Abstract
In comparison with other entomopathogenic Bacillus species, the genome of Brevibacillus laterosporus is poorly characterized. The aim of this study was to examine genetic variability in B. laterosporus by using a range of typing methodologies. Strains of B. laterosporus were examined for variation in 13 chromosomal genes encoding enzymes by multilocus enzyme electrophoresis. Optimal conditions of pulsed-field gel electrophoresis and randomly amplified polymorphic DNA were established that allowed analysis of the genome of B. laterosporus. None of these techniques allowed the identification of a convenient molecular marker for entomopathogenic strains, although one specific primer amplified only DNA from almost all mosquitocidal strains.
Several aspects of the biology of entomopathogenic species of the genus Bacillus have been studied. Bacillus thuringiensis produces a range of toxins encoded principally by plasmids (14). Bacillus sphaericus comprises five genospecies with both entomotoxic and nonentomotoxic strains, which can be differentiated by a variety of molecular methods (2, 11, 26). The toxins produced by this species are not as diverse as those of B. thuringiensis (3, 16). Both species have been used as biological control agents in many countries (5, 18). However, some strains of B. thuringiensis and B. sphaericus present problems, from an economic perspective, such as having low persistence in the environment and restricted targets.
Brevibacillus laterosporus comb. nov. (20), previously classified as Bacillus laterosporus (Laubach 1916b), is an aerobic spore-forming bacterium that can also demonstrate pathogenicity to insects. In common with B. sphaericus and B. thuringiensis, B. laterosporus produces parasporal bodies, which in this species may be canoe-shaped and which serve to cradle the spore (8) or can even be present in different shapes (22). However, these parasporal bodies were not considered to have any entomocidal activity (7) until Orlova et al. (13) demonstrated that some crystals produced during sporulation are highly toxic to Aedes aegypti and Anopheles stephensi larvae.
In common with B. sphaericus, some B. laterosporus strains show no apparent toxic activity to any test organism, and the observed toxicity is not homogeneous among toxic isolates (7, 19, 21). The results of the first bioassays with B. laterosporus demonstrated that some strains presented a larvicidal activity which was 1,000 times lower than that of the B. thuringiensis var. israelensis standard (7, 19). These results discouraged the use of B. laterosporus in biological control. Yet, it is pertinent to note that the first strains found in the B. sphaericus group demonstrated low levels of biocidal activity. Moreover, the observation that some strains demonstrate toxicity to more than one target (21) could be useful, particularly since a number of insects appear to be developing resistance to some of the bioinsecticide treatments used in the field (17). The results of Orlova et al. (13) encourage the search for new strains of B. laterosporus.
Given differences in toxicity levels and in the spectrum of activity, there is a need for molecular markers that could discriminate between strains.
Therefore, in this study, we examined the use of pulsed-field gel electrophoresis (PFGE) and random amplified polymorphic DNA (RAPD) in the analysis of the B. laterosporus genome, because both techniques have been shown to be useful as typing methods because they allow a convenient comparison of the bacterial chromosomes (5, 24). In addition to these techniques, multilocus enzyme electrophoresis (MLEE) was also applied, because it allows the study of the genetic diversity within and between groups of bacteria and because we had previously identified a molecular marker for mosquitocidal strains of B. sphaericus by this methodology (26). In this way, we tried to determine some useful tools with which to generate detailed information on the genetic structure of this species and to identify potential molecular markers for entomopathogenic strains which can assist research groups who are working in screening programs for the isolation of entomopathogenic bacteria.
