompeted these strains to be the dominant colonizer of the pylorus (Fig. 6E). Snodgrassella was the most robust colonizer of the ileum, regardless of which other species were present and appeared to form contiguous biofilms with G. apis near the pylorus and G. apicola along the length of the ileum and near the rectum.
To quantify the abundance of G. apis and G. apicola along the gut, we cut the 11 guts inoculated with engineered G. apis and G. apicola in the middle of the ileum and performed qPCR targeting G. apis-specific ureC and Gilliamella-specific 16S rRNA gene. After normalization based on 16S rRNA gene copy number, G. apis populations with ureC have significantly higher abundance in pylorus than in ileum (Wilcoxon signed-rank test P = 0.014) (SI Appendix, Fig. S8).
In addition to the spatial distribution of selected fluorescent Gilliamella strains, we observed the same pattern in 15 age-controlled bees with a conventional gut microbiome containing the native strain diversity; G. apis possessing ureC is significantly enriched in the pylorus relative to the ileum (Wilcoxon signed-rank test P = 0.012) (Fig. 6F and SI Appendix, Fig. S9). Our results suggest that G. apis and G. apicola colonize the gut in distinct ways: G. apis is strongly localized at the pylorus, and G. apicola colonizes the rest of the ileum and the rectum. Because urea is expected to be most concentrated at the pylorus, this distribution coincides with the difference in abilities of G. apis and G. apicola strains to utilize urea as a potential nitrogen source.
Discussion
The disruption of gene flow is critical for lineage diversification. In host-restricted bacteria, gene flow can be interrupted in two ways: isolation of symbionts in different hosts, following a host shift or divergence of host species, and niche partitioning within individual hosts, causing symbionts to be spatially isolated. The bee gut community provides an unusual opportunity to study the diversification of gut bacteria between and within host species. By analyzing the available 117 Gilliamella genomes and 57 Snodgrassella genomes from social bee species, we reconstructed the diversification of the bee gut microbiome between and within hosts.
Diversification of Gut Bacterial Populations between Hosts.
Consistent with the hypothesis that Gilliamella and Snodgrassella were acquired as gut symbionts in a shared ancestor of eusocial corbiculate bees (5), the phylogeny of Gilliamella and Snodgrassella is largely concordant with their host phylogeny with several exceptions. A. dorsata-derived Gilliamella was placed as the sister group to Gilliamella of other Apis species and Bombus species; Snodgrassella of A. cerana, Apis florea, and Apis andreniformis were placed as the sister group to Snodgrassella of all other A. mellifera- and Bombus-derived Snodgrassella strains based on core-gene trees (Fig. 1). This finding is consistent with previous trees based on core genes (35) but inconsistent with previous trees based on the 16S rRNA gene (38) or fast-evolving protein-coding genes (5). However, based on gene content, Apis-derived strains are more similar to each other than to the Bombus-derived strains (Fig. 2 and SI Appendix, Fig. S1).
Occasional gains and losses of symbionts by hosts can lead to incongruence between symbiont and host phylogenies. We hypothesize that the topology of the core-gene tree reflects gains and losses of Gilliamella and Snodgrassella by particular host lineages, despite overall long-term codiversification. In support of this hypothesis, Kwong et al. (5) have shown that eusocial corbiculate bees have gained and lost their gut associates multiple times. For example, A. dorsata appears to have lost its Snodgrassella, and one group of stingless bees, the genus Melipona, has entirely lost Snodgrassella and Gilliamella (39). A study of Snodgrassella strains in Bombus species showed that one Snodgrassella clade had a broad host range and could colonize hosts in several subgenera, indicating that switches between somewhat divergent host species are possible (40). Experimental transfer experiments showed that Snodgrassella can sometimes switch between Apis species but cannot switch between Apis and Bombus hosts (5, 28).
We further hypothesize that the topology of the gene content tree, which contrasts with the sequence-based tree in coinciding closely with host clades, indicates convergence in accessory gene sets to adapt to Apis hosts. For example, some colonization-beneficial genes of Snodgrassella identified in a mutagenesis study (31) are enriched in Apis-derived Snodgrassella strains. Kwong et al. (5, 28) have also shown that Snodgrassella from Apis species but not from Bombus species can colonize the A. mellifera gut, supporting the idea that Apis-derived Snodgrassella strains share similar functions absent from Bombus-derived strains.
Coexistence of Populations within Hosts.
