Abstract
Purpose
In January 2020, the COVID-19 pandemic started and has severely affected all countries around the world. The clinical symptoms alone are not sufficient for a proper diagnosis. Thus, molecular tests are required. Various institutes and researchers developed real-time PCR-based methods for the detection of the virus. However, the method needs expensive equipment. In the present study, we developed a real-time NASBA assay for the detection of SARS-CoV-2.
Methods
Primers and molecular beacon probes for RdRp and N genes were designed. In silico analysis showed that primers and the probes were specific for SARS-CoV-2. The standard samples with known copy numbers of the virus were tested using the NASBA assay and an FDA-approved real-time PCR kit. A series of standard samples were prepared and tested. Clinical sensitivity, precision analysis, and clinical assessment of the assay were performed.
Results
The limit of detection of the assay was 200 copies/mL. The clinical sensitivity of the assay was 97.64%. The intra-assay and inter-assay for both N and RdRp genes were less than 5% and 10%, respectively. Clinical assessment of the assay showed that the positive agreement rate and negative agreement rate of the assays were determined to be 97.64% and 100%, respectively.
Conclusions
The results of the present study show that the developed real-time NASBA is a sensitive and specific method for the detection of SARS-CoV-2 and is comparable with real-time PCR. NASBA is an isothermal signal amplification method, and if stand-alone fluorescent readers are available, the real-time NASBA can be used without the need for expensive thermocyclers. In addition compared to other isothermal methods like LAMP, the primer design is straightforward. Thus, real-time NASBA could be a suitable method for inexpensive SARS-CoV-2 detection.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11845-022-03046-2.
Keywords: Molecular beacon probe, Real-time NASBA, Real-time PCR, SARS-CoV-2
Introduction
In early January 2020, several cases of pneumonia were observed in Wuhan, China. Consequently, a coronavirus was confirmed to be the cause of the disease and was named SARS-CoV-2 [1, 2]. SARS-CoV-2 is a betacoronavirus from the Coronaviridea family with a single-stranded RNA genome of nearly 30,000 nucleotides [3, 4]. The genome codes 27 proteins including RNA-dependent RNA polymerase (RdRp), nucleocapsid protein (N), envelope protein (E), and spike protein (S). Studies show that the virus enters respiratory cells via S protein, which binds angiotensin-converting enzyme 2 (ACE2) [3, 5]. The virus has been recovered from the nasopharynx, oropharynx, bronchoalveolar lavage (BAL) fluid, and feces of the patients [6]. The symptoms of the disease are not specific and not reliable for diagnosis. Symptoms include fever (44% and 88% on admission and during hospitalization, respectively), and cough (67%). While 56% of the patients showed a ground-glass opacity on admission, no computed tomography (CT) abnormality was observed in 17.9% of the patients [7]. While CT sensitivity is 86–98%, it is not a specific method of detection (25%) [7, 8]. Thus, the most reliable method of confirmation of the disease remains nucleic acid amplification tests (NAAT) [3].
Shortly after the pandemic, molecular diagnostic tests based on quantitative real-time PCR (RT-qPCR) were developed that detected various parts of the viral genome. One of the advantages of RT-qPCR assay is the low limit of detection (LOD). These tests include the USA CDC assay detecting N gene [6], Chinese CDC assay detecting ORF1ab- and N gene [9], an RT-qPCR test developed in Germany based on E, N, and RdRp genes [10], and Japanese CDC assay detecting N gene [11]. However, RT-qPCR-based assays are time-consuming and expensive. Therefore, more rapid testing strategies are required to control the pandemic.
Serological and viral protein testing is an interesting detection option especially, but viral load change during the infection might impair viral protein detection since viral load declines with time [12]. Detecting SARS-CoV-2-specific antibodies have a broader window for detection. However, two obstacles are present. First, they cannot be used for rapid diagnosis and transmission control since antibodies are generated weeks after infection. Second, cross-reactivity is highly possible for antibody testing [13].
