Significance
Unlike fellow nonsegmented negative-strand RNA viruses, exemplified by the devastating Nipah, Ebola, rabies, and measles viruses, vesicular stomatitis virus (VSV) can be considered beneficial, as it is widely used as a vector for anticancer therapy and vaccine development. In these RNA viruses, transcription and replication of the viral genome depend on an RNA-dependent RNA polymerase. Here, we determined the in situ structure of the VSV polymerase complex, consisting of a large protein (L) and a phosphoprotein (P), by cryo-electron tomography and subtomogram averaging. Approximately 55 polymerase complexes are packaged in each bullet-shaped virion through flexible interactions with nucleoproteins. Our results provide insights into the mechanism of L packaging during virus assembly and efficient initiation of transcription during infection.
Keywords: NNS RNA virus, vesicular stomatitis virus, polymerase complex, cryo-electron tomography, subtomogram average
Abstract
The polymerase complex of nonsegmented negative-strand RNA viruses primarily consists of a large (L) protein and a phosphoprotein (P). L is a multifunctional enzyme carrying out RNA-dependent RNA polymerization and all other steps associated with transcription and replication, while P is the nonenzymatic cofactor, regulating the function and conformation of L. The structure of a purified vesicular stomatitis virus (VSV) polymerase complex containing L and associated P segments has been determined; however, the location and manner of the attachments of L and P within each virion are unknown, limiting our mechanistic understanding of VSV RNA replication and transcription and hindering engineering efforts of this widely used anticancer and vaccine vector. Here, we have used cryo-electron tomography to visualize the VSV virion, revealing the attachment of the ring-shaped L molecules to VSV nucleocapsid proteins (N) throughout the cavity of the bullet-shaped nucleocapsid. Subtomogram averaging and three-dimensional classification of regions containing N and the matrix protein (M) have yielded the in situ structure of the polymerase complex. On average, ∼55 polymerase complexes are packaged in each virion. The capping domain of L interacts with two neighboring N molecules through flexible attachments. P, which exists as a dimer, bridges separate N molecules and the connector and C-terminal domains of L. Our data provide the structural basis for recruitment of L to N by P in virus assembly and for flexible attachments between L and N, which allow a quick response of L in primary transcription upon cell entry.
Nonsegmented negative-strand (NNS) RNA viruses are a group of highly diversified eukaryotic viruses that include significant human pathogens such as rabies, measles, Nipah, and Ebola. Our knowledge of the molecular mechanisms of transcription and replication of these viruses comes largely from studies on vesicular stomatitis virus (VSV). VSV has also been used as an excellent vector for anticancer applications (1) and vaccine designs (2), thanks to its lack of integration of the viral genome into host cell DNA, its ability to accommodate large foreign or multiple gene inserts, its strong in vivo immune responses, its fast replication, and its high virus yields (3). Notably, VSV-based pseudotypes displaying SARS-CoV-2 spike proteins have been developed as vaccines against COVID-19, some of which are now on clinical trial (2).
The bullet-shaped VSV particle contains a single-strand RNA genome of 11,161 nucleotides (4), which encodes five viral proteins in the following order from the 3′ to 5′ end: nucleocapsid (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and large protein (L). The highly conserved L protein (5), together with P, forms the viral polymerase complex. L is a multidomain protein that functions as an RNA-dependent RNA polymerase (RdRp) (6), in addition to catalyzing viral messenger RNA (mRNA) capping (7), cap methylation (8, 9) and viral mRNA polyadenylation (10). Instead of naked genomic RNA, L uses the N-RNA complex of the nucleocapsid, which is the RNA genome coated by an N sheath, as the template (11, 12). During transcription and replication of the VSV genome, L does not bind the N-RNA complex directly, but is bridged by its noncatalytic cofactor P, which interacts with L and N oligomers simultaneously (13–15). Additionally, P contains regions required for interaction with the RNA-free N (N0) and serves as an N-specific chaperone to maintain N in a soluble and encapsidation-competent form (16, 17).
Transcription is the first biosynthetic event in the VSV replication cycle, and the polymerase complexes, delivered into host cells along with the nucleocapsid, are used for primary transcription prior to any viral protein synthesis. Accumulation of N0-P complexes, which facilitate encapsidation of the single-stranded RNA (ssRNA) genome, plays a key role in triggering the switch from transcription to replication (18). During replication, the polymerase complex synthesizes an encapsidated complementary strand of the entire ssRNA genome, which is then used as the template for the synthesis of progeny genomes. Normally, VSV rapidly replicates to very high levels within hours and blocks the host mRNA nuclear export, forcing the host to translate only viral mRNA (19, 20). L is vital to this process of transcription and replication; its importance is highlighted by the protein’s extreme sensitivity to mutations. Exchange of different domains between different strains of VSV leads to complete abrogation of polymerase activity (21). ts(G)114, a VSV temperature-sensitive mutant with three substitution mutation sites in L, has its mRNA transcription activity impaired (22). Moreover, most attempts to insert a fluorescent protein into VSV L have resulted in either loss of polymerase activity (21) or temperature sensitivity (23). Notably, sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis has revealed a defect in the packaging of L in the temperature-sensitive mutant, compared to the wild-type VSV (23). Early mass measurements by scanning transmission electron microscopy have determined that ∼50 copies of L and ∼400 copies of P are packaged within each virion (24), but a more recent study via superresolution light microscopy has estimated the number of copies of L to be 20 to 70 (25). This discrepancy aside, there has been no clear information about how L and P are assembled into mature virions and where they are located within the cavity of the bullet-shaped virion, limiting our mechanistic understanding of VSV assembly and replication.
