Significance
The prevailing dogma is that renewed mitogenic signaling is essential to traverse G1 phase of the cell cycle after each division. B lymphocytes undergo multiple mitotic divisions, termed clonal expansion, to expand antigen-specific cells that mediate effective immunity. Here we demonstrate that B cells that have undergone one cell division continue to proliferate even in absence of further mitogenic signals. This mitogen-independent proliferation is accompanied by an altered G1 phase marked by transcriptomic and proteomic features of G2/M. Survivin, a G2/M-specific oncogene, is required in G1 to achieve mitogen-independent proliferation.
Keywords: mitogen-independent proliferation, clonal expansion, B lymphocytes, cell cycle regulation, G1-S
Abstract
B and T lymphocytes of the adaptive immune system undergo proliferative bursts to generate pools of antigen-specific cells for effective immunity. Here we show that in contrast to the canonical view that G1 progression signals are essential after mitosis to reenter S phase, B lymphocytes sustain several rounds of mitogen-independent cell division following the first mitosis. Such division appears to be driven by unique characteristics of the postmitotic G1 phase that has features of S and G2/M phases. Birc5 (survivin), a protein associated with chromosome segregation in G2/M, is expressed in the G1 phase of divided B cells and is necessary for mitogen-independent divisions. The partially active G1 phase and propensity for apoptosis inherited after each division may underlie rapid proliferation and cell death, which are hallmarks of B cell proliferative responses.
Activation of antigen receptors of B and T lymphocytes via antigen engagement is one key signal that initiates responses to specific pathogens (1, 2). Antigen recognition selects a tiny proportion of lymphocytes, which are induced to proliferate and execute an immune response. This proliferative phase, termed clonal expansion, determines effectiveness of subsequent adaptive immune responses. Additionally, antigen nonspecific costimulatory signals provided by innate immune cells contribute to the magnitude of clonal expansion (3, 4). Regulation of lymphocyte proliferation has been studied by treating cells ex vivo with one or more extracellular signals followed by analyses of proliferative responses. In recent years, elegant experimental schemes have been coupled with rigorous computational studies to gain deeper insight into these processes (5–7). Two related questions lie at the heart of these studies. First, what determines the proportion of lymphocytes out of the preexisting pool that will proliferate? Second, how do lymphocytes count the number of divisions so as to have an effective, but limited, proliferative phase?
The prevailing view is that lymphocytes are induced to proliferate when the cumulative response to incoming signals crosses an activation threshold (8). For lymphocytes, the concept of division destiny (DD) has been invoked to signify the number of times a cell divides in response to mitogenic signals (9, 10). DD is related to the mean division number (MDN), which is the average number of cell divisions made over a defined time period. It has been shown that DD is proportionate to levels of Myc that are induced in response to the initiating signal(s) (11). Similarly, the extent of cell division in germinal centers (GCs) has been recently shown to be proportional to Myc levels (12), and combined absence of cMyc and nMyc abrogates B cell proliferation in vitro (13).
The majority of studies of cell division in lymphocytes are carried out with persistent mitogenic stimulation, presumably reflecting our bias that mitogenic restimulation is necessary to incite multiple rounds of cell division for robust clonal expansion. However, it remains unclear whether clonal expansion in vivo requires persistent antigenic stimulation. There is good evidence that signaling and proliferative phases can be spatially segregated. This is most obvious in the GC, where B cell activation via T cell help has been shown to be restricted to the GC light zone (LZ), whereas the bulk of B cell proliferation occurs in the GC dark zone (DZ) (14, 15). Recent studies attribute this to a synergistic effect of B cell receptor (BCR) and CD40 signals in the LZ for optimal Myc induction (16) and cyclin D3–driven CDK4/6 activation in the DZ (17). The simplest interpretation of this distinction is that GC B cells can undergo multiple rounds of division without requiring continued mitogenic signaling. There is also tantalizing evidence that naive B cells can undergo such a signal-independent division ex vivo, which suggests that this form of division may also be involved in clonal expansion (11). Additionally, CD8+ T cells have been shown to undergo several rounds of division in the absence of persistent signaling (18, 19), suggesting that this may be a common feature of cell cycle regulation in lymphocytes. Despite its uniqueness and likely physiological relevance, this form of proliferation has received scant attention.
The decision-making process after mitosis that shunts daughter cells into a G0 state or permits their reentry into G1 has elicited much interest. The classical view, obtained largely from population analyses of fibroblasts in culture, posits that a mitogenic signal is required at the end of each mitosis for cells to progress through the next G1 (20–22). However, analyses of tumor cell lines and, more recently, untransformed mouse embryonic stem (ES) cells, have shown that a subset of cells within a population retain the ability to undergo G1 progression without additional signaling (23, 24). Maintenance of CDK2 activity and loss of p53 have been shown to regulate this form of cell cycle progression (23, 25). Although these players have not been extensively investigated in lymphocytes, there is good evidence that p53 regulates proliferation of murine T lymphocytes (26).
Here we explored mechanisms by which primary murine B lymphocytes make cell division decisions. We demonstrate that commitment to DNA replication (S phase) programs B cells to divide multiple times in the absence of overt mitogenic signaling. The extent of division is limited by cell death rather than by return to quiescence, and circumventing cell death permits up to four rounds of division in 72 h. Mitogen-independent cell cycle progression is driven by unique S- and G2/M-like characteristics of the G1 phase of cells that have divided once, such as, large cell size, low levels of p27, phosphorylated Rb, and expression of G2/M markers survivin, Cenp-A, and Aim1/aurora B. Pharmacologic inhibition of survivin blocked G1 progression in a B cell line and in primary B cells undergoing mitogen-independent division. These observations indicate that, in contrast to textbook models of the cell cycle, B cells inherit a partially active G1 phase after cell division that permits them to move quickly to the next S phase in the absence of exogenous G1 progression signals. Our studies provide direct evidence for Pardee’s hypothesis (20) that retention of features of G2/M in postmitotic cells could trigger a second round of cell cycle progression. We propose that these mechanisms may assist in rapid cell division without differentiation that is required for clonal expansion in response to antigens.
Results
B Cells Undergo Mitogen-Independent Division.
Defranco et al. first demonstrated that B cell mitogenic signals could be removed at a defined time before initiation of DNA synthesis without affecting the ability of cells to enter S phase (27). In modern parlance this observation reflects the need for continuous signaling for cells to progress past the restriction point in G1 (28). The restriction point is characterized by phosphorylation of Rb, release of E2F factors, degradation of cell cycle inhibitors (p21 or p27), and inactivation of Cdh1 (29). Cell cycle studies in fibroblasts indicate that mitogenic signals are again required after traversing S and G2/M to induce G1 progression past the restriction point and reinitiate DNA replication. In contrast to this model, CD8+ T cells undergo several rounds of cell division even if mitogens are removed prior to the first cell division (18, 19). Similar properties have also been attributed to B cells (30); however, mechanisms that permit lymphocytes to deviate from the classical model have not been explored. To better understand this phenomenon, we started our analysis by characterizing the requirement for mitogenic signals for B cell proliferation.
