Significance
B cell clonal selection and expansion from a genetically diverse antibody repertoire guides the immune response to a target antigen. It remains unclear if clonal selection and expansion follow any deterministic rules or are stochastic with regards to phenotypic antibody properties such as antigen-binding, affinity, and epitope specificity. We perform the in-depth genotypic and phenotypic characterization of antibody repertoires following immunization in mice. We identify the degree to which clonal expansion is driven by antibody binding, affinity, and epitope specificity and as such may provide greater insight into vaccine-induced immunity.
Keywords: repertoire, antibody, epitope, specificity, sequencing
Abstract
The capacity of humoral B cell-mediated immunity to effectively respond to and protect against pathogenic infections is largely driven by the presence of a diverse repertoire of polyclonal antibodies in the serum, which are produced by plasma cells (PCs). Recent studies have started to reveal the balance between deterministic mechanisms and stochasticity of antibody repertoires on a genotypic level (i.e., clonal diversity, somatic hypermutation, and germline gene usage). However, it remains unclear if clonal selection and expansion of PCs follow any deterministic rules or are stochastic with regards to phenotypic antibody properties (i.e., antigen-binding, affinity, and epitope specificity). Here, we report on the in-depth genotypic and phenotypic characterization of clonally expanded PC antibody repertoires following protein immunization. We find that clonal expansion drives antigen specificity of the most expanded clones (top ∼10), whereas among the rest of the clonal repertoire antigen specificity is stochastic. Furthermore, we report both on a polyclonal repertoire and clonal lineage level that antibody-antigen binding affinity does not correlate with clonal expansion or somatic hypermutation. Last, we provide evidence for convergence toward targeting dominant epitopes despite clonal sequence diversity among the most expanded clones. Our results highlight the extent to which clonal expansion can be ascribed to antigen binding, affinity, and epitope specificity, and they have implications for the assessment of effective vaccines.
Humoral immunity and successful vaccination require the generation of sustained levels of circulating serum antibodies, which are produced by clonally expanded plasma cells (PCs), a terminally differentiated subset of B cells that reside in lymphoid organs (e.g., bone marrow) for an extended period of time (up to years for mice and humans) (1–5). This dynamic process involves the recombination of germline-encoded genetic elements that encode the antibody (or B cell receptor [BCR]) in single B cells (6); dogma holds that B cell clonal selection, iterative expansion, and differentiation to PCs occur for clones with increased affinity toward the antigen (7–12). While there have been numerous studies describing how this process is orchestrated on the genotypic level in several species (e.g., humans, mice, and zebrafish) (13–21), much less is known about the associated phenotypic antibody repertoire metrics comprising features such as antigen-binding (22–25), quantitative binding affinity, and epitope specificity, which can physically be measured as a consequence of the antibody amino acid (aa) sequence composition. Importantly, most of the studies reporting phenotypic antibody repertoire data were confined to memory B cells or short-lived plasma blasts that express surface BCR, and in contrast to PCs, do not secrete large amounts of antibody proteins (immunoglobulin [Ig]) (22–24, 26–30).
Previous studies on vaccine-induced PC repertoires (murine- and bone marrow-derived) have found that they are dominated by a few (∼3–5) highly expanded clones that are antigen-specific (19, 31), which correlates with the observation that up to 60–90% of the total antigen-specific IgG serum repertoire is comprised of only a few clones (∼4–12) (32–34). However, it remains unclear whether any deterministic factors, such as antigen affinity or epitope specificity, drive the selection of these highly expanded clones and how deep antigen specificity tracks within the PC repertoire. Previous results obtained from adoptive B cell transfer and immunization experiments in BCR transgenic mouse models (monoclonal antibody knockins) revealed that high affinity BCRs promote early splenic B cells to differentiate to PCs (35, 36), a phenomenon that was also reported in a more recent study (37). However, contrasting work showed that instead of PC differentiation, the onset of which was later found to occur during late germinal center reactions (38), higher BCR affinity led to increased overall proliferation of antigen-reactive cells (39). Therefore, it remains unclear how transferable phenotypic antigen binding data from B cells of isolated germinal centers or individual lymph nodes (40, 41) are for the development of long-lived PC repertoires.
Here, we set out to comprehensively address these long-standing questions of clonal selection and expansion of PCs. We developed an integrative genotype-phenotype mapping approach and applied it to the in-depth characterization of clonally expanded PC antibody repertoires across five immunized mice. We used single-cell sequencing and computational analysis of antibody repertoires combined with quantitative antibody-antigen-binding, antigen-affinity, epitope-binning, and antigen-mapping measurements (Fig. 1A) which resulted in the characterization of >230 antibodies (which constitute ∼50% of all captured PCs [IgG] per repertoire).
Fig. 1.
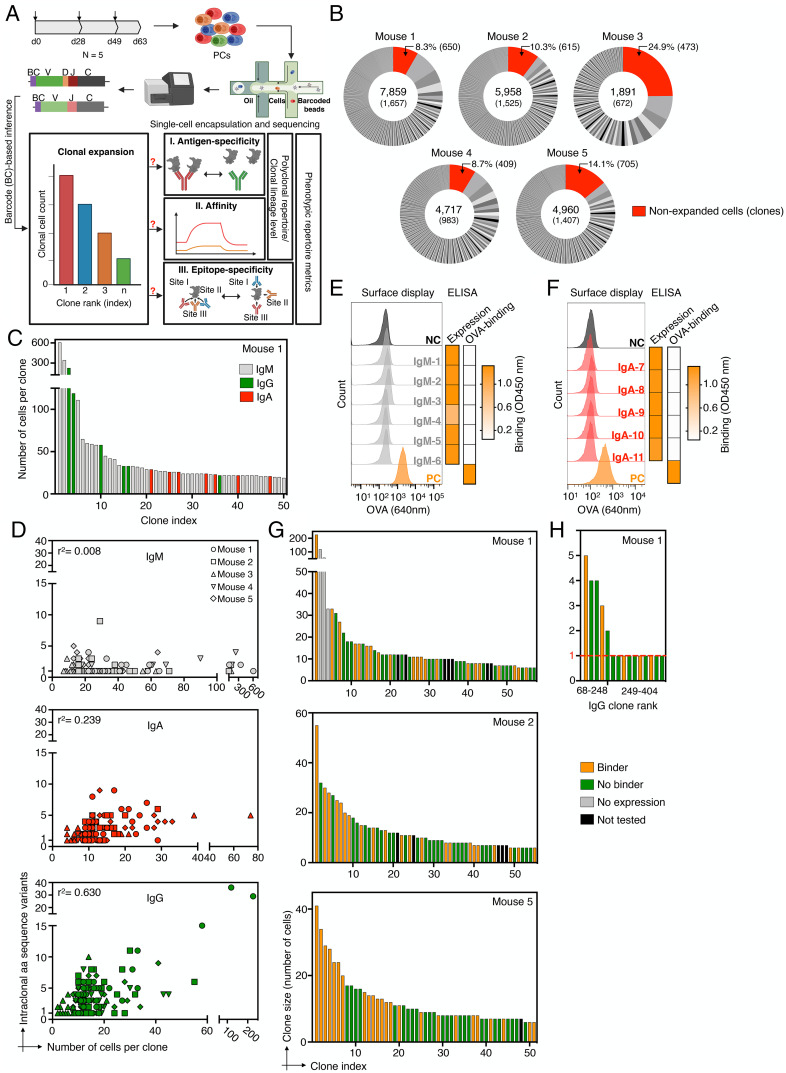
The antigen specificity of clonally expanded plasma cell antibody repertoires. (A) Schematic project outline. Arrows on top of the timeline graph (top left) indicate time points of mouse immunization. (B) Pie charts indicate the fraction of productive plasma cells (PCs) per clone and mouse captured by single-cell sequencing (sc-seq). Numbers in the center indicate the total number of productive cells (Top) and clones (Bottom). Unexpanded clones are shown in red and their cellular percentages and total numbers are indicated on top. Clone definition is based on an unique CDRH3-CDRL3 amino acid (aa) sequence. (C) Representative clonal expansion profile for the 50 most expanded clones of Mouse 1 (MS-1). Clones are colored by isotype majority and color-coded in gray (IgM), green (IgG), and red (IgA). Profiles of all mice are shown in SI Appendix, Fig. S5. (D) Correlation between the number of intraclonal antibody sequence variants (aa) and the number of cells per clone for the 30 most expanded clones per isotype. (E, F) Screening of six and five expanded IgM and IgA clones of MS-1 for antigen binding. Left: flow cytometry histogram plot shows ovalbumin (OVA) labeling of hybridoma cell lines with stable surface expression of selected antibody clones (from IgM or IgA PCs) or expression of positive or negative controls (antibodies with defined binding to OVA [PC, orange] or to hen egg lysozyme [NC, dark gray]). Right: heatmaps indicate antibody expression and binding to OVA based on endpoint ELISA (data obtained from SI Appendix, Figs. S7D and S8B). (G) Antigen-specificity profiling of the top 50–60 expanded clones of MS-1, MS-2, and MS-5. Clones with an endpoint ELISA signal >0.2 (threefold above background) are designated as antigen binders (SI Appendix, Fig. S9A). (H) Antigen specificity of clones showing lower to no clonal expansion (cell count = 1, red dotted line) from MS-1.