Another option is isothermal amplification, which is performed at a single temperature and is faster than RT-qPCR. The most popular methods of isothermal amplification are loop-mediated isothermal amplification (LAMP) and nucleic acid sequence-based amplification (NASBA). LAMP uses 4–6 primers [14] and can be coupled with a reverse transcription step to detect RNA targets [15]. Although LAMP can generate results in nearly 30–40 min, its primer design is complicated, which makes it difficult to develop new assays.
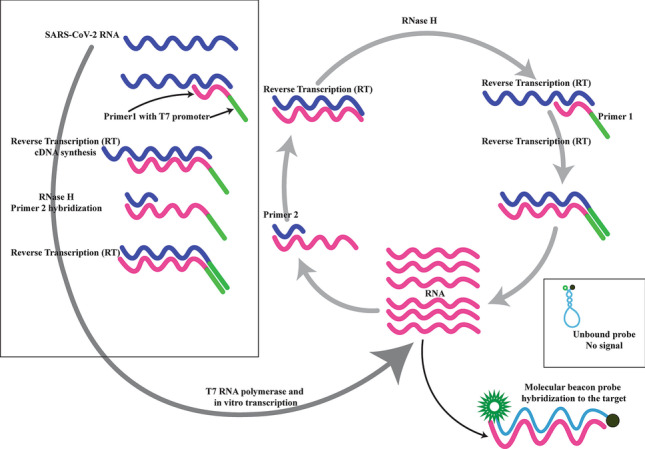
NASBA is another isothermal signal amplification method performed at 37–42 °C. Like LAMP, NASBA can be performed in nearly 30–40 min and can detect both DNA and RNA targets. Unlike LAMP, the NASBA primer design is straightforward in that a T7 RNA polymerase recognition sequence is added to one of the primer pairs. Then, T7 RNA polymerase can rapidly transcribe the target sequence and generate nearly 109 copies of RNA in almost 30 min [16]. NASBA has been developed for the detection of various pathogens such as HIV-1/HCV in serum samples [17, 18] and respiratory viruses using nasal swabs [19]. However, to the best of our knowledge, NASBA and real-time NASBA have not been developed for the detection of SARS-CoV-2 infection.
In the present study, we developed a real-time NASBA assay for the detection of SARS-CoV-2 infection using nasopharyngeal and oropharyngeal swabs (NP/OP). The assay was compared with an FDA-approved RT-qPCR commercial kit for SARS-CoV-2 detection.
Materials and methods
Target gene and primer/probe design
All SARS-CoV-2 sequences were retrieved from GenBank, NCBI, and aligned using ClustalW and MEGA7. Then, the highly conserved regions of the RdRp and N gene were selected to design primers and molecular beacon probes using Beacon Designer 7 (Premier Biosoft, Palo Alto, CA). The specificity and characteristics of the primers and the molecular beacon probe were further analyzed using NCBI BLAST and Oligo7 software, respectively. Table 1 shows the sequence of the primers and probes.
Table 1.
The sequences of the primers and the probe
| RdRp | N | |
|---|---|---|
| Primer1 | AATTCTAATACGACTCACTATAGGGAGGTTAATGTTGTCTACTGTT | AATTCTAATACGACTCACTATAGGG ACTACAGATAGAGACACCAG |
| Primer2 | TGGTTATCTTACTTCTTCTTCTA | GGCTAGACTTTATTATGATTCAA |
| Probe | FAM-CGCGAAATCCTACCACATTCCACCTAGATGGTCGCG-BHQ1 | Yakima Yellow-CGCGAAGCTCTATTCTTTGCACTAATGGCATTCGCG-BHQ1 |
The italics in the P1 primer indicate the T7 promoter
The italics in molecular beacon probe indicate the stem
Standard sample preparation
RNA was extracted from a positive sample (200,000 copies/mL) using an RNA extraction kit (RNJia Virus Kit, Catalogue No. RN983072, RojeTechnologies, Iran). Then, tenfold serial dilutions were prepared from 200,000 to 20 copies/mL.