In this study, we localized and determined the in situ structure of the VSV polymerase complex by cryo-electron tomography (cryoET) and subtomogram averaging. The improved contrast of denoised tomograms enabled direct observation of multiple L molecules attached to the helical nucleocapsid. To resolve the interaction between L and N, we first determined the in situ structure of N and M at 7.5 Å resolution by subtomogram averaging. We then sorted out the subtomograms that had an L molecule attached to N molecules and determined the in situ structure of L complexed with structured domains of P at ∼15 Å resolution. Fitting of the atomic model of L (14, 26) into our in situ structure shows that the capping domain of L interacts with two N molecules at the C-lobe. We also found that the interactions between N and the complex formed by L and P are flexible, a property that meets different demands of the virus at different stages of infection: L needs to be packaged into virions during assembly but to translocate to the 3′ end of the ssRNA genome to initiate transcription after entering the host cells. These results offer insight into the mechanism of transcription initiation in NNS RNA viruses in general and in VSV specifically.
Results
CryoET of Heterogenous VSV Virions and the Visualization of Individual Polymerase Complexes Inside.
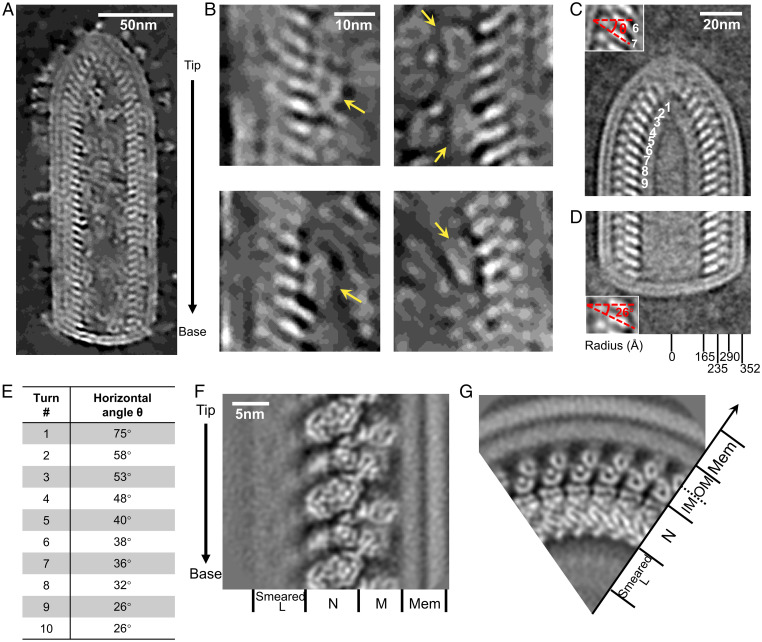
To directly visualize the three-dimensional (3D) organization of VSV virions, we performed cryoET (Movie S1) and reconstructed tomograms (Movie S2), which were then denoised with Noise2map tool in Warp for better observation (27). The bullet-shaped VSV virion has a lipid envelope that is decorated with multiple G trimers and encloses the nucleocapsid (Fig. 1A). In the cavity of the viral particle, which was not reconstructed in the previous cryo-electron microscopy (cryoEM) study (28), we observed ring-like densities attached to the nucleocapsid; based on their size and morphology, which match those of the L structure, we concluded that they are likely L molecules (Fig. 1B). This would be the first time L has been observed in situ in the VSV virion.
Fig. 1.
Architecture of VSV virions resolved by cryoET. (A) A slice through a denoised tomogram showing a section of a representative VSV virion. The arrow on the right denotes the directionality of the virion. (B) Representative L attached to N. Yellow arrows indicate the attached L molecules. (C) A subtomogram averaged density map of VSV conical tip from 88 virions. Inset: zoom in view of layers 6 and 7, with red lines indicating the inclination (horizontal angle θ) of N subunits. (D) A subtomogram averaged density map of VSV blunt base from 90 virions. Inset: zoom in view of two layers, with red lines indicating the inclination of N subunits. (E) Horizontal angles of different helical turn layers in the conical tip in (C). (F) A subtomogram averaged density map of VSV M-N in the trunk. Different proteins of VSV are indicated at the bottom. The arrow on the left indicates the direction from tip to base. (G) The top view of (F), showing radial positions of different VSV proteins. Two layers of M subunits bridge the N layer and the membrane.
For ease of reference, we defined the directionality of the virions from tip to base by following the directional convention established from the direction of its RNA genome, with the 3′ end at the conical tip of the bullet and the 5′ end at the base of the trunk (28). We then obtained the averaged density maps of the tip and base separately (Fig. 1 C and D). The averaged density map of the tip reveals that the tip contains 8 turns of a conical spiral before reaching the helical trunk. This observation can be confirmed by the changing angles of each of those turns: from turns 1 to 8, the horizontal angle θ, measured from the long axis of the N molecule along the horizontal plane of the virion, decreases; from turn 9 on, the horizontal angle θ remains a constant 26° (Fig. 1 D and E).
Previous studies have shown that the helical portion (trunk) of the virion contains two nested left-handed helices: an outer helix, consisting of M, and an inner helix, consisting of N and RNA. These helices have been reported to share identical helical parameters (28): each turn contains exactly 37.5 asymmetric units and rises 50.8 Å along the helical axis, suggesting a rigid size for all VSV virions. However, in our reconstructed tomograms, the radius of the trunk varies from 348 Å-378 Å (SI Appendix, Fig. S1B). This feature of heterogenous radii is also shared by some other NNS viruses, such as Ebola virus (29), and suggests different numbers of asymmetric units per helical turn, as well as different curvatures of the helical turns, both of which would cause misalignment during subtomogram averaging. Thus, we sorted the total 183 VSV virions into seven classes based on their numbers of asymmetric units per helical turn, which ranged from 35.5 per turn to 41.5 per turn. Almost half of the virions consisted of a helix with 38.5 asymmetric units per helical turn (SI Appendix, Fig. S1C), coincident with an earlier estimation based on the number of nucleotides associated with each N subunit visualized in the crystal structure of an N decamer and the radius of the helix in the virus (12). In the following processing to obtain the structure of L, we focused on that majority.