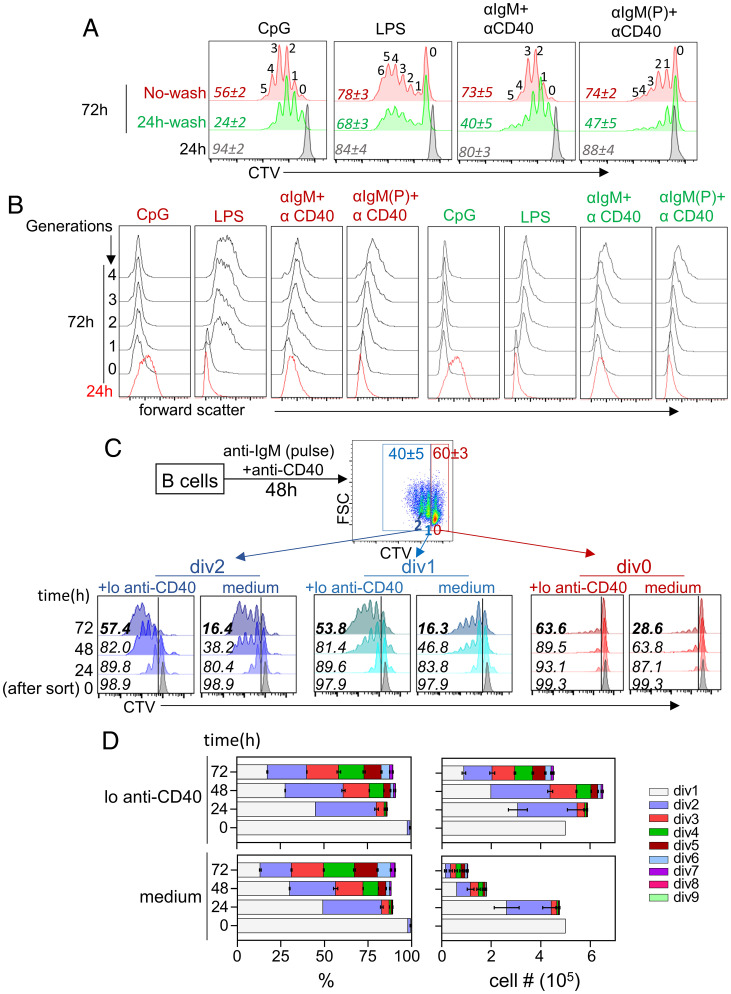
We activated naive splenic B lymphocytes with a variety of mitogenic signals for 24 h and thereafter followed cell division in the absence of overt signaling. We found that none of these treatments induced cell division within 24 h. Yet, multiple rounds of cell division occurred in the next 48 h, regardless of whether mitogen was present continuously or not (Fig. 1A). Cells that lacked continuous mitogenic signals were, however, considerably less viable (Fig. 1A, numbers in italics). As previously noted (9), persistent CpG treatment led to reduced cell size with divisions (Fig. 1B). In contrast, cell size increased with additional divisions for all other stimuli with or without continued stimulation (Fig. 1B, green labels). We conclude that splenic B lymphocytes are programmed to divide multiple times in the absence of continued stimulation. This decision is made during the first cell cycle and is terminated by cell death in vitro.
Fig. 1.
Mitogen-independent proliferation of murine spleen B cells. Splenic B cells were activated with CpG (50 ng/mL), LPS (5 µg/mL), F(ab′)2 anti-IgM (10 µg/mL) + anti-CD40 antibody (5 µg/mL) or pulse BCR stimulation followed by anti-CD40 (31) treatment as indicated. After 24 h, half the cells were washed free of the stimulus and further incubated for an additional 48 h. Remaining cells were continuously stimulated for the entire 72-h period, followed by flow cytometric analysis. (A) Cell division was assessed by CTV dilution at 24 h (gray) and at 72 h (green and red); “no wash” (red) refers to cells that were continuously stimulated. Cells that were washed and incubated further in the absence of mitogen are shown in green. Numbers in italics refer to the percentage of viable cells in each culture; numbers above CTV peaks refer to the cell generations. (B) Cell size was estimated by forward scatter for each cell generation at 24 and 72 h. Red and green labeling refers to cells that had been continuously stimulated or washed free of stimulus at 24 h, respectively. (C) After a 48-h pulse anti-IgM plus anti-CD40 activation, B cells were sorted into populations of cells that had not divided (div0), divided once (div1), and divided twice (div2). Each population was further cultured in medium alone or in medium with 0.125 µg/μL anti-CD40 (lo anti-CD40). CTV dilution after various times of culture is indicated on the Left. Numbers within histogram boxes refer to percentage of viable cells under each condition. (D) Cell percentages (Left) and cell numbers (Right) generated after culture of div1 cells for 48 h in the absence of stimulation. Cell division was determined by CTV dilution as in C (colored bars) using the proliferation modeling tool in FlowJo. (Upper) Cells were treated with low anti-CD40 (lo anti-CD40) and (Lower) cells were incubated in medium alone (medium). Data shown are average of three independent experiments with error bars showing SEM.
To determine when the decision to divide without mitogen was made, we activated cells with a pulse of BCR cross-linking followed by anti-CD40 treatment (31). This regimen led to similar numbers of divided (labeled div1 and div2) and undivided (div0) cells after 48 h, permitting further analyses of both populations (Fig. 1 C, Top). Sorted div1, div2, or div0 cells were cultured for an additional 72 h in the absence of mitogen. We found that very few div0 cells divided, whereas div1 and div2 cells divided robustly despite undergoing extensive cell death (Fig. 1C, labeled “medium”) during culture. Similar results were observed with other mitogens (SI Appendix, Fig. S1A). We conclude that B cells that have completed one mitotic division gain the ability to undergo several additional rounds of mitogen-independent proliferation.
To circumvent cell death, we repeated these experiments by culturing sorted cells in B cell activating factor cytokine (BAFF) containing medium. Though BAFF treatment averted cell death, it also drastically reduced cell division especially at later cell division stages (SI Appendix, Fig. S1B). However, very low concentrations (0.125 µg/mL) of two different clones of anti-CD40 antibodies substantially enhanced cell viability during mitogen-independent proliferation (Fig. 1C, labeled “+lo anti-CD40”), while maintaining proliferation (SI Appendix, Fig. S1B) and cell size (SI Appendix, Fig. S1C). Importantly, this dose of anti-CD40 did not induce proliferation of div0 cells (Fig. 1C, “div0 +lo anti-CD40”). Furthermore, this dose of anti-CD40 did not change the proportions of cells that divided during postsort culture but, instead, increased cell numbers after longer culture periods (Fig. 1D). We conclude that low levels of anti-CD40 provide survival signals without affecting mitogen-independent cell division, suggesting that this form of lymphocyte proliferation can generate substantial cell numbers in the presence of appropriate antiapoptotic activity.
Cell Cycle Characteristics beyond First Mitosis.
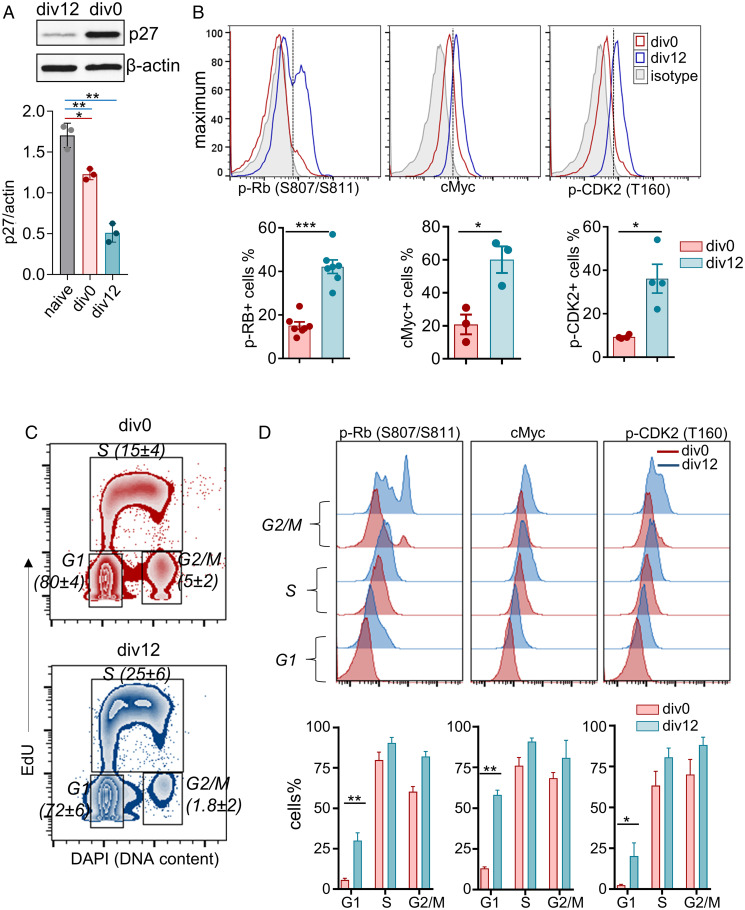
To uncover the basis for mitogen-independent proliferation of div1 and div2 (div12) cells, we first ruled out the possibility that div0 cells were refractory to further division. We found that anti-IgM induced comparable cell division of both div0 and div12 cells (SI Appendix, Fig. S2A), demonstrating that div0 cells could divide with proper mitogenic stimulation. To identify differences between div0 and div12 cells, we assayed biochemical features of the cell cycle. We found that p27 levels were lower in div12 cells (Fig. 2A), whereas hyperphosphorylated Rb (S807/S811), cMyc, and phospho-CDK2 (Thr160) levels were higher in div12 compared to div0 cells (Fig. 2B). Because cells in G1 constituted the majority of cells in both div0 and div12 populations (Fig. 2C), we tentatively concluded that these changes reflected differences in G1 states of the respective populations. This was confirmed by gating directly on cells in G1 (Fig. 2D). Div12 G1 cells were also significantly larger and had increased granularity (SI Appendix, Fig. S2 B and C). These observations demonstrate that the G1 phase of postmitotic B cells is distinct from that of activated but nondivided B cells.
