Significance
Regulatory T cells rely on active processes to maintain a suppressive phenotype inside a tumor, leading to increased tumor burden and worse cancer outcomes. Here, we report a pathway to interfere with regulatory T cell (Treg) stability by disrupting the expression of the interleukin 23 receptor. This approach increases Treg responsiveness to interleukin 12, leading to increased production of gamma-interferon and more efficient antitumor immune responses. The combined engagement of independent pathways to destabilize Treg through the interleukin 23 receptor and the glucocorticoid-induced TNFR-related protein receptor has a synergistic impact on the Treg phenotype and promotes antitumor immune responses. These findings expand our understanding of regulatory T-cell biology and offer tools for cancer immunotherapy.
Keywords: T regulatory cells, Treg conversion, MC38 colon carcinoma, mAb treatment, combination therapy
Abstract
Regulatory T cells (Treg) can impede antitumor immunity and currently represent a major obstacle to effective cancer immunotherapy. Targeting tumor-infiltrating regulatory Treg while sparing systemic Treg represents an optimal approach to this problem. Here, we provide evidence that the interleukin 23 receptor (IL23R) expressed by tumor-infiltrating Treg promotes suppressive activity. Disruption of the IL23R results in increased responsiveness of destabilized Treg to the IL12 cytokine, the production of γ-interferon, and the recruitment of CD8 T cells that inhibit tumor growth. Since the Treg destabilization pathway that is initiated by IL23R blockade is distinct and independent from the destabilization pathway coupled to glucocorticoid-induced TNFR-related protein (GITR) activation, we examined the impact of the coordinate induction of the two destabilization pathways on antitumor immune responses. Combined GITR and IL23R antibody treatment of mice inoculated with MC38 tumors resulted in robust and synergistic antitumor responses. These findings indicate that the delineation of independent Treg destabilization pathways may allow improved approaches to the development of combination immunotherapy for cancers.
Regulatory T cells (Treg) are a critical arm of the adaptive immune response that may prevent autoimmune reactions and resolve ongoing inflammation. Treg also represent a barrier to effective cancer immunity, since recruitment of Treg by growing tumor cells dampens antitumor immune responses and reduces the impact of cancer immunotherapy (1, 2).
Effective targeting of intratumoral Treg has been difficult to accomplish. Gene products that control the function of intratumoral Treg are often intracellular, such as FoxP3, or comprise surface receptors such as CD25 that are also expressed by systemic T cells. We have focused on surface receptors expressed by tumor-infiltrating but not systemic Treg that may allow selective Treg depletion or destabilization within the tumor microenvironment (TME) while sparing tumor-infiltrating systemic Treg (3, 4). For example, targeting the glucocorticoid-induced TNFR-related protein (GITR) receptor by agonistic antibodies (3–7) can reduce FoxP3 expression and destabilize intratumoral Treg, resulting in enhanced interferon gamma (IFNγ) production and increased antitumor immunity. Despite these promising preclinical results, GITR-based protocols have thus far been less effective in humans (8), highlighting the need to identify additional Treg destabilization pathways.
Many tumors produce high levels of interleukin 23 (IL23) (9–11), which potentiate tumor growth and up-regulate IL23 receptor (IL23R) expression by intratumoral T cells, including Treg. The IL23R, a member of the IL12 cytokine receptor family, is composed of an IL23R alpha chain and a signaling chain (IL12Rβ1) that is shared with the IL12 receptor (IL12R) (12, 13). Although the IL23R−STAT3 pathway is the central element in functional T–helper cell 17 (Th17) development (14, 15), expression of the IL23R by tumor-infiltrating Treg has been linked to poor prognosis (16–19).
To more fully characterize the effects of anti-IL23R targeting, we used Treg-specific IL23R knockout models. We found that IL23R marks a subset of highly suppressive tumor-infiltrating Treg and that the ablation or blockade of IL23R expression by Treg promotes protective antitumor immunity. The protective effects of IL23R ablation and blockade reflect increased Treg expression of a functional IL12 receptor, leading to increased IFNγ production and the recruitment of activated CD8 T cells to tumors. We also report that the mobilization of independent Treg destabilization pathways, which combine IL23R blockade with agonistic anti-GITR antibody, resulted in enhanced Treg destabilization and synergistic protection against tumor growth.
Results
IL23R Marks Suppressive, Tumor-Infiltrating Treg.