Structures of M-N by Subtomogram Averaging.
To locate the attachment sites of L within the capsid more precisely by classifying capsid regions (i.e., subparticles) containing M and N, we first obtained a high-resolution structure of M-N by subtomogram averaging. With the helical parameters discovered during the virus sorting above, subtomograms containing M and N were extracted and then locally refined within a limited angular and translational search range. No helical symmetry was imposed during the refinement. Finally, we determined the structure of VSV M-N at a resolution of 7.5 Å (Fig. 1 F and G and SI Appendix, Fig. S2). This averaged structure surprisingly shows two layers of M, not the expected single layer, bridging the N layer and the outermost double-layer membrane. Additionally, inside the cavity of the virion, a 7 nm-thick layer of blurred density attaches to the N layer (Fig. 1F), matching the shortest dimension of the cryoEM structure of L (26). The location of this layer implies the attachment of polymerase L to N. These densities are smeared, possibly due to the flexible attachments of L to N or the asymmetric organization of L.
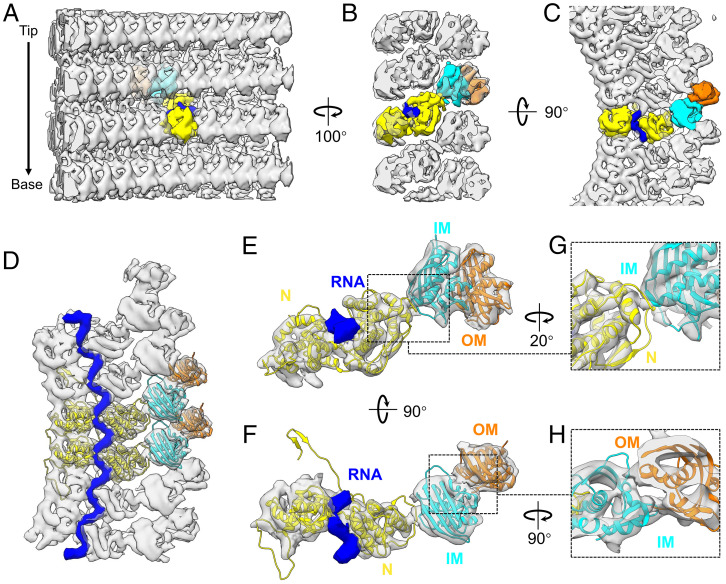
Fitting of available crystal structures of N and M establishes that the stoichiometric ratio of N and M is 1:2 in our reconstruction, which makes M the most abundant protein in the virion; this runs contrary to a previous assertion of a stoichiometric ratio of 1:1, based on a low-resolution (∼11 Å) cryoEM helical reconstruction of the VSV trunk (28). The repeating asymmetric unit, consisting of one copy of N and two copies of M, assembles into three nested left-handed helices (Fig. 2 A–C). The N subunit connects to the inner M (IM) subunit, which connects to the outer M (OM) subunit. Each N subunit closely interacts with the neighboring N subunits in the same helical turn (Fig. 2 C and D), with the genomic RNA tightly encapsidated in a cavity between two lobes of N (Fig. 2 B–D). The IM helix directly interacts with the N helix, each IM subunit located between two turns of the N helix (Fig. 2 B, E, and G), while the OM helix directly interacts with the IM helix, each OM subunit located between two subunits from the same turn of the IM helix (Fig. 2 C, D, F, and H). Given that a purified nucleocapsid can self-organize into a helix (30), the IM helix could thus function to stabilize the spiral structure of the nucleocapsid. The OM helix may bridge the IM-coated nucleocapsid with the membrane and cytoplasmic tail of G trimers, due to M’s ability to associate with the membrane (31). Taken together, our averaged density map complements the architecture of the VSV nucleocapsid in the virion and provides a stage for the classification of L-attached N.
Fig. 2.
Subtomogram averaged structure of proteins M, N, and genomic RNA. (A–C) Three roughly orthogonal surface views of the structure of VSV M-N determined by subtomogram averaging, as viewed from inside (A), side (B) and top (C) of the virion. For ease of visualization, one subunit in each layer is colored: yellow is N, cyan is IM, orange is OM, and blue is a fragment of genomic RNA bound with one N molecule. (D) Top view of the averaged density (semitransparent gray) of six asymmetric units from the same helical turn, with two asymmetric units fitted with the crystal structures (ribbon) of N (PDB: 2GIC) (12) and M (Protein Data Bank [PDB]: 1LG7) (31). The encapsidated RNA is in blue. (E and F) A segmented asymmetric unit of the averaged density (semitransparent gray) fitted with same crystal structures (ribbon), viewed from side (E) and top (F). The cryoEM densities corresponding to the atomic models of the N-arm and the C-loop of N are not included due to insufficient resolution needed for segmentation, since they interact with and are mixed within the densities of neighboring subunits. (G) Detailed view of the interface between N and IM. (H) Detailed view of the interface between IM and OM.
In Situ Structure of Polymerase L in Complex with Cofactor P.