The strains used in this work are listed in Table 1. All strains, except BL01 (supplied by LFB, Laboratório de Fisiologia Bacteriana, IOC/FIOCRUZ, Brazil), were originally supplied by A. A. Yousten (Department of Biology, Virginia Polytechnic Institute and State University, Blacksburg, Va.). More details about these strains are found in the article by Favret and Yousten (7). All strains were maintained at −20°C as spore suspensions in 20% glycerol. Spores were removed from nutrient agar plates which had been incubated at 30°C for at least 2 days. The electrophoretic mobilities of 13 enzymes (EC 1.1.1.3.7, malate dehydrogenase [MDH]; EC 1.4.1.9, lactate dehydrogenase [LDH]; EC 2.4.2.1, nucleoside phosphorylase [NP]; EC 1.4.1.1, alanine dehydrogenase [ADH]; EC 4.2.1.3, aconitase [ACON]; EC 5.3.1.9, glucose phosphate isomerase [GPI]; EC 1.1.1.40, malic enzyme [ME]; EC 3.4.11.1, peptidase [P2]; EC 3.4.11, peptidase [P3]; EC 3.1.1.1, esterase [EST]; EC 3.4.13.9, prolidase [PD]; EC 1.1.1.44, 6-phosphogluconate dehydrogenase [6PG]; and EC 1.1.1.49, glucose-6-phosphate dehydrogenase [G6P]) were determined according to the proximity to the cathode as previously described (26). The methodologies for culture growth, cell lysis, interpretation of the gel, and numerical analysis were as previously described (9, 23, 25, 26).
TABLE 1.
Strains used in this work and data from PFGE, MLEE, and bioassays
| Strain | No. of strains grouped bya:
|
Toxicity forb:
|
||||
|---|---|---|---|---|---|---|
| SRP | RAPD | MLEEc | Lasioderma serricorne | Aedes aegypti | Culex quinquefasciatus | |
| NRS 1642 | 1 | 2 | 117 | + | ||
| NRS 1644 | 1 | 2 | 117 | + | ||
| NRS 1648 | 1 | 2 | 117 | + | − | |
| NRS 1646 | 2 | 6 | 113 | + | + | |
| NRS 1647 | 2 | 8 | 113 | + | + | |
| SHI 1 | 3 | 10 | 115 | + | − | + |
| SHI 2 | 3 | 10 | 111 | + | − | + |
| BON 707 | 4 | 8 | 111 | + | − | + |
| CCEB 342 | 4 | 8 | 118 | + | − | |
| ATCC 6457 | 4 | 9 | 111 | + | + | |
| ATCC 9141 | 4 | 8 | 111 | + | + | + |
| NRS 1267 | 5 | 1 | 113 | + | + | |
| NRS 1264 | 5 | 1 | 116 | |||
| NRS 590 | 6 | 7 | 113 | + | + | + |
| NRS 661 | 7 | 5 | 113 | + | ||
| BON 708 | 7 | 5 | 113 | + | − | |
| BON 712 | 7 | 7 | 113 | + | ||
| NRS 1111 | 7 | 7 | 113 | + | + | |
| NRS 1645 | 8 | 4 | 113 | + | + | |
| SHI 3 | 9 | 12 | + | − | ||
| SHI 4 | 10 | 11 | 114 | + | − | |
| SHI 5 | 11 | 10 | 111 | + | + | + |
| NRS 340 | 12 | 4 | 113 | + | ||
| ATCC 64 | 12 | 7 | 113 | + | + | |
| NRS 1643 | 13 | 10 | 111 | |||
| Montaldi | 14 | 1 | 113 | − | ||
| BON 706 | 4 | 112 | + | |||
| NRS 1338 | 3 | + | ||||
| BL 01 | 110 | |||||
For RAPD analysis, DNA was isolated as reported by Alexander and Priest (1), and its concentration was determined with a model TK0 100 minifluorometer (Hoefer Scientific), and 5-ng/μl dilutions were prepared for RAPD experiments. A series of 14 random sequence decamer primers (OPA-01 to OPA-14), with 60 to 70% G+C content were obtained from Operon Technologies, Inc. Reaction mixtures were made that consisted of 2.5 μl of 10× Pharmacia Taq buffer, each deoxynucleotide triphosphate (dNTP) at a concentration of 100 μM, 5.0 pmol of primer, 1 U of Taq polymerase (Pharmacia), 25 ng of DNA template, and sterile distilled water to bring the final volume to 25 μl. Control reaction mixtures lacking template DNA were also prepared. The optimal concentrations of each component of the reaction mixture were determined by a series of preliminary experiments (not described). All reactions with each primer were performed at least twice by using a Gene Amp PCR system 9600 thermocycler (Perkin-Elmer). The same DNA preparations were studied concurrently with each primer. The amplification reaction involved 45 cycles consisting of denaturation at 94°C for 1 min, annealing at 36°C for 1 min, and extension at 72°C for 2 min. Amplification products were analyzed by separating 12 μl of reaction mixture on a 1.4% agarose gel at 3.2 V cm−1 in TAE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8.0]), followed by staining with 0.5 μg of ethidium bromide ml−1 and examination under UV light.