Fig. 2.
Postmitotic B cells are enriched for markers of G1 progression. Pulse anti-IgM+anti-CD40 activated cells (48 h) were purified into undivided (div0) and divided (div12) populations. Cell cycle features were analyzed biochemically or by flow cytometry. (A) Immunoblotting analysis of p27 and β-actin levels (Top) was performed using whole cell extracts from div0 or div12 cells as indicated. Protein expression was quantified by densitometry (Bottom); error bars show SEM (n = 3). (B) Representative histograms showing intracellular levels of phospho-Rb (pS807/S811), cMyc and phospho-CDK2 (pT160) in div0 (red), and div12 (blue) cells by flow cytometry. Percentages of cells that express indicated target proteins were identified by gating (dotted line) with respect to isotype control (gray). Results from several independent experiments (dots) are shown at Bottom; error bars show SEM. (C) After 48-h activation, cells were pulse labeled with EdU for 1 h followed by Click-iT reaction for Alexa-Fluor labeling. Cell cycle stages in div0 and div12 cells were identified as G1 phase (2n DAPI EdU−), S phase (EdU+), and G2/M phase (4n DAPI EdU−). Representative zebra plots of div0 (Top) and div12 (Bottom) cells are shown. Numbers in parentheses indicate the percentage ± SEM of cells in each cell cycle stage (n = 6). (D) Representative histograms of cell cycle stage–specific intracellular levels of phospho-Rb (pS807/S811), cMyc, and phospho-CDK2 (pT160) in div0 (red) and div12 (blue) cells (Top). Protein positive cells were identified as in B, and % positive cells from two to four independent experiments are shown Lower; error bars represent SEM (standard error of the mean). *P ⪕ 0.05, **P ⪕ 0.01, ***P ⪕ 0.001).
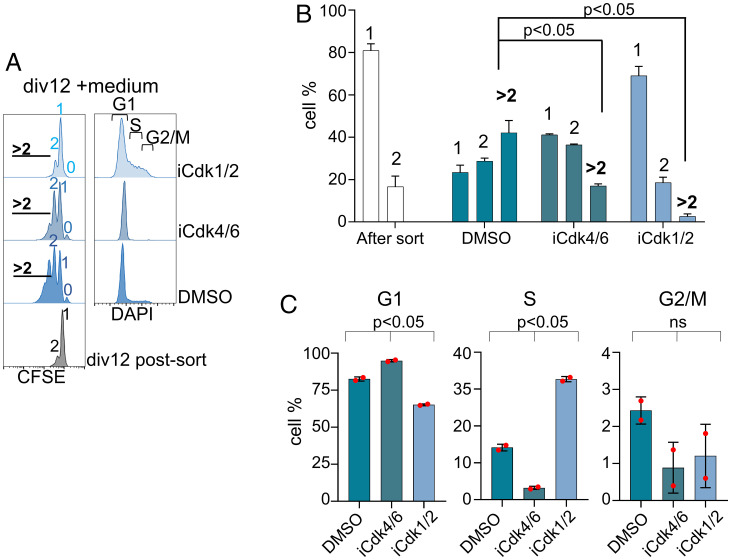
Persistent activity of CDK2 in G1 has been shown to permit a subset of tumor cell lines to circumvent the requirement for mitogen-induced activation of CDK4/6 for G1 progression (23). In contrast, CDK4/6 is required for B cell proliferation in the germinal center light zone (17). To determine whether mitogen-independent B cell proliferation was similarly regulated, we assessed the requirement for CDK1/2 and CDK4/6 using well-established pharmacologic inhibitors. Sorted div12 cells were cultured in the presence of either a CDK4/6 inhibitor (palbociclib, iCdk4/6) or a CDK1/2 inhibitor (CDK1/2 inhibitor III, iCdk1/2) for 48 h, followed by analyses of cell division and cell cycle profiles. At least three additional cell divisions were evident in cells cultured without inhibitors (Fig. 3 A and B, labeled dimethylsulfoxide (DMSO)). By contrast, a fraction of cells treated with iCdk4/6 divided once and then accumulated in G1, whereas CDK1/2 inhibition resulted in cells piling up in S/G2 without division (Fig. 3 A–C). Similar patterns were observed after activation of the div0 population with anti-IgM (SI Appendix, Fig. S2 D and E). We inferred that preexisting S/G2/M phase cells in the div12 population divided once in the presence of iCdk4/6; however, absence of progression to the next S phase demonstrated that mitogen-independent G1 progression required CDK4/6 activity. By contrast, iCdk1/2 prevented mitosis of preexisting S/G2/M cells within div12, resulting in a carboxyfluorescein succinimidyl ester (CFSE) pattern similar to that of div12 cells immediately after sorting (Fig. 3A). We conclude that primary B cells undergoing mitogen-independent proliferation require CDK4/6 kinase activity to progress through G1, paralleling the findings in GC B cells (17).
Fig. 3.
Effect of CDK inhibition on mitogen-independent B cell proliferation. Pulse anti-IgM+anti-CD40 activated cells (48 h) were purified by FACS into undivided (div0) and divided (div12) fractions. After separation, div12 cells were cultured in medium alone in the presence or absence of CDK inhibitors as indicated. CDK4/6 inhibitor (palbociclib, iCdk4/6: 1 µM) and CDK1/2 inhibitor (Cdk1/2 Inhibitor III, iCdk1/2: 10 µM). Proliferation was analyzed by CFSE dilution and cell cycle states by DNA content analysis using DAPI. (A) Representative histograms of cell division (Left) and cell cycle stages (Right) measured after a 48-h culture of purified div12 cells in medium, with no inhibitor (labeled DMSO) or CDK inhibitors as indicated. Cell division numbers are indicated within CFSE profiles. (B and C) Two independent inhibitor experiments from A were quantified to identify percentage of cells present in each division number (B) or in each stage of the cell cycle (C). Error bars represent SEM (unpaired t test).
Differences in Pre- and Postmitotic G1.