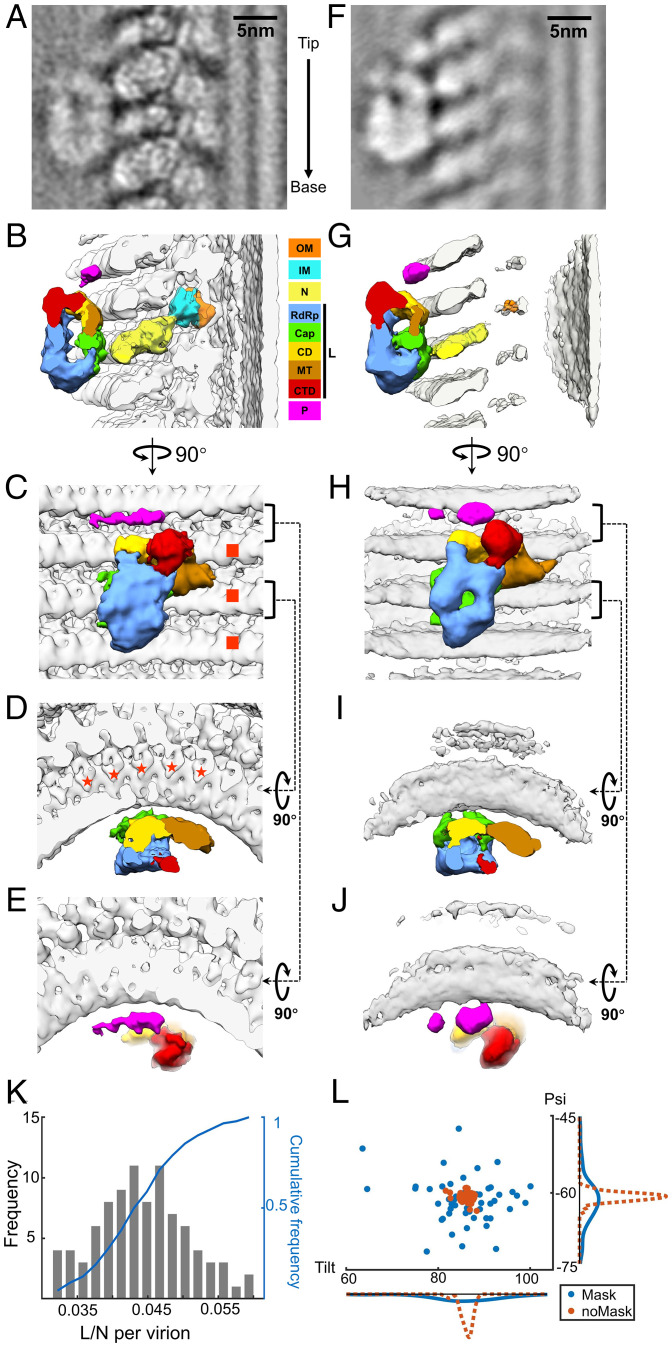
The innermost, blurred layer in our averaged density map of VSV M-N suggests that L attaches to N (Fig. 1 F and G). As N subunits are well-registered, subtomograms containing N with L attached could theoretically be classified out from those containing N without L attached. Thus, a box mask was applied next to the N subunit in the center of the subtomogram, where the L molecules might attach to N, while performing skip-align classification on the previously extracted subtomograms. Eventually, ∼4.5% of the subtomograms, containing L attaching to the central N subunit, were classified out, leading to an averaged structure of L attached to N with a global resolution ∼11 Å (Fig. 3A and SI Appendix, Fig. S2A). The ratios of L molecules to N molecules in individual trunks are not constant, ranging from 3 to 6% (Fig. 3K). Given the number of N subunits (∼1,240 copies) and the averaged ratio of L to N (∼4.5%), each virion therefore contains ∼55 copies of L, which is very close to the value estimated by mass measurements and biochemical quantitation (24).
Fig. 3.
Subtomogram averaged structure of VSV polymerase complex flexibly attached to N. (A) An averaged density map of VSV polymerase L refined without mask around L molecules. The arrow on the right indicates the direction from tip to base. (B–E) 3D segmentations of VSV OM, IM, N, L, and P viewed from side (B), inner (C) and top views (D and E). The interactions of L with N are located between N and the capping domain (Cap) of L. The footprint of an L molecule is across three helical turns of N (marked by red squares in C) and five subunits in each turn (marked by red stars in D). (F) An averaged density map of VSV polymerase L refined with mask around L molecules. N and M subunits are smeared due to the masking around L during subtomogram averaging. (G–J) 3D segmentations of VSV OM, IM, N, L, and P viewed from side (G), inner (H), and top views (I and J). Domains and proteins are colored the same as (B–E). (K) The histogram and cumulative frequency plot of the ratio of L molecules to N molecules per virion (trunk). (L) Distributions of tilt and psi angles of subtomograms from one representative virion. Red: the average without mask; blue: the average with mask. Bottom: frequency distribution of tilt angles; Right: frequency distribution of psi angles. Color keys for OM, IM, N, P, and different domains (following those in ref. 26) of L are shown between panels B and G.
The density assigned to L appears to be quite similar to the previous structure of isolated VSV L solved by cryoEM (14, 26), except for, surprisingly, a thin density that connects L to the nearby N subunits, as well as a thin long density right above L (Fig. 3 B–E). The L molecule is oriented with its ring-like ‘core’ beneath three globular appending domains. Based on the domain assignments from (26), the RdRp domain faces the helical interior while the capping domain faces oppositely, as the capping domain is the contact site with N subunits through the aforementioned thin density (Fig. 3 B and D). Looking from the radial direction, the footprint of the L molecule covers three helical turns of N and five subunits per helical turn (Fig. 3 C and D). This explains why less than 6.7% (1/15) of N subunits have L attached. The thin long density above L is assigned to P, based on its location in the vicinity of the connector domain (CD) and C-terminal domain (CTD) of L (14). Free P molecules have highly flexible regions and exhibit highly heterogenic conformations in solution due to the intrinsically disordered sequences of P (32). Thus, the density observed here may be only its structured regions.