Reproducibility was interpreted as the visualization of the same patterns of bands in different gels after DNA amplification with the same particular primer. For the numerical analysis of RAPD results, photographs of the gels were scanned at 300 dots/in. on an HP Scanjet IIIc/T (Hewlett-Packard, Camas, Wash.), and RAPD patterns obtained by amplification of DNA with primer OPA-2 were analyzed by Gelcompar software Windows version 4.0 (Applied Maths, Kortrijk, Belgium). Patterns were compared through calculation of a similarity matrix with the Dice similarity coefficient. The similarity matrix was transformed into a phenogram by the unweighted pair group method with arithmetic averages (UPGMA) algorithm.
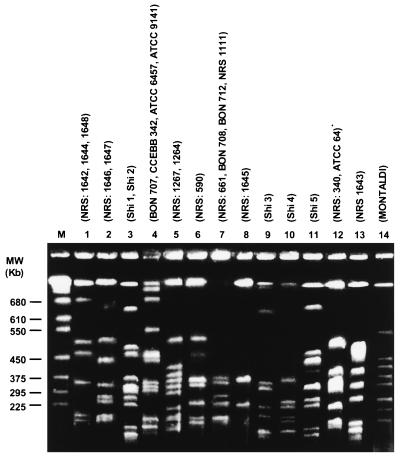
Intact chromosomal DNA was prepared as previously described (24) for use in PFGE. For digestion, the buffer was replaced with 200 μl of restriction enzyme buffer containing 30 U of restriction enzyme, and the insert was incubated for 18 h at the appropriate temperature for the enzymes SmaI, NotI, and SfiI. Among these enzymes, SfiI presented the best results when the pulse time was 50 to 90 s during 42 h at 150 V. SfiI was found to give the most readily interpretable gels, with a large number of clearly separated bands observed for all strains and with no smearing or compression zone at the bottom of the gel. The resulting bands allowed interstrain comparisons to be made (Fig. 1), and the 14 patterns produced were stable and reproducible. Simpson’s index of diversity (10) was calculated. The use of this index allows an objective assessment of how one typing system compares with another. The index ranges from 0 to 1, where a value of 0 indicates no discrimination, while a value of 1 indicates that every strain can be discriminated. The numerical value obtained represents two aspects of discrimination—the number of types and their relative frequency.
FIG. 1.
SfiI-generated PGFE profiles of Brevibacillus laterosporus strains. MW, molecular size. M, molecular size markers.
Among the 13 chromosomal genes encoding enzymes tested by the MLEE technique, those encoding EST, PD, 6PG, and G6P did not present activity for any of the B. laterosporus strains tested, suggesting that the B. laterosporus group apparently does not possess the enzyme G6P (EC 1.1.1.49) and lacks the hexose monophosphate shunt. Data generated with the remaining enzymes grouped the 27 test isolates into 9 zymovars (Table 1). However, the dendrogram based on these data (not shown) did not discriminate entomopathogenic strains from nonpathogenic isolates. MLEE analysis detected only minor differences between strains of B. laterosporus presenting a numerical index of discriminatory ability of D = 0.76. No correlation could be established between the MLEE data recorded in this study and the bioassay results reported in Table 1. This was exemplified by zymovar 113, which contained strains such as 1647 with toxicity to Culex quinquefasciatus (mosquito), Lasioderma serricorne (cigarette beetle), Trichostrongylus colubriformis (zooparasitic nematode), Heterodera glycines (phytoparasitic nematode), Biomphalaria glabrata (snail), and Dreissena polymorpha (zebra mussels) (21), as well as strains like Montaldi, which shows no toxicity to C. quinquefasciatus (23a).