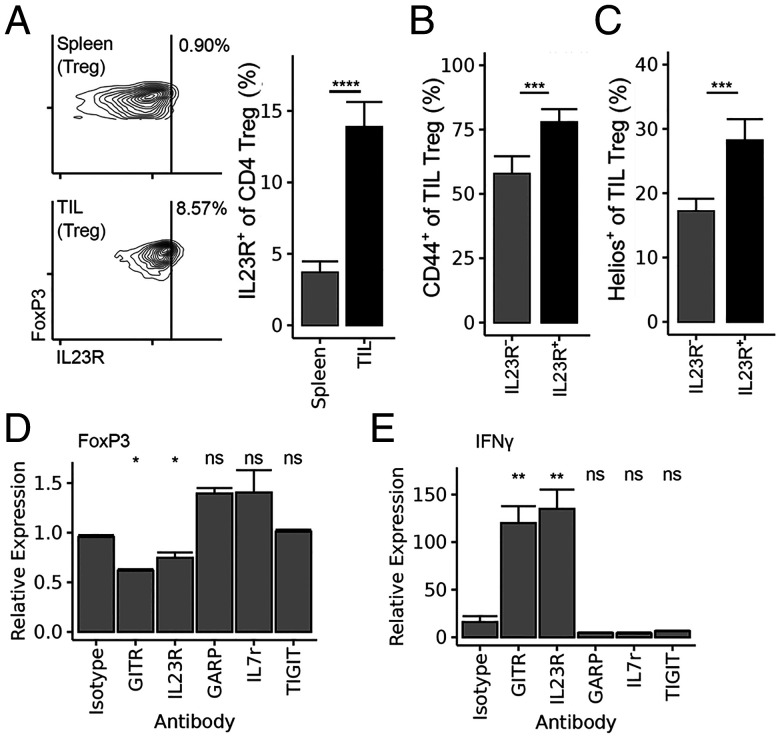
We asked whether IL23R expression by tumor-infiltrating Treg marks a Treg subpopulation with increased stability and suppressive activity. Analysis of tumor-infiltrating cells into MC38 colon carcinoma revealed a significant fraction of IL23R+ tumor-infiltrating Treg (Fig. 1A). A comparison with IL23R− Treg from the same tumors indicated that IL23R+ Treg expressed increased levels of CD44 and the Helios transcription factor (Fig. 1 B and C), which have been correlated with Treg suppressive function and stability (3, 20, 21). To determine whether the expression of IL23R by intratumoral Treg might be linked to a stable, suppressive phenotype, Treg from FoxP3-YFP reporter mice were sorted (SI Appendix, Fig. S1) and incubated with an anti-IL23R antibody. We noted that this antibody and the control anti-GITR antibody, but not other antibodies directed against Treg surface receptors, reduced FoxP3 expression (Fig. 1D) and increased IFNγ expression (Fig. 1E). Taken together, these results suggested that IL23R marked a suppressive subset of tumor-infiltrating Treg and that targeting this receptor in vivo might destabilize Treg and enhance antitumor immune responses.
Fig. 1.
IL23R is expressed on tumor-infiltrating (TIL) Treg. (A) IL23R staining of spleen and tumor-infiltrating Treg (CD4+ CD25+ FoxP3+ lymphocytes) following tumor challenge with 2 × 105 MC38 colon carcinoma cells, as described in Materials and Methods. (B) CD44 and (C) Helios staining on IL23R+ vs. IL23R− TIL Treg identified in A. (D) FoxP3 expression and (E) IFNγ expression of incubated Treg after 5 d of culture in the presence of the indicated antibodies (GITR, glucocorticoid-induced TNFR-related; GARP, glycoprotein A repititions predominant; IL7r, Interleukin 7 receptor; TIGIT, T cell immunoreceptor with Ig and immunoreceptor tyrosine-based inhibitory motif domains). Statistical tests in A, B, and C are paired Wilcoxon tests. Data are pooled from three or four independent experiments (n = 15/group [A–C] or 6/group [D and E]). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, non significant.
IL23R-Deficient Treg Display Increased IL12 Sensitivity and STAT4 Activation.