Results
To this end, we repeatedly immunized five BALB/c mice subcutaneously with the T cell-dependent model antigen ovalbumin (OVA) in monophosphoryl lipid A (MPLA) adjuvant. By performing enzyme-linked immunosorbent assay (ELISA) on serum, we observed robust OVA-specific IgG titers, weak IgM titers, and undetectable IgA titers (SI Appendix, Fig. S1). PCs were isolated (based on CD138+ B220low/− CD19low/− surface expression) from bone marrow 2 wk after the final boost, which represents the peak of PC homing to the bone marrow (42, 43) (Fig. 1A). Next, targeted single-cell sequencing was performed on uniquely barcoded heavy and light chain transcripts, covering all Ig isotypes (10X Genomics V(D)J protocol) (SI Appendix, Fig. S2C). After quality filtering and bioinformatic removal of multiplets, we obtained sequence data on thousands of PCs for each mouse (1891–7859 cells) with one paired full-length variable heavy (VH)-chain and variable light (VL)-chain sequence (SI Appendix, Fig. S2B), which translated to 672–1657 distinct clones (a clone being defined as a unique CDRH3-CDRL3 aa sequence) per mouse (across all isotypes) (Fig. 2B and SI Appendix, Fig. S2C). It should be noted that the captured number of cells only represent a small sample size of the total PC repertoire (3–11%), which makes it difficult to interpret if particular outcomes of this study are representative across the whole repertoire. While total repertoire diversity is under sampled, as indicated by new clones continuing to appear while subsampling, we estimated overall repertoire diversity using both rarefaction analyses and a modified maximum-likelihood method (Recon) (44), which suggests that predictions of clone size distribution was robust (SI Appendix, Fig. S3 A and B). To characterize the state of clonal expansion for each repertoire, we operated under the assumption that since the murine naive BCR sequence diversity (1013) (16) far exceeds the steady-state number of PCs in the bone marrow of a given mouse (∼7.5 × 104) (43), detecting two or more cells with the same clonal CDRH3-CDRL3 sequence could be used to define clonal expansion (irrespective of isotype origin) (45, 46). We found 199–1007 clones to be expanded for each mouse repertoire (30–61% of all clones per repertoire), whereas a substantial proportion of clones was detected only once (Fig. 1B and SI Appendix, Fig. S4). Furthermore, all PC repertoires were reproducibly dominated by highly expanded IgM clones (Fig. 1C and SI Appendix, Fig. S5), some of which made up almost 8% of the total cellular repertoire count (IgM-1). There was a variable degree of highly expanded IgG and IgA clones, depending on the mouse, suggesting usage of these isotypes may not be driven directly by the antigen-adjuvant immunization scheme (SI Appendix, Fig. S5A). As expected, the mutational load between isotypes differed significantly when the 30 most expanded clones per isotype were compared, with most of the IgM clones featuring only a single aa intraclonal variant, whereas the number of intraclonal variants markedly increased for expanded IgA and IgG clones (Fig. 1D and SI Appendix, Figs. S5B and S6). Interestingly, when we tested six of the most expanded IgM clones of a single mouse, some of which were also shared between mice, we did not observe any detectable binding to the OVA antigen (or MPLA adjuvant) (Fig. 1E and SI Appendix, Fig. S7). Likewise, when we screened five of the most expanded IgA clones of the same mouse, no antigen-binding was detected (Fig. 1F and SI Appendix, Fig. S8). Therefore, our data suggests that highly expanded antigen-specific IgM and IgA long-lived PCs (CD19 low) were absent in the investigated repertoires.
Fig. 2.
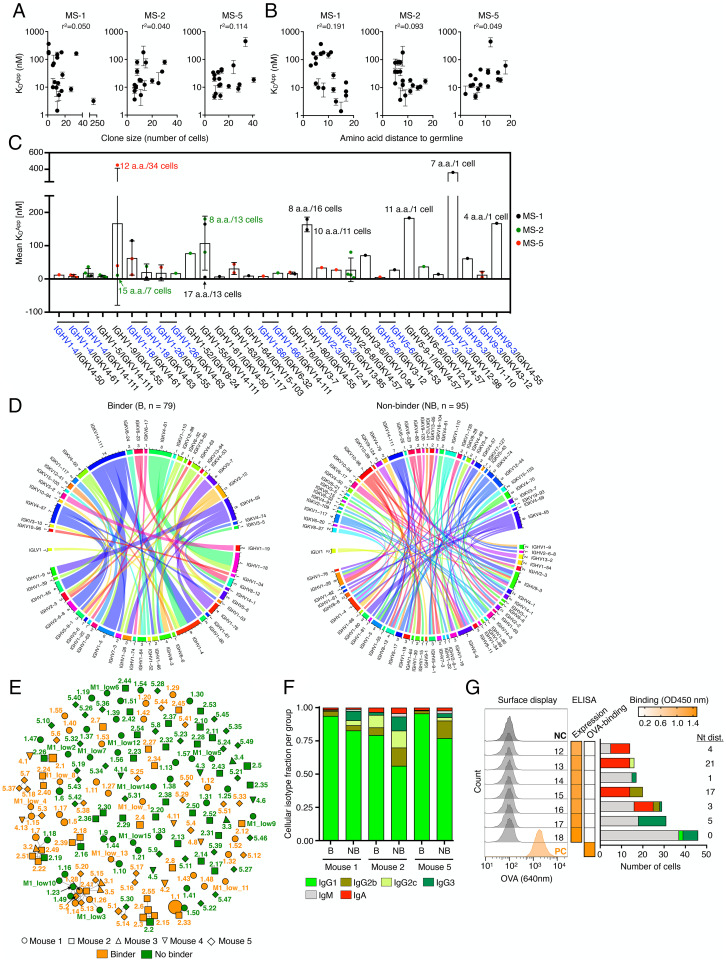
Genotype-phenotype correlations of polyclonal antigen-specific plasma cell repertoires. (A, B) Correlation between clonal apparent dissociation constant (Kd) and clone size (number of cells per clone, A) as well as clonal amino acid (aa) distance to germline (B) for MS-1, MS-2, and MS-5. Error bars indicate SD (n = 2–3 measurements of Kd). (C) Correlation between Kd and VH-VL germline V-gene usage. V-gene pairs featuring shared VH are indicated in blue with a shared horizontal bar on top. Some data points at the extremes are additionally labeled with the aa distance to germline and number of cells for their respective clones. Error bars indicate SD. (D) Circos plots show the diversity of VH-VL-gene pairings in the antigen-binding and nonbinding group of clones. Each line indicates the germline V-gene usage for an individual VH-VL gene pair. The inner track number and the corresponding thickness of the bar indicate the number of clones utilizing a given germline gene. Color corresponds to the respective germline gene. (E) Similarity network plot for all 174 antigen-binders and nonbinders tested across all mice. Edges represent clones separated by edit distance of <4 aa in CDRH3-CDRL3 sequences. Extent of clonal expansion is reflected by the size of the nodes. Annotated labels of each node are according to clone ID in SI Appendix, Tables S1 and S2. (F) Cellular isotype fractions for all binders (B) and nonbinders (NB) per mouse. Increased IgG1 (P adjusted = 0.017) was detected in the binder relative to the nonbinder groups as calculated by ANOVA using Sidak multiple comparison test. (G) Validation of multi-isotype clones by hybridoma surface staining and ELISA screening. Left and Middle: flow cytometry histograms and heatmaps similar to as shown in Fig. 1 E and F (ELISA data shown in SI Appendix, Fig. S14). Right: cellular isotype composition of each clone and associated nt distance from germline.
