Significance
Juvenile nephronophthisis (NPH) is a renal ciliopathy due to a dysfunction of primary cilia for which no curative treatment is available. This paper describes the identification of agonists of prostaglandin E2 receptors as a potential therapeutic approach for the most common NPHP1-associated ciliopathies. We demonstrated that prostaglandin E1 rescues defective ciliogenesis and ciliary composition in NPHP1 patient urine-derived renal tubular cells and improves ciliary and kidney phenotypes in our NPH zebrafish and Nphp1−/− mouse models. In addition, Taprenepag alleviates the severe retinopathy observed in Nphp1−/− mice. Finally, transcriptomic analyses pointed out several pathways downstream the prostaglandin receptors as cell cycle progression, extracellular matrix, or actin cytoskeleton organization. Altogether, our findings provide an alternative for treatment of NPH.
Keywords: kidney, primary cilia, nephronophthisis, drug-screen, prostaglandins
Abstract
Nephronophthisis (NPH) is an autosomal recessive tubulointerstitial nephropathy belonging to the ciliopathy disorders and known as the most common cause of hereditary end-stage renal disease in children. Yet, no curative treatment is available. The major gene, NPHP1, encodes a protein playing key functions at the primary cilium and cellular junctions. Using a medium-throughput drug-screen in NPHP1 knockdown cells, we identified 51 Food and Drug Administration-approved compounds by their ability to alleviate the cellular phenotypes associated with the loss of NPHP1; 11 compounds were further selected for their physicochemical properties. Among those compounds, prostaglandin E1 (PGE1) rescued ciliogenesis defects in immortalized patient NPHP1 urine-derived renal tubular cells, and improved ciliary and kidney phenotypes in our NPH zebrafish and Nphp1 knockout mouse models. Furthermore, Taprenepag, a nonprostanoid prostaglandin E2 receptor agonist, alleviated the severe retinopathy observed in Nphp1−/− mice. Finally, comparative transcriptomics allowed identification of key signaling pathways downstream PGE1, including cell cycle progression, extracellular matrix, adhesion, or actin cytoskeleton organization. In conclusion, using in vitro and in vivo models, we showed that prostaglandin E2 receptor agonists can ameliorate several of the pleotropic phenotypes caused by the absence of NPHP1; this opens their potential as a first therapeutic option for juvenile NPH-associated ciliopathies.
Renal ciliopathies are a group of hereditary diseases caused by mutations in genes encoding proteins playing a role at the primary cilium, a microtubule-based cellular antenna found on almost all vertebrate quiescent cells, which controls key signaling pathways during development and tissue homeostasis (1). In the kidney, as well as in other organs, the primary cilium also acts as an essential mechanosensor through which luminal flow controls tubule diameter and renal epithelium maintenance, explaining the presence of tubular cysts in most renal ciliopathies. Renal ciliopathy spectrum includes autosomal dominant polycystic kidney disease (ADPKD), autosomal recessive polycystic kidney disease, and cystic dysplasia, which are all characterized by enlarged kidneys with numerous cysts (2). It also encompasses nephronophthisis (NPH) that, on the other hand, is generally characterized by small kidneys and progressive tubulointerstitial fibrosis, associated with corticomedullary cysts, leading to end-stage renal disease (ESRD) at a median age of 13 y old. NPH is the first monogenic cause of pediatric ESRD, representing up to 10% of incident cases in developed countries (3, 4). Recent publications have demonstrated that NPH may also occur in adulthood, leading to late-onset ESRD (up to 61 y old) (5). Extrarenal involvements may include retinal degeneration (Senior-Løken syndrome), cerebellar ataxia, skeletal anomalies, and liver fibrosis (6).
To date, more than 23 genes have been linked with NPH, the most frequent being NPHP1 with nearly 25% of affected individuals harboring a homozygous deletion of this gene. NPHP genes encode proteins that form different complexes along the cilium (7). Their mutations are not associated with a complete loss of cilia; they rather lead to subtle phenotypes, such as decreased proportion of ciliated cells, increased cilium length, and ciliary membrane composition defects, which result in defective cilia-dependent signaling (8–10). Besides cilium dysfunction, several NPH-associated gene products, including NPHP1 and its partner NPHP4, have also been involved in the control of cell migration, adhesion and apico-basal cell epithelialization, through the regulation of cytoskeletal organization (8, 11–14).
To date, the only available treatments for NPH patients are dialysis and renal transplantation. As other chronic tubulointerstitial nephropathies characterized by the absence of hypertension or overt albuminuria (except in late stages of the disease), nonspecific nephroprotective treatments by renin-angiotensin system blockade did not delay progression to ESRD (15). Similarly, as enlarged cystic kidneys are uncommon in juvenile NPHP1-associated NPH, molecules developed to limit cyst growth in the context of ADPKD (16, 17)—such as the selective angiotensin-vasopressin type 2 (AVPR2) antagonist Tolvaptan, the CDK inhibitor Roscovitine, or the somatostatin-analog Octreotide—are unlikely to be relevant. Nevertheless, several other molecules were recently pointed out in in vitro and in vivo NPH models including the Hedgehog agonist Purmorphamine, β-catenin inhibitors, ROCK inhibitors, or the flavonoid Eupatilin (4). On the other hand, even though gene therapy has been successfully implemented in early human retinal ciliopathies thanks to local administration of adeno-associated viruses (18), efficient gene therapy in NPH would rather represent a clinical challenge given the lack of effective administration modes for gene delivery in the kidney.
Here, after having performed in vitro screens of small chemical compounds for their ability to lessen phenotypes associated with the loss of NPHP1, we identified prostaglandin E1 (PGE1) and other agonists of prostaglandin E2 (PGE2) receptors as a class of molecules that restore ciliary defects in vitro as well as kidney and retina-associated phenotypes in vivo in both a zebrafish NPH model and Nphp1 knockout (KO) mouse. Also, different transcriptomic approaches showed that these phenotypic restorations can be matched to the effects of these agonists on shared cellular pathways.
Results
Identification of Compounds Able to Rescue NPHP1-Associated Cellular Phenotypes.
We designed an in vitro high-throughput phenotypic drug-screen strategy based on the Prestwick Chemical Library, a collection of 1,120 off-patent small molecules of which 95% are approved by the Food and Drug Agency, European Medicines Agency, or other agencies. The screen was performed on the two most widely used renal cell models to study ciliogenesis and ciliopathies—that is, the mouse inner medullary collecting duct (mIMCD3) and canine Madin-Darby canine kidney (MDCK) cells—in which the expression of Nphp1 was knocked down (KD) through stable expression of short-hairpin RNA (NPHP1_KD).
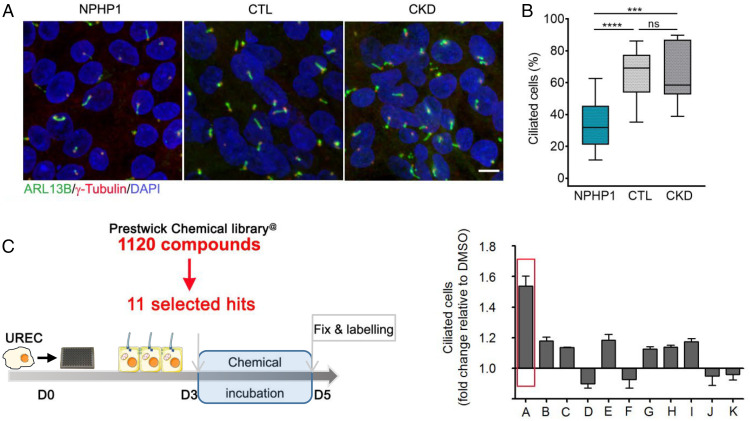
NPHP1_KD mIMCD3 cells display impaired ciliogenesis with a decreased percentage of ciliated cells, as we previously reported for NPHP1_KD MDCK cells (8), as well as abnormally increased cell migration during wound-healing assays (SI Appendix, Fig. S1 A–F). Using fluorescence microscopy-based automated strategies (SI Appendix, Fig. S1G), 51 hits were identified based on their ability to restore cell migration and cilium count defects in either mIMCD3 or MDCK NPHP1_KD cells. Hence, 11 compounds belonging to the different pharmacological classes were selected based on their effects on the analyzed phenotypes, on the absence of obvious toxicity (decrease cell count and/or inhibition of cell migration), and on their chemical action or physicochemical properties that may be relevant in NPH: known or suspected effects on cilium-associated pathways, actin cytoskeleton organization and microtubule polymerization, RhoGTPase activity, cAMP regulation, signaling pathways (e.g., JNK, ERK/MAPK, or Wnt), or their involvement in processes such as fibrosis-related pathways or nephroprotective properties (SI Appendix, Fig. S1G and Table S1).
Rescue of Defective Ciliogenesis in NPHP1 Patients’ Renal Cells by PGE1.
