Significance
Protein aggregates are often toxic, leading to impaired cellular activities and disease. The human HtrA2 trimeric enzyme cleaves such aggregates, and mutations in HtrA2 are causative for various neurodegenerative disorders, such as Parkinson’s disease and essential tremor. The mechanism by which cleavage occurs has been studied using small peptides, but little information is available as to how HtrA2 protects cells from the pathologic effects of aggregation involving protein molecules that can form well-folded structures. Using solution NMR spectroscopy, we investigated the structural dynamics of the interaction between HtrA2 and a model protein substrate, demonstrating that HtrA2 preferentially binds to an unfolded substrate ensemble and providing insights into how HtrA2 function is regulated.
Keywords: mitochondrial proteostasis, ligand-binding thermodynamics, methyl transverse relaxation optimized NMR spectroscopy, conformational selection, protein–protein interaction
Abstract
The human high-temperature requirement A2 (HtrA2) protein is a trimeric protease that cleaves misfolded proteins to protect cells from stresses caused by toxic, proteinaceous aggregates, and the aberrant function of HtrA2 is closely related to the onset of neurodegenerative disorders. Our methyl-transverse relaxation optimized spectroscopy (TROSY)–based NMR studies using small-peptide ligands have previously revealed a stepwise activation mechanism involving multiple distinct conformational states. However, very little is known about how HtrA2 binds to protein substrates and if the distinct conformational states observed in previous peptide studies might be involved in the processing of protein clients. Herein, we use solution-based NMR spectroscopy to investigate the interaction between the N-terminal Src homology 3 domain from downstream of receptor kinase (drk) with an added C-terminal HtrA2-binding motif (drkN SH3-PDZbm) that exhibits marginal folding stability and serves as a mimic of a physiological protein substrate. We show that drkN SH3-PDZbm binds to HtrA2 via a two-pronged interaction, involving both its C-terminal PDZ-domain binding motif and a central hydrophobic region, with binding occurring preferentially via an unfolded ensemble of substrate molecules. Multivalent interactions between several clients and a single HtrA2 trimer significantly stimulate the catalytic activity of HtrA2, suggesting that binding avidity plays an important role in regulating substrate processing. Our results provide a thermodynamic, kinetic, and structural description of the interaction of HtrA2 with protein substrates and highlight the importance of a trimeric architecture for function as a stress-protective protease that mitigates aggregation.
Many proteins must fold into a well-defined three-dimensional (3D) structure in order to properly function. When this process fails, these molecules can form toxic aggregates via exposed hydrophobic regions that are typically sheltered in the properly folded structure. Increasing evidence suggests that the accumulation of toxic aggregates of misfolded proteins is causative for various neurodegenerative disorders, such as Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease (1–3). Cells are equipped with quality-control machinery to counteract these stresses, which either assist protein refolding or target proteins for degradation. High-temperature requirement A (HtrA) proteases are an important class of stress-protective enzymes, widely conserved from bacteria to mammals, whose main function is to cleave toxic proteins in an adenosine 5′-triphosphate (ATP)-independent manner (4–6). Their activities are not limited to the degradation of misfolded proteins, however, but also include roles as protein chaperones and proteases of regulatory proteins involved in various cell-signaling pathways, such as cellular proliferation, mobility, and apoptosis (4).
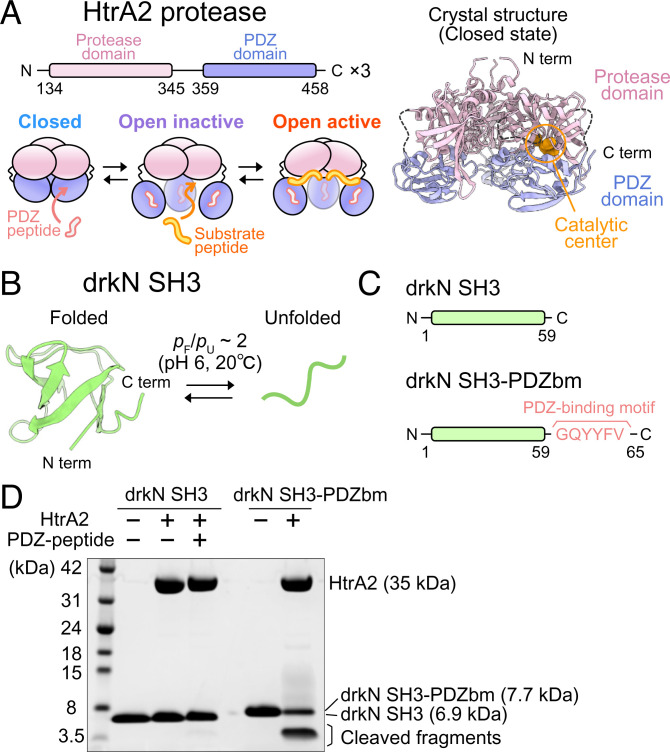
Human HtrA2, also known as the Omi protease, is an HtrA protease that is primarily localized to the mitochondrial intermembrane space (7–9). There is increasing evidence that HtrA2 plays a prominent neuroprotective role by degrading toxic aggregates in mitochondria (9–14) and that its aberrant function results in deregulation of mitochondria and, ultimately, in the onset of neurodegenerative disorders (10, 12–16). This notion is further supported by the finding that missense mutations in the HTRA2 gene are present in patients with Parkinson’s disease or essential tremor (17, 18). The crystal structure of HtrA2 has been solved, revealing a pyramid-shaped homotrimer, in which each HtrA2 protomer is composed of one copy each of serine protease and PDZ (PSD-95, DLG, and ZO-1) domains, with intertrimer interactions mediated by protease–protease contacts (Fig. 1 A, Upper Left and Right) (19). In the crystal structure, the catalytic center in the protease domain is formed by H198, D228, and S306, which is occluded by the PDZ domain, precluding substrate binding. Thus, the crystal structure is thought to represent a closed-inactive conformation that cannot bind and, thus, cleave client molecules (6, 19).
Fig. 1.
drkN SH3-PDZbm, but not drkN SH3, is cleaved by HtrA2. (A) Domain organization of HtrA2 (Left Upper) and cartoon representations of three states of HtrA2 (Left Lower) (31, 32). (A, Right) The crystal structure of trimeric HtrA2 (Protein Data Bank [PDB] ID code 1LCY) in the closed state is shown. The protease (residues 134 to 345) and PDZ (residues 359 to 458) domains are colored pink and light blue, respectively. The region containing residues 282 to 290 (β8–β9 linker) and 344 to 358 (interdomain linker) are shown with dotted lines. The catalytic center residues (residues 198, 228, and 306) are shown as orange spheres in one of the subunits. (B) NMR structure of drkN SH3 T22G mutant (PDB ID code 2A36) (37) and schematic representation of the equilibrium between the folded and unfolded states of the domain. (C) Schematic representations of the drkN SH3 constructs (drkN SH3 and drkN SH3-PDZbm) used in this study. (D) Gel-based proteolytic activity assays of HtrA2 against drkN SH3 or drkN SH3-PDZbm. The assignments of each band are shown to the right.
A mechanistic understanding of HtrA2 function as a stress-protective protease, necessary to establish a therapeutic strategy targeting HtrA2 (5, 20), requires an in-depth study of HtrA2–protein substrate interactions. Molecular biology and proteomics studies have identified an array of physiological substrates of HtrA2, which include regulators of the apoptotic and mitotic pathways (9, 21–26), as well as proteins that can form toxic aggregates, such as amyloid-β and α-synuclein (27–29). However, little is known about the kinetics, thermodynamics, or structural dynamics of the interaction of HtrA2 with protein partners, presumably due to the disordered and/or heterogeneous nature of these clients, which hampers high-resolution structural analyses. Most of the studies so far have been limited, therefore, to biochemical characterizations of the interactions of HtrA2 with short peptide ligands. An early study by Martins et al. (30) determined the optimal sequence for cleavage from a peptide library and also showed that a short hydrophobic peptide that binds to the PDZ domain of HtrA2 (GQYYFV, hereafter referred to as a PDZ-binding motif) greatly enhances the catalytic activity toward substrate peptides. While this study established that at least two cooperative interactions involving protein substrates are expected to be in play (one with the PDZ domain and a second with the protease domain of an HtrA2 protomer), little insight into the binding of protein clients and concomitant enzyme activation was forthcoming, since structures of neither an active nor a substrate-bound state were available.
In a previous set of studies, using solution-state NMR spectroscopy focusing on side-chain methyl groups (31, 32), we elucidated the activation mechanism of HtrA2 by using two different short peptides (30), one that contains the PDZ-binding motif (DDGQYYFV; in what follows, referred to as PDZ-peptide) and a second sequence with an efficiently cleaved site (IRRVSYSF; hereafter referred to as substrate peptide), as illustrated in Fig. 1A. We demonstrated that, in the absence of these peptides, HtrA2 adopts a closed conformation, which is consistent with the known crystal structure (referred to in what follows as the C state). The binding of PDZ-peptide to trimeric HtrA2 occurs in a stepwise manner with positive cooperativity, leading to the formation of an open conformation, whereby the PDZ and protease domains are dissociated from each other (Fig. 1A). Our NMR analyses and peptidase assays indicated that this PDZ-peptide–bound open conformation is not catalytically competent (open inactive [OI] state). Subsequent binding of the substrate peptide to the exposed catalytic center of the OI state leads to a catalytically active conformation, i.e., the open active (OA) state, which involves cooperative structural transitions of the protease domains, revealing an extensive interprotomer network that regulates HtrA2 function (32).