Next, we focused our analysis on the expanded IgG compartment and determined the antigen specificity (by ELISA) of the 50–60 most expanded antibody clones from mouse MS-1, MS-2, and MS-5 (which were chosen based on their respective sequencing depth), along with 15 of the most expanded clones from MS-3 and MS-4 (SI Appendix, Fig. S9 A and B). Together, these clones represented 50–54% of total IgG cells per mouse for MS-1, MS-2, and MS-5, and overall, we found that between 42% and 48% of clones per mouse were antigen-specific (SI Appendix, Fig. S9A; 60% for MS-3 and MS-4). While the most expanded five to ten clones per repertoire were usually antigen-specific, across the rest of the repertoire, antigen-binding and nonbinding clones were evenly distributed (Fig. 1G). In light of this high degree of stochasticity, we determined how deep antigen specificity tracks into the repertoire by testing a set of 15 randomly chosen IgG clones from MS-1, including clones that had a cell count as low as one. Even this deep into the repertoire, several clones corresponding to IgG clone rank 249–404 and total clone rank 1008–1657 showed detectable antigen-binding (Fig. 1H and SI Appendix, Fig. S9 C and D). To further assess the robustness of our antigen-binding classification, we included an additional panel of 10 antibodies with known specificity to human TNFR2 (47) and observed they did not demonstrate any detectable binding to OVA (SI Appendix, Fig. S10).
Next, given the context of a polyclonal, multi-epitope-directed antibody repertoire, we investigated if clonal expansion of PCs is driven by the affinity to the cognate antigen. To this end, we used biolayer interferometry (BLI) to measure the affinity (apparent equilibrium dissociation constants [Kd]) of 55 antigen-specific IgG clones from MS-1, MS-2, and MS-5, resulting in a wide range of affinities (Kd = 2–450 nM) (SI Appendix, Fig. S11 and Table S1). This revealed that there was no direct correlation between clonal expansion (based on cell count of a clone) and antigen affinity, as highly expanded clones (cell count >20) were just as likely to have moderate (Kd ∼100–450 nM) or high (Kd ∼10–100 nM) affinities, including the surprising observation that the highest affinity (Kd < 10 nM) clones often had low cell counts (<10) (Fig. 2A). Likewise, we observed highly expanded clones with significantly disparate affinities, as exemplified by the two most expanded clones of MS-5 (5.1 and 5.2), which displayed Kd values of 18.7 nM and 451.8 nM, respectively (SI Appendix, Table S1). Moreover, we did not observe a direct link between antibody-antigen affinity and the extent of somatic hypermutation, neither on the aa (Fig. 2B) nor on the nucleotide (nt) level (SI Appendix, Fig. S11). In some cases, clones that were closer to germline such as the fourth most expanded clone of MS-5 (5.4, 7 aa mutations) exhibited high affinity (Kd = 8.8 nM), whereas in comparison a highly mutated and highly expanded clone (5.1, 16 aa mutations) had a lower affinity (Kd = 18.7 nM) (SI Appendix, Table S1). For some antigens, such as influenza hemagglutinin and the spike protein of severe acute respiratory syndrome coronavirus 2, it has been shown that certain germlines are structurally predisposed for antigen specificity and high affinity (48–50). Therefore, we determined whether VH-VL germline gene (IGHV-IGKV) combinations impacted the affinity of a given antibody to OVA. For the 31 antigen-specific germline combinations tested, some of which shared the same VH or VL genes, we found that affinity was independent of germline gene usage. Most affinities fell into the same range, and in case of multiple data points per VH-VL combination, the standard deviation for affinity could span a large range (Kd ± 250 nM) (Fig. 2C). Next, we determined if certain VH-VL germline combinations were enriched in the antigen-binding or nonbinding fraction of the tested clones. Analysis of VH-VL germline gene usage by circos plots of the 79 experimentally verified antigen-binding and 95 nonbinding clones did not show enrichment for certain germline combinations in either of the two groups (Fig. 2D). Next, we performed a sequence-similarity network analysis to investigate convergence of IgG clones across all mice (MS-1–MS-5; SI Appendix, Fig. S13) as well as on all experimentally characterized clones. We therefore calculated similar networks using the edit distance of the CDRH3 and CDRL3 sequences and connected those clones that were separated by less than four aa mutations across the summed CDRH3 and CDRL3 distances (51). This revealed the presence of discrete subnetworks (SI Appendix, Fig. S12 and Fig. 2E), which harbored clusters of both antigen-binding and nonbinding clones, highlighting the potential importance of key amino acid sequence motifs beyond the CDRH3-CDRL3 that contribute to antigen specificity (52).
To evaluate whether there were any other features that separate antigen-binding from nonbinding clones, we further analyzed common metrics such as distance to germline, number of intraclonal sequence variants, and CDRH3/CDRL3 length and found that only the number of distinct intraclonal antibody sequence variants was significantly increased for antigen-binding clones (SI Appendix, Fig. S14). Upon closer examination of Ig isotypes (IgM and IgA) and IgG subtypes (IgG1, IgG2b, IgG2c, and IgG3), we observed a consistently higher percentage of IgG1 cells (P adjusted = 0.017) among the antigen-binding fraction of clones (Fig. 2F). When we tested seven clones that were composed of several isotypes (with minimal to no IgG1), none of them showed detectable antigen binding, suggesting that multi-Ig class-switch recombination does not correlate with antigen specificity (Fig. 2G and SI Appendix, Fig. S14).
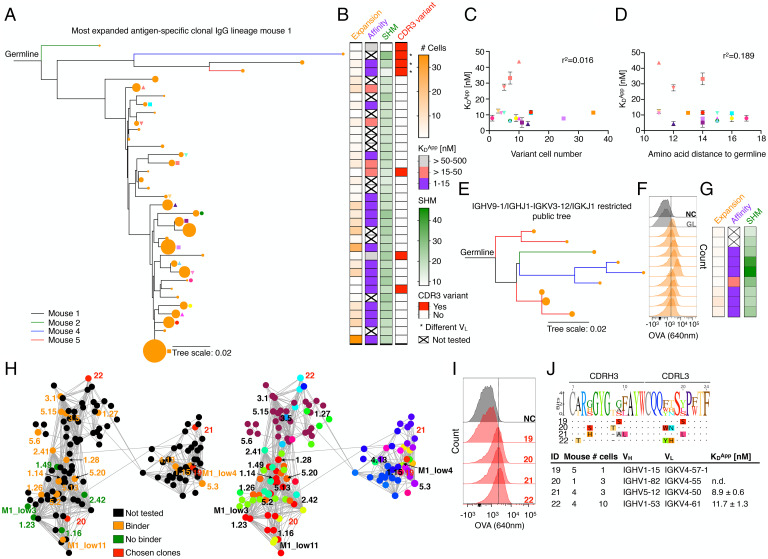
Since we established that within the context of the polyclonal repertoire, selection of highly expanded clones was not driven by antigen binding affinity, we next sought to determine the relationship between antigen affinity and clonal expansion within the evolutionary trajectory of a clonal lineage (PCs sharing identical V- and J-genes and identical CDRH3-CDRL3 sequences). We thus expressed antibodies and measured affinities for the most expanded IgG clonal lineage of MS-1, which featured 29 intraclonal sequence variants distributed across a total of 227 cells (Fig. 3A and SI Appendix, Fig. S15 A and B). We observed no direct correlation between expansion of an intraclonal sequence variant and binding affinity to antigen. Moreover, the number of somatic hypermutations (distance to germline) did not correlate with affinity and intraclonal variants with lower affinity (Kd > 15–50 nM) generally originated from earlier ancestral branch points of the lineage tree (Fig. 3 B–D and SI Appendix, Fig. S16). This provides further evidence for the existence of a clonal physiological affinity ceiling, as has been proposed in previous work (24, 53), and which falls within the range of affinities (Kd = 2–450 nM) measured for polyclonal antibodies (Fig. 2C).
Fig. 3.