The efficiency of the 11 most interesting hits was further validated in urine-derived renal epithelial cells (URECs), a valuable source of cells from NPH individuals, previously described as a model to study ciliogenesis and ciliary functions in the context of ciliopathies (19). Primary URECs were collected either from individuals carrying biallelic mutations in NPHP1 (NPHP1, hereafter Pt1–11), from age-matched control individuals (CTL, hereafter CTL1–5), or from individuals presenting with a nonciliopathy-related chronic kidney disease (CKD, hereafter CKD1–4) (SI Appendix, Table S2), and were immortalized through the expression of a thermosensitive SV40 T-antigen. Transcriptomic analyses (SI Appendix, Fig. S2A) showed that both primary and immortalized URECs exhibit similar expression levels of renal tubular epithelial cell markers, confirming their tubular origin. As expected, the immortalized URECs showed persistent proliferation at permissive temperature (33 °C) but not at nonpermissive temperature (39 °C), as monitored by the loss of SV40 T-antigen and Ki67 nuclear staining in the latter condition (SI Appendix, Fig. S2 B–D).
In order to evaluate ciliogenesis in immortalized URECs, we set up a semiautomated pipeline based on basal-body (γ-Tubulin) (Fig. 1A, red) and ciliary membrane (ARL13B) (Fig. 1A, green) markers to quantify cilium count. At nonpermissive temperature, URECs from NPHP1 individuals presented a significant decrease of cilium count (Fig. 1B and SI Appendix, Table S2), which was associated with an increased proportion of cells presenting with two spots of CP110 per centrosome, indicative of an early blockage of ciliogenesis in NPHP1 URECs (SI Appendix, Fig. S3).
Fig. 1.
Drug screen in NPHP1 urine-derived renal epithelial cells (URECs). (A) Representative images of immunofluorescence of primary cilia (ARL13B, green), basal bodies (γ-Tubulin, red), and nuclei (DAPI, blue) in URECs derived from NPHP1 patients (NPHP1), control individuals (CTL), and CKD patients 5 d after seeding. (Scale bar, 10 µM.) (B) Quantification of ciliogenesis in NPHP1 (n = 11), CTL (n = 6), and CKD (n = 5) URECs expressed as the mean for all individuals and shown using a box-and-whisker plot. n = 1 to 15 experiments (mean of 3.5 experiments per cell line). Mixed linear-regression model with quasibinomial penalization taking into account the correlation of observations coming from the same individuals using a random effect on the cell line: ***P < 0.001, ****P < 0.0001, ns: not significant. (C, Left) NPHP1 URECs (Pt1) were treated for 2 d with the 11 selected compounds from the primary screen and ciliogenesis was evaluated 5 d after seeding. (Right) Quantification of ciliogenesis. A: ALP, 200 nM; B: Cyproheptadine, 1 µM; C: Ethopropazine, 50 nM; D: Fluticasone, 10 nM; E: Methotrexate, 400 nM; F: Mycophenolic acid, 200 nM; G: Paclitaxel, 100 pM; H: Pyrimethamine, 500 nM; I: Simvastatin, 10 nM; J: Tropisetron, 2 µM; K: Verapamil, 80 nM. Mean ± SEM n = 1 to 2 experiments.
Before assessing the effect of the 11 molecules on URECs, we evaluated the nontoxic range of doses at which they could be used (SI Appendix, Table S1). The highest nontoxic concentration, as well as 1-log and 2-log concentrations, were chosen to carry out the ciliogenesis assay on one of the NPHP1 UREC (Pt1). A robust increase of cilium count was observed for Alprostadil (ALP; +1.53), a synthetic analog of the natural occurring PGE1, whereas the other compounds showed modest positive or negative effects on ciliogenesis (Fig. 1C). PGE1 (ALP) is a type-E prostaglandin analog which exerts various effects in inflammation, pain, vascular tone, or renal function, that are mainly mediated by its four receptors, EP1–4 encoded by PTGER1-4 (20). Notably, EP1–4 receptors are expressed in the URECs, predominantly EP2 and EP4, as revealed by qRT-PCR analyses (SI Appendix, Fig. S4A).
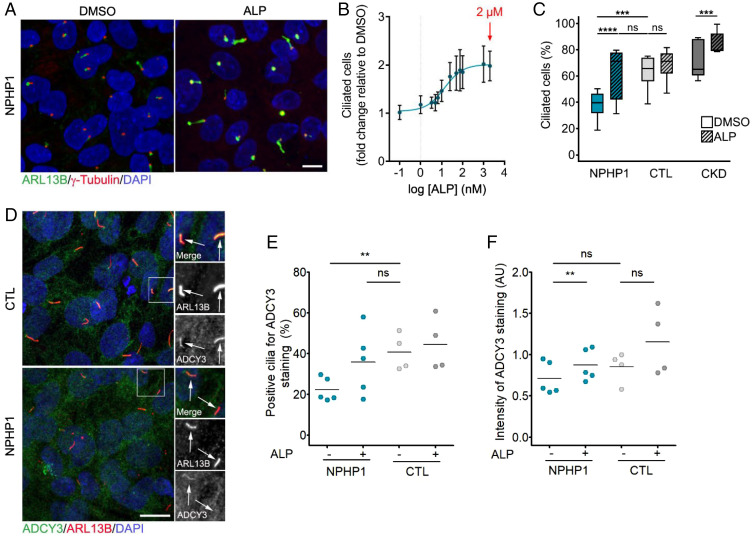
We first performed a dose–response analysis for ALP and determined the half maximal effective concentration (EC50) at 13.75 nM, while the maximal effect on cilium count was observed for concentrations above 1 µM (Fig. 2 A and B). Based on these results, the following experiments were performed using 2 µM ALP. In addition, Dinoprostone (DINO), a synthetic version of the natural occurring PGE2, showed a similar effect as ALP (SI Appendix, Fig. S4B), supporting the positive impact of prostanoids on cilia count in NPHP1 URECs. The positive effect of ALP on ciliogenesis was observed in other NPHP1 URECs (Pt3–4 and 6–11) but not in CTL, even in the one with a low proportion of ciliated cells (Fig. 2C and SI Appendix, Fig. S4 C and D), indicating its specific efficiency toward NPHP1 URECs. It also showed a small but significant positive effect on CKD URECs, indicating that PGE1 could also ameliorate ciliogenesis in a different pathological context. Furthermore, beyond the restoration of ciliogenesis, ALP also restored ciliary composition with an increased percentage of Adenylate Cyclase 3 (ADCY3)+ cilia, associated with an increased intensity of ciliary ADCY3 (Fig. 2 D–F).
Fig. 2.
Validation of prostaglandin signaling as a target in NPHP1-defective renal cells. (A) Representative images of immunofluorescence of primary cilia (ARL13B, green), basal bodies (γ-Tubulin, red), and nuclei (DAPI, blue) in NPHP1 URECs 5 d after seeding treated for 2 d with DMSO (0.04%) or ALP (2 µM). (Scale bars, 10 µM.) (B) Quantification of ciliogenesis in one NPHP1 UREC (Pt1) exposed for 2 d to increasing concentrations of ALP. Mean ± SEM n = 2 to 4 experiments. Nonlinear regression. (C) Quantification of ciliogenesis in URECs derived from NPHP1 patients (NPHP1, n = 11), CTL individuals (n = 6), and CKD patients (n = 5) exposed for 2 d to DMSO (0.04%) and ALP (2 µM). Results are expressed as the mean for all individual categories and are shown using a box-and-whisker plot. n = 1 to 15 experiments per cell line (mean of 3.5 experiments per cell line). Mixed linear-regression model with quasibinomial penalization taking into account the correlation of observations coming from the same individuals using a random effect on the cell line: ***P < 0.001, ****P < 0.0001, ns: not significant. (D) Representative images of immunofluorescence of ADCY3 (green), primary cilia (ARL13B, red), and nuclei (DAPI, blue) in NPHP1 and CTL URECs. Arrows indicate ARL13B+ cilia. (Scale bars, 10 µM.) (E and F) Quantification of the percentage of ADCY3+ cilia (E) and of the intensity of ADCY3 in those cilia (F) in NPHP1 (n = 5) and CTL (n = 4) URECs 5 d after seeding treated for 2 d with DMSO (0.04%) or ALP (2 µM). Each dot represents one individual cell line. Bars indicate mean. n = 3 experiments. Mixed linear-regression model with quasibinomial penalization (E), Mann–Whitney U test (NPHP1 vs. CTL) and paired Student’s t test (DMSO vs. ALP) (F): **P < 0.01, ns: not significant. AU: arbitrary unit.
Generation of an Nphp1 KO Mouse Model Showing Severe Retinopathy and Renal Tubule Dilatation.
One of our next aims was to test the therapeutic potential of PGE1 in a Nphp1 mouse model. Since previously generated models were not reported to exhibit kidney phenotype (21, 22), we generated a Nphp1 KO mouse using the CRISPR/Cas9 system, which harbors a deletion of the start codon within the first exon of Nphp1 resulting in a complete loss of Nphp1 mRNA and protein expression (SI Appendix, Fig. S5), similarly as observed in most NPHP1 patients (23).
The Nphp1−/− mice displayed a retinopathy (SI Appendix, Fig. S6), which was more severe than in the previously described Nphp1 mutant mice (22, 24) with a drastic loss of photoreceptors observed as early as postnatal day (P)21 (SI Appendix, Fig. S6 A and B), associated with a complete loss of the scotopic response (SI Appendix, Fig. S6C).
Interestingly, histological examination of the kidneys revealed the presence of tubular dilatations in the cortex of Nphp1−/− mice as early as 2 mo and more evidently at 5 mo of age (SI Appendix, Fig. S7 A–E). A strong association was observed between the number and size of dilatations per kidney, and the “dilatation index” values for these two parameters showed a progression of kidney damages between 2 and 5 mo (SI Appendix, Fig. S7 F–I). Despite the presence of tubular dilatations, renal function was not impaired in 5-mo-old Nphp1−/− mice (SI Appendix, Fig. S7 J–M), indicating that our mouse model recapitulates only the early stage of the renal disease, consistent with the juvenile form of the disease in humans.