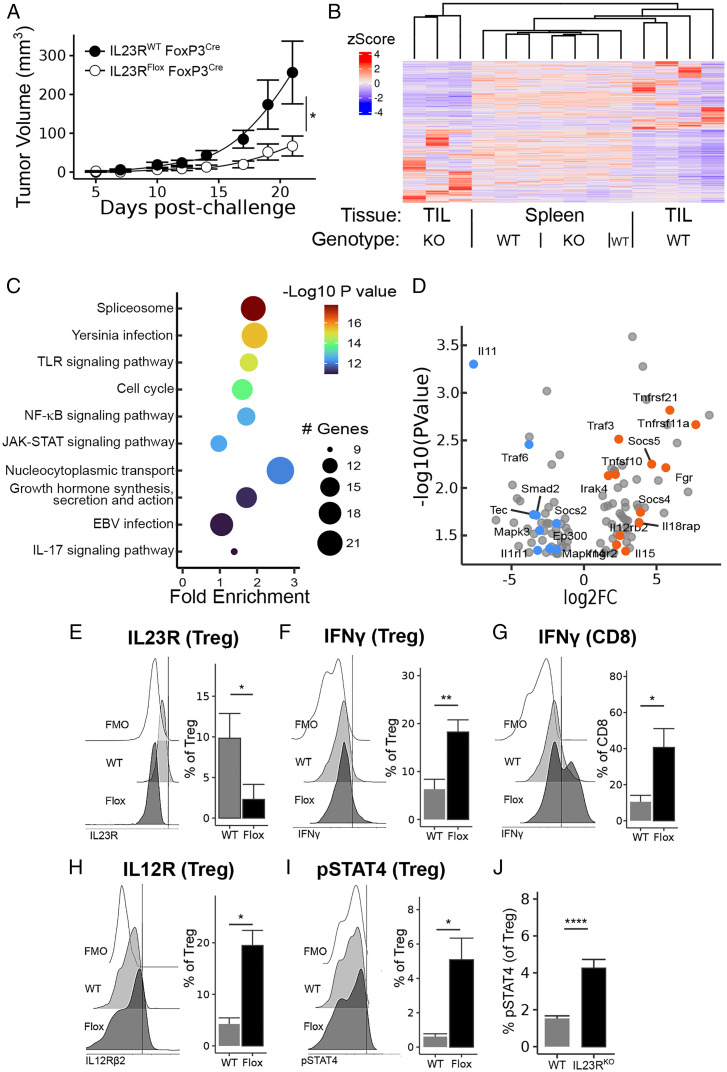
We generated conditional knockout (KO) IL23R-Flox mice (22) after crossing to mice expressing a FoxP3 promoter-driven Cre recombinase (IL23RFlox FoxP3Cre) and backcrossing for 10 generations. Mice with a Treg-specific deletion of IL23R displayed significantly delayed tumor growth compared to congenic age-matched control mice (Fig. 2A). Analysis of tumor-infiltrating Treg from these mice by next-generation RNA sequencing revealed a tumor-specific signature of differentially expressed genes (DEG) (Fig. 2B) that was enriched for immune-relevant pathways (Fig. 2C). Analysis of the genes contributing to these significant immune-relevant pathways revealed a bias toward Th1-like gene expression among IL23R-deficient Treg, while Treg from congenic control mice (IL23R wild-type(WT) FoxP3Cre) displayed a bias toward suppressive and/or Th2-like gene expression (Fig. 2D and SI Appendix, Fig. S2).
Fig. 2.
Treg-specific IL23R ablation protects against MC38 carcinoma. (A) Tumor growth curves after MC38 challenge, as in Fig. 1, in mice bearing a Treg-specific IL23R deletion (IL23RFlox FoxP3Cre) and congenic controls (IL23RWT FoxP3Cre). (B) IL23R-deficient Treg and congenic controls from tumor-infiltrating and spleen samples were subjected to bulk next-generation RNA sequencing. Significant DEGs in tumor samples were not differentially expressed by spleen Treg from the same mice. (C) The top 10 KEGG pathways among these significant DEGs are enriched in inflammatory- or immune-relevant biological processes. TLR, Toll-like receptor; EBV, Epstein–Barr virus. (D) Volcano plot of all genes contributing to any immune-relevant biological process selected from the significant DEGs and manually annotated as inflammatory/Th1-favoring (orange) or regulatory/Th2-favoring (blue). Log2 fold change (log2FC) versus -log10 P value is shown. See also SI Appendix, Fig. S2 for details on this annotation step. (E–I) Flow cytometry analysis of tumor-infiltrating Treg from mice in (A), including (E) Treg IL23R deficiency, (F) Treg and (G) CD8 T-cell IFNγ, (H) Treg IL12Rβ2 expression, and (I) Treg STAT4 activation. Fluorescence minus one (FMO) negative controls are included. (J) Activated STAT4 staining of Treg sorted from C57BL/6 or IL23RKO mice and cocultured with IL12 for 24 h. Tumor growth and flow cytometry data are pooled from three independent experiments (n = 10 to 12/group). *P < 0.05, **P < 0.01, ****P < 0.0001.