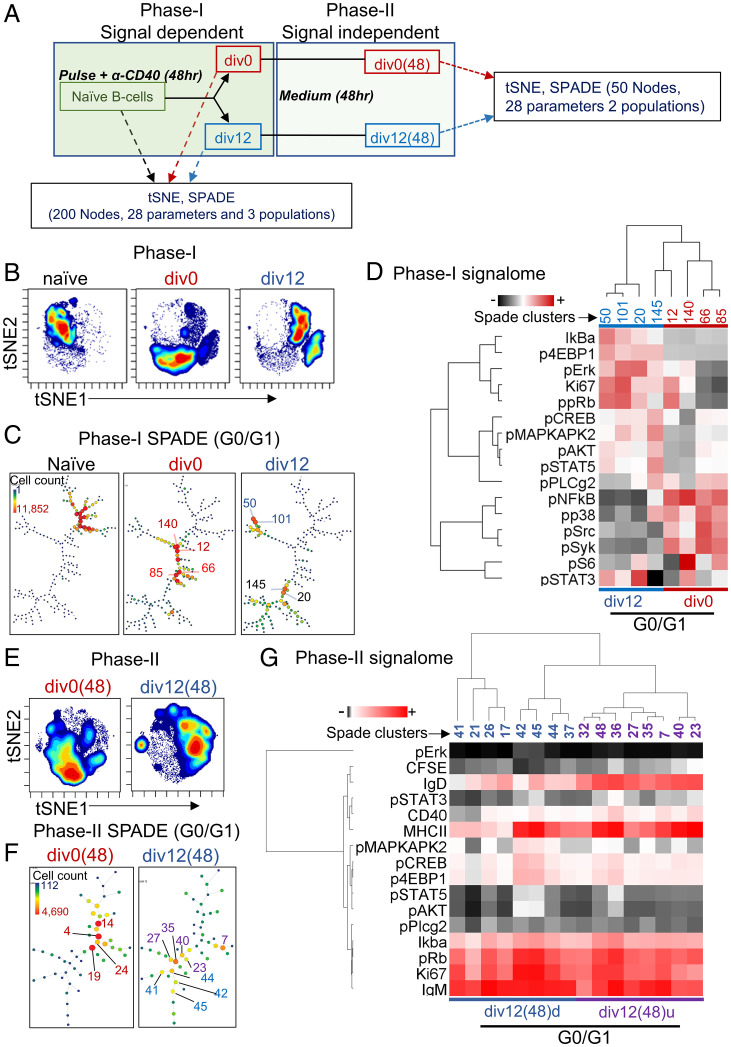
We used mass cytometry by time of flight (CyTOF) to compare G1 states of div0 and div12 cells after 48 h of mitogen-dependent culture (Fig. 4A, phase I). Purified div0 and div12 cells were pulse labeled with iododeoxyuridine (IdU) and cisplatin to identify S phase cells and dead cells, respectively. Following fixation and permeabilization, surface and cell cycle markers and signaling-related phosphoproteins were assayed as previously described (32). Remaining div0 and div12 cells were cultured for an additional 48 h before being similarly processed for CyTOF (Fig. 4A, phase II). t-distributed stochastic neighbor embedding (t-SNE) analysis using 28 parameters (SI Appendix, Fig. S3A) revealed each cell population assayed in phase I was distinct and relatively homogenous (Fig. 4B). The SPADE algorithm distributed three phase I subsets into 200 clusters. We used phospho-H3 (H3 Ser10) to identify M phase cells, cyclin B1 to identify G2 phase cells, and IdU positivity to identify S phase cells as previously described (32). Remaining clusters were attributed to G0/G1 phase cells (SI Appendix, Fig. S3B). We found that G0/G1 phases of naive, div0, and div12 cells differed greatly (Fig. 4C and SI Appendix, Fig. S3B). Clusters of naive cells grouped together, presumably reflecting a homogenous population of G0 cells. The G1 of div0 cells was distinct from naive B cells showing that these cells had responded to stimulation. That they had not divided suggests that these clusters represented different stages of G1 progression induced by continued stimulation during phase I. Why these cells did not progress all the way through the cell cycle remains to be elucidated. Strikingly, div12 cells segregated into two distinct, but well separated, G1 states that were distinct from the late G1 of div0 cells. Four G1 nodes each representing more than 10,000 cells from div0 and div12 subsets were selected for further comparison (numbered in Fig. 4C).
Fig. 4.
CyTOF analysis of B cell division. (A) Experiment design: For analysis of mitogen-dependent B cell division (phase I), CFSE-labeled B cells were activated as indicated for 48 h. IdU was added to cultures for the last 1 h prior to harvest, and undivided (div0) and divided (div12) cells were purified by flow cytometry. Each population was labeled with a panel of antibodies directed against surface and cytosolic proteins (SI Appendix, Fig. S3A). CyTOF analysis was carried out using Cytobank. For analysis of cells that had undergone mitogen-independent division (phase II), div0 and div12 cells purified at the end of phase I were further cultured in the absence of mitogen for an additional 48 h, resulting in the cell populations labeled as div0(48) and div12(48), respectively. CyTOF analysis was carried out following IdU labeling, fixation, and antibody treatments. A total of 28 parameters were used to characterize three cell populations from phase I (naive, div0, and div12) and two cell populations from phase II [div0(48) and div12(48)]. (B) t-SNE profiles of phase I cell populations as indicated. (C) SPADE analysis of G0/G1-stage cells showing distribution of 200 nodes (n = 2) in each of three cell populations from phase I. G0/G1 cells were identified by excluding S phase cells (marked by IdU) and G2/M cells marked by cyclin B and phospho-H3 (SI Appendix, Fig. S3B). Color and size coding represent cell numbers in specific nodes. Numbered nodes were selected for further analysis (D). (D) Analysis of signaling proteins in four SPADE nodes with the largest cell numbers from each of div0 and div12 cell populations. Heatmap representation of hierarchical cluster analysis carried out with the Cluster3.0 program (56), with average linkage. (E) t-SNE profile of phase II cell populations. (F) SPADE analysis phase II G1 cells showing distribution of 50 nodes (n = 2) in each cell population. Cell cycle stages, colors, and size of nodes are as described in C. Numbered nodes were selected for analysis of signaling proteins in G and SI Appendix, Fig. S3F. (G) Analysis of signaling proteins from 16 SPADE nodes of divided (d) (blue) and undivided (u) (purple) B cells of phase II div12(48) cell populations. Heatmap representation of hierarchical cluster analysis as carried out in D.
Each distinct G1 subpopulation of div12 cells was marked by high levels of Ki67, phospho-Rb (pS807/811), phospho-Akt (pT307), phospho-Erk (pT202/pY204), and phospho-STAT5 (pY694) compared to the div0 G1 subset (Fig. 4D and SI Appendix, Fig. S3D). Additionally, div12 G1 cells also contained higher levels of phospho-4EBP1 (pT37/46), a marker for activation of cap-dependent translation. Div0 G1 cells were enriched instead for signaling molecules such as phospho-Src, phospho-p38, phospho-Plcγ2, phospho–NF-κB, and lower levels of IκBα relative to div12 G1 cells (Fig. 4D). These observations demonstrated increased expression of cell cycle progression markers in div12 G1 cells while higher levels of signal-transducing phosphoproteins were evident in div0 G1 cells. We conclude that the G1 state of once divided B cells differs from the state reached after persistent signaling of naive B cells that did not divide.
We next probed the intracellular states of cells that accumulated after incubating div0 and div12 cells for an additional 48 h in the absence of stimulation [Fig. 4A, phase II, div0(48), and div12(48)]. The majority of cells in div0(48) comprised undivided cells, whereas the div12(48) population contained substantial numbers of cells that had undergone mitogen-independent division (SI Appendix, Fig. S3E). Accordingly, t-SNE analysis of div0(48) cells showed one major cell population, whereas div12(48) cells were more heterogenous (Fig. 4E). Because of reduced live cell numbers in phase II cells, we partitioned them into 50 clusters using the SPADE algorithm (Fig. 4F). Div0(48) G1 cells conspicuously lacked Ki67 compared to div12(48) G1 cells despite having comparable levels of phospho-Rb (SI Appendix, Fig. S3F, nodes numbered in Fig. 4F). Receptor-proximal signaling proteins, pSrc and pSyk, were largely inactive in both populations (SI Appendix, Fig. S3F) and a subpopulation of div12(48) G1 cells were enriched for p4EBP1, pCREB, and pMAPKAPK2 (SI Appendix, Fig. S3F).
To understand why only a subset of div12 cells undergoes mitogen-independent proliferation, we further selected 16 nodes comprising cells in G1 within div12(48). Using CFSE levels to distinguish cells that had undergone mitogen-independent proliferation (Fig. 4F, blue) from those that had not (Fig. 4F, purple), we found that surface IgD expression correlated inversely with cell division (Fig. 4G). Lower levels of MHCII expression were also evident on a subset of divided cells. Cells that had divided most (nodes 42 and 45) had relatively higher levels of pErk, pMAPKAPK2, pCREB, p4EBP1, pSTAT5, and pAKT compared to all other subpopulations (Fig. 4G). Many of these phosphoproteins also distinguished div12 from div0 cells during phase I (mitogen-dependent) cell division. We conclude that B cells can undergo at least three to four rounds of mitogen-independent cell division by maintaining and propagating a modified G1 state.
The Postmitotic Transcriptome.