Further characterization of these dilatations revealed that they mainly affect the connecting tubules (CNT) as well as the junction between the CNT and the distal convoluted tubules (DCT2) (SI Appendix, Fig. S8 A and B). Notably, in kidney biopsies from patients carrying homozygous deletion of NPHP1, tubular dilatations were observed in similar tubular segments as well as in the loop of Henlé (SI Appendix, Fig. S8C). These data pointed to a specific role for NPHP1 in the homeostasis of the DCT/CNT region both in mice and humans.
In Vivo Validation of Prostaglandin-Related Molecule in Murine Nphp1 KO Model and NPH Zebrafish Model.
We next investigated whether treatment with the PGE1 analog ALP could have a positive effect on the number and size of tubular dilatations in 5-mo-old Nphp1−/− mice. ALP was administrated intraperitoneally (80 µg/kg, i.p.) daily to 1-mo-old male Nphp1+/+ and Nphp1−/− mice for 4 mo without any impact on growth or survival (SI Appendix, Fig. S9A). Histological analyses revealed that the number and total area of tubular dilatations were reduced in Nphp1−/− mice by ALP treatment, resulting in a significant decrease in the dilatation index with a linear correlation (Fig. 3 A and B and SI Appendix, Fig. S9 B and C). In addition, measurements of cilium length showed that cilia were longer in the dilated regions of Nphp1−/− mouse (Fig. 3 C and D and SI Appendix, Fig. S10A), as observed in other NPHP models (25). We also detected longer cilia in the kidney tubules from individuals with mutations in NPHP1 (SI Appendix, Fig. S10 B and C). Notably, ALP treatment shortened cilia length in Nphp1−/− mice to values similar to those in control mice (Fig. 3 C and D and SI Appendix, Fig. S10A).
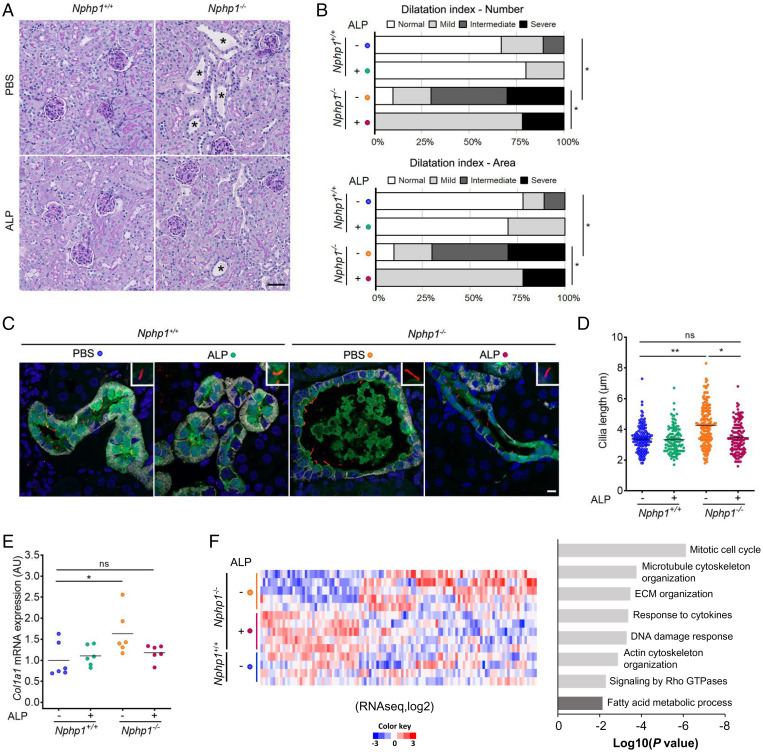
Fig. 3.
In vivo validation of the effect of ALP treatment on Nphp1−/− kidney. (A) Representative images of PAS staining of kidney sections from 5-mo-old Nphp1+/+ and Nphp1−/− mice treated daily with vehicle (PBS) or ALP (80 µg/kg) from month 1 to month 5. (Scale bar, 50 µm.) An asterisk signifies tubular dilatations. (B) Dilatation index for 5-mo-old Nphp1+/+ and Nphp1−/− mice upon ALP treatment. (Upper) Dilatation index for the number of dilatation/10 µm2 (cortex). (Lower) Total area covered by dilatations/cortex area. n = 9 to 10 male mice for each genotype/treatment. χ2 test: *P < 0.05. (C) Representative images of immunostaining of 5-mo-old Nphp1+/+ and Nphp1−/− kidney sections upon ALP treatment with anti-Calbindin (CNT, green), anti-ARL13B (cilia, red), anti-Barttin (TAL, DCT, and CNT, gray) antibodies, and DAPI (nuclei, blue). (Scale bar, 5 µm.) (D) Quantification of cilium length in CNT and DCT2 of 5-mo-old Nphp1+/+ and Nphp1−/− mice upon ALP treatment. n = 3 male mice for each genotype/treatment. Each dot represents one cilium. (E) qRT-PCR analysis of Col1a1 expression from 5-mo-old Nphp1+/+ and Nphp1−/− kidneys upon ALP treatment. Each dot represents one individual mouse. For D and E, bars indicate mean. One-way ANOVA followed by Holm–Sidak’s posttest: *P < 0.05, **P < 0.01, ns: not significant. (F) Significantly differentially expressed genes (DEGs) detected by RNA-seq in 5-mo-old Nphp1−/− mice upon ALP treatment. (Left) Heatmap of the 112 genes from the dataset of Nphp1+/+ (n = 4), Nphp1−/− PBS (n = 5), and Nphp1−/− ALP (n = 5) treated mice using signal log-ratio values. Each row represents a gene with up- (red) and down-regulated (blue) expression. (Right) Pertinent down- (light gray) or up-regulated (dark gray) pathways or processes involving these genes highlighted using Metascape. ECM, extracellular matrix; TAL, thick ascending limb of the loop of Henle.
Despite the fact that interstitial fibrosis is the main characteristic of NPHP1-associated NPH in humans (4), it was not obviously detected by histology in our mouse model. We performed qRT-PCR analysis of several profibrogenic markers in the kidneys of 5-mo-old Nphp1−/− mice (Fig. 3E and SI Appendix, Fig. S11 A–E). Among those markers, expression of Col1a1 was slightly increased in the kidneys of Nphp1−/− mice and attenuated by ALP treatment (Fig. 3E). To gain further insights into the potential beneficial effect of PGE1, we realized a global unsupervised transcriptomic analysis of whole kidneys from control and Nphp1−/− mice (Fig. 3F and SI Appendix, Fig. S11F). This analysis using the gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and DisGeNET, and Reactome datasets revealed that renal fibrosis disease concept (#C0151650) was significantly enriched in Nphp1−/− kidneys (P = 1.0E-10), in addition to several other up-regulated pathways (SI Appendix, Fig. S11F and Dataset S1), including extracellular matrix organization (P = 1.7E-18) and inflammation (response to cytokines, P = 2.3E-07). Furthermore, the up-regulation of most of these pathways was blunted by PGE1 treatment, together with additional ones, such as mitotic cell cycle (P = 7.6E-07) and DNA damage response (P = 5.1E-04) (Fig. 3F and Dataset S2), confirming its global beneficial effect on pathways known to be dysregulated in the context of NPH (4). Interestingly, some of the genes that did not show significant changes by qRT-PCR, were indeed found to be up-regulated in Nphp1−/− mice (Col3a1: +1.54; Acta2: +1.26), with a beneficial effect of PGE1 treatment (Col3a1: +0.68; Acta2: +0.80).
Overall, these data showed that PGE1 treatment is able to attenuate the renal manifestations associated with the loss of Nphp1, including the formation/extension of tubular dilatations, early up-regulation of fibrosis markers and extracellular matrix components, as well as abnormal cilia.
Along with the mouse model, the effect of prostaglandin-related analogs was further examined in zebrafish, a widely used model to study cilia function in context of ciliopathies (26). We used the nphp4 morpholino model, for which we and other have reported cysts in the proximal part of the pronephros, as well as ciliogenesis defects and defects in cloaca formation in its distal end (27, 28). This model is relevant as NPHP1 and NPHP4 play similar roles at the transition zone and cellular junctions; and their loss-of-function in patients, as well as in other animal models, results in identical phenotypes (1, 2). In addition, EP receptors, in particular EP2 and EP4, are expressed in zebrafish pronephros (29). Treatments with PGE1 (ALP) or PGE2 (DINO) molecules resulted in a significant decrease in the proportion of nphp4 morphants with severe pronephric cysts (SI Appendix, Fig. S12 A–C). ALP treatment also reduced the proportion of nphp4 morphants with cloacal dilatations (SI Appendix, Fig. S12D) and partially restored cilium length with a loss of significant difference between control and morphants upon treatment (SI Appendix, Fig. S12E). Overall, these results obtained in zebrafish provide evidence that PGE2 analogs can prevent dilatations of the renal tubules as well as ciliary defects in other in vivo models of NPH.