These studies, and others preceding them (30, 33–35), focused on small peptides. As many known clients of HtrA2 are intact proteins (9, 21–26), it is of considerable interest to examine a protein substrate and to establish whether the same HtrA2 conformations identified using small peptide substrates are at play when processing protein molecules that can assume folded conformations. Further, what are the structures of protein clients when bound to HtrA2, and how does HtrA2 discriminate between substrate and nonsubstrate proteins? Solution NMR spectroscopy is well-suited to these types of investigations, as it offers the ability to analyze dynamic enzyme–substrate interactions at the atomic level. In contrast, the potentially disordered nature of substrate clients and the structural heterogeneity of the resulting complexes challenges similar studies by many other techniques.
Here, we present an NMR-based, thermodynamic, kinetic, and structural analysis of the interaction between HtrA2 and a metastable model protein substrate, the N-terminal Src homology 3 domain from downstream receptor kinase (drkN SH3), to which a PDZ-binding motif has been added to its C terminus, as is present in physiological substrates, to promote docking to HtrA2. We show that HtrA2 binds to this substrate protein via a two-pronged interaction involving 1) the C-terminal PDZ-binding motif of drkN SH3 and the PDZ domain of HtrA2; and 2) a hydrophobic region in the middle of the drkN SH3 molecule and the catalytic center in the protease domain. Based on proteolytic assays of HtrA2 using an oligomeric drkN SH3 substrate, we demonstrate that HtrA2 cleaves oligomeric clients more efficiently than their monomeric counterparts, suggesting that binding avidity plays a significant role in regulating substrate cleavage. Our NMR study establishes that HtrA2 preferentially binds to a transiently sampled unfolded state of drkN SH3, rather than the folded conformation, via a conformational selection mechanism. More generally, it provides insight into how HtrA2 recognizes and processes protein clients and adds to our understanding of the mechanism by which HtrA2 functions as a stress-protective protease.
Results
HtrA2 Cleaves drkN SH3 Possessing a C-Terminal PDZ-Binding Motif.
As described in the introduction, studies of HtrA proteins have largely focused on peptide mimics of protein substrates. In order to provide insight into interactions involving proteins that can assume fully folded structures, we have used drkN SH3 (residues 1 to 59) as a potential substrate of HtrA2. Unlike many other SH3 domains that form stably folded β-barrel structures, drkN SH3 exists in an equilibrium between folded and unfolded states at near-neutral pH and ambient temperature (folded:unfolded ratio of ∼2:1 at pH 6 and 20 °C) (Fig. 1B) (36, 37). Because of this metastability, drkN SH3 has been utilized as a model system to study aspects of protein folding or to investigate chaperone–client interactions (38–41).
We first performed a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE)-based proteolysis assay monitoring the cleavage of drkN SH3 by wild-type (WT) HtrA2. To compare the proteolytic activity between the closed and open forms of HtrA2, we measured the activity in the presence and absence of 1 mM PDZ-peptide (DDGQYYFV) that binds to the PDZ domain of HtrA2, inducing an open conformation that is required for activity (19, 31). In addition, we prepared a fusion protein in which the PDZ-binding motif (bm), GQYYFV, is attached to the C terminus of drkN SH3 (referred to as drkN SH3-PDZbm) (Fig. 1C). Since it has been proposed that HtrA2 recognizes the C-terminal tails of substrates through their PDZ domains, drkN SH3-PDZbm is expected to serve as a mimic of a physiological substrate. Cleavage was not observed, neither in the presence nor absence of PDZ-peptide, upon incubating WT HtrA2 (10 μM as a monomer) with 5 equivalents of drkN SH3 at 37 °C. In contrast, efficient cleavage of drkN SH3-PDZbm was observed, forming ∼4-kDa fragments (Fig. 1D). Liquid chromatography–mass spectrometry (LC-MS) analyses of the cleaved products were performed, identifying cleavage sites at I27–L28, L28–N29, and M30–E31, consistent with cleaved fragments of ∼4 kDa, as observed via SDS-PAGE (SI Appendix, Table S1).
These results establish that drkN SH3 requires a C-terminal PDZ-binding motif for efficient cleavage by HtrA2, suggesting a synergistic binding process involving contacts between 1) the PDZ-binding motif and the PDZ domain; and 2) the substrate cleavage site and the active site on the enzyme. To test whether PDZ-binding motifs are required for the efficient processing of other protein substrates, we performed the same proteolysis assays using the DNA binding domain of the α-helical human telomeric repeat binding factor 1 (hTRF1) and a triple mutant of the β-sheet containing Src homology 3 domain from Gallus gallus Fyn tyrosine kinase (A39V/N53P/V55L Fyn SH3), both of which showed marginal folding stability and have been widely used to analyze chaperone–client interactions (42–49) (SI Appendix, Fig. S1). Similarly, as observed for drkN SH3, these proteins were efficiently cleaved only when the PDZ-binding motif was attached to the C terminus.
Stepwise Binding of drkN SH3-PDZbm Induces the Formation of the Catalytically Active Conformation of HtrA2.
Having established that drkN SH3-PDZbm serves as a model substrate for HtrA2, we next used NMR to analyze the structural changes of HtrA2 that accompany formation of a drkN SH3-PDZbm–protease complex. As the molecular mass of trimeric HtrA2 is 105 kDa, methyl transverse relaxation optimized spectroscopy (methyl-TROSY) was employed, which facilitates the recording of high-resolution and high-sensitivity NMR spectra of protein complexes with molecular masses of hundreds of kilodaltons (50, 51). In the approach used here, samples were prepared with 13CH3 labeling at Ileδ1, Leuδ1 (proR), Valγ1 (proR), and Metε positions in an otherwise deuterated background (referred to as U-2H, proR ILVM-13CH3 labeling in what follows) (52–54) and 13C-1H heteronuclear multiple quantum coherence (HMQC) spectra, which preserve slowly relaxing NMR signals, were recorded. In the following NMR analyses, two background mutations were introduced into HtrA2, including S306A, in which the catalytic Ser residue was replaced with Ala to suppress self-cleavage at high protein concentrations, and I441V, which suppresses the formation of a ligand-binding-incompetent hexamer (31).
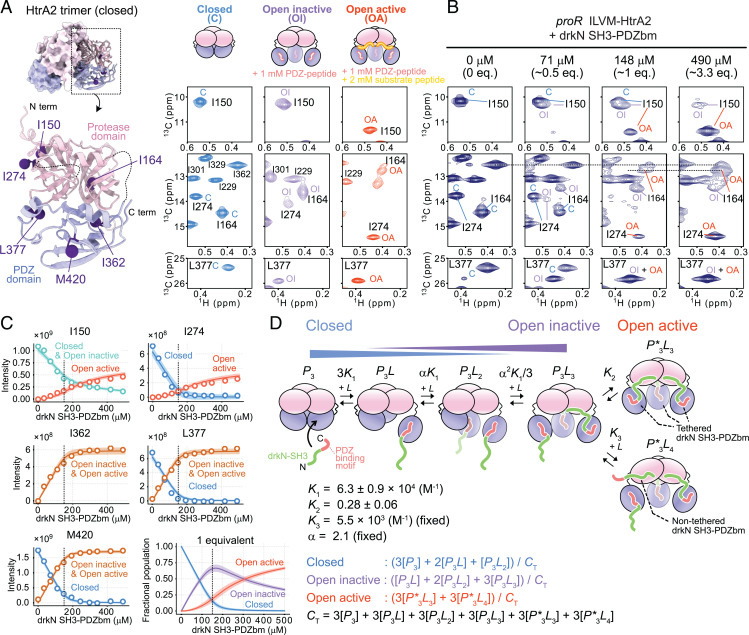
In our previous NMR analyses, we identified several well-resolved methyl probes that are sensitive to different conformational states of HtrA2 (31, 32). Fig. 2A shows spectra with cross-peaks derived from C, OI, and OA states that were obtained with samples containing only HtrA2 (Fig. 2 A, Left), HtrA2 with PDZ-peptide (Fig. 2 A, Center), and HtrA2 with both PDZ and substrate peptides (Fig. 2 A, Right), respectively. The methyl-probes from the PDZ domain (I362, L377, and M420) inform on the binding of PDZ-peptide to generate the OI state, while I150 at the interprotomer interface of each of the three equivalent protease domains is sensitive to the formation of the OA state. Some of the probes from the protease domain (I164 and I274) are sensitive to the presence of both OI and OA states. The assignment of these signals to specific states was confirmed by magnetization-exchange experiments correlating chemical shifts from different conformers that exchange slowly on the NMR chemical-shift timescale (SI Appendix, Fig. S2 B and C). It should be noted that distinct chemical shifts were often observed for methyl probes in each state, consistent with their slow interconversion that is accompanied by significant structural changes. We next prepared a U-2H, proR ILVM-13CH3–labeled HtrA2 sample and recorded 13C-1H HMQC spectra in the presence of various concentrations of U-2H,15N drkN SH3-PDZbm (Fig. 2B and SI Appendix, Fig. S2A). Upon addition of drkN SH3-PDZbm, we observed the successive transition from C to OI states up to an approximate 1:1 drkN SH3-PDZbm:HtrA2 monomer concentration, as well as a transition from OI to OA states when an excess of drkN SH3-PDZbm was added (Fig. 2B). These results establish that binding of drkN SH3-PDZbm to HtrA2 proceeds via a two-pronged mechanism, as previously characterized using short peptides (31, 32), with the first prong involving the C-terminal PDZ-binding motif–PDZ-domain interaction, forming a tethered substrate and responsible for the C to OI transition at the level of an individual HtrA2 protomer, and the second prong resulting from binding of drkN SH3-PDZbm to the protease active site, inducing the OI to OA conformational switch.