To push the resolution of L molecules further, those subtomograms that contained an attached L molecule were locally refined with a tight mask around L. The refinement led to a structure of L and P with more detailed features, while the densities of M and N were smeared (Fig. 3 F–J). The smeared reconstruction of M and N subunits suggests that the orientation of L molecules varies with respect to M and N. Comparing both Tilt and Psi angles of the subtomograms from the refinements with a mask to those from the refinements without a mask results in a scattered distribution within a range less than 20°, indicating that variations of L molecules are in different directions (Fig. 3L). The improved reconstruction of L clearly shows the internal cavities, the substrate channels, and particularly the channel for nucleotides, shown in the foreground of Fig. 3H, compared to the above averaged density map (Fig. 3C). The capping domain of L is still the only domain that interacts with the C-lobe of N. Meanwhile, the improved density map exhibits more predominant connections between L and N: two densities were observed connecting the capping domain to the two neighboring N subunits (Fig. 3I). The contact sites of P with N sit two helical turns above the contact sites of L with N. The density above L was refined into two globules in the vicinity of the CD and CTD domains of L, with the larger one, right above the CD domain of L, connecting to N (Fig. 3 H and J). This is consistent with a previous study that shows that the L binding domain of P (PL) exits L at the CD and CTD domain (14). Since more P molecules are packed into virions than L molecules (24), and the active P molecules are in a dimeric form (33), the smaller globule nearby could belong to the other P molecule in the dimer. Furthermore, a potential connection between the smaller globule and the N subunit is also observed at a lower threshold of the density map (Fig. 3J), structured in the same way as the connection between the larger globule and the N subunit. In addition, the potential connection also appeared between the larger globule and L in the low-threshold density map. Taken together, we determined the in situ structure of the VSV polymerase complex, consisting of an L loosely attaching to N and a P dimer bridging L and N.
Sites of Interactions between L and N and between P and N.
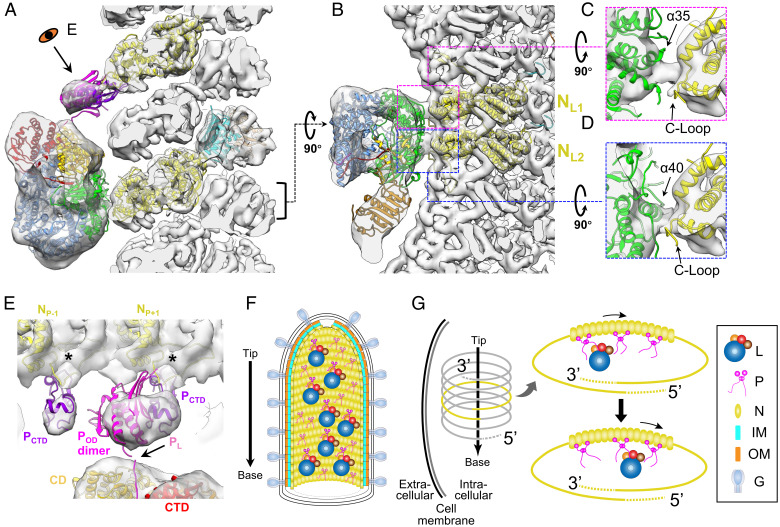
To explore the interactions between the polymerase complex and N further, our reconstruction of L and P (Fig. 3F) and the subnano-resolution reconstruction of M-N (Fig. 1 F and G) were combined, with the atomic models of M, N, L, and structured domains of P fitted in the density (Fig. 4A and Movie S3). The combined density map clearly shows that an L molecule interacts with two nearby N subunits, which are labeled as NL1 and NL2, following the 5′ to 3′ direction of the genome (Fig. 4 B–D). The extended loop in the C-terminal lobe (C-loop, residues 340 to 375) of each N subunit, which has been shown to be involved in the contacts between N subunits (12), is close to the contact point of L. It may also be involved in the interactions between L and N, as may a fragment from residues 376 to 402 (N’s α13 and α14) and the short loop in between (Fig. 4 C and D). Fragments of L from residues 983 to 1004 (L’s α35) and from residues 1081 to 1093 (L’s α40) are in the vicinity of the contacting densities with NL1 and NL2, respectively (Fig. 4 C and D). Potential conformational changes may occur in these structures upon the attachment of L to N to accommodate these interactions.
Fig. 4.
Interactions between VSV L, P, and N, and the models of multiple polymerase complexes in mature virions. (A and B) Combined 3D reconstruction (semitransparent gray) of the subnano averaged structure of VSV M-N and averaged structure of VSV polymerase complex, superimposed with atomic models (ribbon) of VSV L (PDB: 6U1X) (14), POD (PDB: 2FQM) (33), PCTD (PDB: 3HHZ) (13), N (PDB: 3HHZ, 2GIC) (12), and M (PDB: 1LG7) (31) viewed from side (A) and top (B). Following the 5′ to 3′ direction, the two N subunits involved in the interaction with L are numbered NL1, NL2 in (B). (C and D) Detailed side views of the two contacting sites between the capping domain of L and two N subunits. (E) Detailed view of interactions between P and L and between P and N. PL, POD and PCTD are shown here in pink, magenta and purple, respectively; unstructured flexible regions are not shown here. The two N subunits involved in the interaction with PCTD are numbered NP-1 and NP+1 following the 5′ to 3′ direction of the genome. (F) Schematic illustration of a mature virion with, from outer to inner, G decorated membrane, two layers of M helices, N helix, attached L and P dimers. The arrow on the left denotes the directionality of the virion. The L proteins are randomly distributed along the inside of the nucleocapsid, as are the P protein dimers. Some P dimers associate with L proteins, while the others are in unbound state. (G) A model for multiple L molecules’ movement to the 3′ end of the RNA genome by P dimers during infection. After virus entry, the nucleocapsid is released from the virion to serve as the template for subsequent transcription and replication. Multiple polymerase complexes remain attached to N. Driven by the high affinity of L to TIS at the 3′ end of the genome, L molecules are relayed by multiple P dimers all the way to the 3′ end for initiation of primary transcription.