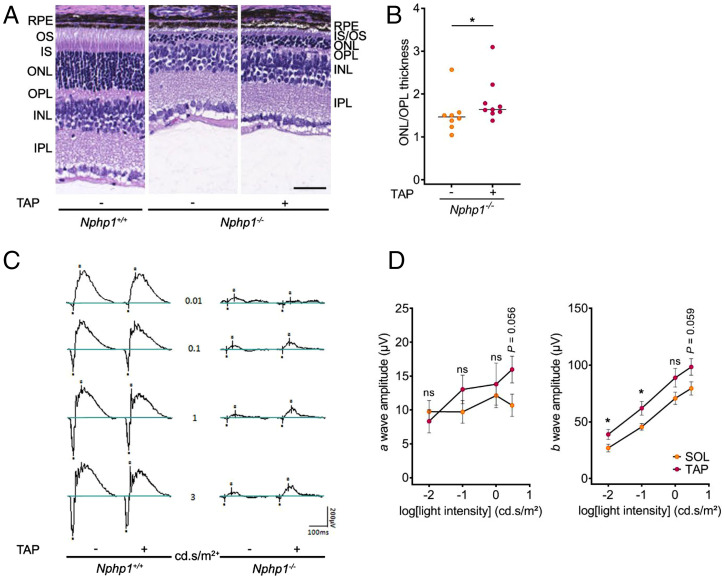
In addition to kidney, we investigated whether EP agonists could hamper retinal degeneration. Notably, EP2 is the most predominantly expressed PGE2 receptor in the eye (30) and the EP2 agonist Taprenepag has been previously used in the context of glaucoma in humans (31). In our Nphp1−/− mouse model, the retinopathy was observed as early as P14 and a loss of light-detection capacity as early as P21 (SI Appendix, Fig. S6). Based on these results, Tapreneprag was injected every 3 d (18 mg/kg, i.p.) from P6 to P21 without deleterious effect on body growth compared to Solutol-treated control mice (SI Appendix, Fig. S13). Treatment with the EP2 agonist led to a partial preservation of the outer nuclear layer of the photoreceptor cells in Nphp1−/− mice at P21 (Fig. 4 A and B) and to an improvement of the light-detection capacity of the photoreceptors with an increased magnitude of b-wave on ERG (Fig. 4 C and D).
Fig. 4.
Effect of Taprenepag, an EP2 agonist, on the Nphp1−/−-associated retinopathy. (A) Representative images of H&E staining of retina sections from P21-old Nphp1+/+ and Nphp1−/− mice treated every 3 d with vehicle (5% Solutol) or Taprenepag (18 mg/kg) from P6 to P21. (Scale bar, 50 µm.) (B) Quantification of ONL/OPL thickness ratio in retinas from P21 Nphp1−/− mice treated with vehicle (n = 8) or Taprenepag (n = 9). Each dot represents one mouse. Bars indicate mean. (C) Scotopic ERG recordings of P21-old Nphp1+/+ or Nphp1−/− retinas upon vehicle or Taprenepag treatment. (D) Quantification of scotopic a- (Left) and b-wave (Right) amplitude versus stimulus intensity (log) in P21 Nphp1−/− mice treated by vehicle (SOL, n = 6) or Taprenepag (n = 7). Mann–Whitney U test: *P < 0.05, ns: not significant. Dots indicate mean ± SEM. INL, inner nuclear segment; IPL, inner plexiform layer; IS, inner segment; ONL, outer nuclear segment; OPL, outer plexiform layer; OS, outer segment; RPE, retinal pigmented epithelium; SOL, Solutol; TAP, Taprenepag.
Altogether, these results indicate that treatment with prostaglandin agonists could partially prevent both renal and retinal ciliopathy-related phenotypes.
Molecular Analysis of PGE1 Treatment in URECs.
Our last aim was to better understand the mechanism underlying the beneficial effect of PGE1 on ciliogenesis. qRT-PCR analyses showed URECs mostly express EP2 and EP4 (SI Appendix, Fig. S4A), which are both coupled to GαS and stimulate cAMP production through activation of adenylate cyclases (AC) in response to agonist stimulation (20). Accordingly, treatment with ALP increased cAMP production in URECs to a level similar to that of forskolin (SI Appendix, Fig. S14A) and increased the phosphorylation of protein kinase A (PKA) substrates (SI Appendix, Fig. S14 B and C). Interestingly, EP4 agonists or increased cAMP production with the AC activator forskolin exert positive effects on ciliogenesis through stimulation of intraflagellar transport (32, 33). Taken together, these results show that ALP treatment activates the cAMP pathway in URECs, which may explain the improved ciliogenesis in NPHP1 conditions. Accordingly with this statement, forskolin treatment showed a similar positive effect on ciliogenesis as that observed with ALP (SI Appendix, Fig. S14 D and E). Finally, in agreement with the down-regulation of cell cycle progression in PGE1-treated kidneys (Fig. 3F), treatment of kidney tubular cells with PGE1 was shown to increase the expression of p27kip1 (34, 35), a key positive regulator of quiescence and ciliogenesis (36–38), which was reported to be positively regulated by cAMP (39). Indeed, treatment of NPHP1 URECs with PGE1 significantly increased the proportion of p27kip1+ nuclei (SI Appendix, Fig. S14 F and G), which likely contributes to its positive effect on ciliogenesis.
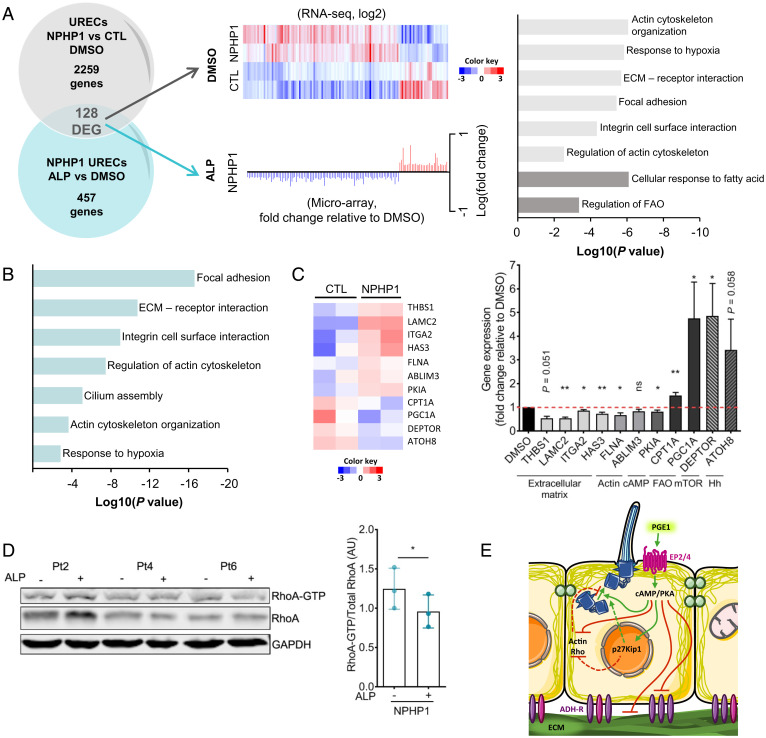
In order to identify additional functional downstream signaling pathways associated with the positive effects of PGE1 in NPHP1 models, we performed comparative transcriptomic profiling analysis in NPHP1 URECs. First, we performed transcriptomics analysis in NPHP1 URECs (Pt1) exposed to either DMSO or 2 µM ALP under the same conditions as for ciliogenesis assays and identified 457 differentially expressed protein-coding genes (150 up-regulated, 307 down-regulated). In parallel, we employed RNA-sequencing (RNA-seq) analysis on NPHP1 URECs (including Pt2 and Pt8) and control lines (including CTL5 and CTL8), and revealed 2,259 dysregulated genes in absence of NPHP1. We then compared the two gene lists and identified 128 dysregulated genes in NPHP1 URECs (31 genes down-regulated and 97 up-regulated) that were inversely modulated upon ALP treatment (Fig. 5A and Dataset S3).
Fig. 5.
Transcriptomic analyses of molecular targets upon ALP treatment in NPHP1 URECs. (A, Left) Venn diagram shows the number of significantly DEGs detected by microarray in URECs derived from NPHP1 patients (NPHP1) and CTL individuals upon ALP treatment versus DMSO (457 regulated genes) in comparison to the number of dysregulated genes detected by RNA-seq in NPHP1 vs. CTL URECs (2,259 regulated transcripts). The 128 genes regulated by ALP inversely modulated in NPHP1 URECs (128 DEG) are underlined. (Center, Upper) Heatmap of the 128 DEGs from the RNA-seq dataset of NPHP1 (n = 2) and CTL (n = 2) URECs using signal log ratio values. Each row represents a gene with up- (red) and down-regulated (blue) expression. (Center, Lower) Fold-change and normalized expression of the 128 DEGs in one NPHP1 UREC (Pt1) selected from the microarray dataset. n = 3 experiments. (Right) Pertinent down- (light gray) or up-regulated (dark gray) pathways or relevant processes involving these genes were highlighted using Metascape. (B) Pertinent pathways or processes involving ciliary modulators dysregulated in NPHP1 URECs upon ALP treatment were highlighted using Metascape. (C, Left) Heatmap of 11 selected genes of the 128 DEGs from RNA-seq dataset of NPHP1 versus CTL URECs. (Left) qRT-PCR validating the positive effect of ALP treatment on the expression of the 11 selected dysregulated genes in NPHP1 URECs (n = 5). Paired Student’s t test: **P < 0.01, *P < 0.05, ns: not significant. Bars indicate mean ± SEM. n = 2 experiments. Hh: Hedgehog. (D, Left) Representative image of Western blot analysis showing the activation of RhoA in NPHP1 URECs (Pt, n = 3) after 24 h of treatment with DMSO (0.04%) or ALP (2 µM). (Right) Quantification of the relative abundance of RhoA-GTP in NPHP1 URECs. Each dot represents one individual cell line. Mean ± SEM. n = 3 experiments. (E) Pathways regulated by ALP/PGE1 in URECs. ADH-R, adhesion receptors. (C and D) Paired Student’s t test: **P < 0.01, *P < 0.05, ns: not significant.