Phenotypic antibody profiling within plasma cell clonal lineages. (A) Phylogenetic lineage tree of the most expanded IgG clone of MS-1 (SI Appendix, Fig. S15). Related clones from different mice are indicated by different colors. The size of the orange nodes at the tip of each branch indicates the number of cells per intraclonal variant. Shapes indicate identity of intraclonal variants plotted in (C) and (D). (B) Left to Right: heatmaps correspond to intraclonal variant expansion (number of cells per variant), binding affinity (Kd), somatic hypermutations (SHM; nucleotide distance to germline), and CDR3 variants (1–6 aa edit distance in CDRH3-CDRL3). Clones featuring a different VL are marked by an asterisk. Intraclonal variants from top to bottom correspond to lineage tree variants, as shown in (A). (C) Correlation between apparent dissociation constant (Kd) and intraclonal variant cell number for all variants indicated in (A). Error bars indicate SD (n = 3–5 measurements of Kd). (D) Correlation between Kd and amino acid distance to germline. Error bars indicate SD (n = 3–5 measurements of Kd). (E) Phylogenetic lineage tree of clones originating from several mice that have similar sequences (identical V- and J-genes, CDRH3-CDRL3 with <5 aa difference) (SI Appendix, Fig. S17 A–C). Branch colors reflect mouse ID from (A) and node sizes reflect clone size. (F) Flow cytometry histograms for OVA binding, similar to Figs. 1E and 2F. GL denotes germline, clones correspond to lineage tree shown in (E). (G) Heatmap shown is similar to Fig. 3B. (H) Network plots of connected IgG sequence nodes from all mice harboring various VH-VL gene combinations. Edges represent clones separated by edit distance of three or less aa based on the concatenated CDR3 sequences. Left: verified binders, nonbinders, not tested clones as well as newly chosen clones are shown. Clone ID according to SI Appendix, Tables S1 and S2. Right: VH-VL gene usage visualization. Color code according to SI Appendix, Fig. S18C. (I) Flow cytometry histograms for OVA binding, similar to Figs. 1E, 2F, and 3F. (J) Concatenated CDRH3-CDRL3 aa sequence logos for clones selected in (H) (red nodes) as well as their antibody characteristics.
Next, we investigated whether clones derived from different mice that shared highly similar sequences (identical V- and J-genes, CDRH3-CDRL3 with <5 aa difference) show similar antigen binding behavior (SI Appendix, Fig. S17). Nine clones were tested, which were similar to a previously identified antigen-specific clone (5.17) and showed consistently similar affinities (Kd = 4–26 nM) (Fig. 3 E–G and SI Appendix, Fig. S17). Next, we examined a major cluster of a sequence similarity network that contained 129 sequence nodes and 32 unique VH-VL germline combinations (CDRH3-CDRL3 sequence nodes connected <4 aa difference). Of those, four clones with different germline genes were randomly selected, and we could confirm antigen binding for three of them (Fig. 3 H–J and SI Appendix, Fig. S18).
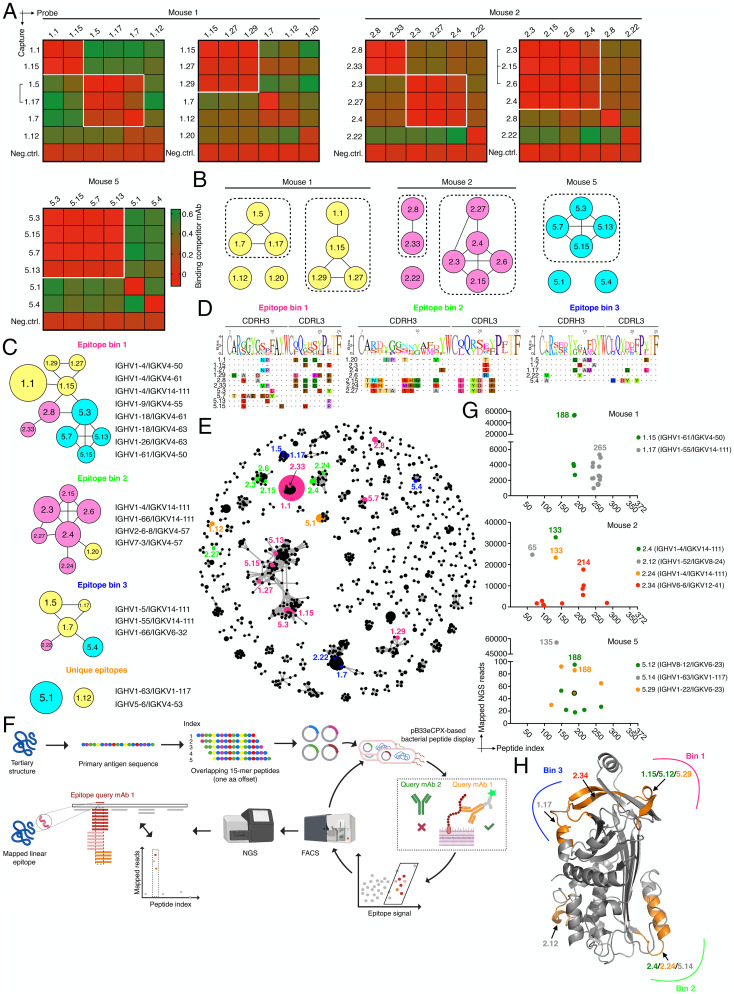
Finally, we aimed to understand the epitope specificity and targeting space of expanded PC clones. Using BLI, we first performed cross-competition epitope binning experiments with highly expanded antibody clones that possessed moderate to high affinity (Kd ∼3–165 nM). This revealed that within a given mouse, a majority of clones could be grouped into discrete epitope bins (Fig. 4 A and B and SI Appendix, Fig. S19). Next, when we tested competitive binding of clones originating from across the different mice (MS-1, MS-2, and MS-5) (SI Appendix, Fig. S19A), three dominant epitope bins emerged, some of which contained clones from all mice (bin 1 and 3), whereas others were mainly occupied by clones from a single mouse (bin 2) (Fig. 4C). For clones 1.12 and 5.1 (the most expanded clone of MS-5), we did not observe any competitive binding suggesting that they target unique epitopes compared to all other clones tested. Interestingly, we observed a number of different VH-VL germline gene combinations for each bin (Fig. 4C and SI Appendix, Fig. S19B). Clonal diversity in each bin was also observed (based on CDRH3-CDRL3 sequences) (Fig. 4D), and when we mapped the bin epitope space of the expanded clones on to the complete IgG clonal sequence similarity network (CDRH3-CDRL3 sequence nodes connected <4 aa difference), we could clearly observe epitope convergence despite a high degree of sequence diversity (Fig. 4E). This was particularly evident for the biggest subnetwork (containing 32 different VH-VL combinations) which exclusively harbored clones of epitope bin 1. Phenotypically, we again did not observe a correlation between clone affinity and epitope specificity when affinity data from different mice per bin were compared (SI Appendix, Fig. S19C).
Fig. 4.
Epitope-targeting space of top expanded clones. (A) Heatmaps show competitive antigen binding based on BLI assays for highly expanded antibody clones in each mouse. Antibodies indicated on the left were captured and probe antibodies on top were used to determine cross-competition for epitope access. Red indicates no binding of the probe antibody as a consequence of epitope blocking by the capture antibody, whereas green denotes binding of the competitor antibody. Groups of antibodies that target the same epitope (epitope bins) are highlighted in white squares. Brackets indicate clonal variants that share the same VH/VL germline V-genes which differed only in CDRH3/CDRL3 aa sequence. An anti-RSVF capture antibody, which does not bind the antigen was used as negative control for all experiments. Clone ID according to SI Appendix, Table S1. (B) Epitope bins with associated clones as determined in (A). Nodes are connected based on observed direct cross-competition. (C) Epitope bins as defined by the cross-competition of clones from different mice. Representative V-gene combinations are shown on the right. Nodes are connected based on direct cross-competition and sizes indicate clone size (number of cells per clone). Colors represent mouse ID as shown in (B). Results are reflective of SI Appendix, Fig. S19. (D) CDRH3/CDRL3 sequence alignment of bin-specific clones. Sequence logo is shown on top and aa residues are highlighted if they are in disagreement with the consensus sequence. (E) Mapping of epitope space as determined in (C) on a sequence similarity network of all IgG clones across all mice (SI Appendix, Fig. S12). Edges represent clones with similar CDR3 sequences based on first calculating separate distance matrices for CDRH3 and CDRL3 amino acid sequences. Following the summation of these two matrices, edges were drawn between those clones separated by <4 aa mutations. Node color according to bin color in (C). Size of clones is reflected by node size. Only those nodes with at least one edge are plotted for visualization purposes. Clone 1.20 is not shown since it was not connected. (F) Linear epitope-mapping workflow using bacterial peptide display. (G) Epitope mapping results of select clones from MS-1, MS-2, and MS-5. For visualization purposes, only data points with >700 mapped reads are shown for MS-1 and MS-2 and clone 5.14 of MS-5; for clones 5.12 and 5.29, only data points with >18 mapped reads are shown. Shared data point between 5.12 and 5.29 is indicated with a circle. Corresponding V-gene combinations are indicated. (H) Mapping of epitope bins from (C) on to the OVA crystal structure using antibody epitope information obtained in (G) (PDB: 1OVA).