Primers OPA-02 (5′-TGCCGAGCTG3′), OPA-04 (5′AATCGGGCTG 3′), and OPA-11 (5′CAATCGCCGT 3′) presented the highest level of polymorphism and gave amplification products for the majority of the B. laterosporus strains tested. Primer OPA-01 (5′CAGGCCCTTC 3′) amplified DNA only from mosquitocidal strains, with the exception of strain 1267 (not shown). In addition, a single band of approximately 900 bp was observed in all B. laterosporus samples when primer OPA-11 was employed in the PCR (not shown). Primer OPA-2 presented the highest level of polymorphism (highest number of bands − Simpson’s index of diversity = 0.93), and for this reason only data produced with that primer were used for the numerical analysis (Fig. 2).
FIG. 2.
(A) Dendrogram showing the relationships between strains of B. laterosporus based on RAPD data obtained with primer OPA-2. Comparisons between the electrophoretic types obtained were made by using the Dice similarity coefficient and clustered by using the UPGMA algorithm (for more details, see the text). (B) RAPD amplification products. (C) Strains of B. laterosporus; the number after the hyphen indicates the grouping according to Table 1.
Interestingly, the spectrum of B. laterosporus pathogenicity is more complex than for Bacillus species, with some strains (e.g., NRS 1647) demonstrating toxicity to a range of target organisms (21). In the present study, the primer OPA-01 generated amplification products from the majority of mosquitocidal strains, with no products observed for the other strains tested. This observation indicates that this primer could be used to identify sequences associated with strains which demonstrate mosquitocidal activity. However, this will only be substantiated through the examination of additional strains. The use of a higher number of strains will be necessary, given that no amplification product was generated from the mosquitocidal strain NRS 1267. In this way, we agree with Singer’s opinion regarding the biological activity of B. laterosporus: “We have only scratched the surface of our investigation (21).” MLEE data obtained by Singer (21) and in this study revealed that mosquitocidal strains of B. laterosporus are phenotypically very closely related.
Very similar PFGE patterns (differing in only one or two bands) were recorded for the majority of strains used in this study, indicating that they were clonally related. A correlation between MLEE, PFGE, and RAPD data was apparent for some strains. Thus, strains from SRP1 all belong to zymovar 117 and RAPD group 1. These strains not only shared the same molecular structure but have the same targets for pathogenicity, strongly suggesting that these strains belong to the same clone. Some strains (e.g., NRS 707 and ATCC 9141 [SRP4]) gave the same MLEE and PFGE patterns as well as very similar RAPD profiles, strongly suggesting that they are representatives of the same clone. An examination of the banding patterns obtained by PFGE revealed that the majority of samples differ in one (SRP1 and SRP14) to six (SRP3 and SRP6) bands. It is well documented that this kind of variation can be the result of a single simple genetic event, while differences in four to six bands are probably the result of two genetic events. This finding suggests that the genetic events that cause heterogeneity are not widespread in B. laterosporus.
RAPD and PFGE had almost the same discriminatory power D = 0.93 and D = 0.94, respectively), but it should be emphasized that the patterns obtained by PFGE were more stable and reproducible. Like all PCR techniques, some bands on RAPD had different intensities in different gels. PFGE does not present that characteristic. However, the use of the RAPD technique resulted not only in the possibility of the visualization of the fingerprinting of each strain, but in the promising existence of one molecular marker for B. laterosporus at the species level and another for mosquitocidal strains.
Microbial screening programs are most effective if based on a sound taxonomic framework (4). Such frameworks allow the development of effective selective isolation strategies and the accurate recognition of the required organism or of novel microbes (15).