Gut microbial communities harbor genetic variation within individual hosts (41–43). One basis of this variation may be the presence of different populations adapted to specific ecological niches (8, 44). Such niche partitioning, if associated with spatial separation, could lead to the formation of distinct populations with reduced gene flow (20, 21). We showed that a gene flow-based approach identifies distinct populations without setting a gANI threshold (Fig. 3). In support of the niche-partitioning hypothesis, specific accessory genes with different functions are enriched in different populations. Gilliamella populations differ in gene sets related to pectin degradation (Fig. 4) and sugar metabolism (SI Appendix, Fig. S3), supporting that G. apicola populations are the major degraders of pectin (30). These functional differences are supported by in vitro assays (27). G. apicola populations differ in genes for using the monosaccharide products of pectin degradation, and these differences are also supported by in vitro assays (35). In contrast, G. apis populations are enriched with only the transporters and the genes related to pectin-derived galacturonide metabolism. Potentially, they utilize different substrates from pollen and therefore can coexist in the same host, as documented for Lactobacillus species in bee guts (45).
Our results also support that different Gilliamella populations could interact differently with the host environment. Some populations of Gilliamella have lost T6SSs (SI Appendix, Fig. S6), which may benefit the colonization of bee guts (31), suggesting distinct colonization processes.
Nitrogen Utilization and the Spatial Distribution of Gut Bacterial Populations.
We found that G. apis populations harbor an 11-kb region containing 12 genes related to urea metabolism. On the symbiont side, amino acid nitrogen is limiting in the hindgut as it is absorbed by the host upstream in the midgut; this was verified by a mutagenesis study showing that Snodgrassella requires genes for biosynthesis of all protein amino acids in order to colonize bee guts (31). But, in contrast to the mammalian large intestine where overall nitrogen is likely limiting (46), insect hindguts have a supply of nitrogen. The Malpighian tubules deposit nitrogenous wastes into the gut immediately following the pyloric sphincter (Fig. 6). We hypothesized that G. apis populations utilize urea derived from host waste, and therefore colonize most abundantly near the pylorus. Using engineered fluorescent bacteria, we showed that a G. apis strain possessing urea utilization genes localized to the pylorus region of the honey bee gut. We observed the same pattern in bees containing native strain diversity: G. apis strains with ureC genes are enriched in the pylorus region. The urea transporters and urease genes possessed by G. apis are predicted to allow rapid acquisition and use of urea, a capability lacking in G. apicola. We hypothesize that members of the gut community unable to utilize urea instead depend on nitrogen-containing compounds, such as amino acids, produced by members such as G. apis and Snodgrassella that are able to use it. This dependence may explain why G. apicola colonizes more readily in the presence of Snodgrassella than alone. Similarly, members of G. apis population 1 show variations in the presence of urea transporters and urease genes (Fig. 5A), although their genomes show high similarity (Fig. 1) and gene flow (Fig. 3). This finding suggests that members of population 1 could utilize urea differently. G. apis population 1 strains without urease genes could utilize nitrogen-containing compounds produced by other members in the population. The selective pressure on utilizing urea is not strong enough for the urea metabolism-related genes to quickly spread through the population (47).
Potentially, urea utilization also results in nitrogen recycling at the level of the host. Nitrogen acquisition is critical for herbivorous insects, as plant material is nitrogen-poor. In some other insects—including ants, termites, cockroaches, and aphids—symbionts are able to recycle nitrogenous waste to improve the host nitrogen budget (48, 49), but this has not yet been demonstrated in bees.
In Snodgrassella, the distribution of urea utilization genes across strains also matches the host distribution (Fig. 5B), suggesting that nitrogenous waste utilization is critical for host adaptation. Consistent with this hypothesis, the ureDG genes were found to be beneficial to Snodgrassella colonization in honey bee guts (31).
By using engineered fluorescent bacteria, we were able to image bacteria during colonization, without fixation or processing protocols that may distort tissue morphology or disrupt biofilms. Because sectioning is also not required, it is feasible to inspect multiple gut regions or even the whole gut for colonization, rather than thin sections. The engineered strains of Snodgrassella and G. apicola appear to colonize similarly to wild-type strains within native gut communities, based on microscopy using fluorescent in situ hybridization (19, 37). The results of qPCR assays based on bees containing native strain diversity (Fig. 6) further confirm the spatial distribution of natural Gilliamella populations. Our experiments captured a single snapshot of G. apis and G. apicola colonization. Future studies could investigate how G. apis, and other gut bacterial strains, spatially organize on different timescales or under different conditions.