Pathway enrichment analysis using the GO, KEGG, and Reactome datasets revealed that, in agreement with the analysis of the Nphp1−/− mice kidneys (Fig. 3F and SI Appendix, Fig. S11F), the 128 dysregulated genes were predominantly involved in the regulation of actin cytoskeleton (P = 1.0E-06), focal adhesion (P = 7.9E-06), extracellular matrix–receptor interaction (P = 2.5E-06), and integrin cell surface interactions (P = 4.7E-05). In addition, several pathways crucial for mitochondrial function commonly impaired in CKDs were also modulated, including positive regulators of fatty acid oxidation (FAO; P = 4.4E-04) (40) and the response to hypoxia (P = 9.1E-07) (41). Several dysregulated genes were also associated with renal fibrosis, including the renal profibrotic factors GREM1 and THBS1 (42, 43), as well as negative regulators of the mammalian target of rapamycin (mTOR) pathway (DEPTOR) (44) or cyclic AMP signaling (PDE7, PKIA, ANX3), consistent with known signaling pathways downstream of EPs (45) and with our observations. Furthermore, in agreement with the positive effect of ALP on ciliogenesis, 65 of the 457 ALP-regulated genes were found to be cilium-related (Fig. 5B and Dataset S4). Among those 65 genes, ABLIM3 encoding an actin modulator, which negatively regulates ciliogenesis (46), was down-regulated after PGE1 treatment, whereas ATOH8, encoding an homolog of the positive regulator of ciliogenesis ATOH1 (47, 48), was up-regulated.
To validate our transcriptomic analyses, we then selected 11 dysregulated genes belonging to the main pathways/cellular processes identified, including genes associated with cell adhesion (LAMC2, ITGA2), renal fibrosis (THBS1, HAS3, LAMC2, ITGA2), regulation of cAMP (PKIA), mTOR (DEPTOR), and FAO (CPT1A, PGC1Α), as well as potential modulators of ciliogenesis (ABLIM3, ATOH8). qRT-PCR analyses in several NPHP1 URECs confirmed the effect of ALP treatment on both the up- and down-regulation of most of the genes tested (Fig. 5C). Those results showed that, similarly as in Nphp1−/− mouse kidneys, PGE1 treatment down-regulated expression of almost all extracellular matrix components, integrin signaling, and actin cytoskeleton regulators. The up-regulation of Rho GTPase activity, which could be linked to a global up-regulation of actin and adhesion-related processes, was evidenced in several ciliopathy conditions and was involved in the associated ciliary defects (49). Notably, we confirmed that PGE1 treatment also decreased the amount of active RhoA in NPHP1 URECs (Fig. 5D).
Altogether, our results suggest that a prostaglandin compound would be able to restore ciliary and profibrogenic pathway in NPHP1-deficient cells through modulation of cell cycle progression, actin, adhesion, and extracellular matrix cues (Fig. 5E).
Discussion
There is approximatively less than 5% of treatment available for the more than 7,000 orphan diseases so far identified, most of which have genetic causes (50). The work presented here exemplifies the principle of identification of small pharmacological molecules to compensate genetic defects as a source for potential therapeutic agents. A phenotypic screening strategy using a reference kidney cell line derived from an NPHP1 patient led to the rapid hit identification of a prostaglandin (PGE1, ALP) that reverted whole-locus NPHP1 deletion-associated ciliary phenotypes, an effect confirmed using a panel of NPHP1 patient-derived cell lines. In vivo, PGE1 lessened the renal phenotypes observed in both NPH zebrafish model and novel Nphp1 KO murine model. In addition, another PGE2 receptor agonist (Taprenepag) with an orthogonal chemical structure ameliorates the severe retinopathy found in Nphp1−/− mice.
Although ciliary defects observed in the different NPHP1 in vitro and in vivo models are variable, they are all rescued by PGE1 treatment. Our present and past analyses consistently indicate that the loss of NPHP1 in kidney epithelial cells results in decreased ciliogenesis, which was linked to epithelialization delay in MDCK grown in two-dimensional conditions (8). It is noteworthy that when NPHP1_KD MDCK cells are grown in three-dimensions (spheroids), cilia get longer, as similarly observed in vivo in the kidneys of both Nphp1−/− mice and patients. As for NPH zebrafish and other ciliopathy models, ciliogenesis is decreased and cilia are shorter in the distal pronephros (26). Importantly, the length of primary cilia depends on various factors, including microtubule and actin cytoskeleton dynamics, epithelialization, and signaling pathways such as ROS, mTOR, cAMP, and cell cycle, which may be differentially regulated in different tissues and altered in NPH models (1, 4). This diversity of processes that influence ciliogenesis and cilium length could explain why, depending on the cellular or animal models, we observed different ciliary phenotypes.
Thus, the decreased ciliogenesis observed in vitro in NPHP1-deficient kidney tubular cells grown in standard culture conditions is a robust proxy, which led to the identification of PGE1 as a molecule that restores ciliogenesis defects in all other NPH models.
Several studies support the principle of our findings linking PGE2 receptor stimulation to ciliogenesis, which is mediated by downstream cAMP signaling (32, 33). Interestingly, ciliopathies associated with mutations in NPHP or IFT genes have been independently linked to a down-regulation of cAMP/PKA signaling associated with a decrease of ciliary localization of ADCY3 (9, 10, 51). In addition, increasing cAMP using forskolin was shown to ameliorate cilium count and length in CEP290-defective fibroblasts (10). In our study, we show that PGE1 treatment rescues ciliogenesis as well as the percentage of ADCY3+ cilia in NPHP1 URECs, and that this effect is linked to downstream cAMP production.
While the positive effect of EPs/cAMP on ciliogenesis was previously linked to a stimulation of intraflagellar transport (32, 33), our data also point to additional contributors. As reported for PGE2 (34, 35), we showed that PGE1 treatment increased the proportion of cells expressing p27kip1, a main positive regulator of quiescence (G0), a stage of the cell cycle during which ciliogenesis occurs (37). The expression of p27kip1 is tightly controlled during the cell cycle but mainly at the protein level by ubiquitin-dependent degradation (52), explaining why it did not appear in our various transcriptomic analyses. Interestingly, negative regulators of p27Kip1, such as CKS1B (53), were down-regulated (+0.69) along with cell cycle progression pathways upon PGE1 treatment in Nphp1−/− kidneys. As p27kip1 is also a negative regulator of Rho GTPases (54), its increased expression might therefore contribute to the inhibition of RhoA activation observed upon PGE1 treatment together with modulation of the extracellular matrix/adhesion pathways (see below). In summary, our data, together with previously published studies, indicate that the robust and strong positive effect of PGE1 on ciliogenesis in NPHP1 cells is the consequence of convergent pathways downstream of prostaglandin E2 receptors.
Besides the direct positive impact of EP receptor-mediated cAMP signaling on ciliogenesis, other prostaglandin-dependent pathways or cellular processes are susceptible to also modulate ciliation. It is now well documented that actin cytoskeleton contractility and extracellular matrix rigidity are major determinants of ciliogenesis (55, 56). Interestingly, fibrosis and thickening of the basal membrane surrounding kidney tubules are two main characteristics of the NPHP1-associated juvenile form of NPH. In agreement with these pathological observations, our transcriptomic analyses showed that renal fibrosis disease was one of the main up-regulated pathways in Nphp1−/− mice kidneys and that extracellular matrix organization (collagens, LAMC2), actin-cytoskeleton regulation, and integrin were among the most enriched GO terms up-regulated in both URECs and Nphp1−/− mice kidneys. Strikingly, PGE1 treatment had a beneficial effect on these profibrotic pathways, as well as on other processes, including cell cycle progression.
In accordance with our results, EP2 and EP4 activation was described to exert antifibrotic properties in several models of kidney fibrosis (57, 58), and was shown to play a major role in the differentiation of myofibroblasts and expression of extracellular matrix proteins (59). Altogether, our results suggest that the NPHP1-deficient background (KO mice and URECs) is sensitive to PGE1 treatment, which, in addition to its positive effects on ciliogenesis, has a protective effect on profibrotic gene expression and renal lesions.
Conclusion.
Our results stress that PGE2/EP signaling is likely to tackle NPH-associated phenotypes through a convergent mode of actions, including direct effects on cilia linked to cAMP signaling but also indirect effects through cell cycle and adhesion/matrix regulation also relevant to counteract the up-regulation of profibrotic genes observed in the pathological context. These combined effects are likely to explain the beneficial impact of agonists of PGE2 receptors observed on cilia, in vitro (URECs) and in vivo (kidney, retina), as well as on renal tubule dilatation. These restoration effects on preclinical NPHP1-associated manifestations offer exciting new potential therapeutic targets. Finally, this study provides evidence for a potential use of a small-molecule–based therapy in the specific context of the NPHP1 locus deletion-associated NPH, the most frequent form of NPH. More importantly, our study demonstrates that phenotype changes induced by a complete locus deletion are amenable to be reversed, and they can be functionally complemented with an indirect therapeutic molecular point of intervention using small molecules.