Treatment of STAT3KO mice with the anti-IL23R antibody displayed similar protection to antibody-treated WT mice (SI Appendix, Fig. S3), suggesting that reduced IL23R-linked signaling through its canonical STAT3 signaling pathway did not account for the protection noted above. The formation of functional IL23R and IL12R reflects, in part, competition for the shared IL12Rβ1 signaling chain (13). Reduced levels of IL23R or IL23R destabilization secondary to reduced IL23 ligand binding may increase the association of the IL12Rβ1 chain with IL12Rβ2. We noted that Il12rb2 was one of the transcripts significantly overexpressed in IL23R-deficient tumor-infiltrating Treg (Fig. 2D), resulting in the formation of high-affinity IL12 receptor complexes (13). We hypothesized that Treg-specific IL23R loss or blockade might enhance the IL12 responsiveness of Treg and thus alter their functional phenotype (SI Appendix, Fig. S4).
Flow cytometric analysis of tumor-infiltrating Treg from WT and IL23RFlox FoxP3Cre mice confirmed their IL23R-deficient status (Fig. 2E) and supported the hypothesis that these IL23R-deficient Treg display increased IL12 sensitivity. We also noted increased IFNγ expression, a downstream effector of IL12 signaling, by both Treg (Fig. 2F) and tumor-infiltrating CD8 (Fig. 2G) from IL23RFlox FoxP3Cre tumors. Moreover, we observed increased IL12R expression (Fig. 2H) and increased phosphorylated STAT4 (pSTAT4) activation (Fig. 2I) by IL23R-deficient Treg. Finally, we noted increased responsiveness to IL12 by activated IL23RKO compared to IL23WT Treg, as judged by increased pSTAT4 after incubation in IL12 for 24 h (Fig. 2J). Together, these results suggest that tumor protection mediated by IL23R-deleted Treg may reflect increased IL12 responsiveness and associated IFNγ secretion by tumor-infiltrating Treg, leading to the recruitment of activated CD8 T cells.
Impact of Enhanced Treg IL12 Signaling on the TME.
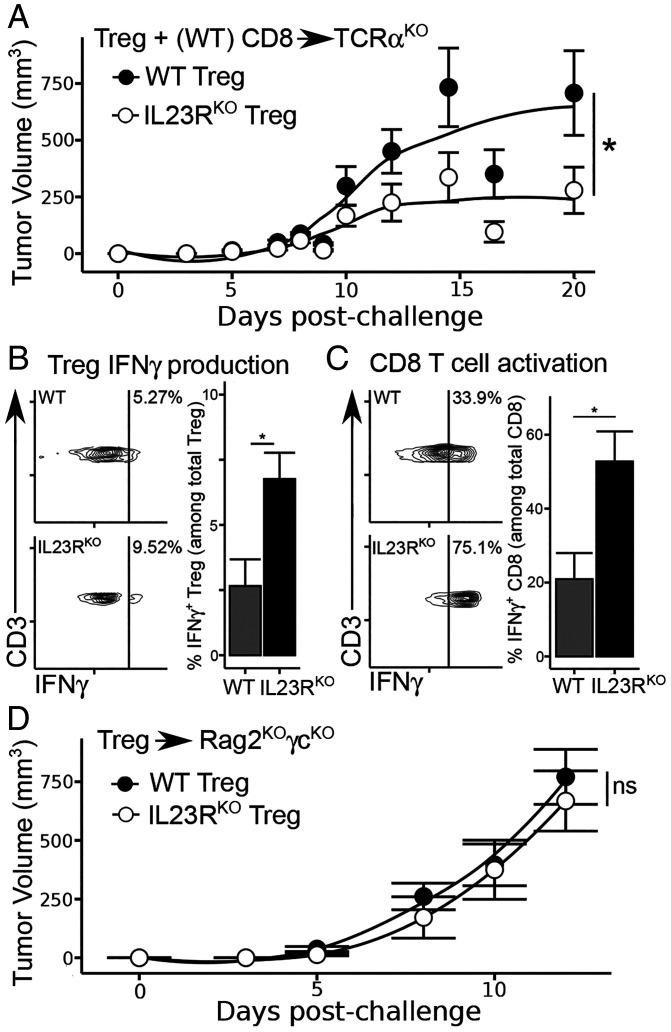
We asked whether the increased tumor protection from IL23RKO Treg depended in part on their recruitment of activated CD8 T cells. The adoptive transfer of 5 × 105 Treg from IL23RKO or IL23WT mice with 1 × 106 CD8 and CD25− CD4 T cells from WT donors into TCRαKO mice indicated that the stronger antitumor responses of IL23RKO Treg hosts (Fig. 3A) was associated with increased IFNγ production by tumor-infiltrating IL23RKO Treg (Fig. 3B), as well as increased activation of intratumoral CD8 T cells (Fig. 3C). Enhanced CD8 T-cell activation was required for tumor protection since the transfer of IL23RKO Treg alone into Rag2−/−γc−/− hosts did not reduce tumor burden (Fig. 3D). Taken together, these results suggest that Treg destabilization after the disruption of surface IL23R expression promotes IL12R/STAT4–dependent IFNγ responses (SI Appendix, Fig. S4) and improved antitumor immune responses.