Real-time NASBA and RT-qPCR on standard samples
The standard samples were tested using Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (Sansure Biotech Inc.) according to the manufacturer’s instruction.
The standards were also tested using the developed real-time NASBA assay (RT-NASBA). RT-NASBA reactions contained 0.4 µM of each primer and 0.2 µM of each probe. Five microliters of purified RNA extracted from each sample was added to 15 µL of multiplex amplification mixture in a 0.2-mL microcentrifuge tube. The tubes were then incubated at 65 °C to disrupt the secondary structures of the target RNA. Each tube was immediately cooled to 41 °C for 5 min, after which 2 µL of the enzyme mixture containing 2.6 µg of bovine serum albumin (in 50% glycerol; Roche Diagnostics Corp., Indianapolis, Ind.), 40U of T7 RNA polymerase, 8 U of avian myeloblastosis virus reverse transcriptase, 0.2 U of RNase H, and 12.5 U of RNasin (All enzymes were purchased from Fermentas, except AMV-reverse transcriptase from Roche) were added. Development of fluorescence was followed in closed tubes for 90 min at 41 °C. Fluorescence intensity data were recorded every minute of the RT-NASBA reaction. All tests were performed in triplicates using the RotorGene Q instrument (Qiagen, Germany).
Analytical sensitivity
The NASBA primer/probe set was queried using NCBI BLASTn against nr/nt database to find the level of similarity with sequences other than SARS-CoV-2.
Limit of detection
Analytical sensitivity was assessed using the American FDA guidelines (Version July 28, 2020). A nasopharyngeal/oropharyngeal clinical sample of SARS-CoV-2 with known viral load was used. The sample was diluted to 200, 100, and 50 copies/mL, and 20 replicates of each one were tested. LOD is defined as the lowest concentration sample that returns a positive result in 95% of 20 replicates of positive samples.
Clinical sensitivity
Eighty-five positive samples (Cq range, 14–36), which were tested using Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (Sansure Biotech Inc), were also tested using the developed RT-NASBA assay. In addition, the standard sample was used to prepare 2000 (1 log10 > LOD), 20,000 (2 log10 > LOD), and 200,000 (3 log10 > LOD) copies/mL in a negative sample matrix (nasopharyngeal/oropharyngeal). These samples were extracted and tested in triplicate using the developed RT-NASBA assay.
Analytical specificity
To determine the analytical specificity of the RT-NASBA assay, the primer/probe sets were queried using NCBI BLASTn against nr/nt database to find the level of similarity SARS-CoV-2 sequences and the primer/probe sets.
Clinical specificity
Ten pooled human genome and 104 copies/mL the nucleic acids of respiratory pathogens (Vircell, AmpliRun®DNA/RNA) were spiked in a nasopharyngeal/oropharyngeal matrix. The prepared samples were then extracted and tested using the RT-NASBA assay. Table 2 shows the tested respiratory pathogens and 10 pooled human genomes.
Table 2.
Investigation of the cross-reaction of the new coronavirus (2019-nCoV) with NASBA real-time multiplex method
| Virus/bacteria/parasite | Source/sample type | Concentration | Ct value (ORF1ab gene/N gene) |
|---|---|---|---|
| Adenovirus | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| Influenza A | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| Influenza B | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| Legionella pneumophila | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| Cryptococcus neoformans | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| Chlamydia pneumonia | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| Streptococcus pneumoniae | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| Respiratory Syncytial Virus | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| Mycoplasma pneumoniae | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| Streptococcus pyogenes | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| Mycobacterium tuberculosis | AmpliRun®DNA/RNA Vircell | 104 copies/mL | -/- |
| 10 pooled human genome | Clinical sample | 10 ng/µL | -/- |
Precision analysis
The precision analysis includes intra-assay and inter-assay. Intra-assay refers to the variability of the result of the replicate of samples in the same run. To do so, each standard sample was tested in triplicates. Inter-assay refers to the variability of the result of replicates of samples in different runs/days. Each standard sample was tested five times on three different days to determine the precision of the RT-NASBA assay.