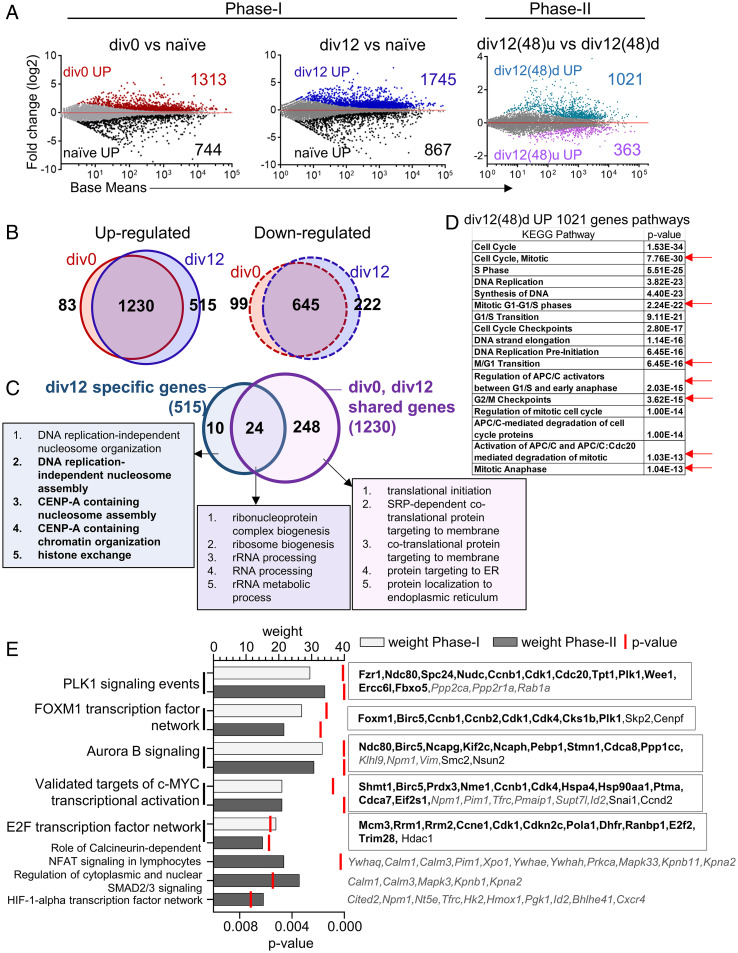
To search for possible mechanisms by which such different G1 states were inherited or maintained in div0 versus div12 cells, we carried out total RNA sequencing (RNA-seq) from different cell subsets (SI Appendix, Fig. S4A). Most genes, and associated biological pathways (gene ontologies [GO]), that were either up- or down-regulated in div0 or div12 cells compared to their naive precursors during the first 48 h culture were common to both subsets (Fig. 5 A and B, phase I and SI Appendix, Fig. S4 B and C). Fewer genes were uniquely down- (222) or up- regulated (515) in div12 cells compared to div0 cells, that might confer the unique proliferative properties of div12 compared to div0 cells. Div12 selective up-regulated genes revealed enrichment of pathways related to centromeric chromatin and cellular metabolism (Fig. 5C and SI Appendix, Fig. S4D). Several of the identified chromatin-associated pathways were related to processes that occur in G2/M and postmitotic early G1 phases of the cell cycle (33). These observations suggested either that 1) G2/M phase RNA dominated the transcriptome of div12 cells or 2) G2/M-related RNAs were also present in G1 phase cells that comprised the largest subpopulation within div12. Because only 2% of div12 cells were in G2/M (Fig. 2C), we favor the interpretation that div12 G1 cells express several G2/M-selective RNAs. GO analysis for div12-selective down-regulated genes were enriched for lymphocyte activation, differentiation, and adhesion pathways (SI Appendix, Fig. S4E).
Fig. 5.
Transcriptomes of B cells undergoing mitogen-dependent and mitogen-independent division. Total RNA was prepared from three cell populations of mitogen-dependent phase I (naive, div0, and div12) and two cell populations of mitogen-independent phase II [div12(48)d and div12(48)u corresponding to div12 cells that had divided or not during mitogen-independent culture]. Following RNA-seq differentially expressed genes were identified using DESeq2 with false discovery rate (FDR) ≤0.05 for phase I and ≤0.1 for phase II (n = 2). (A) MA plots showing differentially expressed genes during phase I and phase II proliferation. For phase I analysis, gene expression was compared between div0 and naive B cells or between div12 and naive B cells (Left). For phase II analysis, gene expression was compared between cells that had further divided in absence of signals [div12(48)d) and cells that had not divided further (div12(48)u] (Right). (B) Venn diagrams showing overlap of up-regulated (Left) and down-regulated (Right) genes in div0 and div12 when compared to naive cells. Numbers indicate genes in each category. (C) GO analysis was carried out using up-regulated genes that were shared by both div0 and div12 populations (1,230 genes in B, corresponding to 272 GOs) or genes that were uniquely up-regulated in div12 cells (515 genes in B, corresponding to 34 GOs). Venn diagram shows the overlap between shared and unique GO categories. Top 5 GO categories (based on P ≤ 0.05; SI Appendix, Fig. S4 B and D) are shown. (D) Top Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched from up-regulated (1,021) genes of phase II divided B cells div12(48)d. Red arrow indicate the mitosis specific pathways. (E) PID (57) analysis was carried out with genes that were up-regulated during phase I (shared between div0 and div12 cells) or up-regulated during phase II [div12(48)d compared to div12(48)u]. Enriched PIDs were scored by P value and weight (proportion of PID genes identified in the differential gene expression analysis) (Left). Genes shared between phase I and phase II cells are shown in bold (Right). Genes restricted to phase II are shown in italics while phase I–specific genes are nonbold.
We further compared the transcriptomes of cells that had or had not undergone additional (mitogen-independent) cell division during 48 h of phase II culture and found 1,021 genes to be expressed at higher levels in cells that divided during this phase (Fig. 5 A, Right and SI Appendix, Fig. S4A). These genes enriched for biological processes such as mitotic cell cycle, protein modification, and biosynthesis (SI Appendix, Fig. S4F), including G2/M-specific pathways (Fig. 5D, red arrows). By contrast, genes whose expression was higher in cells that did not divide during phase II culture were conspicuously devoid of cell cycle–related biological pathways (SI Appendix, Fig. S4G). Thus, cells permissive to undergo mitogen-independent division express a cell cycle–related transcriptome.
To search for possible networks that drive mitogen-dependent (phase I) or independent (phase II) cell cycle progression, a pathway interaction database (PID) analysis was performed. PID analysis identified five pathways that were common to phases I and II and three that were unique to phase II (Fig. 5E). All five shared pathways corresponding to processes involved in cell cycle progression. Of these, PLK1 signaling, the FOXM1 transcription factor network, and aurora B signaling (SI Appendix, Fig. S4 H–J, respectively) are associated with G2/M stages of the cell cycle (34, 35). PID networks uniquely present in phase II cells were related to calcium signaling via calmodulin, nucleocytoplasmic transport via karyopherins, and 14-3-3 proteins and HIF-1α transcription factor network genes (Fig. 5E). The evidence of G2-like characteristics of div12 cells drew our attention to G2/M-specific RNAs that dominated the transcriptome of dividing B cells.
G2/M Features in G1 of Divided B Cells.