Fig. 3.
Induced Treg-specific IFNγ leads to improved CD8 T-cell response. (A) Tumor growth curves (as in Fig. 2) in TCRαKO mice following the adoptive transfer of 5 × 105 CD4 Treg from WT or IL23RKO donors, as well as 1 × 106 CD4 non-Treg and 1 × 106 CD8 from WT donors. (B) IFNγ expression in tumor-infiltrating Treg and (C) CD8 T cells isolated from TCRαKO hosts. (D) Tumor growth curves, as in (B), but only 5 × 105 CD4 Treg were transferred and Rag2KOγcKO hosts were used. Data are pooled from three independent experiments (n = 9 to 12/group). *P < 0.05; ns, nonsignificant.
Full Deletion of IL23R Mimics the Treg-Specific Deletion Phenotype.
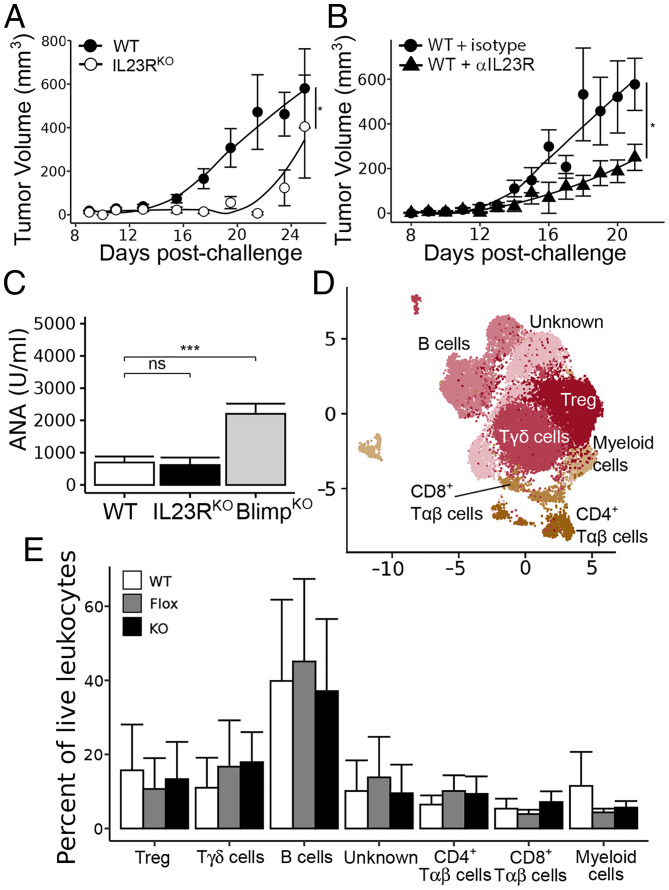
In addition to tumor-infiltrating Treg, IL23R may also be expressed by gut-associated Th17 cells and possibly Treg. We therefore asked whether full IL23R targeting would phenocopy Treg-specific targeting. Analysis of IL23RKO mice (23) (Fig. 4A) or mice treated with the anti-IL23R antibody(Fig. 4B) indicated that protection against MC38 growth was retained in both cases. In the latter case, the protective effects of the anti-IL23R antibody were similar to those observed in IL23RFlox FoxP3Cre mice (Fig. 4B).
Fig. 4.
Impact of full IL23R deletion on antitumor response. (A) Tumor growth curves, as in Fig. 2, in WT vs. IL23RKO mice. (B) Tumor growth curves of WT mice treated with 100 μg anti-IL23R antibody or isotype control by intraperitoneal injection once tumors were palpable (∼7 d) and every 48 h after for a total of three injections. (C) Serum antinuclear antibody (ANA) levels detected by enzyme-linked immunosorbent assay of aged (> 1 y) WT, IL23RKO, and Blimp1KO mice, which serve as a positive control for Treg destabilization. (D) Dimensional reduction of immune cells isolated from combined lamina propria and Peyer’s patches of IL23RWT FoxP3Cre (WT), IL23RFlox FoxP3Cre (Flox), and IL23RKO (KO) mice. (E) No statistically significant differences were found in any gut immune population identified by unsupervised clustering analysis. Data are pooled from two or three independent experiments (n = 8 to 10 mice/group; n = 6 mice/group for high-dimensional flow cytometric analysis). *P < 0.05, ***P < 0.001.