In order to more precisely define epitope specificity in relation to the sequence and structure of the OVA antigen, we used a bacterial linear peptide display system (54), where an overlapping 15-mer (aa) peptide library of the primary antigen sequence is expressed on the surface of Escherichia coli. Highly expanded antibody clones are then used to label the antigen peptide library and sequential rounds of fluorescence-activated cell sorting (FACS) is performed to enrich for peptide binding, followed by targeted deep sequencing of peptide encoding regions. Subsequently, epitopes are identified using sequence-alignment and read count statistics (Fig. 4F). To benchmark the system, we used two commercial monoclonal antibodies with well-defined OVA specificity for which we could successfully confirm their respective epitopes (SI Appendix, Fig. S20). We then determined the epitopes of nine highly expanded clones across the different mice (MS-1, MS-2, and MSaa-5) (Fig. 4G and SI Appendix, Fig. S21). For all clones (except 2.4 and 5.14, for which only a single highly enriched peptide was found to bind the antibody), FACS enrichment followed by read count analysis resulted in a small number (9 to 18) of overlapping peptide sequences that were highly enriched compared to the background. Within those, we defined the peptide sequence with the highest read count enrichment (observed 86- to 56,000-fold above background) as the corresponding epitope. Importantly, we identified the same epitope for two clones (2.4 and 2.24) sharing highly similar sequences (identical VHJH-VLJL germline, CDRH3-CDRL3 edit distance of 4 aa). Finally, several of the clones were also representative of the identified epitope bins (Fig. 4C) (e.g., clones 1.15, 1.17, and 2.4/2.24), allowing us to precisely map these bins onto the structure of the OVA antigen, which revealed that they were indeed occupying distinct areas of the antigen (Fig. 4H).
Discussion
Here, we set out to determine if clonal selection and expansion of PC repertoires follow any deterministic rules or is stochastic with regards to their phenotypic antibody properties (i.e., antigen-binding, antigen-affinity, and epitope specificity). First, we discovered that clonal expansion is largely driving antigen specificity for the most expanded clones (∼top 10) in each repertoire. However, for the remaining proportion of the repertoire, antigen specificity was found to be largely stochastic and could not be predicted based on solely the number of expanded cells (Fig. 1 G and H). It should be mentioned, however, that antigen specificity was only assessed in the context of a model protein antigen (ovalbumin) in its native conformation. Therefore, we are not able to draw definitive conclusions on the extent of actual “non-binding clones” (vs. by-stander activation) since some B cells might have been targeting nonnative quasi antigen species that arise following extended exposure to body temperature and/or degradation by extracellular proteases. Next, we determined if PC repertoire affinities allowed for accommodating the widely accepted germinal center B cell selection model: selective expansion of B cells is driven by an avidity-based selection mechanism in germinal centers involving high-affinity BCRs expressed on B cells that consequentially present the highest levels of peptide-MHC to T follicular helper cells (41, 55, 56), which in turn controls the rate of B cell proliferation (57). In contrast to this model, we did not observe any deterministic correlation between antibody affinity and clonal expansion of PCs, furthermore affinity was also not correlated with the number of somatic hypermutations; these stochastic findings were consistent both on the polyclonal repertoire level (Fig. 2 A and B) and the clonal lineage level (Fig. 3 C and D).
While we acknowledge that our experimental setup reflects the collective PC-differentiated integration of multiple germinal center outputs that to some extent mirror ongoing (primarily) naive B cell engagement (58), similar results have been observed recently for differentially expanded germinal center B cells (40). Therefore, we speculate that over time avidity-based selection may not only be a consequence of affinity alone but, for example, can also lead to autoantibody redemption via somatic hypermutation (59). Furthermore, from a B cell’s perspective, capturing of antigen may further be modulated by differential BCR stability and surface expression levels, which can also be fine-tuned by somatic hypermutation. To illustrate, recent work has suggested that somatic hypermutation is associated with decreased conformational antibody stability (60). Furthermore, highly differential antibody secretion rates from single plasma cells have been reported, which can span up to 3–4 log units (61). It remains to be determined how these findings translate to surface expression levels of germinal center B cells and subsequent clonal selection, expansion and differentiation to PCs.
It is also critical to appreciate that B cell selection and expansion depend on a multitude of different extrinsic and intrinsic parameters (62) that only in concert can explain the driving forces behind PC repertoire formation. Therefore, all of these parameters collectively introduce a high-level of stochasticity when the impact of individual parameters or of a small subset thereof on clonal expansion are analyzed separately.
Finally, our results also indicate that despite a high level of antibody sequence diversity, the selection of PCs has a deterministic component with regards to specificity toward a few dominant epitopes, which may be attributed to epitope accessibility and availability during B cell maturation and PC differentiation (Fig. 4 C–E). This could also provide a rationale for the shifting serum epitope immunodominance hierarchies over time, as observed following influenza A virus infection (63).
Collectively, our findings highlight the complex interplay between genotypic and phenotypic features which only in concert drive clonal selection and expansion of differentiated PC repertoires. While the results presented in this study are confined to a single highly immunogenic antigen, it remains to be seen how transferable these findings are with antigens that differ in key parameters such as protein size, stability, valency, immunogenicity and the like. This work has implications for the assessment of vaccine-induced immunity, as understanding how to selectively expand PC clones with a desired activity profile may support the development of vaccines that provide broad and long-lasting protection by the humoral immune system.
Materials and Methods
Mouse Immunizations.
Mouse immunizations were performed under the guidelines and protocols approved by the Basel-Stadt cantonal veterinary office (Basel-Stadt Kantonales Veterinäramt Tierversuchsbewilligung #2582). Five female BALB/c mice (Janvier Laboratories France, 9 wk old) were housed under specific pathogen-free conditions and maintained on a standard chow diet. Mice were repeatedly immunized subcutaneously on day 0, 28, and 49 into the flank with 150 μL of a phosphate-buffered saline (PBS)-based immunization mixture containing 100 μg ovalbumin (Sigma, A5503) and 20 μg monophosphoryl lipid A (MPLA) adjuvant (Sigma, L6895). Blood samples were collected from the tail vein on day 0 and by heart-puncture on day 63. On day 63, mice were euthanized by CO2 asphyxiation and cervical dislocation, and femurs and tibias were collected.
Isolation of Plasma Cells from Bone Marrow.
Harvested femurs and tibias were clipped with surgical scissors at both ends and the bone marrow was flushed out with ∼5 mL of a chilled and sterile filtered solution of PBS pH 7.2, 0.5% bovine serum albumin (BSA) and 2 mM ethylenediamine tetraacetic acid (EDTA) using a 30-gauge BD Micro-Fine+ insulin needle. Bone marrow cells were filtered through a 40-μm nylon cell strainer (FALCON, 352340). PCs defined as CD138+ B220low/− CD19low/− antibody-secreting plasma cells were isolated by magnetic activated cell sorting (MACS) using the CD138+ Plasma Cell Isolation Kit mouse (Miltenyi Biotech, 130-092-530) following the manufacturer’s instructions. Briefly, nonplasma cells were first magnetically depleted using a mixture of biotinylated antibodies against CD49b (DX5) and CD45R (B220) and anti-Biotin microbeads. Then, plasma cells were positively selected from the pre-enriched cell fraction by direct labeling with CD138 microbeads.