In this work, we used three different techniques in an attempt to analyze genetic polymorphisms in B. laterosporus and look for molecular markers associated with pathogenicity. Such markers would facilitate the identification of potential candidates for use as bioinsecticides. The results indicate that both RAPD and PFGE have the potential to detect polymorphism in this species.
This study represented the first attempt to employ a range of molecular techniques in the analysis of genetic variability in B. laterosporus. These results could be of assistance to research groups which are working in screening programs for the isolation of entomopathogenic bacteria.
Acknowledgments
We thank Douglas McIntosh for reviewing the manuscript and Allan A. Yousten for suggestions. We also thank Heloisa M. N. Diniz for computer drawings and help with the figures.
REFERENCES
- 1.Alexander B, Priest F G. Numerical classification and identification of Bacillus sphaericus including some strains pathogenic for mosquito larvae. J Gen Microbiol. 1990;136:367–376. doi: 10.1099/00221287-136-2-367. [DOI] [PubMed] [Google Scholar]
- 2.Aquino de Muro M, Mitchell W J, Priest F G. Differentiation of mosquito pathogenic strains of Bacillus sphaericus from non-toxic varieties by ribosomal RNA gene restriction patterns. J Gen Microbiol. 1992;138:1159–1166. doi: 10.1099/00221287-138-6-1159. [DOI] [PubMed] [Google Scholar]
- 3.Bravo A. Phylogenetic relationships of Bacillus thuringiensis δ-endotoxin family proteins and their functional domains. J Bacteriol. 1997;179:2793–2801. doi: 10.1128/jb.179.9.2793-2801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bull A T, Goodfellow M, Slater H. Biodiversity as a source of innovation in biotechnology. Annu Rev Microbiol. 1992;46:219–252. doi: 10.1146/annurev.mi.46.100192.001251. [DOI] [PubMed] [Google Scholar]
- 5.Carlson C R, Caugant D A, Kolstø A-B. Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl Environ Microbiol. 1994;60:1719–1725. doi: 10.1128/aem.60.6.1719-1725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consoli R A G B, Santos B S, Lamounier M A, Secundino N F C, Rabinovitch L, Silva C M B, Alves R S A, Carneiro N F F. Efficacy of a new formulation of Bacillus sphaericus 2362 against Culex quinquefasciatus (Diptera: Culicidae) in Montes Claros, Minas Gerais, Brazil. Mem Inst Oswaldo Cruz. 1997;92:571–573. doi: 10.1590/s0074-02761997000400025. [DOI] [PubMed] [Google Scholar]
- 7.Favret M E, Yousten A A. Insecticidal activity of Bacillus laterosporus. J Invertebr Pathol. 1985;45:195–203. doi: 10.1016/0022-2011(85)90009-6. [DOI] [PubMed] [Google Scholar]
- 8.Hanney C L. The parasporal body of Bacillus laterosporus var Laubach. J Biophys Biochem Cytol. 1957;3:1001–1010. doi: 10.1083/jcb.3.6.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris H, Hopkinson D A. Handbook of enzyme electrophoresis in human genetics. New York, N.Y: Elsevier Sciences Publishers; 1976. [Google Scholar]
- 10.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krych V, Johnson J L, Yousten A A. Deoxyribonucleic acid homologies among strains of Bacillus sphaericus. Int J Syst Bacteriol. 1980;30:476–484. [Google Scholar]