Fig. 2.
Binding model of drkN SH3-PDZbm to HtrA2. (A, Left) Close-up view of HtrA2 monomer structure showing methyl probes used in the titration analyses as purple spheres (PDB ID code 1LCY). (A, Right) The 13C-1H HMQC correlation maps of the methyl signals that show distinct chemical shift differences between the C (blue), OI (purple), and OA (red) states. The cartoon representations of each state are shown on top of the spectra. The spectra of OI and OA states were recorded in the presence of 1 mM PDZ-peptide (OI) or both 1 mM PDZ-peptide and 2 mM substrate peptide (OA) (40 °C and 23.5 Tesla). (B) The 13C-1H HMQC correlation maps of 150 μM (monomer concentration) U-2H, proR ILVM-13CH3 S306A/I441V HtrA2 with varying concentrations of U-2H,15N drkN SH3-PDZbm (40 °C and 18.8 Tesla). (C) Plots of the intensities of methyl correlations as a function of the concentration of drkN SH3-PDZbm. The solid lines are the fitted curves, and the 95% CI of each fitted curve is contained within the thick line estimated from Monte Carlo error analyses (84). The fractional populations of the C, OI, and OA states calculated from the fitted parameters are shown in C, Right Bottom. In each plot, the concentration of one equivalent added ligand is indicated as a dotted vertical line. (D) Thermodynamic binding model along with cartoon representations of each state used in fits of the titration data. The best-fit values and the estimated errors of the parameters are shown below the model (see Stepwise Binding of drkN SH3-PDZbm Induces the Formation of the Catalytically Active Conformation of HtrA2 for details). Equations for fractional populations of the C, OI, and OA states and the total protein concentration are also listed. Eq., equivalent; N term, N terminus; C term, C terminus.
Prior to a quantitative analysis of the titration profile (Fig. 2 B and C) by model fitting, a number of important points can be made through a simple inspection of the data. First, the population of the C state decreases rapidly with increasing ligand and is largely depleted when an equimolar amount of ligand is added, while the OA state increases relatively slowly (I150 and I274 of Fig. 2C). This suggests that the OA state is only substantially formed after the PDZ domains are almost fully occupied by the C-terminal PDZ-binding motifs of drkN SH3-PDZbm, consistent with our previous observations when separate PDZ- and substrate-peptides were used (31, 32). Second, in the presence of an equimolar amount of drkN SH3-PDZbm, where the PDZ domains of HtrA2 were almost fully occupied, signals from both OI and OA states were observed for I150 and I164. Thus, despite the high local concentration of substrate available, not all of the active sites on HtrA2 are bound (second prong of the interaction, triggering formation of OA), consistent with a relatively weak contact involving the substrate and protease domains. Finally, the signal intensities reporting on the OA state continue to increase, even after the addition of more than one equivalent of drkN SH3-PDZbm, suggesting that additional substrate molecules can access HtrA2’s active sites without first binding to the PDZ domain.
We have fit a thermodynamic model to the NMR titration profiles, taking into account the observations noted above (Fig. 2 C and D). The model, motivated by previous peptide studies (31, 32), includes free (P3), partially ligated (P3L and P3L2, where one or two ligand molecules are tethered to a trimer, respectively), and fully bound (P3L3) trimeric states describing the stepwise binding of the C-terminal PDZ-binding motif of drkN SH3-PDZbm to the PDZ domains of HtrA2. As highlighted in Fig. 2D, the protomers in these states range from all closed (P3) to all open (P3L3), but the HtrA2 molecules are inactive. In addition, two distinct catalytically active states, P*3L3 and P*3L4 (asterisk denotes the OA state), are included, with substrates bound at the protease active sites. These substrates can either be tethered to HtrA2 via their PDZ domains (P*3L3) or one of the four untethered (three of the four ligands in P*3L4 are tethered), accounting for the fact that the population of the OA state increases at substrate concentrations beyond where the PDZ domains of the enzyme are saturated. Following our previous analyses of the binding of PDZ-peptides to HtrA2 (31), we assume that the unbound and bound protomers in each P3Lj (j ∈ 0–3) conformer contribute to signals from the C and OI states, respectively, as (state C) and (state OI), and that successive binding events occur with positive cooperativity (α factor in scheme of Fig. 2D), as observed previously in peptide titrations (31, 32). Due to the inherent complexity of the binding scheme, we chose to fix some of the equilibrium constants. First, the cooperativity factor α was set to 2.1, a value estimated from cooperativity values measured from a previous titration using PDZ-peptides (31). Second, the association constant for the P3L3 + L → P*3L4 reaction (K3), corresponding to the binding of a nontethered drkN SH3-PDZbm substrate to the protease domain of HtrA2, was estimated from a second titration experiment and then fixed in fits of the model of Fig. 2D. In this second experiment, described in detail in SI Appendix, SI Materials and Methods, PDZ-peptides were covalently attached to PDZ domains of HtrA2 via cysteine-based chemistry to create the OI state, as described (32), and the modified HtrA2 was subsequently titrated with drkN SH3-PDZbm. A value of K3 = 5,470 M−1 was obtained in this manner (SI Appendix, Fig. S3). The NMR titration profiles (Fig. 2C) could be well explained by the branched model of Fig. 2D, with the equilibrium constants for each binding process obtained as shown. Notably, binding of substrate to the PDZ domains (prong 1), forming tethered client molecules, occurs with higher affinity than the subsequent association of client with the protease domains (prong 2). Alternative models lacking the branched path from the P3L3 state (i.e., only one of P*3L3 and P*3L4 present) were poorly fit to the data (SI Appendix, Fig. S4).
It is important to stress that, although the population of each state can be established from this titration, information is not available as to the number of protease-bound substrates in each conformer. The present titration is focused exclusively on HtrA2, and the methyl probes are localized to the intertrimer interface that is sensitive to transitions between OI and OA conformers (32), and not to the number of protease-bound ligands per se. Thus, from these data alone, it is not possible to ascertain whether P3L3, for example, has zero, one, or two substrates engaged with the three active sites of the trimer, although, by definition, all three PDZ domains are occupied. It is also not possible to distinguish a scenario in which binding of a single substrate to a protease-domain active site triggers formation of OA from one where all three substrates are required to bind for the OI to OA transition to occur. Nevertheless, in combination with a related NMR analysis, in which signals from drkN SH3-PDZbm were observed in a relaxation-based experiment, as described in drkN SH3-PDZbm Binds to HtrA2 via a Conformational Selection Pathway, it is possible to estimate that all three protease domain–substrate interactions are formed in state OA (P*3L3 and P*3L4) and that, likely, two such interactions also exist in the OI state P3L3 (Discussion). This situation is indicated in Fig. 2D. Thus, according to our model, the transition from the OI to the OA HtrA2 conformation occurs only when all active sites are bound, with the unimolecular equilibrium between OI and OA states favoring OI (K2 = 0.33 ± 0.05). Details of the model fitting are described in SI Appendix, SI Materials and Methods.
Synergistic Binding of Substrates Enhances the Proteolytic Activity of HtrA2.
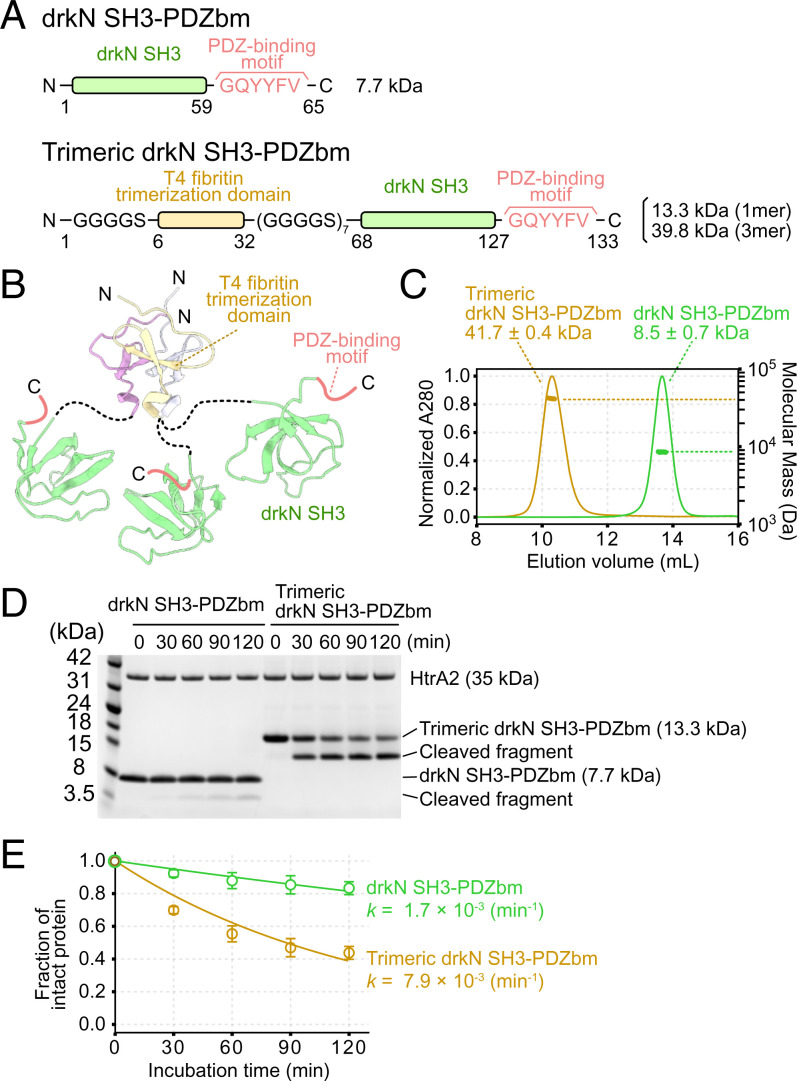
The thermodynamic model described in the previous section assumes that formation of the OA state occurs only after all PDZ domains in a trimer are bound with substrate, consistent with results from our previously described protomer-mixing experiments (32), where preventing substrate binding to a single protomer was sufficient to eliminate activity. In this case, it might be predicted, therefore, that substrates able to form a tripartite interaction involving tethering to all three PDZ domains at once would be more efficiently cleaved by HtrA2. In order to test this, we performed a proteolytic activity assay against an engineered oligomeric substrate that is expected to exhibit strong binding avidity.