Clinical assessment of the RT-NASBA
The clinical performance of the assay was evaluated using 185 nasopharyngeal/oropharyngeal swab samples from COVID-19-suspected individuals. The samples were also evaluated using Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (Sansure Biotech Inc), which is an American-FDA-approved diagnostic kit.
Results
RT-NASBA and RT-qPCR on standard samples
The standard samples were tested using RT-qPCR (Table 3) and the developed RT-NASBA (Table 4) in triplicate. Both N and RdRp genes were detectable using both assays. However, RT-NASBA could not detect any signal at 20 copies/mL concentration (Table 3).
Table 3.
Detection of standard samples using Sansure RT-qPCR assay
| Assay | Target gene | Concentration (copies/mL) | Mean Cq for triplicates |
|---|---|---|---|
|
2019-nCoV positive specimen Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (Sansure Biotech Inc.) |
N | 200000 | 22.9587 |
| 20000 | 26.61745 | ||
| 2000 | 30.65462 | ||
| 200 | 36.17117 | ||
| 20 | 38.99604 | ||
| RdRp | 200000 | 24.41095 | |
| 20000 | 27.98925 | ||
| 2000 | 31.68987 | ||
| 200 | 37.21188 | ||
| 20 | 38.9026 |
Table 4.
Detection of standard samples using the developed RT-NASBA assay
| Assay | Target gene | Concentration (copies/mL) | Result of triplicates |
|---|---|---|---|
| RT-NASBA | N | 200000 | Positive |
| 20000 | Positive | ||
| 2000 | Positive | ||
| 200 | Positive | ||
| 20 | Undetermined | ||
| RdRp | 200000 | Positive | |
| 20000 | Positive | ||
| 2000 | Positive | ||
| 200 | Positive | ||
| 20 | Undetermined |
Analytical sensitivity
The RT-NASBA primer/probe sets were queried against NCBI SARS-CoV-2 sequences using BLASTn. The result showed that the primer/probe sets were 100% identical to the SARS-CoV-2 corresponding regions. Supplementary Table 1 shows the result for the N gene primer/probe set.
Limit of detection of the RT-NASBA
According to FDA guidelines (Version July 28, 2020), LOD is defined as the lowest concentration sample that returns positive results in 95% positive samples. Figure 1 shows the result for 20 replicates of each standard sample using the RT-NASBA assay. Only in 200 copies/mL standard were 95% of the samples positive. Therefore, the LOD of the assay was determined to be 200 copies/mL.
Fig. 1.
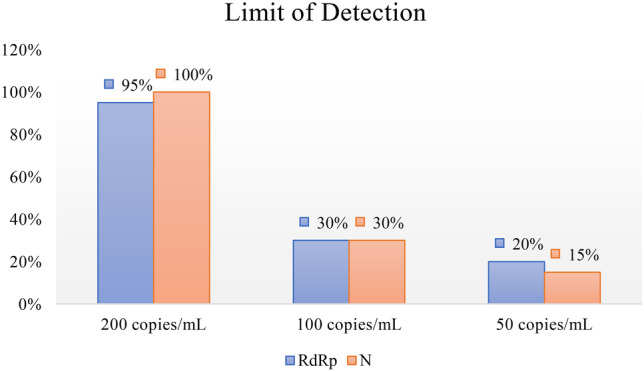
The LOD of the RT-NASBA assay. The RT-NASBA assay could detect 95%, 30%, and 20% of samples with 200, 100, and 50 copies/mL of SARS-CoV-2 RdRp gene, respectively. The assay could detect 100%, 30%, and 15% of samples with 200, 100, and 50 copies/mL of SARS-CoV-2 N gene, respectively. Thus, the LOD of the assay is 200 copies/mL
Clinical sensitivity
Eighty-five samples with a Cq range of 14–36 were tested using Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (Sansure Biotech Inc). Using the RT-NASBA assay, 83 of these samples tested positive and 2 tested negative. Therefore, the clinical sensitivity of the assay was determined to be 97.64%.