Single-Cell Sequencing of Antibody Repertoires.
Single-cell sequencing libraries were constructed from the isolated bone marrow plasma cells following the demonstrated 10X Genomics’ protocol: “Direct target enrichment-Chromium Single Cell V(D)J Reagent Kits” (CG000166 REV A). Briefly, single cells were co-encapsulated with gel beads (10X Genomics, 1000006) in droplets using 5 lanes of one Chromium Single Cell A Chip (10X Genomics, 1000009) with a target loading of 13,000 cells per reaction. V(D)J library construction was carried out using the Chromium Single Cell 5′ Library Kit (10X Genomics, 1000006) and the Chromium Single Cell V(D)J Enrichment Kit, Mouse B Cell (10X Genomics, 1000072) according to the manufacturer’s instructions. All of the reverse transcribed complementary DNA was used as input for VDJ library construction. Final libraries were pooled and sequenced on the Illumina NextSEq. 500 platform (mid output, 300 cycles, paired-end reads) using an input concentration of 1.8 pM with 5% PhiX.
Repertoire Analysis.
Raw sequencing files arising from multiple Illumina sequencing lanes were merged and supplied as input to the command line program cell ranger on a high-performance cluster. Reads were aligned to the germline using cellranger (v3.1.0) segments from the murine VDJ reference (vdj_GRCm38_alts_ensembl-3.1.0) and subsequently assigned into clonal families based on identical combinations of CDRH3+CDRL3 amino acid sequences. Only those clones containing exactly one productive heavy chain and one productive light chain were retained in the analysis. Isotype majority was determined based on the constant region alignment containing the within each clonal family, with all IgG subtypes merged. Full-length variable sequences (spanning from FR1 to FR4) for each cell were obtained by using the call_MIXCR function in Platypus (64), which relies upon aligning full-length contig sequences from the all_contig.fasta file output from cellranger using MiXCR (65). To quantify unique variants, full-length VDJRegion (defined as FR1-FR4) for both heavy and light chains were appended together for each cell. Similarity networks were created by calculating the pairwise edit distance using the stringdist package (66) in R and subsequently creating an adjacency matrix, where each entry (node) corresponded to a unique CDRH3+CDRL3 nucleotide sequence (network shown in Fig. 2E was created based on unique CDRH3+CDRL3 aa sequence). Edges were drawn between nodes with an edit distance of three or less amino acid mutations and following networks were created by R package igraph. Edit distances were calculated using the stringdist package. Circos plots demonstrating germline gene usage were created using the circlize package (67) in R. The unmutated germline reference gene was determined by Cellranger’s alignment and set as the outgroup for the phylogenetic tree. Nucleotide and amino acid distance to germline was determined via IMGT (68). Rarefaction analysis (species richness, Shannon diversity, and Simpson diversity) was performed using the iNEXT package. The iNEXT function was used with default parameters (knots = 40, se = TRUE, conf = 0.95, number of bootstraps = 50) on the extracted abundance data (number of cells per clone). The resampling endpoint (maximum sample size) was selected as 50,000 cells. Recon was performed using the recon_v3.0 python script, with the default parameters (parameter limit = 20, sampling error threshold = 30). Resampling from the inferred parent distribution was performed for a maximum clone size of 10 and then plotted against the original sample.
Antibody Expression.
Antibodies were either transiently expressed at small scale in HEK 293 Expi cells using the ExpiFectamine 293 Transfection Kit (Thermo, A14524) and the pFUSE2ss vector system for both, IgM/IgK and IgG/IgK/L (Invitrogen) according to previous protocols (69), or stable hybridoma cell lines were engineered by CRISPR/Cas9 genome editing as described before (70, 71). Of note, these hybridoma cell lines are able to surface display as well as secrete antibody of the IgG2c isotype, which allows for both, FACS-based as well as ELISA-based specificity profiling.
Hybridoma Cell Culture.
Hybridoma cell lines were cultivated in high-glucose DMEM (Thermo, 61965-026), supplemented with 10% (vol/vol) of ultra-low IgG FBS (Thermo, 16250078), 100 U/mL Pen/Strep (Thermo, 15140-122), 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Thermo, 15630-056), and 50 μM 2-mercaptoethanol (Thermo, 31350-010). Cell lines were maintained at 37 °C, 5% CO2, and passaged every 72 h.
Antibody Validation by ELISA.
Sterile-filtered cell culture supernatant (0.2 μm) of a 6-d culture was used to confirm both, antibody expression as well as OVA specificity. ELISA plates were coated with the capturing reagent in PBS (OVA [Sigma, A5503] for antigen ELISAs, anti-mouse IgM [γ-chain specific; Sigma, M8644] and anti-mouse IgG [light chain specific; Jackson ImmunoResearch, 115-005-174] for IgM and IgG expression ELISAs) at 4 μg/mL, blocked with PBS supplemented with 2% (wt/vol) milk (AppliChem, A0830), and incubated with (serial dilutions of) cell culture supernatant (supernatant of a hen-egg lysozyme/OVA specific cell line served as negative/positive controls respectively). IgM and IgG binding was detected using anti-mouse kappa light chain-HRP (Abcam, ab99617) or anti-mouse IgG (Fc-specific)-HRP (Sigma, A2554) secondary antibody, respectively. Binding was quantified using the 1-Step Ultra TMB-ELISA substrate solution (Thermo, 34028) and 1M H2SO4 for reaction termination. Absorbance at 450 nm was recorded on an Infinite 200 PRO (Tecan). All commercial antibodies were used according to manufacturer’s recommendations. For heatmaps shown in Figs. 1 and 2, each of the steps was timed to last equally long. For isotype-specific serum ELISAs, serum was incubated with 1:5 serial dilutions of prediluted (1:100) serum (naïve serum served as control). Either total IgG or the isotypes were detected by incubation with biotinylated IgA (RMA-1), IgG1 (RMG-1), IgG2b (RMG2b-1), IgG2c (RMG2a-62), IgG3 (RMG3-1), IgM (RMM-1, all BioLegend), or IgK (Abcam, ab99617), washed, and incubated with streptavidin-HRPO (BD). Binding was quantified using ABTS development (Sigma).
Antibody Validation by Surface Staining of Stable Hybridoma Cell Lines.
Flow cytometry scanning of hybridoma cells was performed on a BD FACS Aria III. Typically, 5 × 105 cells were stained for 30 min on ice in 50 μL of a labeling mix consisting of anti-IgG2c-AlexaFluor488 (Jackson ImmunoResearch, 115-545-208), anti-IgK-Brilliant Violet421 (BioLegend, 409511), and OVA-AlexaFluor647 (0.86 mg/mL) at 1:100, 1:80, and 1:50, respectively. Before scanning, cells were washed twice.
Antibody Affinity Measurements.
Supernatants of Expi cultures and monoclonal hybridoma populations were collected, concentrated (Amicon, UFC810008) and filtered through a 0.2-μm filter (Sartorius, 16534-K). Affinities were then measured on an Octet RED96e machine (FortéBio) with the following parameters: anti-mouse IgG Fc Capture (AMC) biosensors (FortéBio, 18-5088) were hydrated in conditioned media diluted 1 in 2 with kinetics buffer (1×KB) (FortéBio, 18-1105) for at least 10 min before conditioning through 4 cycles of regeneration consisting of 10 s incubation in 10 mM glycine, pH 1.52, and 10 s in 1×KB. Conditioned sensors were then loaded with concentrated conditioned medium diluted 1 in 2 with 1×KB (reference sensor) or cell culture supernatant diluted 1 in 2 with 1×KB. Loaded sensors were then equilibrated in 1×KB and typically incubated with various concentrations of OVA antigen in 1×KB ranging from 0 nM (reference sample) up to 200 nM and 1 μM, respectively, for polyclonal and intraclonal variants mAbs. Finally, sensors were incubated in 1×KB to allow antigen dissociation. Kinetics analysis was performed in analysis software Data Analysis HT v11.0.0.50 and Kd-values were calculated from fits with an association R2 > 0.85.
Epitope Binning.
Transiently expressed antibodies were purified from 30 mL of HEK 293 Expi cultures using Protein G GraviTrap columns (Sigma, GE28-9852-55) and the Ab Buffer Kit (Sigma, GE28-9030-59) according to the manufacturer instructions. Before epitope binning, purified antibodies were confirmed for OVA-positivity by ELISA and OVA-binding kinetics were reconfirmed by BLI measurements prior to each experiment.