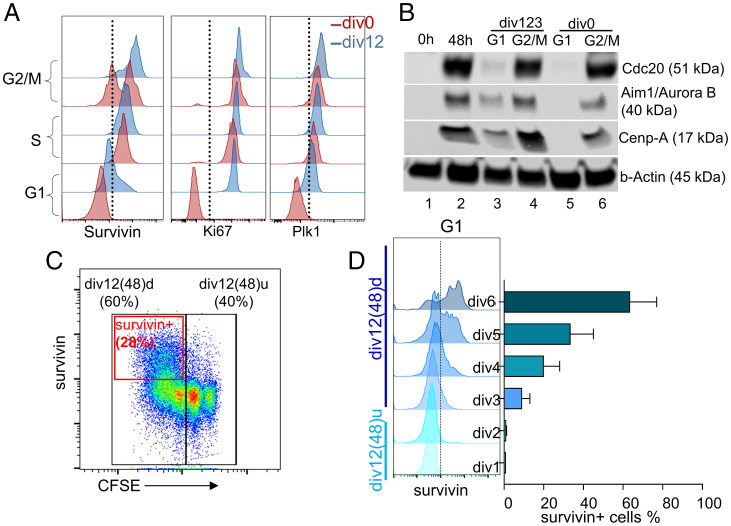
To further substantiate G2/M features in the G1 phase of cells competent to undergo mitogen-independent division, we reexamined the CyTOF data. We found several G2/M phase proteins were selectively expressed in G1 phase clusters of divided (div12) compared to nondivided (div0) cells (SI Appendix, Figs. S5A and S3D). The predominance of CENP-related pathways in the div12 transcriptome prompted us to query the expression of Birc5 (also known as survivin), a G2/M-specific factor in transformed cells, where it assists mitotic chromosome segregation via interaction with centromeric proteins (36, 37). Survivin is also an integral part of FOXM1 transcription factor and aurora B signaling networks that featured in our PID analyses (SI Appendix, Fig. S4 I and J). Consistent with this mechanism, survivin has been shown to be important for G2/M progression in developing B cells and in proliferating germinal center B cells (38). We found that Birc5 mRNA and protein were expressed at higher levels in div12 compared to div0 cells and maintained thereafter in cells that had undergone three to four rounds of mitogen-independent division (SI Appendix, Fig. S5 B and C). Additionally, Birc5 mRNA expression in phase I and phase II correlated with Foxm1, Aurkb, and Plk1 mRNA (SI Appendix, Fig. S5D). To identify the cell cycle stage at which survivin was expressed, we costained phase I cells with 5-Ethynyl-2′-deoxyuridine (EdU) and anti-survivin antibody for flow cytometry. As expected, survivin was expressed at high levels in S and G2/M stages of both div0 and div12 cells (Fig. 6A); however, only div12 cells expressed survivin at the G1 stage (Fig. 6A and SI Appendix, Fig. S5E). Two other S/G2/M markers Ki67 and Plk1 showed the same pattern of expression as Birc5 (Fig. 6A).
Fig. 6.
G2/M features in G1 phase of dividing B cells. (A) Representative histograms demonstrating cell cycle stage–specific expression of survivin, Ki67, and Plk1. For survivin, after the first 48 h of B cell activation (pulse anti-IgM+anti-CD40), cell cycle stages were identified as in Fig. 2C, using pulse EdU labeling and DAPI (n = 3). Ki67 and Plk1 expression is after 48 h of LPS-stimulated B cells; cell cycle stages were identified by DNA content analysis as in Fig. 3A (n = 2). (B) Immunoblots for G2/M-specific proteins from whole cell extracts of FUCCI mouse splenic B cells (representative of two independent experiments). Resting (lane 1) and 48 h LPS-activated B cells (lane 2), G1 (lane 3), and G2/M (lane 4) stages of divided B cells (div123) and G1 (lane 5) and G2/M (lane 6) stages of undivided B cells (div0) after 48 h of LPS activation. (C) Survivin expression after 48 h of mitogen-independent division of div12 cells was assayed by flow cytometry. Black boxes represent cell division states; div12(48)u cells did not divide during culture, div12(48)d cells divided during culture. Red box marks survivin-positive cells. (D) G1 stage–specific survivin expression in cells that have undergone different numbers of divisions from div12 cells during 48 h of mitogen-independent culture. Representative histograms (Left) and quantification (Right; mean ± STD, n = 2).
To further examine expression of G2/M markers in B cells that underwent mitogen-induced division, we isolated G1 and SG2M stage cells from div0 and div123 B cell populations using fluorescence ubiquitination cell cycle indicator (FUCCI) mice (39) (SI Appendix, Fig. S5F). Whole cell extracts derived from these cells were probed for expression of G2/M markers that were not readily evaluated by flow cytometry. Expression of Cdc20, Aim1/aurora B, and Cenp-A was readily apparent by immunoblotting extracts obtained from G1 phase of div12 cells but not of div0 cells (Fig. 6B). All three proteins were expressed at higher levels in S/G2/M cells from both div12 and div0 populations. Taken together, we conclude that B lymphocytes that have completed one division express several G2/M markers in the G1 phase. Additionally, survivin expression in G1 persisted over the course of mitogen-independent division, especially in cells that had divided most during culture (Fig. 6 C, red box and D). We hypothesized that survivin is a key determinant of mitogen-independent progression to S phase.
The Role of Survivin in G1–S Progression in Divided B Cells.
A role for survivin at the G2/M stage of mitogen-dependent B cell proliferation has previously been described (38). Evaluation of its role in signal-independent G1 progression required a different experimental design than use of survivin-deficient B cells. We first tested our hypothesis in a cell-line model. For this we generated FUCCI-BJAB cells in which different cell cycle stages could be purified based on red and green fluorescence (39) (SI Appendix, Fig. S6A). Survivin expression was apparent throughout G1 (SI Appendix, Fig. S6B). To evaluate the role of survivin at different stages of the cell cycle, we sorted G2/M or early G1 FUCCI-BJAB cells and cultured them for an additional 18 h in the presence or absence of LLP3 (a blocker of survivin–Ran interaction) (40) or S12 (a pseudoallosteric survivin antagonist) (41). Consistent with its known function in mitosis, both survivin inhibitors reduced cell cycle progression in G2/M cells (SI Appendix, Fig. S6 C, Upper). However, both inhibitors also markedly reduced S phase reentry of early G1 cells (SI Appendix, Fig. S6 C, Lower), indicating an essential role for survivin in late G1.
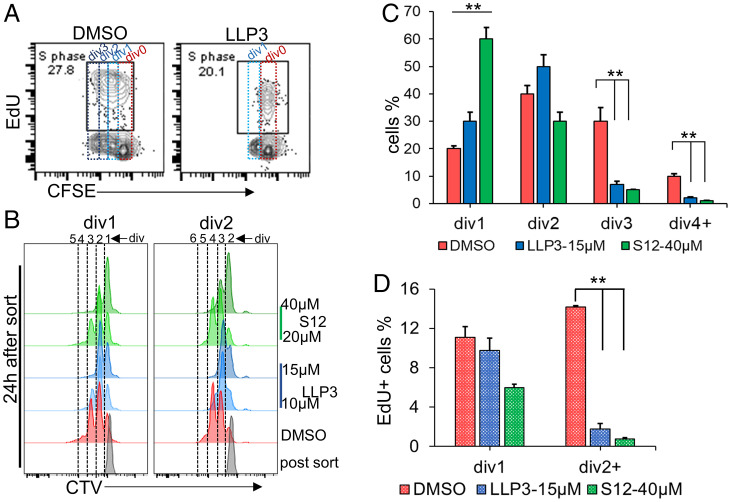
To test the role of survivin during phase I (mitogen-dependent) proliferation of primary B cells, LLP3 (15 µM) or vehicle (DMSO) was added just after anti-IgM pulse + anti-CD40 activation and cells were analyzed for division by CFSE dilution and S phase by EdU incorporation at 54 h. DMSO-treated cells showed three divisions and 27% EdU+ S phase cells (Fig. 7A), while LLP3-treated cells showed only one division and 20% EdU+ S phase cells (Fig. 7A). EdU+ S phase cells were present after each division of DMSO-treated cells (SI Appendix, Fig. S7 A, Top) while in LLP3-treated cultures, S phase cells were only observed in div0 population (SI Appendix, Fig. S7 A, Bottom). These observations indicate that pharmacological inhibition of survivin blocks G1 to S transition beyond the first mitotic division, but not progression through the first cell cycle.
Fig. 7.