Our findings implicate spatial barriers within host guts in limiting gene flow in modern populations of Gilliamella. We note that G. apicola and G. apis routinely occur with the same individual bees and that analyzed genomes of both were sampled from multiple continents. Thus, neither geographic separation of host populations nor confinement to different host individuals contributes to the maintenance of these genetically distinct populations. Potentially, divergence of G. apicola and G. apis began during a period when their ancestors were restricted to geographically separate host populations, although this possibility would not explain why they have acquired distinct ecological niches within individual guts. A plausible scenario is that acquisition of ecologically impactful genes, such as those for urea utilization, caused a Gilliamella strain to localize to a new gut location, such as the pylorus. This shift would then impose spatial barriers to DNA exchange, resulting in genome-wide sequence divergence as mutations are fixed in each lineage. At some divergence level, gene flow due to homologous recombination ceases because of requirements of recombination processes, such as that mediated by RecA (50).
In summary, combining comparative genomics and microscopy with engineered fluorescent bacteria, we showed population-level diversification of gut bacterial species between and within corbiculate bee host species. In Gilliamella and Snodgrassella, two clades of core bee symbionts, both host speciation and niche partitioning have contributed to diversification of gut bacterial species. Acquisition of functional genes can help with adapting to specific hosts and ecological niches. Niche partitioning can further lead to spatial differentiation in the gut, potentially leading to disruption of gene flow and speciation.
Materials and Methods
Detailed materials and methods are available in SI Appendix, Supplementary Materials and Methods.
Data Collection, Genome Annotation, and Phylogenetic Reconstruction.
We downloaded available Gilliamella and Snodgrassella genomes from the NCBI. After removing low quality and contaminated genomes, we annotated the genomes and assigned genes into orthologous groups using anvi’o (51). We performed functional annotations using GhostKoala (52) for biological pathways, dbCan2 (53) for CAZyme genes, and CRISPRCasFinder (54) for CRISPR spacers. To investigate functional genes enriched in different lineages and populations, we used a generalized linear model with the logit linkage function implemented in anvi’o to compute an enrichment score and P value for each biological function and orthologous group.
We defined core-gene families as single-copy gene families that existed in at least 80% of the species (i.e., 94 of the 117 Gilliamella genomes or 46 of the 57 Snodgrassella genomes). We aligned and concatenated nucleotide sequences of core genes and constructed the phylogenetic trees using IQ-TREE (55).
Population Delineation Based on Gene Flow.
To delineate populations in Gilliamella and Snodgrassella, we used PopCOGenT (21) to estimate the amount of recombination among strains. Recombination between divergent genomes with low gANI was verified by investigating identical regions between genomes and gene-specific sweeps in the core and flexible genome using PopCOGenT (SI Appendix, Supplementary Materials and Methods and Fig. S11). To assess whether vertical descent substitutions could give a false signal of population delineation, we simulated genomes with the same divergence but without homologous recombination using Seq-Gen (56). The results of PopCOGenT showed that simulated genomes (with ≥0.0355% divergence) do not have evidence of homologous recombination with each other (SI Appendix, Fig. S10). Thus, the observed populations in the Gilliamella and Snodgrassella genomes reflect homologous recombination.
Gut Colonization Experiment, Microscopy, and qPCR Analyses.
We first confirmed the location of G. apis and G. apicola populations along with Snodgrassella using engineered strains expressing three different fluorescent proteins (37). We performed three independent experiments by collecting 280 bees emerged overnight from brood frames and inoculating them with different strain combinations (57). After 7 d, we imaged 42 whole guts using a Zeiss 710 Laser Scanning Confocal Microscope.
We also tested the spatial distributions of G. apis and G. apicola in bees with a conventional gut microbiome. In order to control for age, we collected around 80 bees that emerged overnight from one brood frame in the laboratory. We exposed these newly emerged bees to gut homogenates from 12 forager bees, a method previously shown to establish a large and diverse gut community indistinguishable from a native gut community (34). At 7 d after inoculation, we dissected the bee guts to quantify the abundance of G. apis and G. apicola in different gut regions. We divided guts into pylorus and distal ileum samples by cutting in the middle of the ileum on 11 guts inoculated with engineered strains and 24 guts inoculated with the conventional community. We performed DNA extraction and qPCR to quantify ureC genes of G. apis and 16S rRNA genes of both G. apis and G. apicola. To control for the abundance of Gilliamella between pylorus and ileum, we normalized the ureC copy number by dividing it by total Gilliamella 16S rRNA gene copy number. For bees with the conventional community, we only used guts colonized by both G. apis and G. apicola, including 15 bees with ureC gene copy number ≥ 1,000 for ileum and 16S gene copy number ≥ 1,000 for both pylorus and distal ileum. We then performed a Wilcoxon signed-rank test between pylorus and ileum samples using wilcox.test function in R.