Epitope binning following a classical sandwich protocol was performed on an Octet RED96e machine (FortéBio), using anti-mouse IgG Fc Capture (AMC) biosensors (FortéBio, 18-5088) with the following steps. (0) Hydration of biosensors in 1×KB for 30 min. (1) Baseline equilibration in 1×KB for 60 s. (2) First loading of capture antibody at 40–60 μg/mL in 1×KB for 240 s. (3) Quenching of biosensors in 50 μg/mL polyclonal mouse IgG (Rockland, 010-0102) for 300 s. (4) Baseline in 1×KB for 240 s. (5) Second loading of capture antibody at 40–60 μg/mL in 1×KB for 240 s. (6) Baseline in 1×KB for 200 s. (7) Loading of OVA at 150 nM in 1×KB for 600 s. (8) Baseline in 1×KB for 60 s. (9) Loading of probe antibody at 25 μg/mL in 1×KB for 600 s. (10) Regeneration of sensors. Analysis was performed in analysis software Data Analysis HT v11.0.0.50.
Library Generation for Epitope Mapping.
Bacterial epitope mapping was based on the pB33eCPX plasmid, which was previously established (54) to fuse a peptide to the enhanced outer membrane protein X (OmpX) for extracellular peptide presentation. First, we modified the plasmid using primers EpMap_1 and EpMap_2 along with ssODN EpMap1_ssODN (SI Appendix, Table S3) using the NEBuilder Mastermix (NEB, E2621S) following the manufacturer’s instructions in order to create hairpin-free overlap regions that could be used for homology-based library cloning. Next, the epitope library was generated using MC1061 E. coli cells, NEBuilder Mastermix, primers EpMap_3 and EpMap_4 (SI Appendix, Table S3) along with 0.078 pmol of a ssODN library of 105 nt in length (30 bp homology overhangs on each side [GGAACTTCTGTAGCTGGACAATCTGGACAA and GGAGGGCAGTCTGGGCAGTCAGGTGATTAC] flanking a 45-bp stretch encoding a 15mer aa peptide sequence; ssODN library was ordered as a Tier 1 oligo pool from Twist Bioscience). The library was designed to cover the whole OVA amino acid sequence, with a window size of 45 bp and a cutoff of one codon, resulting in a library size of 372. The final library size was determined to contain 268,900 transformants using serial dilutions (722x oversampled), and when we analyzed 20 clones by Sanger sequencing using primer EpMap_5, we found that 95% contained a correct sequence of which 95% were unique. Glycerol stocks were subsequently stored at −80 °C until further use.
Epitope Mapping by Bacterial Surface Display.
Bacterial display-based epitope mapping was carried out as described before (72). Briefly, 500 mL of LB-medium supplemented with chloramphenicol (34 μg/mL) and 0.2% (wt/vol) 0.2 μm sterile filtered glucose were inoculated with the transformant library and grown for 12 h at 37 °C. Next, cells were subcultured 1:50 into 5 mL of LB supplemented with chloramphenicol and grown for 2 h at 37 °C, before protein expression was induced for 1 h at 37 °C using 0.04% (wt/vol) L-arabinose. Typically ∼1.1E7 cells were subsequently harvested at 3,000 × g for 5 min, washed and stained for 30 min on ice in 100 μL of a labeling mix consisting of 20 μL of concentrated and extensively PBS buffer-exchanged hybridoma cell culture supernatant (containing antibody protein of the top expanded clones) and 80 μL of PBS pH 7.2, 0.5% BSA, and 2 mM EDTA (for assay establishment, 0.5 μg of control antibodies Clone 4B4E6 [Chondrex, 7096] and Clone 2322 [Chondrex, 7094] were spiked into concentrated and buffer exchanged cell culture supernatant of a hybridoma cell line, that did not express antibody). Cells were washed twice and stained in 100 μL for 30 min on ice using an anti-mouse IgG-Brilliant Violet421 secondary antibody at 1:90 (BioLegend, 405317). Cells were again washed twice, resuspended in 500 μL of PBS, and sorted into sterile SOC media using a BD FACS Aria III. Typically, between 10,000 and 50,000 cells were sorted on 2–3 consecutive days to enrich for pure antigen-binding populations, before plasmid was extracted and sequencing libraries were generated.
Generation of Epitope Mapping Sequencing Libraries.
Bacterial plasmid DNA was extracted from a 3 mL overnight culture using the QIAprep Spin Miniprep Kit (Qiagen, 27104). Next, NGS libraries were generated following a two-step primer extension protocol (73). 30 μg of plasmid DNA were amplified using Kapa Hifi HotStart Ready mix (Kapa Biosystems, KK2602) in a 50 μL reaction using primers EpMap_7 and EpMap_8 (which bound to regions that were ∼70 bp away from the peptide encoding region) with the following cycling parameter: 95 °C for 3 min, 21 cycles of 98 °C for 20 s, 59 °C for 15 s, 72 °C for 20 s, and 72 °C for 30 s final extension.
After gel purification, the final library was constructed the same way using 20 μg of PCR1 product along with primer EpMap_9 and one of 20 Illumina index primers (EpMap_idx) using the following cycling parameter: 95 °C for 3 min, 21 cycles of 98 °C for 20 s, 56 °C for 15 s, 72 °C for 25 s, and 72 °C for 30 s final extension. Final libraries of the correct length (∼400 bp) were gel-purified on a 2% (wt/vol) agarose-gel and subjected to fragment analyzer analysis (Advanced Analytical Technologies) using DNF-473 Standard Sensitivity NGS fragment analysis kit prior to sequencing. High-quality library pools were sequenced on the Illumina MiSeq platform using the reagent kit v3 (2 × 300 cycles, paired-end) with 10% PhiX.
Bioinformatic Epitope Extraction.
Forward and reverse reads were merged in Geneious v10.2.6 and subsequently read into R (v.4.0.4) using the “read.fasta” function from the “seqinr” package. The number of occurrences of each epitope within these reads was determined using the “str_count” function from the “stringr” package.
Data Visualization.
FACS plots were created using FlowJo v10 (BD). Sequence alignments, phylogenetic trees, and logo plots were exported from Geneious v10.2.6. Structural epitope visualization was performed using PyMol v2.4.2. BLI affinity traces were exported from Data Analysis HT v11.0.0.50 (FortéBio). Figs. 1A and 4F were created with BioRender.com. The generation of all other figures was either already described or they were produced using Prism v8 (GraphPad).
Supplementary Material
Acknowledgments
We acknowledge the ETH Zurich D-BSSE Single Cell Unit and the Genomics Facility Basel for excellent support and assistance, in particular I. Nissen, T. Schär, E. Burcklen, T. Horn, and C. Beisel. We also would like to thank M.D. Hussherr and G. Camenisch for assistance with animal authorization and experiments. pB33eCPX was a gift from Patrick Daugherty (Addgene plasmid #23336; http://n2t.net/addgene:23336; RRID:Addgene_23336). This work was supported by the European Research Council (Starting Grant 679403 to S.T.R.) and ETH Zurich Research Grants (to S.T.R.). The professorship of S.T.R. is supported by an endowment from the S. Leslie Misrock Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. G.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113766119/-/DCSupplemental.
Data Availability
Sequencing data have been deposited in European Bioinformatics Institute database.