We designed a trimeric version of drkN SH3-PDZbm and compared proteolysis rates relative to a drkN SH3-PDZbm monomer (Fig. 3 A and B). To stabilize the trimeric structure, a trimerization domain from the T4 bacteriophage fibritin protein was fused to the N terminus of drkN SH3-PDZbm (55–58), with a flexible linker (seven repeats of GGGGS) inserted between the trimerization domain and drkN SH3-PDZbm to ensure that all three drkN SH3-PDZbm copies had enough flexibility to simultaneously interact with trimeric HtrA2. The formation of the client trimer was confirmed by using multiangle light scattering coupled with size-exclusion chromatography (SEC-MALS), where the molecular mass of the fusion protein (41.7 ± 0.4 kDa) was about threefold larger than that of the monomer (13.3 kDa) (Fig. 3C). We then performed proteolysis assays using the monomeric and trimeric drkN SH3-PDZbm substrates by incubating the same monomeric concentrations of client with a fixed amount of HtrA2. The cleavage rate of trimeric drkN SH3-PDZbm was ∼4.6-fold faster than for the drkN SH3-PDZbm monomer (Fig. 3 D and E), consistent with a stimulation of HtrA2 proteolytic activity through binding avidity mediated by the tripartite substrate–enzyme interaction.
Fig. 3.
Trimeric drkN SH3-PDZbm is cleaved more readily than the monomeric form. (A) Schematic representations of drkN SH3-PDZbm (Upper) and trimeric drkN SH3-PDZbm (Lower) constructs used in this study. (B) Cartoon model of trimeric drkN SH3-PDZbm. NMR-based structures of the trimerization domain (PDB ID code 1RFO) and drkN SH3 (PDB ID code 2A36) are shown. (C) SEC-MALS profiles of drkN SH3-PDZbm (green) and trimeric drkN SH3-PDZbm (gold). Curves show absorbance at 280 nm (left y axis) and the dots indicate molecular masses (right y axis, log scale). (D) Gel-based proteolytic activity assays monitoring the degradation of drkN SH3-PDZbm (Left) or trimeric drkN SH3-PDZbm (Right) by WT HtrA2. The time course of the reaction was monitored by SDS-PAGE. (E) Plots of the fraction of intact protein, as measured by intensities on an SDS-PAGE gel, as a function of incubation time. The intensities were normalized to those obtained at 0 min, and the profiles were fit to a single exponential decay function. The apparent cleavage rates (k) are shown. Error bars correspond to one SD based on triplicate measurements.
drkN SH3-PDZbm Is Unfolded When Bound to HtrA2 via Both Prongs.
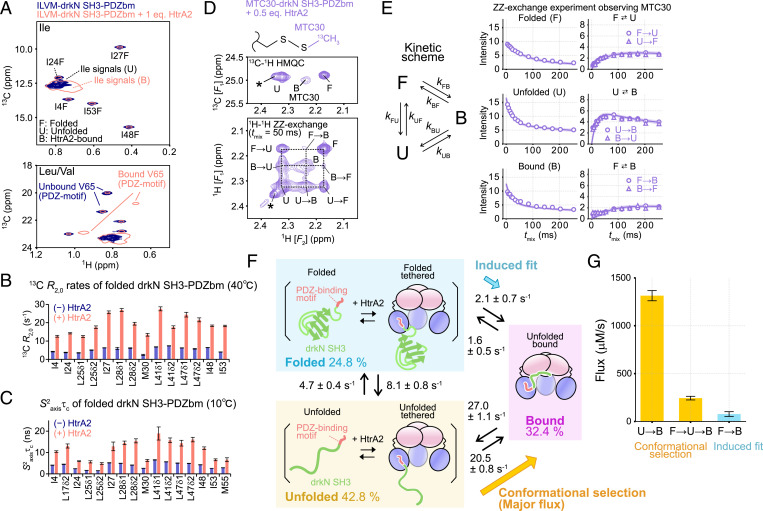
To further explore the binding mechanism of drkN SH3-PDZbm to HtrA2, we sought to characterize the structural dynamics of the substrate in the HtrA2-bound state. To this end, we prepared a U-2H, ILVM-13CH3 drkN SH3-PDZbm [only one of the pair of isopropyl methyls of Leu and Val was 13CH3-labeled (52, 53)] and recorded 13C-1H HMQC spectra with and without U-2H, S306A/I441V HtrA2. As drkN SH3-PDZbm is a metastable protein, we observed two separate sets of signals in the absence of HtrA2, one for each of the folded (F) and unfolded (U) conformations. The relative populations of these two states were estimated to be ∼35% and ∼65%, respectively, at 40 °C. In the presence of HtrA2, we observed a new set of signals whose chemical shifts were similar, but distinct, particularly in the 1H dimension, from the unfolded state, which we refer to as the HtrA2-bound (B) state in what follows (Fig. 4A). The binding of drkN SH3-PDZbm could also be directly monitored from the methyl signals of V65, a residue at the C terminus of the PDZ-binding motif of drkN SH3-PDZbm, that showed large chemical-shift changes in the presence of HtrA2, presumably reflecting the direct association of substrate with enzyme. Although most of the B- and U-state signals were severely overlapped, assignments could be established through 13C[t1]-tmix-1H[t2] ZZ-exchange experiments (59), with the methyl chemical shifts of the B state unambiguously read out in many cases from exchange cross-peaks connecting B signals with the well-resolved F-state signals (SI Appendix, Fig. S5 A and B). The obtained B-state chemical shifts are consistent with random coil values (60), so that the B-state structure of drkN SH3-PDZbm (two binding prongs are engaged) is largely unfolded (SI Appendix, Fig. S5C).
Fig. 4.
Structural characterization of drkN SH3-PDZbm bound to HtrA2. (A) The 13C-1H HMQC correlation maps of Ile (Upper) and Leu/Val (Lower) regions of 150 μM U-2H, ILVM drkN SH3-PDZbm with (pink, single contour) and without (navy, multiple contours) 150 μM (monomer concentration) U-2H, S306A/I441V HtrA2. The folded, unfolded, and HtrA2-bound states are denoted as F, U, and B, respectively. (B) Plots of 13C single-quantum transverse relaxation rates using a 2-kHz Carr–Purcell–Meiboom–Gill field, with (pink) and without (navy) U-2H, S306A/I441V HtrA2 (40 °C and 23.5 Tesla). (C) Plots of the order parameter squared multiplied by the rotational correlation time (S2axisτc) of methyl threefold symmetry axes (folded state of drkN SH3-PDZbm) with (pink) and without (navy) U-2H, S306A/I441V HtrA2 (10 °C and 23.5 Tesla). (D) The 13C-1H HMQC (Upper) and 13C-edited 1H[t1]-tmix-1H[t2] ZZ-exchange [Lower (68)] datasets recorded on 150 μM U-2H, ILV-13CH3, M30C 13C-MMTS-labeled drkN SH3-PDZbm in the presence of 75 μM (monomer concentration) U-2H S306A/I441V HtrA2 (50-ms mixing time, 23.5 Tesla, and 40 °C). The asterisk denotes a signal from an impurity. The chemical structure of the MTC group is shown above the spectra. (E, Left) Triangular kinetic scheme showing the interconversion between F, U, and B states of drkN SH3-PDZbm. (E, Right) ZZ-exchange profiles of diagonal and exchange cross-peaks as a function of mixing time. The solid lines are the fitted curves, and the 95% CI of each fitted curve is contained within the thick line estimated from Monte Carlo error analyses. The two symmetric cross-peaks (e.g., U→F and F→U) are plotted separately as circles and triangles. (F) Cartoon representations of the kinetic scheme describing the binding of drkN SH3-PDZbm to HtrA2. The best-fit values and the estimated errors of the rate constants of each transition and the associated equilibrium populations of the F (blue background), U (yellow background), and B (pink background) states are shown (see drkN SH3-PDZbm Binds to HtrA2 via a Conformational Selection Mechanism for details). The flux directions corresponding to induced fit (blue) and conformational selection (orange) are shown. (G) Flux values from U to B (Left), F to U to B (Center), and F to B (Right). Errors were calculated from the SD of parameter distributions obtained from Monte Carlo error analyses of the ZZ-exchange profiles. Eq., equivalent.