In addition to confirming efficacy, we tested for potential side effects of IL23R disruption using WT and IL23RKO mice aged 12 mo. We did not observe evidence of autoimmunity in IL23RKO mice compared to age-matched WT controls, as judged by the production of antinuclear antibodies (Fig. 4C) and comparisons of Peyer’s patches and lamina propria of WT, IL23RFlox FoxP3Cre, and IL23RKO mice aged 12 mo, which did not uncover alterations in their gut immune populations (Fig. 4 D and E). In contrast, mice carrying a Treg-specific Blimp deletion displayed impaired Treg function (24).
Synergy between Antibody-Mediated Targeting of IL23R and GITR.
Since the pathway leading to Treg destabilization following IL23R blockade is distinct from the Treg destabilization pathway initiated by agonistic GITR treatment (3, 4), we asked whether the combined targeting of both pathways might have a synergistic impact on Treg and improve tumor protection. We analyzed tumor growth after separate and combined administration of anti-IL23R and agonistic GITR antibody. The administration of IL23R or GITR antibodies alone slowed tumor growth, while the combined administration of lower doses of each antibody completely protected mice from tumor growth (Fig. 5A). We then carried out a dose–response analysis of each antibody alone and in combination. We observed reduced efficacy of both monotherapies compared to combined treatment, consistent with a synergistic effect (Fig. 5B). From this point onward, we used a larger tumor inoculum to allow characterization of the TME (Fig. 5C) and to investigate overall tumor survival (Fig. 5D). We observed a significant improvement in tumor clearance and survival in combination-treated animals compared to either monotherapy.
Fig. 5.
IL23R antibody treatment combined with GITR activation to promote tumor immunity. (A) Tumor growth curves in C57BL/6 mice treated intraperitoneally with anti-IL23R (100 μg/treatment), anti-GITR (100 μg/treatment), isotype controls (100 μg/treatment), or a combination of both IL23R and GITR antibodies (50 μg each/treatment), as in Fig. 4B. (B) Tumor inhibition of each antibody monotherapy and combination therapy, shown as a percentage of isotype-treated mice, where 0% inhibition indicates tumors that grew equal to isotype treatment and 100% inhibition indicates no tumor growth. (C) Tumor growth curves as in (A) using a higher dose (5 × 105) of MC38. (D) Survival analysis of high-dose–challenged mice as in (C). (E–H) Flow cytometry analysis of tumor-infiltrating Treg high-dose challenged mice including those with (E) Treg IFNγ expression, (F) Treg IL12Rβ2 expression, (G) CD8 T-cell IFNγ expression, and (H) CD8 T-cell Fas expression. Data are pooled from three independent experiments (n = 14 to 15/group). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, non significant. Iso, isotype.
Investigation of the TME at an early timepoint (to allow for tumor-infiltrating cells to be harvested from combination-treated animals) revealed a Treg destabilization and CD8 T-cell activation phenotype similar to that observed in Fig. 2, including enhanced IFNγ (Fig. 5E) and IL12R (Fig. 5F) expression by Treg and IFNγ expression by CD8 T cells (Fig. 5G). We also noted increased Fas expression by tumor-infiltrating CD8 T cells (Fig. 5H), with the highest Fas expression observed after GITR monotherapy, suggesting an induction of CD8 T-cell exhaustion. Although GITR monotherapy alone may activate Treg and CD8 IFNγ expression, as previously described (5–8), this evidence of CD8 T-cell exhaustion may explain the incomplete tumor immunity provoked by GITR monotherapy, compared to combination therapy. These findings support a synergistic protective mechanism achieved by combined GITR and IL23R treatment.
Discussion
Here we show that IL23R is a tumor-selective marker of suppressive Treg and that targeting it either alone or in combination with agonistic GITR treatment offers tumor protection that reflects increased IL12R-coupled STAT4 activation, IFNγ production by intratumoral Treg, and recruitment of activated CD8 T cells to growing tumors. This IL23R-dependent Treg destabilization pathway is distinct from GITR-linked inhibition of Helios-dependent STAT5 activation, leading to reduced FoxP3 expression by destabilized Treg (3, 4, 21). These considerations led us to determine whether the engagement of both the IL23R−STAT4 and GITR−STAT5 pathways might have a synergistic impact on Treg destabilization and tumor immunity, which proved to be the case (Fig. 5).