Materials and Methods
Human URECs.
Patients and control individuals were recruited at Necker Hospital in the frame of the approved NPH_1 protocol and urine samples were collected upon written informed consent and anonymized together with personalized data. URECs were isolated from urine collected from patients suffering from NPH carrying biallelic NPHP1 mutations, healthy age-matched donors, and patients suffering from ciliopathy-unrelated CKD (stages 2 to 5) (SI Appendix, Table S2). URECs were cultured as previously described (60) with the following modifications. Briefly, peeled renal tubular epithelial cells were cultured in primary medium (DMEM/F12, 10% FBS, 100 U/mL penicillin, 100 mg/mL streptomycin, amphotericinB, 1× REGM SingleQuots [CC-4127, Lonza]) in six-well plates at 37 °C in 5% CO2, and medium (0.5 mL) was renewed every 24 h. At 96 h, the medium was replaced by proliferation UREC medium (REBM Basal Medium [CC-3191, Lonza], 1× REGM SingleQuots, 10 ng/mL rhEGF [R&D system], 2% FBS certified [Invitrogen]). At 80% confluence, cells were trypsinized (TrypsinE select, Invitrogen) and plated in 12-well plates (2 × 104 cells). After 24 h, cells were transduced by retroviral gene transfer of thermosensitive SV40 T antigen (LOX-CW-CRE, Addgene) at a multiplicity of infection of 5 with 8 μg/mL polybrene (61). After 24 h, medium was changed and cells were grown at permissive temperature (33 °C).
Antibodies.
The following antibodies were used: ADCY3 (1/100e; Invitrogen, PA5-35382), ARL13B (1/800e; ProteinTech, 17711-1-AP), Barttin (1/400e; Santa Cruz, sc365161), Calbindin D-28K (1/500e; Novus, NBP2-50028SS), GAPDH (1/1,000e; Millipore, MAB374), γ-Tubulin (1/5,000e; Sigma, T6557), PKAS (1/1,000e; Cell Signaling, 9624), RhoA (1/1,000e; Cytoskeleton, ARH05). Secondary Alexa-conjugated secondary antibodies were from Invitrogen.
Immunofluorescence-Based Assays in URECs.
Cell culture and compound treatments.
For ciliogenesis assays, URECs (105 cells per well) were seeded in 96-well glass microplates (Sensoplates, Greiner) at 39 °C. For other immunofluorescence experiments, URECs (4 × 105 cells per well) were seeded in 24-well plates at 39 °C. On the third day of culture, cells were incubated for 48 h in the presence of the compounds, all dissolved in DMSO (0.04 to 0.1%).
Immunofluorescence.
Cells were fixed after 1 to 5 d of culture in either cold methanol for 5 min or either with 4% PFA for 20 min, quenched in 50 mM NH4Cl, and permeabilized 15 min with 0.1% Triton if fixed with 4% PFA, then treated 30 min with PBS, 0.1% Tween 20, 3% BSA, before being incubated 1 h with primary antibodies, and then 1 h with appropriate fluorescent secondary antibody and DAPI (1/2,000e; Thermo Fisher Scientific, 62247).
Data acquisition, quantification, and analysis.
For ciliogenesis assays, images were acquired using the Opera Phenix (40×, Perkin-Elmer). Automated acquisition of 41 z-stack per well was performed. The number of ciliated cells was measured using a semiautomated analysis using Harmony software (Perkin-Elmer). Briefly, images were analyzed using the building blocks approach to detect in this order, nuclei (DAPI staining) and, with a 20-pixel enlarged region, the basal body (γ-Tubulin signal), and the primary cilia (ARL13B staining). With conventional filters (intensity, size, and so forth), the software segmented candidate primary cilium. Hundreds of phenotypic parameters were calculated for every candidate cilium using signal enhancement ratio texture (intensity patterns) and advanced STAR morphology parameters (distribution of either texture features or fluorescence intensities inside a region of interest). Using the PhenoLOGIC machine-learning option of Harmony, the parameters best suited to discriminate cilia were defined and used to obtain final detection and counting.
For other assays, confocal images were acquired using a Spinning Disk microscope (40× or 63×, Zeiss). All quantifications were performed using ImageJ software.
RNA Extraction for Transcriptomic Analyses.
URECs were seeded in ciliogenesis conditions: primary cells were seeded (2 × 104 cells per well) in 2 wells of 12-well plates and incubated 1 wk at 37 °C; immortalized patients and controls cells were seeded (105 cells per well) in 8 wells of 96-well glass microplates at 39 °C and incubated for 4 d. To evaluate the impact of ALP treatment on transcriptome, cells were incubated for 24 h on the third day of culture with DMSO or 2 μM of ALP. Kidneys from control and Nphp1−/− mice, treated or not treated with PGE1, were lysed in RLT buffer using the T18 digital ULTRA-TURRAXX (Ika). Total mRNA was isolated using an Extraction Mini Kit (Qiagen) following the recommendations of the manufacturer.
RNA-seq Analysis.
For each sample, 50-million read pair-end sequencing was performed on NovaSEq. 6000 Illumina. FASTQ files were mapped to the ENSEMBL [Human(GRCh38/hg38)/Mouse GRCm38/mm10] reference using Hisat2 and counted by featureCounts from the Subread R package (http://www.r-project.org/). Read-count normalizations and group comparisons were performed by the Deseq2 statistical method. Flags were computed from counts normalized to the mean coverage. All normalized counts <20 were considered as background (flag 0) and ≥20 as signal (flag = 1). P50 lists used for the statistical analysis regroup the genes showing flag = 1 for at least half of the compared samples. The results were filtered at P ≤ 0.05 and fold-change 1.2 for URECs and 1.1 for mice kidneys. Cluster analysis was performed by hierarchical clustering using the Spearman correlation similarity measure and ward linkage algorithm. Heatmaps were made with the R package ctc: Cluster and Tree Conversion, and imaged by Java Treeview software (Java Treeview, extensible visualization of microarray data) (62). Functional analyses were carried out using Metascape (http://metascape.org/).
qRT-PCR Analysis.
Total RNA was reverse-transcribed using SuperScript II Reverse Transcriptase (LifeTechnologies) according to the manufacturer’s protocol. qRT-PCR was performed with iTaq Universal SYBR Green Supermix (Bio-Rad) on the ViiA 7 Real-Time PCR System (Thermo Fisher Scientific). Each biological replicate was measured in technical duplicates. The primers used for quantitative real-time PCR are listed in SI Appendix, Table 3.
Mice Experiments.
Generation of Nphp1−/− mice by CRISPR/Cas9.
Nphp1−/− mice were generated using a CRISPR/Cas9 system, as previously described (63). Guide RNAs targeting exon1 of Nphp1 were designed using CRISPOR (crispor.tefor.net/). Single-guide RNA and Cas9 protein (TACGEN) were injected into the pronucleus of the C57BL/6J zygotes. All exonic off-targets were tested to ascertain absence of undesired mutations. Two founders carrying indel mutations were back-crossed with C57BL/6J mice and the Nphp1+/− offspring were further confirmed by PCR genotyping with appropriate primers. We focused on the mouse line carrying a 73-bp deletion (c.-33_40del) in exon1 encompassing the ATG.
Treatment of mice with prostaglandin receptor agonists.
For kidney experiments, ALP (80 µg/kg) or PBS were administrated intraperitoneally daily to 1-mo-old Nphp1+/+ and Nphp1−/− male mice for 4 mo. For retina experiments, Taprenepag (18 mg/kg) or Solutol (5%) were administrated intraperitoneally every 3 d from P6 to P21 to Nphp1+/+ and Nphp1−/− male and female mice.
Histology and immunohistochemistry.
Kidneys and eyes were fixed overnight at 4 °C in 4% PFA and embedded in paraffin. The retinal and kidney structures were assessed by H&E and periodic acid Schiff staining of 4-µm sections, respectively. For immunofluorescence, after paraffin removal and antigen retrieval treatment (10 mM Tris pH9, 1 mM EDTA, 0.05% Tween 20, 87 °C, 50 min), sections were blocked with PBS, 0.1% Tween 20, 1% BSA for 1 h and incubated overnight at 4 °C with the indicated primary antibodies. After two washes, sections were incubated for 1 h with appropriate secondary antibodies and DAPI. Confocal images were acquired using a Spinning Disk microscope (40× or 63×, Zeiss). Count and area of kidney dilatations and cilia length was measured using ImageJ. Retina layer thickness was measured with QuantaCell’s costum scripts.
Dilatation index.
In order to quantify more accurately the severity of the disease, a dilatation index, taking into account the dilatation number and dilatation size, was established. For each calculated ratio—the 1) dilatation surface area to cortex area and 2) dilatation number to cortex area—a score was defined as follows: score = 1 (no lesions) for all values between zero and the third quartile of all values for Nphp1+/+; score = 2 (mild) for all values between score 1 and the median value for Nphp1−/−; score = 3 (intermediate) for all values between score 2 and the third quartile of all values for Nphp1−/−; and score = 4 (severe) for all values above score 3. Scores are protocol-specific and calculated based on all the animals included in the protocol.