Our titration results (Stepwise Binding of drkN SH3-PDZbm Induces the Formation of the Catalytically Active Conformation of HtrA2) suggested a two-pronged client–enzyme interaction, with contacts involving protease domains formed less frequently than those associated with PDZ moieties. We therefore wondered whether an intermediate PDZ-tethered, protease-domain unbound complex (i.e., only one prong formed) could be observed directly, providing support for our binding model. To test for this state, we recorded a pair of experiments that are sensitive reporters of methyl-group dynamics and focused on the F-state signals of drkN SH3-PDZbm (61, 62), with and without one equivalent of HtrA2 (Fig. 4 B and C and SI Appendix, Fig. S6). The experiments included 1) measurement of apparent transverse relaxation rates of methyl 13C nuclei, with potential exchange contributions suppressed by applying a train of closely spaced chemical-shift refocusing pulses (2-kHz Carr–Purcell–Meiboom–Gill experiment) (61, 63); and 2) quantifying S2axisτc values, where S2axis and τc are the order parameter squared of the methyl threefold axis and the effective rotational correlation time of the methyl, respectively (62). Notably, the apparent 13C transverse relaxation rates at 40 °C (Fig. 4B) and S2axisτc values at 10 °C (Fig. 4C) were larger in the presence of HtrA2, indicating that the overall tumbling of the F state of drkN SH3-PDZbm was significantly reduced with added enzyme. This reduction was similarly observed at low temperature (10 °C; SI Appendix, Fig. S6A), where the exchange between each conformer is expected to be slow, so that the increased relaxation rates do not arise from rapid relaxation in the B state coupled with chemical exchange (i.e., B to F). Also, the effect is not due to increased viscosity associated with the addition of HtrA2, as the dynamics of drkN SH3 did not change when the native drkN SH3 domain without the PDZ-binding motif, which does not form a stable complex, was mixed with HtrA2 (SI Appendix, Fig. S6 B and C). These results paint a picture of a binding intermediate, whereby the folded conformation of drkN SH3-PDZbm binds to HtrA2 through its PDZ domain (tethered), but does not unfold, so that one of the two prongs of the binding interaction is formed. In this model, drkN SH3-PDZbm exchanges between F, U, and B structural ensembles, where F and U are not solely unbound states, but also include contributions from C-terminal–tethered conformers that are bound to PDZ domains. Thus, there is a distinction in terms of the SH3 domain client structure between a bound (and unfolded) conformation, where the core of drkN SH3 interacts with the HtrA2 protease domain, and a tethered state, in which only PDZ contacts are made.
drkN SH3-PDZbm Binds to HtrA2 via a Conformational Selection Mechanism.
Although the B state of drkN SH3-PDZbm is unfolded, it remains an open question as to whether HtrA2 binds first to the F state of drkN SH3-PDZbm to unfold the structure or preferentially to an already-existing U state. In the context of ligand–protein interactions, these two limiting cases are referred to as induced fit (IF) and conformational selection (CS), respectively (64, 65). Discrimination between these two possibilities requires measurement of fluxes between states, as described (41, 66). With this in mind, the interconversion between the F, U, and B states of drkN SH3-PDZbm, which occurs in the slow-exchange regime on the NMR chemical-shift timescale, was quantified by using ZZ-exchange experiments.
In order to minimize signal overlap, we made use of residue-specific 13C-methanethiosulfonate (MMTS) labeling of an engineered Cys side chain in drkN SH3-PDZbm (67). As drkN SH3-PDZbm does not have intrinsic Cys residues, Met30 was mutated to Cys, followed by labeling with 13C-MMTS to form an S-methylthiocysteine (MTC) group that serves as a site-specific NMR probe (Fig. 4 D, Top). The advantage of using a single MTC probe is that cross-peaks fall in an “empty” region of the 13C-1H spectrum, so that overlap with signals from other residues is not an issue. Three distinct peaks from the MTC group, corresponding to F, U, and B states of U-2H, ILV-13CH3, and M30C 13C-MMTS–labeled drkN SH3-PDZbm, were observed in the presence of 0.5 equivalent of U-2H S306A/I441V HtrA2, which could be used to probe the thermodynamics and kinetics of the interconversion between these three states (Fig. 4D).
The 13C-edited ZZ-exchange (1H[t1]-tmix-1H[t2]) experiments (68) were recorded as a function of mixing time (Fig. 4 D and E). In the spectrum recorded with tmix = 50 ms, the exchange cross-peaks between the F and B states were much weaker than those between U and B, unequivocally demonstrating that the U state of drkN SH3-PDZbm predominantly interacts with HtrA2 to form the B state. The ZZ-exchange profiles were fit to a triangular kinetic scheme in which F, U, and B states interconvert with each other (Fig. 4E). The obtained six rate constants, as well as the populations of each state, are indicated in Fig. 4F, where the F and U states are depicted to include both unbound and C-terminally tethered states, as described in drkN SH3-PDZbm Is Unfolded when Bound to HtrA2 via Both Prongs. In order to determine which of the IF and CS pathways is dominant, it is important to compare the fluxes between the two pathways (41, 66). With this in mind, fluxes were calculated (FF→B = kFBpF[drkN SH3-PDZbm] for the IF pathway and FU→B= kUBpU[drkN SH3-PDZbm] for the CS pathway, where pF and pU are the equilibrium populations of the F and U states) with FU→B ∼17-fold larger than FF→B. We also calculated the flux for the three-state pathway {FF→U→B = (FF→U−1 + FU→B−1)−1, where FF→U = kFUpF[drkN SH3-PDZbm]}, which was also larger than FF→B (Fig. 4G). These results establish a dominant CS pathway for the binding of drkN SH3-PDZbm to HtrA2.
The Hydrophobic Region of drkN SH3-PDZbm Interacts with the Catalytic Center of HtrA2.
Our studies of the drkN SH3-PDZbm/HtrA2 binding mechanism (Fig. 2D) and the structural dynamics of the enzyme–substrate complex (Fig. 4) made use of side-chain methyl probes, as the size of the complex prohibited recording high-quality backbone amide data. However, by working with drkN SH3 without the C-terminal PDZ-binding motif, which has a much lower affinity for HtrA2 than drkN SH3-PDZbm, and exploiting its rapid on/off binding kinetics, it is possible to “read out” structural properties of the SH3 bound state in high-quality spectra of the free form of the molecule and to identify interaction sites by inspecting residue-specific differential broadening or intensity reductions that inform on the transient binding to HtrA2 (69). Fig. 5A illustrates schematically the exchanging system that was used, in which the OI HtrA2 state was created through the addition of saturating amounts of PDZ-peptide.
Fig. 5.
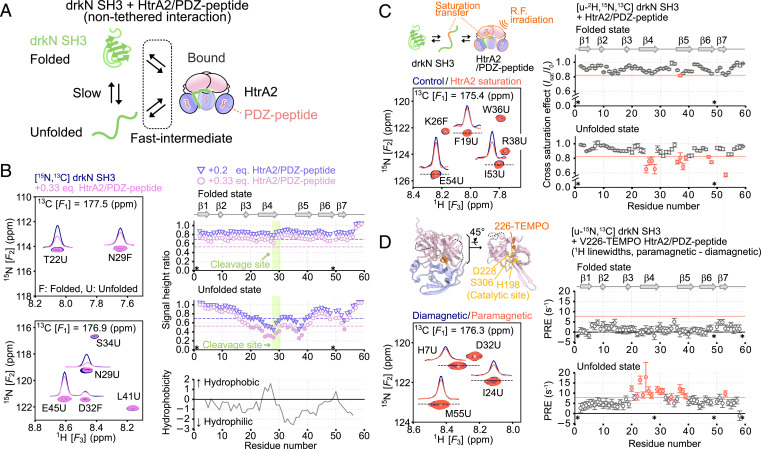
Mapping of HtrA2 interaction sites on drkN SH3. (A) Schematic cartoon describing the nontethered interaction between drkN SH3 without the C-terminal PDZ-binding motif and the open, inactive state of HtrA2 bound to PDZ-peptide. (B, Left) The 15N-1H planes from 3D HNCO spectra of U-15N,13C drkN SH3 with (pink) or without (navy) unlabeled S306A/I441V HtrA2 bound to PDZ-peptide. F and U denote the folded and unfolded states of drkN SH3. (B, Right Top and Middle) Plots of signal height ratios of drkN SH3 correlations in the presence of 0.2 (triangle, purple) or 0.33 (pink circle) equivalents of HtrA2. The signal heights were normalized against those in the absence of HtrA2. The secondary structure elements in the folded state are shown above the plots. The average − SD values are indicated as dotted horizontal lines (average over all residues, both from F and U states). Residues with ratios below the average − SD are shown as filled symbols. (B, Right Bottom). Plot of hydrophobicity calculated based on the Kyte–Doolittle scale using an averaging window size of 7 (71). (C, Left Upper) Schematic representation of the TCS experiment. (C, Left Lower) The 15N-1H planes from 3D TROSY-HNCO spectra of U-2H,15N,13C drkN SH3 with (orange) or without (navy) 1H radio-frequency (R.F.) irradiation of S306A/I441V HtrA2 bound to PDZ-peptide. A ratio of drkN SH3:HtrA2/PDZ-peptide of 2:1 (monomer) was used. (C, Right) Plots of the cross-saturation effects for the folded and unfolded states of drkN SH3. The average − SD values are indicated by orange horizontal lines. Residues with a ratio below average − SD are colored orange. (D, Left Upper) A close-up view of the structure of the catalytic center of HtrA2 (PDB ID code 1LCY). The catalytic residues (residues 198, 228, and 306) are shown as yellow sticks, and the beta carbon of V226 where the TEMPO-modification was introduced is shown as an orange sphere. (D, Left Lower) The 15N-1H planes from 3D HNCO spectra of U-15N,13C drkN SH3 with a diamagnetic (navy) or paramagnetic (orange) label on HtrA2 (drkN SH3:HtrA2 monomer concentrations in a ratio of 2:1). (D, Right) Plots of PRE rates of the folded and unfolded states of drkN SH3. Average + SD values are indicated as orange horizontal lines. Residues with PRE rates larger than average + SD are colored orange. All of the measurements were performed at 10 °C and 14.0 Tesla. Asterisks indicate the Pro residue (P49) or residues that were not analyzed due to signal overlap or broadening. Eq., equivalent.