The observation that combined antibody targeting of IL23R and GITR provides synergistic protection against tumor growth may reflect a complementary impact on antitumor immune responses. We have shown previously that GITR activation destabilizes Treg through diminished Helios expression (3), resulting in reduced STAT5 activation and an impaired Treg suppressive phenotype (4). We have also noted that decreased Helios leads to a Th1-like phenotype and the expression of high-affinity, self-reactive T-cell receptors (4). Conversely, IL23R interference responds to environmental IL12 and leads to increased STAT4 activation by Treg, which can polarize the immune microenvironment toward type 1 immunity through the production of cytokines, including IFNγ. Since the TME that develops in IL23R-deficient mice is associated with enhanced effector CD8 T-cell activity (Fig. 3), we suggest that the activation of a STAT4−IFNγ pathway secondary to IL23R blockade may promote a type 1 immune microenvironment. It is likely that Treg → Th1-like conversion through a GITR−Helios−STAT5 pathway combined with changes in the TME secondary to the engagement of an IL23R−STAT4−IFNγ pathway may exert a synergistic impact on the TME, leading to increased CD4 and CD8 effector responses and more efficient tumor clearance.
This may also explain why IL23R monotherapy was less effective against high doses of MC38 in our experiments. Agonistic GITR antibodies directly destabilize Treg through the loss of Helios (3), while IL23R blockade sensitizes Treg to be destabilized by environmental signals such as IL12. Therefore, altering the tumor microenvironment by altering the tumor inoculum could be expected to have a stronger impact on IL23R monotherapy than on GITR monotherapy, which we observed. In addition, the Th1-supporting microenvironmental changes induced by IL23R monotherapy may be insufficient to keep pace with the more aggressive MC38 regimen unless it is supported by additional Treg destabilization and CD8 activation through IL23R and GITR combination therapy.
We have shown previously that Helios-deficient Treg induced by GITR treatment up-regulate the expression of the Il12rb2 gene but not Il12rb1 gene (4) (SI Appendix, Fig. S5). Increased IL12Rβ2 expression in GITR-treated Treg may not suffice for enhanced IL12R formation without a commensurate increase in IL12Rβ1 availability provided by IL23R blockade. Although IL23R blockade does not lead to increased Il12rb1 transcription, the release of the IL12Rβ1 receptor chain may promote high-affinity IL12 receptor expression when combined with free IL12Rβ2. Coordinate treatment with anti-GITR and anti-IL23R Abs yielding increased availability of both components of the high-affinity IL12 receptor may account for improved IL12 sensitivity of Treg in combination therapy compared to either monotherapy (Fig. 5F).
The combination approach summarized above leading to Treg destabilization and establishment of a type 1 immune microenvironment is relevant to ongoing attempts to harness Treg therapies in the clinic. Although GITR agonism is exceptionally effective at improving cancer outcomes in murine cancer models, it has been less impressive in human trials to date. Although ongoing clinical studies in human cancers may be more promising (8, 25), GITR monotherapy may induce activation as well as CD8 T-cell exhaustion in humans (8). This outcome is similar to our observation that although GITR monotherapy strongly induces CD8 T-cell IFNγ production, it also induces CD8 T-cell exhaustion (Fig. 5). In contrast, GITR/IL23R combination therapy induced robust CD8 T-cell IFNγ production by activated CD8 T cells that expressed lower levels of T-cell exhaustion markers. We hypothesize that the improved CD8 T-cell response to combination treatment reflects IFNγ production by intratumoral Treg that potentiates effector CD8 T-cell responses without substantial CD8 T-cell exhaustion.
Collectively, the mechanism of Treg destabilization described here in the context of tumor immunotherapy has broader implications for our understanding of both Treg and Th17 biology. The finding that IL23R blockade can influence IL12 signaling expands our understanding of the role of the receptor in Treg and Th17 biology. These findings open the possibility that the IL23−IL17 axis that regulates gut immunity may be sensitive to IL12 as well as IL23 targeting and suggest approaches that target the IL12R and IL23R in the context of inflammatory bowel disease and cancers. In view of the current unrealized potential of Treg destabilization secondary to antibody monotherapy, approaches that utilize complementary pathways to improve cancer outcomes through Treg targeting may open avenues of research into reshaping the tumor microenvironment through Treg intervention.