- 12.Montaldi F A, Roth I L. Parasporal bodies of Bacillus laterosporus sporangia. J Bacteriol. 1990;172:2168–2171. doi: 10.1128/jb.172.4.2168-2171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlova M V, Smirnova T A, Ganushkina L A, Yacubovich V Y, Azizbekyan R R. Insecticidal activity of Bacillus laterosporus. Appl Environ Microbiol. 1998;64:2723–2725. doi: 10.1128/aem.64.7.2723-2725.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter A G. Mosquitocidal toxins, genes and bacteria: the hit squad. Parasitol Today. 1996;12:175–179. doi: 10.1016/0169-4758(96)10013-2. [DOI] [PubMed] [Google Scholar]
- 15.Priest F G, Aquino de Muro M, Kaji D. Systematic of insect pathogenic bacilli: uses in strain identification and isolation of novel pathogens. In: Priest F G, Ramos-Cormenzana A, Tindall B, editors. Bacterial diversity and systematics. New York, N.Y: Plenum Press; 1994. pp. 275–296. [Google Scholar]
- 16.Priest F G, Ebdrup L, Zahner V, Carter P E. Distribution and characterization of mosquitocidal toxin genes in some strains of Bacillus sphaericus. Appl Environ Microbiol. 1997;63:1195–1198. doi: 10.1128/aem.63.4.1195-1198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao D R, Mani T R, Rajendran R, Joseph A S, Gajanana A, Reuben R. Development of a high level of resistance to Bacillus sphaericus in a field population of Culex quinquefasciatus from Kochi India. J Am Mosq Control Assoc. 1995;11:1–5. [PubMed] [Google Scholar]
- 18.Régis L, Silva-Filha M H N L, Oliveira C M F, Rios E M, Silva S B, Furtado A F. Integrated control measures against Culex quinquefasciatus, the vector of filariasis in Recife. Mem Inst Oswaldo Cruz. 1995;90:115–120. doi: 10.1590/s0074-02761995000100022. [DOI] [PubMed] [Google Scholar]
- 19.Rivers D B, Vann C N, Zimmack H L, Dean D H. Mosquitocidal activity of Bacillus laterosporus. J Invertbr Pathol. 1991;58:444–447. doi: 10.1016/0022-2011(91)90191-r. [DOI] [PubMed] [Google Scholar]
- 20.Shida O, Takagi H, Kadowaki K, Komagata K. Proposal for two new genera, Brevibacillus gen. nov. and Aneurinibacillus gen. nov. Int J Syst Bacteriol. 1996;46:939–946. doi: 10.1099/00207713-46-4-939. [DOI] [PubMed] [Google Scholar]
- 21.Singer S. The utility of strains of morphological group II Bacillus. Adv Appl Microbiol. 1996;42:219–261. doi: 10.1016/s0065-2164(08)70374-5. [DOI] [PubMed] [Google Scholar]
- 22.Smirnova T A, Minenkova I B, Orlova M V, Lecadet M M, Azizbekyan R R. The crystal-forming strains of Bacillus laterosporus. Res Microbiol. 1996;147:343–350. doi: 10.1016/0923-2508(96)84709-7. [DOI] [PubMed] [Google Scholar]
- 23.Vieira V V, Teixeira L, Zahner V, Momen H, Fackland R R, Steigerwalt A G, Brenner D J, Castro A C D. Genetic relationships among the different phenotypes of Streptococcus dysgalactiae strains. Int J Syst Bacteriol. 1998;48:1231–1243. doi: 10.1099/00207713-48-4-1231. [DOI] [PubMed] [Google Scholar]
- 23a.Yousten, A. A. Personal communication.
- 24.Zahner V, Momen H, Priest F G. Serotype H5a5b is a major clone within mosquito-pathogenic strains of Bacillus sphaericus. Syst Appl Microbiol. 1998;21:162–170. doi: 10.1016/S0723-2020(98)80020-9. [DOI] [PubMed] [Google Scholar]
- 25.Zahner V, Momen H, Rabinovitch L. A comparative study of enzyme variation in Bacillus cereus and Bacillus thuringiensis. J Appl Bacteriol. 1989;67:275–282. doi: 10.1111/j.1365-2672.1989.tb02496.x. [DOI] [PubMed] [Google Scholar]
- 26.Zahner V, Rabinovitch L, Cavados C F G, Momen H. Multilocus enzyme electrophoresis on agarose gel as an aid to the identification of entomopathogenic Bacillus sphaericus. J Appl Bacteriol. 1994;76:327–335. doi: 10.1111/j.1365-2672.1994.tb01636.x. [DOI] [PubMed] [Google Scholar]