Precision analysis
Intra-assay
The standard samples were tested in triplicates to determine the intra-assay of the RT-NASBA. According to the College of American Pathologists (CAP) guidelines, the CV of intra-assay should be less than 5%. The highest and lowest CV of the RT-NASBA assay for the N gene was 1.2% and 0.51%, respectively. The highest and lowest CV of the intra-assay for RdRp was 1.19% and 0.29%, respectively. The results are acceptable according to CAP guidelines.
Inter-assay
The standard samples were tested using the RT-NASBA in quintuplicate on three different days. According to CAP guidelines, the CV of inter-assay should be less than 10%. The highest and lowest CV of the RT-NASBA assay for the N gene was 2.27% and 0.8%, respectively. The highest and lowest CV of inter-assay for RdRp was 1.99% and 0.44%, respectively. The results are acceptable according to CAP guidelines.
Clinical assessment of the RT-NASBA
One hundred and eighty-five NP/OP clinical samples from COVID-19-suspected individuals were tested using both Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (Sansure Biotech Inc) and the RT-NASBA.
Using the Sansure RT-qPCR kit, 100 of the samples were determined to be negative and the remaining 85 samples were positive. The RT-NASBA assay result showed that 83 samples were positive, and 102 samples were negative. Therefore, the positive agreement rate (PPA) and negative agreement rate (NPA) of the two assays were determined to be 97.64% and 100%, respectively (Table 5).
Table 5.
Clinical assessment of the RT-NASBA assay and agreement rate of RT-NASBA and RT-qPCR
| Assay | RT-qPCR (Sansure Biotech Inc.) | Total | ||
|---|---|---|---|---|
| Positive | Negative | |||
| RT-NASBA | Positive | 83 | 0 | 83 |
| Negative | 2 | 100 | 102 | |
| Total | 85 | 100 | 185 | |
Discussion
In the present study, we developed an RT-NASBA assay using molecular beacon probes based on N and RdRp genes to detect SARS-CoV-2 and compared it with Sansure RT-qPCR FDA-approved kit. Using the standard samples, the RT-NASBA assay was comparable to the RT-qPCR kit, and the LOD of the RT-NASBA assay was 200 copies/mL. In silico analysis showed that the RT-NASBA primer/probe sets were 100% compatible with SARS-CoV-2 sequences. Compared to Sansure RT-qPCR kit, the clinical sensitivity of the RT-NASBA assay was determined to be 97.64%. The precision of the assay was also acceptable based on CAP guidelines since the intra-assay and inter-assay of the assay were less than 5% and 10%, respectively. Clinical assessment of the assay showed that the PPA and NPA of the assay were 97.65% and 100% compared to Sansure RT-qPCR assay.
Since the start of the pandemic, various researchers and organizations developed RT-qPCR-based assays for the detection of SARS-CoV-2 [6, 9–11]. However, RT-qPCR needs specialized equipment, which is expensive and may not be available to all laboratories. On the contrary, isothermal methods are performed at a single temperature and are simpler to perform. The most well-known examples of isothermal methods are LAMP and NASBA. Several LAMP-based assays have been developed for the detection of SARS-CoV-2. For example, Yu et al. developed an RT-LAMP with a visual readout. They claimed that the limit of detection of the assay was 60 copies/mL of SARS-CoV-2 ORF1ab using 2 µL sample [20]. In another study, Yang and colleagues developed an RT-LAMP that had a limit of detection of 1000 copies/mL [21]. Broughton et al. developed an interesting RT-LAMP-based lateral flow assay for SARS-CoV-2 detection which is coupled with CRISPR-Cas-12 [22]. However, primer design and the optimization of LAMP reaction are not straightforward and, in the case of Broughton’s assay, need CRISPR-Cas-12, which is not readily available.