In addition to the crystal structure of N decamer, there is also a reported crystal structure of a decameric complex (N-PCTD decamer) that contains N and the C-terminal domain of P (PCTD). In this crystal structure, PCTD interacts with and lies between the C-lobes of two adjacent N subunits (13). Based on the amino acid sequence, PCTD (residues 193 to 265) is connected to PL (residues 49 to 105) through the oligomerization domain of P (POD, residues 109 to 170) and an unstructured sequence (residues 171 to 192). Thus, with the sequence information and the protein interactions observed in our reconstruction, we assigned the large globular density, which contacts a nearby N subunit and may potentially contact L (Fig. 3 H and J), to be one PCTD and a dimer of POD. The small globular density is assigned to a second PCTD from the other P molecule of the dimer. The connecting density between the large globular density and the N subunit is weak, suggesting conformational flexibility of N in the P-bound state.
Therefore, although precise fitting of the atomic models of PCTD or POD in the cryoET map is not possible due to limited resolution of the latter, we were able to position a P dimer interacting with each L in the virion by taking advantage of the cryoEM structure of L with PL (14), and the crystal structure of N-PCTD decamer (13), which already established the positions of PL and PCTD relative to L and N, respectively. Because different radii between N decamer and virion trunk, the relative orientations between adjacent N subunits are slightly different in the crystal structure of N-PCTD decamer from that in our cryoET structure of the virion trunk (SI Appendix, Fig. S3 A and B). As a result, after directly placing the atomic models of two N subunits (NP and NP+1) and their associated PCTD into the cryoET density map, we need to rotate PCTD ∼30° away from NP (SI Appendix, Fig. S3C) such that only the interactions between PCTD and the adjacent NP+1 remain. Notably, in this placement, the C-loops of N subunits, involving in the interactions between N and PCTD in the crystal structure of N-PCTD decamer, point to the connecting densities between the N subunits and the globular densities (Fig. 4E and SI Appendix, Fig. S3D). The placement of POD dimer into the remaining portion of the large globular density was guided by ensuring the N-terminus of one subunit in the POD dimer proximal to the C-terminus of PL bound in L (Fig. 4E and SI Appendix, Fig. S3E) previously established in the cryoEM structure (14). Our positioning of P as described above points to a role of P in bridging L to N for the packaging of L during virus assembly.
Discussion
This paper reports the in situ structure of the polymerase complex and its structural organization with respect to the nucleocapsid and matrix proteins in the virion of VSV, a prototype NNS RNA virus. The averaged density maps of regions containing L, P, N, and M clearly show that L, together with its cofactor P, binds to specific sites of nearby N subunits through loose attachments and P binding.
Like other NNS RNA viruses, viral polymerases are assembled into VSV virions to initiate viral reproductive cycle. These polymerases participate in early transcription after virus entry, yielding at least one cycle of transcription within the first hour after infection (34, 35). This high efficiency leads to ∼10K progeny VSV virions from a single cell within 10 h postinfection (36). Multiple polymerases in a mature virion may be responsible for this efficient transcription during the early stages of infection. We estimated that ∼55 L molecules are attached to the nucleocapsid in each virion; this large number of polymerases may explain why the VSV nucleocapsid, having to make room for these vital proteins, needs to be assembled within a spacious trunk. Our cryoET results show that these L molecules bind to N molecules specifically but are almost randomly distributed along the nucleocapsid, coincident with observations from immunogold electron microscopy (37). During transcription and replication, L gains access to the N-embedded RNA genome via P, which engages L and N simultaneously (15, 38). P has been shown to be a nonglobular molecule containing both structured domains and disordered regions (13, 33, 39). In our reconstruction, the structured domains of P, two copies of PCTD and a POD dimer, are assigned to the globular density above L, one copy of PCTD and the POD dimer to the large globular density in the vicinity of the CD domain of L and another copy of PCTD to the small globular density (Fig. 4E). PL, reported to be structured and bound with L (14), was not directly resolved here due to limited resolution. Therefore, our reconstruction suggests that P, already bound to N, bridges L and N during assembly, similar to the bridging role it plays during transcription and replication (38). Additionally, L remains associated with the nucleocapsid in the mature virion through relatively flexible interactions between its capping domain and the two C-lobes from two N subunits located in a different helical turn (Fig. 3). Such L-N association would ensure all L molecules within the virion to have similar orientations with respect to the axis of the virion. Similarly orientated, flexibly associated polymerase complexes would promote their directional movements toward the 3′ end of the genome to initiate transcription.
Our results show that an intact VSV virion has multiple polymerase complexes, attached to N through loose attachments and randomly distributed along the helical nucleocapsid inside the trunk (Fig. 4F). According to the stop-start VSV transcription model, the polymerase enters the template at a single site, the 3′ end of the encapsidated genome (40, 41). The transcription of VSV’s five genes is in a sequential manner along the template (42), with an ∼30% transcriptional attenuation at each gene junction (34). Due to the random distribution of L molecules on the template, these attached L molecules need to translocate along the template and eventually engage the 3′ end to initiate synthesis of mRNA in primary transcription after cell entry. Although translocation may be bidirectional, movement to the transcription initiation site (TIS) at the 3′ end of the genome may be more statistically likely due to the strong affinity of L to the TIS (43). We propose that the flexible interactions between L and N observed in mature VSV virions may allow for L molecules to slide along the template before initiation of primary transcription. In the current model, P proteins, intrinsically flexible, have been depicted as an "ensemble of continuously exchanging conformers" (32): during assembly, P dimers bind to N subunits directly through PCTD, and the remaining part of P swings to ‘grab’ L molecules randomly through PL interactions. Those "grabbed" L molecules are introduced to the nucleocapsid, and once the nucleocapsid is wound up in a helix, L molecules loosely bind to the nearby N subunits at the C-lobe specifically to enhance attachment (Fig. 4G). P molecules, which exist two times more in quantity than L inside the virion (24), may also function as carriers facilitating the translocation of L molecules after virus entry. Driven by the high affinity of L to TIS (43), relay between the additional P dimers may facilitate sliding of L molecules to the 3′ end to engage TIS (Fig. 4G). With the organization of multiple polymerase complexes on the nucleocapsid, therefore, L molecules could quickly initiate primary mRNA transcription for all five genes, especially the gene of L, which is located furthest downstream. The de novo synthesis of L, in turn, ensures the repetitive synthesis of mRNA, the antigenome and the genome, leading to highly efficient virus replication. Visualization of this dynamic process inside infected cells awaits future in vivo studies.