Supplementary Material
Acknowledgments
We thank Kim Hammond for taking care of the hives used in this work and for her assistance with figure preparation; John Jones and Sydney Jones for use of property in Driftwood, Texas; and Daniel Weaver for helping to set up the Driftwood hives. This work was supported by NIH Grant R35GM131738 (to N.A.M.).
Footnotes
Competing interest statement: S.P.L. and N.A.M. have filed a patent application on the commercial use of engineered gut bacteria to improve honey bee health. J.E.P., S.P.L., and N.A.M. have filed a patent application on the commercial use of probiotics to improve honey bee health.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115013119/-/DCSupplemental.
Data Availability
Data and code used in this study, including genome assemblies, gene annotations, orthologous groups, and functional annotations are available on GitHub (https://github.com/lyy005/bee_gut_bacteria) and Zenodo (https://zenodo.org/record/5209528).
References
- 1.Ley R. E., Peterson D. A., Gordon J. I., Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Moeller A. H., et al. , Cospeciation of gut microbiota with hominids. Science 353, 380–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flint H. J., Bayer E. A., Rincon M. T., Lamed R., White B. A., Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6, 121–131 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Brune A., Dietrich C., The gut microbiota of termites: Digesting the diversity in the light of ecology and evolution. Annu. Rev. Microbiol. 69, 145–166 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Kwong W. K., et al. , Dynamic microbiome evolution in social bees. Sci. Adv. 3, e1600513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain C., Rodriguez-R L. M., Phillippy A. M., Konstantinidis K. T., Aluru S., High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mark Welch J. L., Hasegawa Y., McNulty N. P., Gordon J. I., Borisy G. G., Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl. Acad. Sci. U.S.A. 114, E9105–E9114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereira F. C., Berry D., Microbial nutrient niches in the gut. Environ. Microbiol. 19, 1366–1378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coyte K. Z., Rakoff-Nahoum S., Understanding competition and cooperation within the mammalian gut microbiome. Curr. Biol. 29, R538–R544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldford J. E., et al. , Emergent simplicity in microbial community assembly. Science 361, 469–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Rossum T., Ferretti P., Maistrenko O. M., Bork P., Diversity within species: Interpreting strains in microbiomes. Nat. Rev. Microbiol. 18, 491–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rayssiguier C., Thaler D. S., Radman M., The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342, 396–401 (1989). [DOI] [PubMed] [Google Scholar]
- 13.Dykhuizen D. E., Green L., Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 173, 7257–7268 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayr E., “The biological species concept” in Species Concepts and Phylogenetic Theory: A Debate, Wheeler Q. D., Meier R., Eds. (Columbia University Press, New York, 2000), pp. 17–29. [Google Scholar]
- 15.Funk D. J., Helbling L., Wernegreen J. J., Moran N. A., Intraspecific phylogenetic congruence among multiple symbiont genomes. Proc. Biol. Sci. 267, 2517–2521 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moeller A. H., et al. , Social behavior shapes the chimpanzee pan-microbiome. Sci. Adv. 2, e1500997 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hongoh Y., et al. , Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol. Ecol. 15, 505–516 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Sabree Z. L., et al. , Genome shrinkage and loss of nutrient-providing potential in the obligate symbiont of the primitive termite Mastotermes darwiniensis. Appl. Environ. Microbiol. 78, 204–210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinson V. G., Moy J., Moran N. A., Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 78, 2830–2840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bobay L.-M., Ochman H., Biological species are universal across life’s domains. Genome Biol. Evol. 9, 491–501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arevalo P., VanInsberghe D., Elsherbini J., Gore J., Polz M. F., A reverse ecology approach based on a biological definition of microbial populations. Cell 178, 820–834. e14 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Bobay L.-M., Ochman H., Biological species in the viral world. Proc. Natl. Acad. Sci. U.S.A. 115, 6040–6045 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., O’Donnell A. C., Ochman H., Discriminating arboviral species. J. Gen. Virol. 102, 001572 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bobay L.-M., Ellis B. S.-H., Ochman H., ConSpeciFix: Classifying prokaryotic species based on gene flow. Bioinformatics 34, 3738–3740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludvigsen J., Porcellato D., Amdam G. V., Rudi K., Addressing the diversity of the honeybee gut symbiont Gilliamella: Description of Gilliamella apis sp. nov., isolated from the gut of honeybees (Apis mellifera). Int. J. Syst. Evol. Microbiol. 68, 1762–1770 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Ellegaard K. M., Suenami S., Miyazaki R., Engel P., Vast differences in strain-level diversity in the gut microbiota of two closely related honey bee species. Curr. Biol. 30, 2520–2531.