Amide 15N-1H spectra of drkN SH3 were recorded in the presence and absence of PDZ-peptide–bound HtrA2 (10 °C to minimize hydrogen exchange) (70). Although separate bound-state signals were not observed, owing to the fast–intermediate exchange regime, individual correlations for residues in the drkN SH3 F and U states, which interconvert slowly, were present. This hampers the residue-specific analysis of two-dimensional spectra due to cross-peak overlap. We, therefore, recorded 3D HNCO spectra of U-13C,15N, drkN SH3, to which varying amounts (0, 0.2, or 0.33 equivalents) of unlabeled S306A/I441V HtrA2 in complex with PDZ-peptide was added. Fig. 5 B, Left shows site-specific intensity reductions in many of the signals relative to the isolated SH3 domain, and, notably, these reductions were more prominently observed in the U state (Fig. 5 B, Left), consistent with the preferential binding of HtrA2 to the U state as discussed in drkN SH3-PDZbm Binds to HtrA2 via a Conformational Selection Mechanism. Significant intensity reductions in the U state were observed primarily in the β4–β5 region, and to correlations from residues in β6–β7 to a lesser extent, where we annotate the U state according to secondary structure formed in the folded conformation (Fig. 5 B, Right Top and Middle). Not surprisingly, one of these regions includes the cleavage sites (residues I27–L28, L28–N29, and M30–E31 of strand β4), identified in our LC-MS analysis, so that the interactions observed in the HNCO spectra very likely correspond to those involving the protease domain that are required for proteolysis. The cleaved region is predicted to lie in a hydrophobic stretch based on the Kyte–Doolittle scale (71), suggesting that HtrA2 may recognize hydrophobic regions of unfolded substrates for cleavage (Fig. 5 B, Right Bottom).
To further validate that the intensity reductions in spectra of drkN SH3 reflect direct binding to HtrA2, we carried out a transferred cross-saturation (TCS) experiment (72–74). In the TCS experiment, the intensities of amide correlations from a uniformly deuterated drkN SH3 sample are quantified in datasets where the aliphatic protons of HtrA2 are either saturated by an applied radio-frequency field or unperturbed. As saturation transfer from HtrA2 to drkN SH3 occurs efficiently for amide protons on drkN SH3 that are proximal to the binding interface, a binding site can be ascertained by monitoring site-specific intensity reductions of the drkN SH3 amide signals in the unbound state (Fig. 5 C, Left Upper). The 3D TROSY-HNCO spectra of U-2H,13C,15N drkN SH3 were acquired with and without saturation of unlabeled S306A/I441V HtrA2 (drkN SH3:HtrA2 = 2:1, monomer to monomer; Fig. 5 C, Left Lower). Significant TCS effects were observed in β4–β5 and β6–β7 in the unfolded state (Fig. 5 C, Right), consistent with the regions identified from intensity reductions (Fig. 5B) and providing further evidence that these are the interaction sites on drkN SH3. We repeated the experiment on a sample containing only drkN SH3 and confirmed that the observed intensity reductions are not from intramolecular spin diffusion caused by direct irradiation of residual protons of the U-2H,13C,15N drkN SH3 sample (SI Appendix, Fig. S7).
As a final validation of the interaction site on drkN SH3, we measured intermolecular paramagnetic relaxation enhancements (PRE) (75), with a spin label attached to HtrA2 V226C that is located close to the catalytic center residue (D228) (Fig. 5 D, Left Upper). Intermolecular PREs were quantified from increases in 1H linewidths in HNCO spectra of U-13C,15N drkN SH3, to which V226C 2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPO)-labeled S306A/I441V HtrA2 was added (drkN SH3:HtrA2 = 2:1, monomer to monomer; Fig. 5 D, Left Lower and Right). Significant PREs were again observed in β4–β5 and β7 of drkN SH3 in the unfolded state. Taken together, our data demonstrate that the hydrophobic stretch of residues, mainly comprising β4–β5, as well as β6–β7, are the interaction sites on drkN SH3 and that the catalytic centers of the HtrA2 protease domains are involved in drkN SH3 binding (second prong).
Discussion
HtrA2 is a key stress-protective protease that cleaves misfolded proteins in an ATP-independent manner, with aberrant functions of the enzyme closely linked to pathogenic neurological conditions. Most biochemical and structural studies have involved short peptide fragments, such as 1) PDZ-peptides that bind to the PDZ domains of HtrA2 to form an open, albeit inactive, structure; or 2) substrate mimics that bind to the active sites of HtrA2 when PDZ-peptides have first been added. These studies have elucidated important roles of oligomeric assembly and interprotomer allostery (19, 30–32), but are limiting because HtrA2 substrates are proteins, not short peptides, and many are capable of forming folded structures. Physiological substrates, thus, include PDZ-binding regions, localized to their C termini (5, 30), as well as an additional segment that is bound and cleaved by the protease domains of the enzyme. Understanding the structural dynamics of binding when there are two prongs in the same molecule, as well as the binding mechanism in this case, is therefore of considerable interest.
In this study, we used the metastable protein substrate drkN SH3-PDZbm and studied its interaction with HtrA2 by using a combination of solution-based NMR methods and proteolytic activity assays. We show that a similar activation scheme is in play for the SH3 domain as that proposed on the basis of our previous NMR studies involving PDZ-peptides and substrate peptides (31, 32). The two-pronged drkN SH3-PDZbm/HtrA2 interaction, occurring via conformational selection (Fig. 4), involves three distinct conformational states of HtrA2. The C-terminal PDZ-binding motifs of drkN SH3-PDZbm clients successively bind to the PDZ domains of HtrA2 trimers to induce the structural transition from state C to state OI, followed by the binding of a hydrophobic segment of drkN SH3 containing the sites of hydrolysis to the catalytic center of the enzyme, leading to the OI to OA transition and catalytic activity (Fig. 2). Notably, the second prong of the interaction, involving the hydrophobic region of drkN SH3, is weak. Proteolysis therefore likely only occurs when substrates are tethered to the enzyme via PDZ domains and their local concentration is high, so that all PDZ-binding sites are occupied. In this context, it may well be that anchoring of substrates close to the enzyme’s protease domains allows the active sites to rapidly “search” for cleavable sequences within the tethered substrate. This is consistent with the results of the proteolytic assay of Fig. 1, where HtrA2 efficiently cleaved drkN SH3 only when the PDZ-binding motif was attached to its C terminus. The requirement for linking the two interacting sites on the same substrate molecule is similarly observed for the bacterial HtrA protease DegP (76), suggesting that the mechanism proposed here may be applicable to the whole HtrA family.
Our study highlights the importance of carrying out experiments on each component of a complex, multistep binding reaction to build a reliable model of association (Fig. 2D). For example, in the drkN SH3-PDZbm–HtrA2 titration, only HtrA2 was NMR-active (Fig. 2), providing information about the evolution of the HtrA2 conformational states, from C to OI to OA as a function of added substrate. No insight into the status of the substrate—for example, at what stage it goes from tethered (single-prong interaction involving only the PDZ domain) to bound (double-prong association with PDZ and protease domains)—could be obtained. Our previous studies, involving the use of separate peptides as activators and substrates, allows separation of the individual enzyme conformations (C, OI, and OA) since the OA state can only form when substrate is added (after addition of PDZ-peptide), indicating that hydrolysis does not occur until all PDZ domains on the enzyme are saturated (31, 32). However, it is unclear whether binding of a single drkN SH3 domain to the protease domain (second prong) is sufficient for triggering formation of OA or whether additional substrates must be bound in a similar way to HtrA2 prior to activation. Correspondingly, our relaxation studies focusing on drkN SH3 in the presence of NMR invisible HtrA2 allow the separation of unfolded, folded, and bound substrate conformations (Fig. 4), showing that an intermediate exists in which substrates are tethered to PDZ domains, but not interacting with protease domains, but they do not inform on the conformational status of HtrA2. However, by combining results from these two sets of experiments, it is possible to generate a more nuanced model of substrate binding, as we show in Fig. 2D. In particular, it becomes possible to estimate the number of protease domain–substrate interactions that are formed when the enzyme is saturated with substrate (i.e., HtrA2 in the P3L3, P*3L3, and P*3L4 states). Consider monomer concentrations of 150 μM for drkN SH3-PDZbm and 75 μM for HtrA2, as used in the experiments of Fig. 4 D and E. The molar concentrations of each species depicted in Fig. 2D can be calculated from the parameters extracted from the drkN SH3-PDZbm–HtrA2 titration, along with the concentration of free drkN SH3-PDZbm, to give: [P3] ∼ 0 μM, [P3L] = 0.2 μM, [P3L2] = 2.0 μM, [P3L3] = 13.6 μM, [P*3L3] = 3.9 μM, [P*3L4] = 5.4 μM, and [L] = 72.1 μM. Assuming that only one molecule of drkN SH3-PDZbm interacts with the protease domain of HtrA2 in each of the P*3L3 and P*3L4 states to give rise to the B-state signal of Fig. 4D (i.e., only a single protease subunit must be occupied to form OA), the fractional population of the B state is expected to be ([P*3L3] + [P*3L4])/LT = 0.062 (LT = 150 μM, the total concentration of drkN SH3-PDZbm), which is much smaller than the experimentally observed fractional population of the B state derived from magnetization exchange (pB = 0.324; Fig. 4F). When three molecules of drkN SH3-PDZbm are required to interact with the protease domain to form P*3L3 and P*3L4 (i.e., the active state), the fractional population of the B state is (3[P*3L3] + 3[P*3L4])/LT = 0.184, more similar to the experimental value (Fig. 4F). Interestingly, if it is further assumed that either one or two of the active sites are occupied already in P3L3, in addition to full occupancy in the P*3L3 and P*3L4 states, the fractional population of the B state is calculated to be ([P3L3] + 3[P*3L3] + 3[P*3L4])/LT = 0.275 or (2[P3L3] + 3[P*3L3] + 3[P*3L4])/LT = 0.366, closer still to the observed fractional population of the B state (Fig. 4F). This combined analysis suggests that all three protease domains within the HtrA2 trimer are occupied by drkN SH3-PDZbm in the OA state, so that the formation of the catalytically active OA state is a highly cooperative process that occurs only after all of the catalytic sites within a trimer are engaged. The picture that emerges is consistent with our previous HtrA2 protomer mixing experiments showing that formation of the OA state involves a concerted structural transition of all three protease domains (32), regulating enzyme activity so that cleavage of substrates only occurs when their local concentrations are sufficiently high, so as to saturate the binding sites on HtrA2.
