Fig. 1 |. Discovery of a 4EP biosynthetic pathway, strain engineering of gut bacteria, and colonization of mice to produce 4EPS.
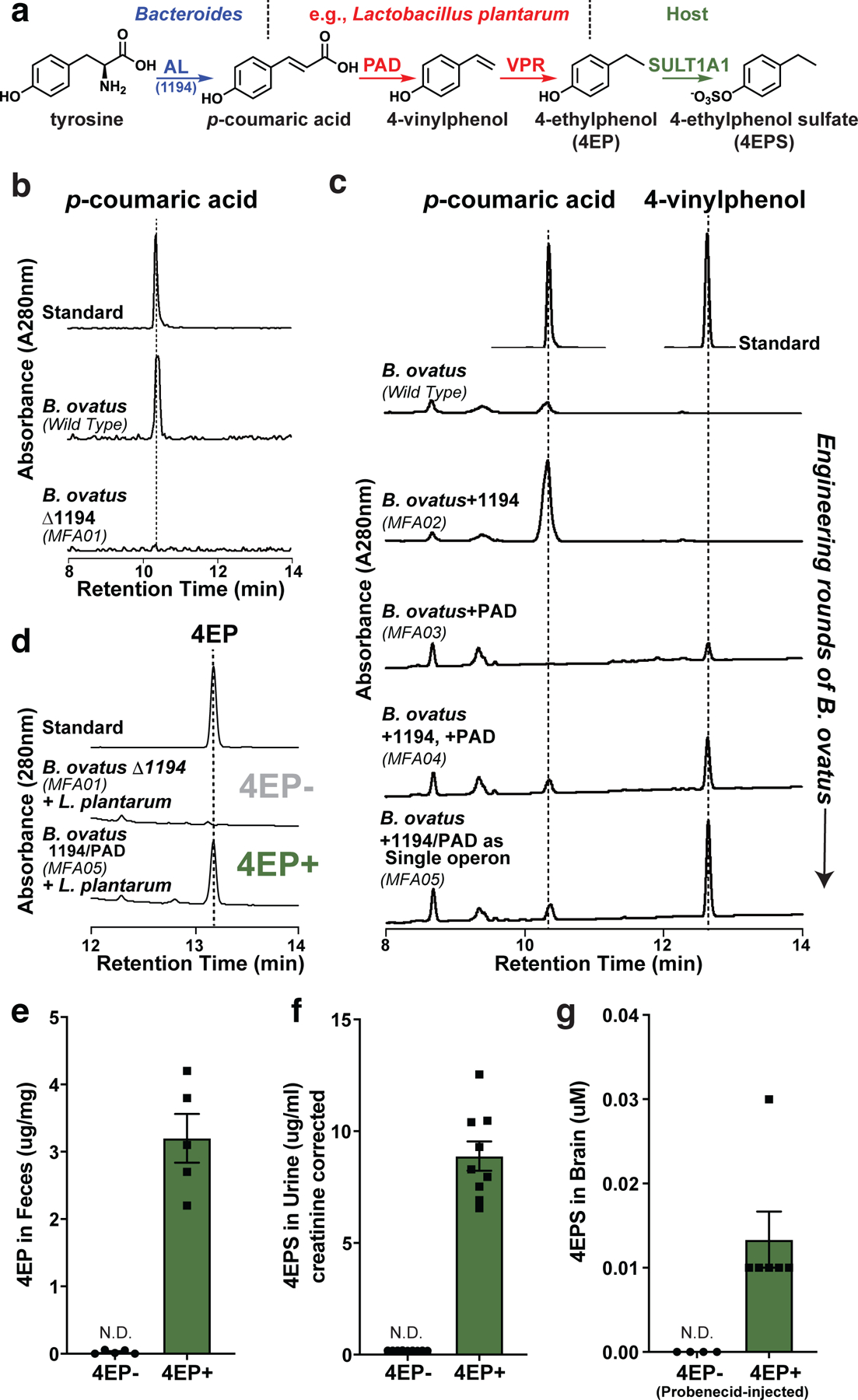
a, Proposed biosynthetic pathway of 4EP in gut bacteria via tyrosine ammonia lyase (AL), phenolic acid decarboxylase (PAD), and vinyl phenol reductase (VPR) enzymes, and subsequent sulfation to 4EPS by host sulfotransferase (SULT1A1). The genes identified and cloned for bacterial strain engineering can convert tyrosine to 4EP via two chemical intermediates. b and c, 4EP precursors, p-coumaric acid and 4-vinylphenol from in vitro cultures of Bacteroides ovatus strains provided tyrosine as substrate. b, Conversion, or lack thereof, of tyrosine to p-coumaric acid by BACOVA_01194 gene (AL/1194 in figure) and deletion mutant (MFA01), respectively. c, The BACOVA_01194 gene (AL/1194 in figure) was introduced separately (MFA02), in addition to the pad gene (MFA04), or both genes as a single, highly expressed operon (MFA05). Standards are shown at top. Further details in Extended Data Figure 1, Methods, and Supplementary Information Tables 1-3. d, 4EP levels from in vitro cultures (provided with tyrosine) of 4EP- strain pair (B. ovatus Δ1194 (MFA01) and Lactobacillus plantarum) or the highest 4-vinylphenol producing engineered strain, B. ovatus 1194/PAD (MFA05) with L. plantarum (4EP+ strain pair). A standard is shown at the top. e, Levels of 4EP in feces (μg/mg) of mice colonized with bacterial strains lacking (4EP-) and producing (4EP+) 4EP (n=5). f, Levels of 4EPS in urine of mice colonized with 4EP- or 4EP+ (n=9) bacterial strains. Additional urine data, Extended Data Fig. 1k. g, Brain levels of 4EPS (after injection with probenecid) in mice colonized with 4EP- (n=4) and 4EP+ (n=6) bacterial strains. Additional controls, Extended Data Fig. 1f. At least two biologically independent trials were performed for each experiment. Abbreviations: N.D., not detectable. Data in panels e-g represent mean ± SEM.