Effects of pharmacological inhibition of survivin on B cell division. (A) Splenic B cells were activated with pulse anti-IgM+anti-CD40 in the presence of 15 µM LLP3 or vehicle (DMSO). Cell division and S phase were analyzed by CFSE dilution and EdU incorporation, respectively, at 54 h. CFSE dilution and EdU-positive (S phase) cells after 54 h, DMSO (Left) LLP3 (Right), cell divisions are shown by dotted boxes. (B) Survivin inhibition in LPS-stimulated B cells. After 48 h, cells that had divided once (div1) or twice (div2) were sorted and further cultured for 24 h in the presence or absence of indicated dosages of two survivin inhibitors, S12 or LLP3. CTV dilution histograms of phase II proliferation after 24 h in div1 and div2 populations (B) and quantified over two independent experiments for div1 population for indicated dosages of inhibitors (C). (D) S phase cells from div1 cultures (C) were identified by pulsing with EdU for 1 h. Bar graphs represent EdU-positive cells after a 24-h phase II culture. Cells that had divided during culture (div2, div3, and div4) are labeled as div2+. *P ≤ 0.05 and **P ≤ 0.001 in Student’s t test, respectively.
We further assessed the effect of survivin inhibitors in mitogen-independent B cell proliferation. Divided (div1 or div2) cells obtained after 48 h lipopolysaccharide (LPS) activation underwent an additional two to three divisions upon 24 h of secondary culture in the absence of mitogen (Fig. 7B, DMSO). Both LLP3 and S12 significantly reduced mitogen-independent proliferation in a dose-dependent manner (Fig. 7B and quantified for div1 cells in Fig. 7C). EdU pulse labeling of div1 cells during secondary culture revealed that divided cells, div2+ (consisting of div2, div3, and div4) had significantly reduced numbers of cells in S phase (Fig. 7D). EdU+ cells that did not divide during mitogen-independent proliferation (div1) presumably reflected the well-known role of survivin during G2/M and possibly S phase. Similar observations were made for the divided (div12) B cells obtained after pulse BCR + anti-CD40 activation. Div12 cells that had not been treated with inhibitor underwent one to two additional rounds of mitogen-independent division. By contrast, additional cell division was inhibited in the transition to EdU+ cells by LLP3 (SI Appendix, Fig. S7 B–D). These observations are consistent with an essential role for survivin in S phase reentry during mitogen-independent B cell proliferation.
Discussion
Humoral responses are characterized by two critical proliferative phases. The first occurs during clonal expansion of a tiny fraction of antigen responsive cells. The second occurs during the germinal center reaction that results in generation of effector cells and memory. The extent of lymphocyte proliferation has been linked to the concept of division destiny, a cell autonomous condition that is proposed to be the cumulative outcome of multiple signals that impinge on responding cells (11). Mechanisms that underly multiple rounds of cell division during clonal expansion remain unclear. Much more is known about B cell proliferation in the GC, including the puzzling observation that signaling occurs in the GC light zone, whereas the bulk of cell division occurs in the dark zone in the apparent absence of cell stimulation (14). Proliferation in the GC is controlled by Myc (12) and recently shown to be driven by cyclin D3 and Cdk4/6 (17). Here we show that the G1 state of B lymphocytes after the first cell division is distinct from the G1 state of undivided activated B cells. This state is characterized by phosphoproteins and RNA that are hallmarks of G2/M (42). We propose that, contrary to prevailing dogma, B cells carry over hallmarks of G2 into the postmitotic G1 phase. This state permits G1 progression in a CDK4/6-dependent manner without requiring renewed mitogenic stimulation. Importantly, B cells undergoing mitogen-independent proliferation maintained their size over several divisions, likely due to enhanced metabolic activity (43). During the time course of our in vitro studies, mitogen-independent proliferation of B cells was limited only by accentuated cell death, which could be attenuated via nonmitogenic CD40 stimulation. It is quite possible that the extent of mitogen-independent proliferation in vivo is further regulated by other survival signals, such as those resulting from Cxcl12/Cxcr4 interactions in the GC dark zone (44, 45). We propose that the ability of B cells to undergo multiple cell divisions with limited stimulation is an important determinant of clonal expansion and cell proliferation in GCs.
These observations raised two related questions. First, how do features of G2/M phases persist in the postmitotic G1 phase and second, what drives continued mitogen-independent cell division? The prevailing view is PP1 and PP2A activity in late G2/M dephosphorylates key proteins, such as pRb, p130 (42), and AKT (46) and also inactivates Ras signaling (47), thereby resetting the clock in the newly generated G1. Because many of these targets remained phosphorylated in postmitotic G1, we infer that phosphatase activity is attenuated in G2/M of primary B lymphocytes, thereby leading to inheritance of a partially activated G1 after cell division. Reduced PP2A activity may underlie elevation of the Ca2+/calcineurin pathway in postmitotic cells noted in our PID analysis (48, 49). Two other pathways that were highlighted by the PID analysis, SMAD signaling and HIF-1α network, also connect to GC biology (50, 51). However, their role in clonal expansion, if any, remains unknown. We also observed elevated Myc protein levels in postmitotic G1 and our transcriptomic analyses revealed up-regulation of Myc target genes (52) during mitogen-independent cell division. We hypothesize that persistent Myc expression contributes to mitogen-independent cell division by maintaining an elevated metabolic state. Attempts to evaluate the role of PI3K or downstream effectors such as mTOR using pharmacologic inhibitors were unsuccessful due to excessive cell death in cultures treated with inhibitors.
We identified genes relevant for G2/M by comparing transcriptomes and annotated pathway interaction database of cells that were permissive for mitogen-independent proliferation. Among these genes, Birc5 (survivin) caught our attention as one with prior connection to multiple cell cycle stages. Though primarily known as a factor important for chromosome segregation and survival in G2/M (38, 53), survivin has also been proposed to activate CDK4/6 (54) and for G1 progression of primed CD4+ T cells (55). We found that survivin expression in G1 correlated with the propensity for mitogen-independent cell division and was highest in cells that had divided multiple times without exogenous signals. Importantly, modulating survivin function with two highly selective, but mechanistically distinct, small molecule inhibitors of survivin reduced mitogen-independent cell division, thereby identifying it as an essential component of this form of G1-to-S progression. Whether survivin expression is sufficient for mitogen-independent cell divisions remains to be established.
In summary, we provide evidence that primary B cells undergo several rounds of cell division without requiring renewed G1 progression signals after each mitosis. Thus, requirements for B cell proliferation are distinct from current views of the cell cycle and more in line with properties of tumor cells, a proportion of which have been shown to be capable of G1 progression in the absence of growth factor signals. However, primary B cells undergoing such mitogen-independent proliferation lack survival signals that are a hallmark of transformed cells. It is possible that microenvironmental signals in secondary lymphoid organs cooperate with this property of B cells to tune the extent of expansion during immune responses.
Materials and Methods
Mice and Cell Cultures.
All experimental mice were 8- to 12-wk-old C57BL/6J and FUCCI mice. Splenic primary B cells were isolated using negative magnetic enrichment. B cells were labeled with cell trace dyes either CFSE or CellTrace Violet (CTV) according to manufacturer protocol. Mitogenic activations were done using goat anti-mouse IgM F(ab′)2 (Jackson Immunoresearch) and anti-CD40 as described earlier (31) or with LPS and CpG. Details of inhibitors and other reagents are provided in SI Appendix, Table S1.
FACS, Flow Cytometry, CyTOF, and Western Blotting.
Divided and nondivided B cells were sorted based on cell trace dye levels using fluorescence-activated cell sorting (FACS) Aria II or FACS Aria Fusion (BD Biosciences). Antibodies used for flowcytometry and CyTOF are listed in SI Appendix, Table S2. Intracellular staining, EdU Click-iT, and cell cycle analysis details are available in method sections of SI Appendix, Materials and Methods. Whole cell extracts were made using radioimmunoprecipitation assay (RIPA) buffer and proteins were separated by electrophoresis through 4 to 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane. Membranes were incubated with respective antibodies, and blots were developed using the enhanced chemiluminescence (ECL).
RNA-Seq and Analysis.
Total RNA from 1 × 106 (phase I) and 5 × 105 (phase II) cells was isolated using the Qiazol (Qiagen) kit according to the manufacturer’s protocol. Ribosomal RNA was depleted from 100 ng of total RNA and a barcoded library was made and 50-bp single end reads were generated from each sample using standard protocol for NextSEq 500 sequencer. Reads were analyzed using the online server Galaxy RNA-seq pipeline (usegalaxy.org). RNA-seq data are deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession no. GSE195698.
Statistical Analysis and Graphs.