Electroretinography.
For electroretinography, 21-d-old mice were dark-adapted overnight before the experiments and anesthetized (intraperitoneal ketamine [66.7 mg/kg] and xylazine [11.7 mg/kg]). Eyes were exorbitated with neosynephrine and the pupils were dilated with mydriatycum eye drops. The reference electrode was placed under the back skin, just above the tail, and the measuring electrodes on the cornea on which a drop of ophthalmic gel was added (Lacrigel, Europhia). For the scotopic electroretinograms, the flash duration varied from 3 to 5 ms, with the final flash output ranging from 0.001 to 3 cd⋅s/m2. Responses were amplified, filtered, digitized, and a- and b-waves were measured (CelerisTM Diagnosys Espion Visual Electrophysiology System).
For mice studies, all image analyses and quantifications were done blinded.
Active RhoA Pull-Down Assay.
Cells were incubated during 15 min with 0.04% DMSO or 2 µM ALP. RhoA-GTP was measured using the RhoA Pull-down Activation Assay Biochem Kit (BK036, Cytoskeleton) following the recommendations of the manufacturer. Briefly, cell lysates were incubated with 50 µg of GST-tagged Rhotekin-RBD beads for 1h at 4 °C. Lysates and precipitates were run on polyacrylmid gels (12%) under reducing conditions and analyzed by Western blotting using indicated primary antibodies (RhoA, GADPH) and the Odyssey CLx imaging system (LI-COR Biosciences). Active RhoA was normalized to total RhoA, which was itself normalized on GAPDH using Image Studio Lite software (LI-COR Biosciences).
Statistics.
Statistical calculations were performed with R or GraphPad Prism v8. Figure representation was performed using GraphPad Prism v8 and is presented as dot plots or box-and-whisker plots. The Shapiro–Wilk test was used to test the normality. Differences between two groups that do not meet the normal distribution were compared using two-tailed Mann–Whitney U test. If not possible or if the two groups meet the normal distribution, differences were compared using unpaired two-tailed Student’s t test. When comparing more than two groups, a one-way ANOVA followed by Holm–Sidak’s multiple comparisons test was used. To analyze the effect of ALP treatment on ciliogenesis or ADCY3+ cilia staining in cells coming from the same individuals, we used a mixed linear-regression model with quasibinomial penalization (R software: qlmmPQL function of MASS package), taking into account the correlation of observations coming from the same individuals using a random effect on the cell line. To analyze the effect of ALP treatment in cells coming from the same individuals, we used a paired two-tailed Student’s t test. To compare the correlation between two parameters, we used a Pearson’s test. To determine if there is a significant difference between two proportions, we used Fisher’s exact test. To determine if there is a significant difference between more than two proportions, we used a χ2 test. P < 0.05 was considered statistically significant. Statistical tests and exact sample sizes used to calculate statistical significance are stated in the figure legends.
Study Approval.
Human studies.
The NPH_1 protocol on the research of therapeutic targets in the frame of NPH and renal-associated ciliopathies has been approved by the French National Committee for the Protection of Persons under the ID-RCB no. 2016-A00541-50 and is kept in full accordance with the principles of the Declaration of Helsinki and Good Clinical Practice guidelines. The human renal biopsies belonging to the Imagine Biocollection are declared to the French Minister of Research under the number DC-2020-3994 and this biocollection was approved by the French Ethics Committee for Research at Assistance Publique-Hôpitaux de Paris under the Institutional Review Board registration no. 00011928.
Animal studies.
All animals were handled in strict accordance with good animal practice as defined by the national animal welfare bodies, and all animal procedures were performed in accordance with protocols approved by the ethical committee of the Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation (APAFIS#22608-2018042715366694 v4).
Supplementary Material
Acknowledgments
We thank the patients and their families for their participation; Olivia Boyer, Aurélie Hummel, and Guillaume Lezmi (Pediatric Nephrology, Necker Enfants Malades, Assistance Publique–Hôpitaux de Paris, Paris, France), Amélie Lezmi Ryckewaert and Sophie Taque (Pediatric Hematology and Oncology, Hôpital Universitaire, Rennes, France), Odile Boespflug-Tanguy (Centre de Compétence des Leucodystrophies et Leucoencéphalopathies de Cause Rare, Pôle Femme et Enfant, Hôpital Estaing, Centre Hospitalier Universitaire de Clermont-Ferrand, Clermont-Ferrand, France), and Jérôme Harambat and Brigitte Llanas (Department of Pediatrics, Bordeaux University Hospital, Bordeaux, France) who helped with the follow-up of patients; the Department of Clinical Research (Salma Kotti, Elisabeth Hulier-Ammar) at Imagine Institute with the sponsorship team (Didier Beudin, Olivier Freund, Isabelle Buffet, Haizia Hammoudi, Ahlam Ramdani, Nicholas Renaud) that facilitates and structures the set-up of the clinical research projects, and the investigation team (Pauline Touche, Marc Melesan, Aurélie Jospin, Joséphine Cornet, Kenza Bensari, Maxime Douillet) that prepares and ensures the follow-up of the clinical trials; Romain Marlange and Nicolas Doulet (Innovation & Technology Transfer Department, Imagine Institute), who helped us to establish the collaboration with industrial partners Alexion and Medetia; Cécile Jeanpierre for critical reading of the manuscript; the Biophenics core facilities (Curie Institute, Paris), in particular Aurianne Lescure for her technical assistance; the zebrafish, mice, transgenesis, histology, and imaging facilities of Structure Fédérative de Recherche Necker, especially Pierre David for his help in establishing the Nphp1 knockout CRISPR-Cas mouse model, and Nicolas Cagnard (Bioinformatics Platform, Imagine Institute) for his help on RNA-sequencing analysis; Victor Racine (QuantaCELL) for the design of the semiquantitative analysis of zebrafish body curvature and retina layers thickness; Xavier Gérard and Jean-Michel Rozet (Imagine Institute) for their precious and helpful advice for phenotypic analysis of retinal degeneration; and Michael Perricone, who initiated the collaboration between the Imagine Institute and Alexion. This work was supported by INSERM, the Ministère de l’Education Nationale de la Recherche et de la Technologie, by a State funding from the Agence Nationale de la Recherche (ANR) under the “Investissements d’avenir” program (ANR-10-IAHU-01), and by a public grant “RHU-C’IL-LICO” overseen by the ANR as part of the second “Investissements d’Avenir” program (ANR-17-RHUS-0002). We acknowledge the Imagine Institute for the purchase of Leica SP8 STED and Zeiss Spinning Disk microscopes, and the Fondation ARC (EML20110602384) for the purchase of the Leica SP8 confocal microscope.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115960119/-/DCSupplemental.
Data Availability
The datasets generated are available in the ArrayExpress database at the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI), https://www.ebi.ac.uk/arrayexpress/ (accession nos. E-MTAB-11582 (64), E-MTAB-11593 (65), E-MTAB-11594 (66), and E-MTAB-11595 (67)). All other study data are included in the main text and supporting information.