Taken together, our in situ structure of the polymerase complex reveals the flexible attachments of L to N via P dimers, facilitating L packaging into the virion during assembly and translocating to the 3′ end of the ssRNA genome to initiate transcription after virus entry. Such flexible attachment would allow VSV to efficiently undergo multiple stages of the viral life cycle and may be vital to both primary transcription and efficient replication. The results not only provide insights into VSV transcription and replication but also are directly relevant to ongoing engineering efforts in anticancer and vaccine development, including those urgently needed to mitigate the fast-emerging COVID-19 virus variants.
Materials and Methods
VSV virions (Indiana serotype, San Juan strain) were produced as previously described (44). Briefly, inoculum was plaque-purified after multiple passages in HeLa cells with a very low multiplicity of infection to suppress truncated defective-interference particles. Full virions were then isolated through three rounds of centrifugations and a 10 mL density gradient. To prepare cryoET grids, 2.5 μL of the virus suspension, which had already been mixed with 5 nm diameter colloidal gold in a 20:1 ratio, was applied onto glow-discharged 200 mesh copper grids (3.5/1, Quantifoil), and then plunged into liquid ethane. Grids were loaded into a Titan Krios (FEI) 300 kV transmission electron microscope equipped with a Gatan imaging filter Quantum LS and a Gatan K3 detector. Tomographic tilt series were collected between -60° and 60° with a 3° angular increment in dose-symmetry scheme (45) by SerialEM 3.8.0 (46) at a nominal magnification of 64,000× (pixel size 0.69 Å).
In total, 40 tilt series were collected and then reconstructed in IMOD (47). Due to heterogeneity of VSV virions, virion particles were classified into seven classes according to the numbers of asymmetric units per helical turn of the trunk part of the virion, ranging from 35.5 to 41.5 with an interval of 1. Of 183 manually selected virions, 91 virions with 38.5 asymmetric units per turn comprised the major class (SI Appendix, Fig. S2) and were used for subsequent averaging of M-N and M-N-L subtomograms. A total of 82,991 subtomograms containing M and N were extracted, aligned, and averaged with RELION 3.0.8 (48), resulting a 7.5 Å-resolution averaged density map on the basis of the gold-standard FSC = 0.143 criterion. To classify out the subtomograms with an L subunit attached to N, we applied a box mask close to the central N in the averaged volume and performed RELION 3D classification on the previously aligned subtomograms with the skip-align option. Only one of eight classes showed a ring-shaped L density next to the central N. After removing duplicates, 3,699 subtomograms were retained and locally refined either with or without a tight mask around L. More features appeared for the L density while M and N smeared in the averaged density map refined with a mask, due to flexible attachment between L and N.
Please refer to SI Appendix for more detailed descriptions of cryoET imaging and data processing.
Supplementary Material
Acknowledgments
We thank Titania Nguyen for editing the manuscript and Dr. Peng Ge for valuable suggestions on data processing. This project is supported partly by grants from the US NIH (AI094386 and GM071940 to Z.H.Z.). We acknowledge the use of resources at the Electron Imaging Center for Nanomachines supported by UCLA and grants from the NIH (1S10OD018111 and 1U24GM116792) and the NSF (DBI-1338135 and DMR-1548924).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. H.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111948119/-/DCSupplemental.
Data Availability
The cryoET averaged density maps of VSV M-N and M-N-L are deposited to the Electron Microscopy Data Bank, under the accession numbers EMD-26465 and EMD-26468, respectively.