NASBA, on the other hand, is a simple isothermal signal amplification method. NASBA primers are easily designed by adding a T7 promoter to one of the primers and rapidly amplifies RNA targets [23]. In addition, it does not need specialized equipment like RT-qPCR thermocycler. The result can be visualized using gel electrophoresis [16]. In the case of using fluorescent probes, the signal can be detected using either RT-qPCR thermocycler or stand-alone fluorescent detectors.
NASBA has been used for the detection of infectious agents. Paryan et al. developed a NASBA assay for the detection of HIV-1 and HCV. The LOD of the assay was 100 copies/mL, and its sensitivity and specificity were 93% and 100%, respectively. Mohammadi-yeganeh et al. also used RT-NASBA for the detection of HIV-1 and HCV using molecular beacon probes. The LOD of the assay was 1000 copies/mL. The sensitivity and specificity of the assay were 98% and 100%, respectively. Keightly et al. developed an RT-NASBA assay for the detection of SARS-CoV in 2005 [24]. They found that their RT-NASBA assay was comparable to Triple-Target CDC TaqMan RT-PCR assay for SARS-CoV.
During COVID-19 pandemics, NASBA-based methods have been developed for SARS-CoV-2 detection. Chakravarthy and colleagues developed an RNA biosensor using NASBA with a sensitivity of 100 copied. The assay uses NASBA to amplify RNA targets. When the target RNA is present, the biosensor turns on a reporter gene translation, and the result is colorimetrically or luminometrically detected [25]. Cao et al. also developed an RNA-based riboswitch that uses NASBA to amplify the trigger RNA [26]. Wu and colleagues developed a NASBA-based high-throughput sequencing assay (INSIGHT), with a detection limit of 50 copies/mL. The NASBA product in this study can also be detected using portable fluorescent detectors or lateral flow strips [27]. This is an interesting strategy since the system can be used as point-of-care (POC) testing. This is mainly due to the capacity of NASBA to produce enormous amounts of RNA.
Conclusions
To the best of our knowledge, our RT-NASBA method is the only NASBA method for the detection of SARS-CoV-2 that uses molecular beacon probes. Although we used a regular real-time thermocycler for the detection of signals, the RT-NASBA results can be used to detect signals using stand-alone fluorescent readers. Therefore, the whole procedure will be less expensive than other methods. Furthermore, since the primer design is straightforward, the sensitivity and specificity of the assay are acceptable, and it is reproducible, the RT-NASBA assay can be a suitable method for the detection of SARS-CoV-2.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The author thanks Cellular and Molecular Biology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran, for providing technical support.
Author contribution
The conceptualization was done by MP and SMY. The formal analysis and interpretation were done by VK and AT. The resource and writing-original draft preparation were carried out by VK and AT. The supervision was done by SMY. The whole manuscript was read and approved by all authors.
Funding
The project was funded by Shahid Beheshti University of Medical Sciences, Tehran, Iran (grant number: 23650). This project was under the supervision of ethics committe of Shahid Beheshti University of Medical Sciences, Tehran, Iran( Ethics code:IR.SBMU.RETECH.REC.1399.503).
Availability of supporting data
The data achieved and analyzed during this study are available from the corresponding author on reasonable request.
Declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mahdi Paryan, Email: mparyan@gmail.com, Email: m_paryan@pasteur.ac.ir.
Samira Mohammadi-Yeganeh, Email: smyeganeh@gmail.com, Email: s.mohammadiyeganeh@sbmu.ac.ir.