Statistical tests and graphs were created using Prism (version 7.0) (GraphPad) or Excel (Microsoft). Student’s t test (two tailed) was used to determine the statistical significance and the results are represented as *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
Detailed method descriptions are in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank all the members of the R.S. laboratory in the Gene Regulation Section for valuable discussion. We also thank Dr. Ananda Roy and Dr. Anindya Dutta for critical suggestions during manuscript preparation. We thank Dr. Isabel Beerman (National Institute on Aging [NIA]) for sharing the FUCCI mice. This work was supported in part by the Intramural Research Program of the NIH, NIA.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115567119/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in Gene Expression Omnibus (GEO) (GSE195698).
References
- 1.Sablitzky F., Wildner G., Rajewsky K., Somatic mutation and clonal expansion of B cells in an antigen-driven immune response. EMBO J. 4, 345–350 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langhoff E., Steinman R. M., Clonal expansion of human T lymphocytes initiated by dendritic cells. J. Exp. Med. 169, 315–320 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenschow D. J., Walunas T. L., Bluestone J. A., CD28/B7 system of T cell costimulation. Annu. Rev. Immunol. 14, 233–258 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Liu Y., C. A. Janeway, Jr, Cells that present both specific ligand and costimulatory activity are the most efficient inducers of clonal expansion of normal CD4 T cells. Proc. Natl. Acad. Sci. U.S.A. 89, 3845–3849 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duffy K. R., et al. , Activation-induced B cell fates are selected by intracellular stochastic competition. Science 335, 338–341 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Duffy K. R., Hodgkin P. D., Intracellular competition for fates in the immune system. Trends Cell Biol. 22, 457–464 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Dowling M. R., et al. , Stretched cell cycle model for proliferating lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 111, 6377–6382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossman Z., Paul W. E., Dynamic tuning of lymphocytes: Physiological basis, mechanisms, and function. Annu. Rev. Immunol. 33, 677–713 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Turner M. L., Hawkins E. D., Hodgkin P. D., Quantitative regulation of B cell division destiny by signal strength. J. Immunol. 181, 374–382 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Marchingo J. M., et al. , T-cell stimuli independently sum to regulate an inherited clonal division fate. Nat. Commun. 7, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinzel S., et al. , A Myc-dependent division timer complements a cell-death timer to regulate T cell and B cell responses. Nat. Immunol. 18, 96–103 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Finkin S., Hartweger H., Oliveira T. Y., Kara E. E., Nussenzweig M. C., Protein amounts of the MYC transcription factor determine germinal center B cell division Capacity. Immunity 51, 324–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habib T., et al. , Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J. Cell Biol. 179, 717–731 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Victora G. D., et al. , Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesin L., Ersching J., Victora G. D., Germinal center B cell dynamics. Immunity 45, 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo W., Weisel F., Shlomchik M. J., B cell receptor and CD40 signaling are rewired for synergistic induction of the c-Myc transcription factor in germinal center B cells. Immunity 48, 313–326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pae J., et al. , Cyclin D3 drives inertial cell cycling in dark zone germinal center B cells. J. Exp. Med. 218, e20201699 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaech S. M., Ahmed R., Memory CD8+ T cell differentiation: Initial antigen encounter triggers a developmental program in naïve cells. Nat. Immunol. 2, 415–422 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Stipdonk M. J., Lemmens E. E., Schoenberger S. P., Naïve CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat. Immunol. 2, 423–429 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Pardee A. B., G1 events and regulation of cell proliferation. Science 246, 603–608 (1989). [DOI] [PubMed] [Google Scholar]
- 21.Zetterberg A., Larsson O., Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc. Natl. Acad. Sci. U.S.A. 82, 5365–5369 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldin V., Lukas J., Marcote M. J., Pagano M., Draetta G., Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 7, 812–821 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Spencer S. L., et al. , The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L., et al. , G1 cyclins link proliferation, pluripotency and differentiation of embryonic stem cells. Nat. Cell Biol. 19, 177–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H. W., Chung M., Kudo T., Meyer T., Competing memories of mitogen and p53 signalling control cell-cycle entry. Nature 549, 404–408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe M., Moon K. D., Vacchio M. S., Hathcock K. S., Hodes R. J., Down modulation of tumor suppressor p53 by T cell receptor signaling is critical for antigen-specific CD4(+) T cell responses. Immunity 40, 681–691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Defranco A. L., Raveche E. S., Asofsky R., Paul W. E., Frequency of B lymphocytes responsive to anti-immunoglobulin. J. Exp. Med. 155, 1523–1536 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pardee A. B., A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 71, 1286–1290 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cappell S. D., Chung M., Jaimovich A., Spencer S. L., Meyer T., Irreversible APC(Cdh1) inactivation underlies the point of no return for cell-cycle entry. Cell 166, 167–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rush J. S., Hodgkin P. D., B cells activated via CD40 and IL-4 undergo a division burst but require continued stimulation to maintain division, survival and differentiation. Eur. J. Immunol. 31, 1150–1159 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Damdinsuren B., et al. , Single round of antigen receptor signaling programs naive B cells to receive T cell help. Immunity 32, 355–366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behbehani G. K., Bendall S. C., Clutter M. R., Fantl W. J., Nolan G. P., Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A 81, 552–566 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bui M., et al. , Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell 150, 317–326 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu Z., et al. , Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat. Cell Biol. 10, 1076–1082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lampson M. A., Cheeseman I. M., Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21, 133–140 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altieri D. C., Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer 8, 61–70 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Carmena M., Wheelock M., Funabiki H., Earnshaw W. C., The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13, 789–803 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miletic A. V., et al. , Essential role for survivin in the proliferative expansion of progenitor and mature B cells. J. Immunol. 196, 2195–2204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakaue-Sawano A., et al. , Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Guvenc H., et al. , Impairment of glioma stem cell survival and growth by a novel inhibitor for Survivin-Ran protein complex. Clin. Cancer Res.J. Am. Assoc. Cancer Res. 19, 631–642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berezov A., et al. , Disabling the mitotic spindle and tumor growth by targeting a cavity-induced allosteric site of survivin. Oncogene 31, 1938–1948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolupaeva V., Janssens V., PP1 and PP2A phosphatases--cooperating partners in modulating retinoblastoma protein activation. FEBS J. 280, 627–643 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Lee Y., et al. , Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature 510, 547–551 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barinov A., et al. , Essential role of immobilized chemokine CXCL12 in the regulation of the humoral immune response. Proc. Natl. Acad. Sci. U.S.A. 114, 2319–2324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teicher B. A., Fricker S. P., CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res.J. Am. Assoc. Cancer Res. 16, 2927–2931 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Kuo Y. C., et al. , Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J. Biol. Chem. 283, 1882–1892 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Naetar N., et al. , PP2A-mediated regulation of Ras signaling in G2 is essential for stable quiescence and normal G1 length. Mol. Cell 54, 932–945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruvolo P. P., The broken “Off” switch in cancer signaling: PP2A as a regulator of tumorigenesis, drug resistance, and immune surveillance. BBA Clin. 6, 87–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wlodarchak N., et al. , Structure of the Ca2+-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation. Cell Res. 23, 931–946 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albright A. R., et al. , TGFβ signaling in germinal center B cells promotes the transition from light zone to dark zone. J. Exp. Med. 216, 2531–2545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho S. H., et al. , Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature 537, 234–238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iritani B. M., Eisenman R. N., c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. U.S.A. 96, 13180–13185 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li F., et al. , Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396, 580–584 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Suzuki A., et al. , Survivin initiates cell cycle entry by the competitive interaction with Cdk4/p16(INK4a) and Cdk2/cyclin E complex activation. Oncogene 19, 3225–3234 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Song J., Salek-Ardakani S., So T., Croft M., The kinases aurora B and mTOR regulate the G1-S cell cycle progression of T lymphocytes. Nat. Immunol. 8, 64–73 (2007). [DOI] [PubMed] [Google Scholar]
- 56.de Hoon M. J., Imoto S., Nolan J., Miyano S., Open source clustering software. Bioinformatics 20, 1453–1454 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Schaefer C. F., et al. , PID: The Pathway Interaction Database. Nucleic Acids Res. 37, D674–D679 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in Gene Expression Omnibus (GEO) (GSE195698).