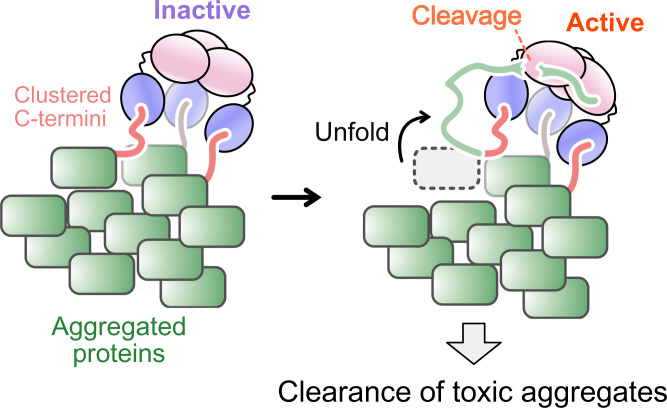
One of the hallmark features of HtrA2 as a stress-protective protease is its ability to cleave aggregated proteins (11, 28, 29). Our binding model provides insight into how this might occur and how HtrA2 would distinguish protein aggregates that must be cleared from isolated proteins that are required for cellular function. HtrA2 is predicted to become catalytically active when all of its PDZ domains are occupied by C termini of substrates (Fig. 2D). Thus, clustering of client C termini in an aggregated state would facilitate association with HtrA2 via a tripartite interaction, easily forming the fully substrate-bound state required for catalytic activity. Monomeric or nonaggregated proteins, on the other hand, would be less efficiently cleaved unless at high concentrations, since the likelihood of forming the OA HtrA2 conformation would be reduced (Fig. 6). The significantly increased hydrolysis rates of trimeric drkN SH3-PDZbm relative to monomeric drkN SH3-PDZbm (Fig. 3) supports this notion.
Fig. 6.
Proposed mechanism for the clearance of toxic aggregates by HtrA2. HtrA2 binds to the clustered C termini of aggregated proteins via a tripartite interaction formed through its three PDZ domains. Hydrophobic regions of unfolded proteins forming the aggregate are recognized by the exposed catalytic center of HtrA2, leading to degradation. In our model (Fig. 2D), the active HtrA2 state is only formed when all three catalytic sites are occupied by substrates.
Thus, the trimeric architecture of HtrA2 functions both as a scaffold to select toxic aggregates, while at the same time suppressing uncontrolled cleavage of nonsubstrate proteins. The trimeric HtrA2 structure also supports an extensive interprotomer network that regulates catalytic activity, while also functioning as the basic unit to form an auto-inhibited hexameric state (31, 32). Therefore, the oligomeric assembly of HtrA2 plays a variety of nuanced roles in selecting substrates and in regulating its catalytic activity. Other families of HtrA proteases are also integral for the removal of toxic aggregates. These include HtrA1, involved in the degradation of toxic tau fibrils (77, 78), which shares a similar domain architecture with HtrA2, suggesting a similar mechanism of function potentially for many HtrA enzymes and explaining the evolutionary conservation of oligomeric structures across these families.
Our spin-relaxation studies of drkN SH3-PDZbm side-chain methyl groups demonstrate a highly dynamic substrate–enzyme complex. When the C terminus of drkN SH3-PDZbm is tethered to the HtrA2 PDZ domain, the core region of drkN SH3 is in equilibrium between folded, unfolded, and unfolded protease-bound states, reflecting the weak and transient second prong of the binding interaction. This explains why structural studies of HtrA–substrate complexes have focused on small peptides, with models of HtrA in complex with protein substrates limited to oligomeric cages where the bound substrate is poorly resolved (76, 79–82). In this regard, solution NMR spectroscopy offers a unique window into studies of protein client–HtrA interactions, demonstrating how the oligomeric architecture of the HtrA family plays a critical role in regulating enzyme function.
Materials and Methods
HtrA2 and drkN SH3 proteins were expressed in Escherichia coli and purified by Ni-affinity chromatography, hydrophobic interaction chromatography (for HtrA2), and SEC. All NMR measurements were performed at 23.5 Tesla (1-GHz 1H frequency), 18.8 Tesla (800-MHz 1H frequency), or 14.0 Tesla (600-MHz 1H frequency). Structures of proteins for visualization were generated by using UCSF ChimeraX (83). Details of protein expression, purification, NMR experiments, and data analysis are provided in SI Appendix.
Supplementary Material
Acknowledgments
SEC-MALS experiments were conducted with the help of the Hospital for Sick Children Structural and Biophysical Core Facility. Y.T. is supported through a Japan Society for the Promotion of Science Overseas Research Fellowship and an Uehara Memorial Foundation postdoctoral fellowship. R.W.H. is grateful to the Canadian Institutes of Health Research (CIHR) and the SickKids Research Institute for postdoctoral fellowships. This research is funded through grants from the CIHR and the Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2203172119/-/DCSupplemental.
Data Availability
All relevant data are included in the paper and in SI Appendix. Backbone NMR chemical shifts of drkN SH3 have been deposited in the Biological Magnetic Resonance Bank, https://bmrb.io/ (entry 51327).
References
- 1.Dobson C. M., Getting out of shape. Nature 418, 729–730 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Gregersen N., Bross P., Vang S., Christensen J. H., Protein misfolding and human disease. Annu. Rev. Genomics Hum. Genet. 7, 103–124 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Chiti F., Dobson C. M., Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Clausen T., Kaiser M., Huber R., Ehrmann M., HTRA proteases: Regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell Biol. 12, 152–162 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Skorko-Glonek J., et al. , HtrA protease family as therapeutic targets. Curr. Pharm. Des. 19, 977–1009 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Zurawa-Janicka D., et al. , Structural insights into the activation mechanisms of human HtrA serine proteases. Arch. Biochem. Biophys. 621, 6–23 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Faccio L., et al. , Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J. Biol. Chem. 275, 2581–2588 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Gray C. W., et al. , Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur. J. Biochem. 267, 5699–5710 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Suzuki Y., et al. , A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell 8, 613–621 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Radke S., et al. , Mitochondrial protein quality control by the proteasome involves ubiquitination and the protease Omi. J. Biol. Chem. 283, 12681–12685 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moisoi N., et al. , Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 16, 449–464 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Martins L. M., et al. , Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol. Cell. Biol. 24, 9848–9862 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plun-Favreau H., et al. , The mitochondrial protease HtrA2 is regulated by Parkinson’s disease-associated kinase PINK1. Nat. Cell Biol. 9, 1243–1252 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Desideri E., Martins L. M., Mitochondrial stress signalling: HTRA2 and Parkinson’s disease. Int. J. Cell Biol. 2012, 607929 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin M. T., Beal M. F., Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Fitzgerald J. C., et al. , Phosphorylation of HtrA2 by cyclin-dependent kinase-5 is important for mitochondrial function. Cell Death Differ. 19, 257–266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strauss K. M., et al. , Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson’s disease. Hum. Mol. Genet. 14, 2099–2111 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Unal Gulsuner H., et al. , Mitochondrial serine protease HTRA2 p.G399S in a kindred with essential tremor and Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 111, 18285–18290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W., et al. , Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 9, 436–441 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Chien J., Campioni M., Shridhar V., Baldi A., HtrA serine proteases as potential therapeutic targets in cancer. Curr. Cancer Drug Targets 9, 451–468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srinivasula S. M., et al. , Inhibitor of apoptosis proteins are substrates for the mitochondrial serine protease Omi/HtrA2. J. Biol. Chem. 278, 31469–31472 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Yang Q. H., Church-Hajduk R., Ren J., Newton M. L., Du C., Omi/HtrA2 catalytic cleavage of inhibitor of apoptosis (IAP) irreversibly inactivates IAPs and facilitates caspase activity in apoptosis. Genes Dev. 17, 1487–1496 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki Y., Takahashi-Niki K., Akagi T., Hashikawa T., Takahashi R., Mitochondrial protease Omi/HtrA2 enhances caspase activation through multiple pathways. Cell Death Differ. 11, 208–216 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Cilenti L., et al. , Regulation of HAX-1 anti-apoptotic protein by Omi/HtrA2 protease during cell death. J. Biol. Chem. 279, 50295–50301 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Kuninaka S., et al. , The tumor suppressor WARTS activates the Omi/HtrA2-dependent pathway of cell death. Oncogene 24, 5287–5298 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Vande Walle L., et al. , Proteome-wide identification of HtrA2/Omi substrates. J. Proteome Res. 6, 1006–1015 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Park H.-J., Seong Y.-M., Choi J.-Y., Kang S., Rhim H., Alzheimer’s disease-associated amyloid beta interacts with the human serine protease HtrA2/Omi. Neurosci. Lett. 357, 63–67 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Kooistra J., Milojevic J., Melacini G., Ortega J., A new function of human HtrA2 as an amyloid-β oligomerization inhibitor. J. Alzheimers Dis. 17, 281–294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung H.-J., Jamal M. A. H. M., Hong S.-T., The function of bacterial HtrA is evolutionally conserved in mammalian HtrA2/Omi. Sci. Rep. 10, 5284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martins L. M., et al. , Binding specificity and regulation of the serine protease and PDZ domains of HtrA2/Omi. J. Biol. Chem. 278, 49417–49427 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Toyama Y., Harkness R. W., Lee T. Y. T., Maynes J. T., Kay L. E., Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR. Proc. Natl. Acad. Sci. U.S.A. 118, e2025022118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toyama Y., Harkness R. W., Kay L. E., Dissecting the role of interprotomer cooperativity in the activation of oligomeric high-temperature requirement A2 protein. Proc. Natl. Acad. Sci. U.S.A. 118, e2111257118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bejugam P. R., et al. , Allosteric regulation of serine protease HtrA2 through novel non-canonical substrate binding pocket. PLoS One 8, e55416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zurawa-Janicka D., et al. , Temperature-induced changes of HtrA2(Omi) protease activity and structure. Cell Stress Chaperones 18, 35–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh N., D’Souza A., Cholleti A., Sastry G. M., Bose K., Dual regulatory switch confers tighter control on HtrA2 proteolytic activity. FEBS J. 281, 2456–2470 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E., A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR 4, 727–734 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Bezsonova I., Singer A., Choy W.