References
- 1.Reiter J. F., Leroux M. R., Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 18, 533–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun D. A., Hildebrandt F.. Ciliopathies. Cold Spring Harb. Perspect. Biol. 9, a028191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salomon R., Saunier S., Niaudet P.. Nephronophthisis. Pediatr. Nephrol. 24, 2333–2344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stokman M. F., Saunier S., Benmerah A., Renal ciliopathies: Sorting out therapeutic approaches for nephronophthisis. Front. Cell Dev. Biol. 9, 653138 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snoek R., et al. , NPHP1 (Nephrocystin-1). J. Am. Soc. Nephrol. 29, 1772–1779 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.König J., et al. , Phenotypic spectrum of children with nephronophthisis and related ciliopathies. Clin. J. Am. Soc. Nephrol. 12, 1974–1983 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sang L., et al. , Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145, 513–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delous M., et al. , Nephrocystin-1 and nephrocystin-4 are required for epithelial morphogenesis and associate with PALS1/PATJ and Par6. Hum. Mol. Genet. 18, 4711–4723 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bizet A. A., et al. , Mutations in TRAF3IP1/IFT54 reveal a new role for IFT proteins in microtubule stabilization. Nat. Commun. 6, 8666 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molinari E., et al. , Targeted exon skipping rescues ciliary protein composition defects in Joubert syndrome patient fibroblasts. Sci. Rep. 9, 10828 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benzing T., et al. , Nephrocystin interacts with Pyk2, p130Cas, and tensin and triggers phosphorylation of Pyk2. Proc. Natl. Acad. Sci. U.S.A. 98, 9784–9789 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donaldson J. C., Dise R. S., Ritchie M. D., Hanks S. K., Nephrocystin-conserved domains involved in targeting to epithelial cell-cell junctions, interaction with filamins, and establishing cell polarity. J. Biol. Chem 277, 29028–29035 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Veland I. R., et al. , Inversin/Nephrocystin-2 is required for fibroblast polarity and directional cell migration. PLoS One 8, e60193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez-Hernandez V., et al. , Bardet–Biedl syndrome proteins control the cilia length through regulation of actin polymerization. Hum. Mol. Genet. 22, 3858–3868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Locatelli F., Del Vecchio L., Cavalli A., Inhibition of the renin-angiotensin system in chronic kidney disease: A critical look to single and dual blockade. Nephron. Clin. Pract. 113, c286–c293 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Torres V. E., et al. , Tolvaptan in patients with autosomal dominant polycystic kidney disease. N. Engl. J. Med. 367, 2407–2418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Husson H., et al. , Reduction of ciliary length through pharmacologic or genetic inhibition of CDK5 attenuates polycystic kidney disease in a model of nephronophthisis. Hum. Mol. Genet. 25, 2245–2255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bainbridge J. W. B., et al. , Long-term effect of gene therapy on Leber’s congenital amaurosis. N. Engl. J. Med. 372, 1887–1897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molinari E., et al. , Human urine-derived renal epithelial cells provide insights into kidney-specific alternate splicing variants. Eur. J. Hum. Genet. 26, 1791–1796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasrallah R., Hassouneh R., Hébert R. L., Chronic kidney disease: Targeting prostaglandin E2 receptors. Am. J. Physiol. Renal. Physiol. 307, F243–F250 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Jiang S.-T., et al. , Targeted disruption of Nphp1 causes male infertility due to defects in the later steps of sperm morphogenesis in mice. Hum. Mol. Genet. 17, 3368–3379 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Louie C. M., et al. , AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat. Genet. 42, 175–180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunier S., et al. , Characterization of the NPHP1 locus: Mutational mechanism involved in deletions in familial juvenile nephronophthisis. Am. J. Hum. Genet. 66, 778–789 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang S.-T., et al. , Essential role of nephrocystin in photoreceptor intraflagellar transport in mouse. Hum. Mol. Genet. 18, 1566–1577 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Ramsbottom S. A., et al. , Targeted exon skipping of a CEP290 mutation rescues Joubert syndrome phenotypes in vitro and in a murine model. Proc. Natl. Acad. Sci. U.S.A. 115, 12489–12494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Z., Zhang X., Jia S., Yelick P. C., Zhao C., Zebrafish as a model for human ciliopathies. J. Genet. Genomics 43, 107–120 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Burcklé C., et al. , Control of the Wnt pathways by nephrocystin-4 is required for morphogenesis of the zebrafish pronephros. Hum. Mol. Genet. 20, 2611–2627 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Slanchev K., Pütz M., Schmitt A., Kramer-Zucker A., Walz G., Nephrocystin-4 is required for pronephric duct-dependent cloaca formation in zebrafish. Hum. Mol. Genet. 20, 3119–3128 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Poureetezadi S. J., Cheng C. N., Chambers J. M., Drummond B. E., Wingert R. A., Prostaglandin signaling regulates nephron segment patterning of renal progenitors during zebrafish kidney development. eLife 5, e17551 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biswas S., Bhattacherjee P., Paterson C. A., Prostaglandin E2 receptor subtypes, EP1, EP2, EP3 and EP4 in human and mouse ocular tissues—A comparative immunohistochemical study. Prostaglandins Leukot. Essent. Fatty Acids 71, 277–288 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Schachar R. A., Raber S., Courtney R., Zhang M., A phase 2, randomized, dose-response trial of taprenepag isopropyl (PF-04217329) versus latanoprost 0.005% in open-angle glaucoma and ocular hypertension. Curr. Eye Res. 36, 809–817 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Jin D., et al. , Prostaglandin signalling regulates ciliogenesis by modulating intraflagellar transport. Nat. Cell Biol. 16, 841–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Besschetnova T. Y., et al. , Identification of signaling pathways regulating primary cilium length and flow-mediated adaptation. Curr. Biol. 20, 182–187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasrallah R., et al. , Prostaglandin E2 increases proximal tubule fluid reabsorption, and modulates cultured proximal tubule cell responses via EP1 and EP4 receptors. Lab. Invest. 95, 1044–1055 (2015). [DOI] [PubMed] [Google Scholar]
- 35.May-Simera H. L., et al. , Primary cilium-mediated retinal pigment epithelium maturation is disrupted in ciliopathy patient cells. Cell Rep. 22, 189–205 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komatsu Y., Kaartinen V., Mishina Y., Cell cycle arrest in node cells governs ciliogenesis at the node to break left-right symmetry. Development 138, 3915–3920 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izawa I., Goto H., Kasahara K., Inagaki M., Current topics of functional links between primary cilia and cell cycle. Cilia 4, 12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yukimoto H., et al. , A novel CDK-independent function of p27Kip1 in preciliary vesicle trafficking during ciliogenesis. Biochem. Biophys. Res. Commun. 527, 716–722 (2020). [DOI] [PubMed] [Google Scholar]
- 39.New D. C., Wong Y. H., Molecular mechanisms mediating the G protein-coupled receptor regulation of cell cycle progression. J. Mol. Signal. 2, 2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Console L., et al. , The link between the mitochondrial fatty acid oxidation derangement and kidney injury. Front. Physiol. 11, 794 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faivre A., Scholz C. C., de Seigneux S., Hypoxia in chronic kidney disease: Towards a paradigm shift? Nephrol. Dial. Transplant. 36, 1782–1790 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Church R. H., et al. , Gremlin1 plays a key role in kidney development and renal fibrosis. Am. J. Physiol. Renal. Physiol. 312, F1141–F1157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy-Ullrich J. E., Thrombospondin 1 and its diverse roles as a regulator of extracellular matrix in fibrotic disease. J. Histochem. Cytochem. 67, 683–699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C., et al. , Loss of DEPTOR in renal tubules protects against cisplatin-induced acute kidney injury. Cell Death Dis. 9, 441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y., et al. , Prostaglandins in the pathogenesis of kidney diseases. Oncotarget 9, 26586–26602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao J., et al. , miR-129-3p controls cilia assembly by regulating CP110 and actin dynamics. Nat. Cell Biol. 14, 697–706 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Schroeder N., Wuelling M., Hoffmann D., Brand-Saberi B., Vortkamp A., Atoh8 acts as a regulator of chondrocyte proliferation and differentiation in endochondral bones. PLoS One 14, e0218230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang C.-H., et al. , Atoh1 controls primary cilia formation to allow for SHH-triggered granule neuron progenitor proliferation. Dev. Cell 48, 184–199.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Smith C. E. L., Lake A. V. R., Johnson C. A., Primary cilia, ciliogenesis and the actin cytoskeleton: A little less resorption, a little more actin please. Front. Cell Dev. Biol. 8, 622822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pogue R. E., et al. , Rare genetic diseases: Update on diagnosis, treatment and online resources. Drug Discov. Today 23, 187–195 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Halbritter J., et al. , Defects in the IFT-B component IFT172 cause Jeune and Mainzer-Saldino syndromes in humans. Am. J. Hum. Genet. 93, 915–925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pagano M., et al. , Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269, 682–685 (1995). [DOI] [PubMed] [Google Scholar]
- 53.Shi W., et al. , CKS1B as drug resistance-inducing gene-a potential target to improve cancer therapy. Front. Oncol. 10, 582451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Besson A., Gurian-West M., Schmidt A., Hall A., Roberts J. M., p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 18, 862–876 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Failler M., et al. , Whole-genome screen identifies diverse pathways that negatively regulate ciliogenesis. Mol. Biol. Cell 32, 169–185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brücker L., Kretschmer V., May-Simera H. L., The entangled relationship between cilia and actin. Int. J. Biochem. Cell Biol. 129, 105877 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Vukicevic S., et al. , Role of EP2 and EP4 receptor-selective agonists of prostaglandin E2 in acute and chronic kidney failure. Kidney Int. 70, 1099–1106 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Jensen M. S., et al. , Activation of the prostaglandin E 2$EP 2$receptor attenuates renal fibrosis in unilateral ureteral obstructed mice and human kidney slices. Acta Physiol. 227, e13291 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li K., et al. , The roles of various prostaglandins in fibrosis: A review. Biomolecules 11, 789 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ajzenberg H., et al. , Non-invasive sources of cells with primary cilia from pediatric and adult patients. Cilia 4, 8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salmon P., et al. , Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Mol. Ther. 2, 404–414 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Saldanha A. J., Java Treeview—Extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Ucuncu E., et al. , MINPP1 prevents intracellular accumulation of the chelator inositol hexakisphosphate and is mutated in Pontocerebellar Hypoplasia. Nat. Commun. 11, 6087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.H. Garcia et al., Transcriptional analysis of the effect of Alprostadil on NPHP1 URECs. ArrayExpress. https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-11582. Deposited 15 March 2022. [Google Scholar]
- 65.H. Garcia et al., Transcriptional analysis of NPHP1 URECs. ArrayExpress. https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-11593. Deposited 15 March 2022. [Google Scholar]
- 66.H. Garcia et al., Transcriptional analysis of Nphp1-/- KO mouse model kidneys. ArrayExpress. https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-11594. Deposited 14 March 2022. [Google Scholar]
- 67.H. Garcia et al., Transcriptional analysis of Nphp1-/- KO mouse model kidneys in response to Alprostadil treatment. ArrayExpress. https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-11595. Deposited 14 March 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated are available in the ArrayExpress database at the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI), https://www.ebi.ac.uk/arrayexpress/ (accession nos. E-MTAB-11582 (64), E-MTAB-11593 (65), E-MTAB-11594 (66), and E-MTAB-11595 (67)). All other study data are included in the main text and supporting information.