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W et al (2020) A novel coronavirus from patients with pneumonia in China. N Engl J Med 382(8):727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed]
- 3.Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamming I, Timens W, Bulthuis ML et al (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203(2):631–637. 10.1002/path.1570 [DOI] [PMC free article] [PubMed]
- 6.Holshue ML, DeBolt C, Lindquist S et al (2020) First case of 2019 novel coronavirus in the United States. N Engl J Med 382(10):929–936. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed]
- 7.Guan WJ, Ni ZY, Hu Y et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu P, Lu R, Zhao L, et al. Three novel real-time RT-PCR assays for detection of COVID-19 virus. China CDC Wkly. 2020;2(25):453–457. doi: 10.46234/ccdcw2020.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman VM, Landt O, Kaiser M et al (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RTPCR. Euro Surveill 25(3):2000045. 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed]
- 11.Nao N, Shirato K, Katano H et al (2020) Detection of second case of 2019-nCoV infection in Japan. National Institute of Infectious Diseases, Japan
- 12.To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv H, Wu NC, Tsang OT et al (2020) Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. bioRxiv. 10.1101/2020.03.15.993097
- 14.Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong Y-P, Othman S, Lau Y-L et al (2018) Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms. 124(3):626–643. 10.1111/jam.13647 [DOI] [PMC free article] [PubMed]
- 16.Malek L, Sooknanan R, Compton J. Nucleic acid sequence-based amplification (NASBA) Methods Mol Biol. 1994;28:253–260. doi: 10.1385/0-89603-254-x:253. [DOI] [PubMed] [Google Scholar]
- 17.Paryan M, Forouzandeh Moghadam M, Kia V et al ( 2013) A simple and rapid method for the detection of HIV-1/HCV in co-infected patients. Iran J Biotechnol 11(2):74–79. 10.5812/IJB.10717
- 18.Mohammadi-Yeganeh S, Paryan M, Mirab Samiee S, et al. Molecular beacon probes-base multiplex NASBA real-time for detection of HIV-1 and HCV. Iran J Microbiol. 2012;4(2):47–54. [PMC free article] [PubMed] [Google Scholar]
- 19.Wat D, Gelder C, Hibbitts S, et al. The role of respiratory viruses in cystic fibrosis. J Cyst Fibros. 2008;7(4):320–328. doi: 10.1016/j.jcf.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu L, Wu S, Hao X, et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem. 2020;66(7):975–977. doi: 10.1093/clinchem/hvaa102%JClinicalChemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W, Dang X, Wang Q et al (2020) Rapid Detection of SARS-CoV-2 Using Reverse transcription RT-LAMP method. MedRxiv. 10.1101/2020.03.02.20030130
- 22.Broughton JP, Deng X, Yu G et al (2020) CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol 38(7):870–874. 10.1038/s41587-020-0513-4 [DOI] [PMC free article] [PubMed]
- 23.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350(6313):91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 24.Keightley MC, Sillekens P, Schippers W, et al. Real-time NASBA detection of SARS-associated coronavirus and comparison with real-time reverse transcription-PCR. J Med Virol. 2005;77(4):602–608. doi: 10.1002/jmv.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakravarthy A, Nandakumar A, George G et al (2021) Engineered RNA biosensors enable ultrasensitive SARS-CoV-2 detection in a simple color and luminescence assay. Life Sci Alliance 4(12). 10.26508/lsa.202101213 [DOI] [PMC free article] [PubMed]
- 26.Cao M, Sun Q, Zhang X et al (2021) Detection and differentiation of respiratory syncytial virus subgroups A and B with colorimetric toehold switch sensors in a paper-based cell-free system. Biosens Bioelectron 182:113173. 10.1016/j.bios.2021.113173 [DOI] [PubMed]
- 27.Wu Q, Suo C, Brown T et al (2021) INSIGHT: a population-scale COVID-19 testing strategy combining point-of-care diagnosis with centralized high-throughput sequencing. Sci Adv 7(7). 10.1126/sciadv.abe5054 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data achieved and analyzed during this study are available from the corresponding author on reasonable request.