-Y., Tollinger M., Forman-Kay J. D., Structural comparison of the unstable drkN SH3 domain and a stable mutant. Biochemistry 44, 15550–15560 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Zhang O., Forman-Kay J. D., Structural characterization of folded and unfolded states of an SH3 domain in equilibrium in aqueous buffer. Biochemistry 34, 6784–6794 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E., Comparison of the backbone dynamics of a folded and an unfolded SH3 domain existing in equilibrium in aqueous buffer. Biochemistry 34, 868–878 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Lee J. H., et al. , Heterogeneous binding of the SH3 client protein to the DnaK molecular chaperone. Proc. Natl. Acad. Sci. U.S.A. 112, E4206–E4215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekhar A., et al. , Conserved conformational selection mechanism of Hsp70 chaperone-substrate interactions. eLife 7, e32764 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishikawa T., Nagadoi A., Yoshimura S., Aimoto S., Nishimura Y., Solution structure of the DNA-binding domain of human telomeric protein, hTRF1. Structure 6, 1057–1065 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Gianni S., et al. , Unifying features in protein-folding mechanisms. Proc. Natl. Acad. Sci. U.S.A. 100, 13286–13291 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neudecker P., et al. , Identification of a collapsed intermediate with non-native long-range interactions on the folding pathway of a pair of Fyn SH3 domain mutants by NMR relaxation dispersion spectroscopy. J. Mol. Biol. 363, 958–976 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Libich D. S., Tugarinov V., Clore G. M., Intrinsic unfoldase/foldase activity of the chaperonin GroEL directly demonstrated using multinuclear relaxation-based NMR. Proc. Natl. Acad. Sci. U.S.A. 112, 8817–8823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sekhar A., Rosenzweig R., Bouvignies G., Kay L. E., Hsp70 biases the folding pathways of client proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E2794–E2801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Libich D. S., Tugarinov V., Ghirlando R., Clore G. M., Confinement and stabilization of Fyn SH3 folding intermediate mimetics within the cavity of the chaperonin GroEL demonstrated by relaxation-based NMR. Biochemistry 56, 903–906 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wälti M. A., Libich D. S., Clore G. M., Extensive sampling of the cavity of the GroEL nanomachine by protein substrates probed by paramagnetic relaxation enhancement. J. Phys. Chem. Lett. 9, 3368–3371 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harkness R. W., et al. , Competing stress-dependent oligomerization pathways regulate self-assembly of the periplasmic protease-chaperone DegP. Proc. Natl. Acad. Sci. U.S.A. 118, e2109732118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tugarinov V., Hwang P. M., Ollerenshaw J. E., Kay L. E., Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 125, 10420–10428 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Rosenzweig R., Kay L. E., Bringing dynamic molecular machines into focus by methyl-TROSY NMR. Annu. Rev. Biochem. 83, 291–315 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Tugarinov V., Kanelis V., Kay L. E., Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 1, 749–754 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Gelis I., et al. , Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131, 756–769 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gans P., et al. , Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew. Chem. Int. Ed. Engl. 49, 1958–1962 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Tao Y., Strelkov S. V., Mesyanzhinov V. V., Rossmann M. G., Structure of bacteriophage T4 fibritin: A segmented coiled coil and the role of the C-terminal domain. Structure 5, 789–798 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Frank S., et al. , Stabilization of short collagen-like triple helices by protein engineering. J. Mol. Biol. 308, 1081–1089 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Yang X., et al. , Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76, 4634–4642 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meier S., Güthe S., Kiefhaber T., Grzesiek S., Foldon, the natural trimerization domain of T4 fibritin, dissociates into a monomeric A-state form containing a stable β-hairpin: Atomic details of trimer dissociation and local β-hairpin stability from residual dipolar couplings. J. Mol. Biol. 344, 1051–1069 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Wagner G., et al. , Exchange of two-spin order in nuclear magnetic resonance: Separation of exchange and cross-relaxation processes. J. Am. Chem. Soc. 107, 6440–6446 (1985). [Google Scholar]
- 60.Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D., 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 5, 67–81 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Lundström P., Vallurupalli P., Religa T. L., Dahlquist F. W., Kay L. E., A single-quantum methyl 13C-relaxation dispersion experiment with improved sensitivity. J. Biomol. NMR 38, 79–88 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Sun H., Kay L. E., Tugarinov V., An optimized relaxation-based coherence transfer NMR experiment for the measurement of side-chain order in methyl-protonated, highly deuterated proteins. J. Phys. Chem. B 115, 14878–14884 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Loria J. P., Rance M., Palmer A. G., A relaxation-compensated Carr–Purcell–Meiboom–Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332 (1999). [Google Scholar]
- 64.Boehr D. D., Nussinov R., Wright P. E., The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Changeux J.-P., Edelstein S., Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep. 3, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hammes G. G., Chang Y.-C., Oas T. G., Conformational selection or induced fit: A flux description of reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 13737–13741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Religa T. L., Ruschak A. M., Rosenzweig R., Kay L. E., Site-directed methyl group labeling as an NMR probe of structure and dynamics in supramolecular protein systems: Applications to the proteasome and to the ClpP protease. J. Am. Chem. Soc. 133, 9063–9068 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Religa T. L., Sprangers R., Kay L. E., Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science 328, 98–102 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Matsuo H., et al. , Identification by NMR spectroscopy of residues at contact surfaces in large, slowly exchanging macromolecular complexes. J. Am. Chem. Soc. 121, 9903–9904 (1999). [Google Scholar]
- 70.Y. Toyama, R. W. Harkness, L. E. Kay, Backbone 1H, 13C and 15N chemical shift assignments for the N-terminal SH3 domain of Drk. Biological Magnetic Resonance Data Bank. https://bmrb.io/data_library/summary/index.php?bmrbId=51327. Deposited 13 February 2022. [Google Scholar]
- 71.Kyte J., Doolittle R. F., A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982). [DOI] [PubMed] [Google Scholar]
- 72.Takahashi H., Nakanishi T., Kami K., Arata Y., Shimada I., A novel NMR method for determining the interfaces of large protein–protein complexes. Nat. Struct. Biol. 7, 220–223 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Nakanishi T., et al. , Determination of the interface of a large protein complex by transferred cross-saturation measurements. J. Mol. Biol. 318, 245–249 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Ueda T., et al. , Cross-saturation and transferred cross-saturation experiments. Q. Rev. Biophys. 47, 143–187 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Battiste J. L., Wagner G., Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear Overhauser effect data. Biochemistry 39, 5355–5365 (2000). [DOI] [PubMed] [Google Scholar]
- 76.Kim S., Grant R. A., Sauer R. T., Covalent linkage of distinct substrate degrons controls assembly and disassembly of DegP proteolytic cages. Cell 145, 67–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tennstaedt A., et al. , Human high temperature requirement serine protease A1 (HTRA1) degrades tau protein aggregates. J. Biol. Chem. 287, 20931–20941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poepsel S., et al. , Determinants of amyloid fibril degradation by the PDZ protease HTRA1. Nat. Chem. Biol. 11, 862–869 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Krojer T., et al. , Structural basis for the regulated protease and chaperone function of DegP. Nature 453, 885–890 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Jiang J., et al. , Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 11939–11944 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krojer T., Sawa J., Huber R., Clausen T., HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues. Nat. Struct. Mol. Biol. 17, 844–852 (2010). [DOI] [PubMed] [Google Scholar]
- 82.Truebestein L., et al. , Substrate-induced remodeling of the active site regulates human HTRA1 activity. Nat. Struct. Mol. Biol. 18, 386–388 (2011). [DOI] [PubMed] [Google Scholar]
- 83.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Press W. H., Flannery B. P., Teukolsky S. A., Vetterling W. T., Numerical Recipes in C. The Art of Scientific Computing (Cambridge University Press, Cambridge, UK, ed. 2, 1992). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the paper and in SI Appendix. Backbone NMR chemical shifts of drkN SH3 have been deposited in the Biological Magnetic Resonance Bank, https://bmrb.io/ (entry 51327).