References
- 1.Slifka M. K., Antia R., Whitmire J. K., Ahmed R., Humoral immunity due to long-lived plasma cells. Immunity 8, 363–372 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Manz R. A., Hauser A. E., Hiepe F., Radbruch A., Maintenance of serum antibody levels. Annu. Rev. Immunol. 23, 367–386 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Amanna I. J., Carlson N. E., Slifka M. K., Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 357, 1903–1915 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Turner J.S., et al. , SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature 595, 421–425 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Akkaya M., Kwak K., Pierce S. K., B cell memory: Building two walls of protection against pathogens. Nat. Rev. Immunol. 20, 229–238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hozumi N., Tonegawa S., Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc. Natl. Acad. Sci. U.S.A. 73, 3628–3632 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisen H. N., Siskind G. W., Variations in affinities of antibodies during the immune response. Biochemistry 3, 996–1008 (1964). [DOI] [PubMed] [Google Scholar]
- 8.Weigert M. G., Cesari I. M., Yonkovich S. J., Cohn M., Variability in the lambda light chain sequences of mouse antibody. Nature 228, 1045–1047 (1970). [DOI] [PubMed] [Google Scholar]
- 9.Jacob J., Kelsoe G., Rajewsky K., Weiss U., Intraclonal generation of antibody mutants in germinal centres. Nature 354, 389–392 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Burnet F. M., A modification of Jerne’s theory of antibody production using the concept of clonal selection. CA Cancer J. Clin. 26, 119–121 (1976). [DOI] [PubMed] [Google Scholar]
- 11.Victora G. D., Nussenzweig M. C., Germinal centers. Annu. Rev. Immunol. 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Nutt S. L., Hodgkin P. D., Tarlinton D. M., Corcoran L. M., The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 15, 160–171 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Weinstein J. A., Jiang N., R. A. White, 3rd, Fisher D. S., Quake S. R., High-throughput sequencing of the zebrafish antibody repertoire. Science 324, 807–810 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang N., et al. , Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc. Natl. Acad. Sci. U.S.A. 108, 5348–5353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elhanati Y., et al. , Inferring processes underlying B-cell repertoire diversity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greiff V., et al. , Systems analysis reveals high genetic and antigen-driven predetermination of antibody repertoires throughout B cell development. Cell Rep. 19, 1467–1478 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Briney B., Inderbitzin A., Joyce C., Burton D. R., Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 566, 393–397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soto C., et al. , High frequency of shared clonotypes in human B cell receptor repertoires. Nature 566, 398–402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy S. T., et al. , Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat. Biotechnol. 28, 965–969 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Laserson U., et al. , High-resolution antibody dynamics of vaccine-induced immune responses. Proc. Natl. Acad. Sci. U.S.A. 111, 4928–4933 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeKosky B. J., et al. , Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. U.S.A. 113, E2636–E2645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellebedy A. H., et al. , Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol. 17, 1226–1234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Setliff I., et al. , High-throughput mapping of B cell receptor sequences to antigen specificity. Cell 179, 1636–1646.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horns F., Dekker C. L., Quake S. R., Memory B cell activation, broad anti-influenza antibodies, and bystander activation revealed by single-cell transcriptomics. Cell Rep. 30, 905–913.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gérard A., et al. , High-throughput single-cell activity-based screening and sequencing of antibodies using droplet microfluidics. Nat. Biotechnol. 38, 715–721 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Goodwin E., et al. , Infants infected with respiratory syncytial virus generate potent neutralizing antibodies that lack somatic hypermutation. Immunity 48, 339–349.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Y., et al. , Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 182, 73–84.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilchuk P., et al. , Integrated pipeline for the accelerated discovery of antiviral antibody therapeutics. Nat. Biomed. Eng. 4, 1030–1043 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers T. F., et al. , Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wec A. Z., et al. , Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 369, 731–736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B., et al. , Facile discovery of a diverse panel of anti-Ebola virus antibodies by immune repertoire mining. Sci. Rep. 5, 13926 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wine Y., et al. , Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc. Natl. Acad. Sci. U.S.A. 110, 2993–2998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavinder J. J., et al. , Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc. Natl. Acad. Sci. U.S.A. 111, 2259–2264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voss W. N., et al. , Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science 372, 1108–1112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paus D., et al. , Antigen recognition strength regulates the choice between extrafollicular plasma cell and germinal center B cell differentiation. J. Exp. Med. 203, 1081–1091 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phan T. G., et al. , High affinity germinal center B cells are actively selected into the plasma cell compartment. J. Exp. Med. 203, 2419–2424 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor J. J., Pape K. A., Steach H. R., Jenkins M. K., Humoral immunity. Apoptosis and antigen affinity limit effector cell differentiation of a single naïve B cell. Science 347, 784–787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisel F. J., Zuccarino-Catania G. V., Chikina M., Shlomchik M. J., A temporal switch in the germinal center determines differential output of memory B and plasma cells. Immunity 44, 116–130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan T. D., et al. , Antigen affinity controls rapid T-dependent antibody production by driving the expansion rather than the differentiation or extrafollicular migration of early plasmablasts. J. Immunol. 183, 3139–3149 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Tas J. M. J., et al. , Visualizing antibody affinity maturation in germinal centers. Science 351, 1048–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gitlin A. D., Shulman Z., Nussenzweig M. C., Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature 509, 637–640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slifka M. K., Matloubian M., Ahmed R., Bone marrow is a major site of long-term antibody production after acute viral infection. J. Virol. 69, 1895–1902 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manz R. A., Thiel A., Radbruch A., Lifetime of plasma cells in the bone marrow. Nature 388, 133–134 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Kaplinsky J., Arnaout R., Robust estimates of overall immune-repertoire diversity from high-throughput measurements on samples. Nat. Commun. 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han A., Glanville J., Hansmann L., Davis M. M., Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gate D., et al. , Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agrafiotis A., et al. , B cell clonal expansion is correlated with antigen-specificity in young but not old mice. bioRxiv [Preprint] (2021). 10.1101/2021.11.09.467876 (Accessed 12 April 2022). [DOI]
- 48.Lingwood D., et al. , Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnes C. O., et al. , SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreer C., et al. , Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell 182, 1663–1673 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miho E., Roškar R., Greiff V., Reddy S. T., Large-scale network analysis reveals the sequence space architecture of antibody repertoires. Nat. Commun. 10, 1321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedensohn S., et al. , Convergent selection in antibody repertoires is revealed by deep learning. bioRxiv (2020) doi: 10.1101/2020.02.25.965673. [DOI]
- 53.Batista F. D., Neuberger M. S., Affinity dependence of the B cell response to antigen: A threshold, a ceiling, and the importance of off-rate. Immunity 8, 751–759 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Rice J. J., Daugherty P. S., Directed evolution of a biterminal bacterial display scaffold enhances the display of diverse peptides. Protein Eng. Des. Sel. 21, 435–442 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Victora G. D., et al. , Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shulman Z., et al. , Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science 345, 1058–1062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gitlin A. D., et al. , HUMORAL IMMUNITY. T cell help controls the speed of the cell cycle in germinal center B cells. Science 349, 643–646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mesin L., et al. , Restricted clonality and limited germinal center reentry characterize memory B cell reactivation by boosting. Cell 180, 92–106.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burnett D. L., et al. , Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination. Science 360, 223–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shehata L., et al. , Affinity maturation enhances antibody specificity but compromises conformational stability. Cell Rep. 28, 3300–3308.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Eyer K., et al. , Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring. Nat. Biotechnol. 35, 977–982 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Angeletti D., Yewdell J. W., Understanding and Manipulating Viral Immunity: Antibody Immunodominance Enters Center Stage. Trends in Immunology 39, 549–561 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Angeletti D., et al. , Defining B cell immunodominance to viruses. Nat. Immunol. 18, 456–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yermanos A., et al. , Platypus: An open-access software for integrating lymphocyte single-cell immune repertoires with transcriptomes. NAR Genom. Bioinform. 3, l (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bolotin D. A., et al. , MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Loo M., The stringdist Package for Approximate String Matching. The R Journal 6, 111 (2014). [Google Scholar]
- 67.Gu Z., Gu L., Eils R., Schlesner M., Brors B., Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Brochet X., Lefranc M.-P., Giudicelli V., IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503-8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vazquez-Lombardi R., et al. , Transient expression of human antibodies in mammalian cells. Nat. Protoc. 13, 99–117 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Pogson M., Parola C., Kelton W. J., Heuberger P., Reddy S. T., Immunogenomic engineering of a plug-and-(dis)play hybridoma platform. Nat. Commun. 7, 12535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parola C., et al. , Antibody discovery and engineering by enhanced CRISPR-Cas9 integration of variable gene cassette libraries in mammalian cells. MAbs 11, 1367–1380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Getz J. A., Schoep T. D., Daugherty P. S., Peptide discovery using bacterial display and flow cytometry. Methods Enzymol. 503, 75–97 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Menzel U., et al. , Comprehensive evaluation and optimization of amplicon library preparation methods for high-throughput antibody sequencing. PLoS One 9, e96727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in European Bioinformatics Institute database.