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel P., Martinson V. G., Moran N. A., Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. U.S.A. 109, 11002–11007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong W. K., Engel P., Koch H., Moran N. A., Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl. Acad. Sci. U.S.A. 111, 11509–11514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steele M. I., Kwong W. K., Whiteley M., Moran N. A., Diversification of type VI secretion system toxins reveals ancient antagonism among bee gut microbes. MBio 8, e01630-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng H., et al. , Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. U.S.A. 116, 25909–25916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell J. E., Leonard S. P., Kwong W. K., Engel P., Moran N. A., Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc. Natl. Acad. Sci. U.S.A. 113, 13887–13892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellegaard K. M., Engel P., Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 10, 446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carpenter M. H., Harpur B. A., Genetic past, present, and future of the honey bee (Apis mellifera) in the United States of America. Apidologie (Celle) 52, 63–79 (2021). [Google Scholar]
- 34.Zheng H., Powell J. E., Steele M. I., Dietrich C., Moran N. A., Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. U.S.A. 114, 4775–4780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng H., et al. , Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. MBio 7, e01326-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makarova K. S., et al. , Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leonard S. P., et al. , Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth. Biol. 7, 1279–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koch H., Abrol D. P., Li J., Schmid-Hempel P., Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 22, 2028–2044 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Cerqueira A. E. S., et al. , Extinction of anciently associated gut bacterial symbionts in a clade of stingless bees. ISME J. 15, 2813–2816 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powell E., Ratnayeke N., Moran N. A., Strain diversity and host specificity in a specialized gut symbiont of honeybees and bumblebees. Mol. Ecol. 25, 4461–4471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaiber A., et al. , Functional and genetic markers of niche partitioning among enigmatic members of the human oral microbiome. Genome Biol. 21, 292 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilbert S. A., Mark Welch J. L., Borisy G. G., Spatial ecology of the human tongue dorsum microbiome. Cell Rep. 30, 4003–4015.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olm M. R., et al. , inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat. Biotechnol. 39, 727–736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louca S., et al. , Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2, 936–943 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Brochet S., et al. , Niche partitioning facilitates coexistence of closely related honey bee gut bacteria. eLife 10, e68583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reese A. T., et al. , Microbial nitrogen limitation in the mammalian large intestine. Nat. Microbiol. 3, 1441–1450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cordero O. X., Polz M. F., Explaining microbial genomic diversity in light of evolutionary ecology. Nat. Rev. Microbiol. 12, 263–273 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Hu Y., et al. , Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome. Nat. Commun. 9, 964 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen A. K., Pers D., Russell J. A., “Symbiotic solutions to nitrogen limitation and amino acid imbalance in insect diets” in Advances in Insect Physiology, Oliver K. M., Russell J. A., Eds. (Elsevier, 2020), vol. 58, pp. 161–205. [Google Scholar]
- 50.Watt V. M., Ingles C. J., Urdea M. S., Rutter W. J., Homology requirements for recombination in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 82, 4768–4772 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eren A. M., et al. , Anvi’o: An advanced analysis and visualization platform for ’omics data. PeerJ 3, e1319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M., Sato Y., Morishima K., BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Zhang H., et al. , dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46 (W1), W95–W101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Couvin D., et al. , CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46 (W1), W246–W251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minh B. Q., et al. , IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rambaut A., Grassly N. C., Seq-Gen: An application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 13, 235–238 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Leonard S. P., et al. , Engineered symbionts activate honey bee immunity and limit pathogens. Science 367, 573–576 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code used in this study, including genome assemblies, gene annotations, orthologous groups, and functional annotations are available on GitHub (https://github.com/lyy005/bee_gut_bacteria) and Zenodo (https://zenodo.org/record/5209528).