References
- 1.Hastie E., Grdzelishvili V. Z., Vesicular stomatitis virus as a flexible platform for oncolytic virotherapy against cancer. J. Gen. Virol. 93, 2529–2545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yahalom-Ronen Y., et al. , A single dose of recombinant VSV-ΔG-spike vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 11, 6402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majid A. M., Barber G. N., “Recombinant vesicular stomatitis virus (VSV) and other strategies in HCV vaccine designs and immunotherapy” in Hepatitis C Viruses: Genomes and Molecular Biology, Tan S.-L., Ed. (Horizon Bioscience, Norfolk, UK, 2006), pp. 423–450. [PubMed] [Google Scholar]
- 4.Huang A. S., Wagner R. R., Comparative sedimentation coefficients of RNA extracted from plaque-forming and defective particles of vesicular stomatitis virus. J. Mol. Biol. 22, 381–384 (1966). [DOI] [PubMed] [Google Scholar]
- 5.Poch O., Blumberg B. M., Bougueleret L., Tordo N., Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: Theoretical assignment of functional domains. J. Gen. Virol. 71, 1153–1162 (1990). [DOI] [PubMed] [Google Scholar]
- 6.Emerson S. U., Wagner R. R., L protein requirement for in vitro RNA synthesis by vesicular stomatitis virus. J. Virol. 12, 1325–1335 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogino T., Banerjee A. K., Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 25, 85–97 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Hercyk N., Horikami S. M., Moyer S. A., The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology 163, 222–225 (1988). [DOI] [PubMed] [Google Scholar]
- 9.Grdzelishvili V. Z., et al. , A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J. Virol. 79, 7327–7337 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt D. M., Mehta R., Hutchinson K. L., The L protein of vesicular stomatitis virus modulates the response of the polyadenylic acid polymerase to S-adenosylhomocysteine. J. Gen. Virol. 69, 2555–2561 (1988). [DOI] [PubMed] [Google Scholar]
- 11.Emerson S. U., Wagner R. R., Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J. Virol. 10, 297–309 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green T. J., Zhang X., Wertz G. W., Luo M., Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313, 357–360 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Green T. J., Luo M., Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc. Natl. Acad. Sci. U.S.A. 106, 11713–11718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenni S., et al. , Structure of the vesicular stomatitis virus l protein in complex with its phosphoprotein cofactor. Cell Rep. 30, 53–60.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emerson S. U., Schubert M., Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 84, 5655–5659 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leyrat C., et al. , Structure of the vesicular stomatitis virus N0-P complex. PLoS Pathog. 7, e1002248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M., Ogino T., Banerjee A. K., Interaction of vesicular stomatitis virus P and N proteins: Identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J. Virol. 81, 13478–13485 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wertz G. M., Howard M. B., Davis N., Patton J., The switch from transcription to replication of a negative-strand RNA virus. Cold Spring Harb. Symp. Quant. Biol. 52, 367–371 (1987). [DOI] [PubMed] [Google Scholar]
- 19.Petersen J. M., Her L. S., Varvel V., Lund E., Dahlberg J. E., The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol. Cell. Biol. 20, 8590–8601 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faria P. A., et al. , VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17, 93–102 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Ruedas J. B., Perrault J., Putative domain-domain interactions in the vesicular stomatitis virus L polymerase protein appendage region. J. Virol. 88, 14458–14466 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galloway S. E., Wertz G. W., A temperature sensitive VSV identifies L protein residues that affect transcription but not replication. Virology 388, 286–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruedas J. B., Perrault J., Insertion of enhanced green fluorescent protein in a hinge region of vesicular stomatitis virus L polymerase protein creates a temperature-sensitive virus that displays no virion-associated polymerase activity in vitro. J. Virol. 83, 12241–12252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas D., et al. , Mass and molecular composition of vesicular stomatitis virus: A scanning transmission electron microscopy analysis. J. Virol. 54, 598–607 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodges J. A., et al. , A polymerase super complex within vesicular stomatitis virus. Biophys. J. 104, 415a (2013). [Google Scholar]
- 26.Liang B., et al. , Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162, 314–327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tegunov D., Cramer P., Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge P., et al. , Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 327, 689–693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bharat T. A. M., et al. , Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc. Natl. Acad. Sci. U.S.A. 109, 4275–4280 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desfosses A., et al. , Self-organization of the vesicular stomatitis virus nucleocapsid into a bullet shape. Nat. Commun. 4, 1429 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Gaudier M., Gaudin Y., Knossow M., Crystal structure of vesicular stomatitis virus matrix protein. EMBO J. 21, 2886–2892 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leyrat C., et al. , Ensemble structure of the modular and flexible full-length vesicular stomatitis virus phosphoprotein. J. Mol. Biol. 423, 182–197 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Ding H., Green T. J., Lu S., Luo M., Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J. Virol. 80, 2808–2814 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iverson L. E., Rose J. K., Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23, 477–484 (1981). [DOI] [PubMed] [Google Scholar]
- 35.Flamand A., Bishop D. H., Primary in vivo transcription of vesicular stomatitis virus and temperature-sensitive mutants of five vesicular stomatitis virus complementation groups. J. Virol. 12, 1238–1252 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timm A., Yin J., Kinetics of virus production from single cells. Virology 424, 11–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harmon S. A., Robinson E. N. Jr., Summers D. F., Ultrastructural localization of L and NS enzyme subunits on vesicular stomatitis virus RNPs using gold sphere-staphylococcal protein A-monospecific IgG conjugates. Virology 142, 406–410 (1985). [DOI] [PubMed] [Google Scholar]
- 38.Mellon M. G., Emerson S. U., Rebinding of transcriptase components (L and NS proteins) to the nucleocapsid template of vesicular stomatitis virus. J. Virol. 27, 560–567 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jamin M., Yabukarski F., Nonsegmented negative-sense RNA viruses—Structural data bring new insights into nucleocapsid assembly.Adv. Virus Res. 97, 143–185 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Testa D., Chanda P. K., Banerjee A. K., Unique mode of transcription in vitro by vesicular stomatitis virus. Cell 21, 267–275 (1980). [DOI] [PubMed] [Google Scholar]
- 41.Emerson S. U., Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell 31, 635–642 (1982). [DOI] [PubMed] [Google Scholar]
- 42.Abraham G., Banerjee A. K., Sequential transcription of the genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 73, 1504–1508 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang X., Bendjennat M., Saffarian S., Vesicular stomatitis virus polymerase’s strong affinity to its template suggests exotic transcription models. PLOS Comput. Biol. 10, e1004004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holland J. J., Villarreal L. P., Breindl M., Factors involved in the generation and replication of rhabdovirus defective T particles. J. Virol. 17, 805–815 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagen W. J. H., Wan W., Briggs J. A. G., Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197, 191–198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Mastronarde D. N., Dual-axis tomography: An approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343–352 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Bharat T. A. M., Scheres S. H. W., Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat. Protoc. 11, 2054–2065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryoET averaged density maps of VSV M-N and M-N-L are deposited to the Electron Microscopy Data Bank, under the accession numbers EMD-26465 and EMD-26468, respectively.