Materials and Methods
Animal Models and Cell Isolation.
C57BL/6 (WT), B6.129S1-Stat3tm1Xyfu/J (Stat3Flox), and B6.129(Cg)-FoxP3tm4(YFP/icre)Ayr/J (FoxP3Cre) mice were obtained from Jackson Laboratories. IL23RKO mice were a gift from Dr. Vijay Kuchroo. IL23RFlox mice were a gift from Dr. Philip Rosentiel. Stat3Flox FoxP3Cre mice were generated by interbreeding Stat3Flox and FoxP3Cre mice. IL23RFlox FoxP3Cre mice were generated by backcrossing IL23RFlox mice onto FoxP3Cre mice for 10 generations, then intercrossing F1 progeny to generate both IL23RFlox/Flox FoxP3Cre mice (IL23RFlox FoxP3Cre) and IL23RWT/WT FoxP3Cre mice (congenic controls). All animal husbandry and manipulations were performed under protocols approved by the Dana–Farber Cancer Institute (DFCI) Institutional Animal Care and Use Committee (IACUC) guidelines.
For in vitro analysis or flow cytometry, cells were isolated from spleens by mechanical dissociation. Tumor-infiltrating leukocytes were isolated by treatment with collagenase, dispase, and DNase I for 1 h at 37 °C, followed by mechanical dissociation and density centrifugation. Cells from the lamina propria and Peyer’s patches were isolated using the OctoMACS tissue dissociator (Miltenyi) following the manufacturer’s instructions.
Flow Cytometry.
For conventional cytometry, single-cell suspensions were prepared as above and stained using Zombie fixable viability dyes (Biolegend) in 1× PBS. Surface antibody staining using antibodies indicated in Table 1 was performed in 1× PBS supplemented with 1% FBS and 1 mM EDTA. Intracellular staining was performed using the FoxP3 Staining Buffer set (Thermo) following the manufacturer’s instructions. All samples were acquired on a Fortessa X20 (BD) and analyzed using the R statistical programming language. Samples for Treg sorting were first depleted of red blood cells, B cells, and CD8 T cells by magnetic selection (StemCell), then stained as above and sorted on an Aria III (BD). All sorted Treg came from mice with a FoxP3-YFP reporter, so Treg were sorted based on FoxP3 expression (SI Appendix, Fig. S1).
Table 1.
Antibodies used for flow cytometry
| Target | Fluorochrome | Source |
|---|---|---|
| CD45 | BV785 | Biolegend 103149 |
| TCRb | BV605 | Biolegend 109241 |
| CD4 | BUV395 | BD 563790 |
| F4/80 | FITC | Biolegend 123108 |
| CD11b | PerCP-Cy5.5 | Biolegend 108706 |
| FoxP3 | BV421 | Biolegend 126419 |
| CD8a | BV650 | Biolegend 100742 |
| B220 | BV711 | Biolegend 103255 |
| CD3 | BUV737 | BD 741788 |
| NK1.1 (CD161) | APC | Biolegend 108710 |
| CD25 | APC-Cy7 | Biolegend 101918 |
| CD62L | APC-R700 | BD 553219 |
| CD11c | PEDazzle594 | Biolegend 117348 |
| TCRgd | PE-Cy7 | Biolegend 118124 |
| pSTAT3 | APC | BD 560392 |
| pSTAT4 | AlexaFluor 647 | BD 558137 |
| IL12Rb1 | PE | R&D FAB1998P |
| CD6 | PE | Biolegend 146403 |
| CCR6 | PE | Biolegend 129804 |
| Fas (CD95) | PE | Biolegend 152607 |
| Ly108 | PE | Biolegend 134606 |
| GITR | PE | Biolegend 126310 |
| CD48 | PE | Biolegend 102707 |
| PD-1 | PE | Biolegend 135206 |
| CD54 | PE | Biolegend 116108 |
| IL23R | PE | Biolegend 150904 |
| TNF | PE | Biolegend 506306 |
| Perforin | PE | Biolegend 154406 |
| IL17A | PE | Biolegend 506904 |
| IL12Rb2 | PE | Miltenyi 130–125-221 |
| CD130 | PE | Biolegend 149404 |
| CD98 | PE | Biolegend 128208 |
| CD69 | PE | BD 553237 |
| Ki67 | PE | Biolegend 151209 |
| Blimp1 | PE | BD 564268 |
| IFNg | PE | Biolegend 505808 |
| IL10 | PE | Biolegend 505008 |
For high-dimensional cytometry, samples were stained as above for viability and surface dyes, then split into 24 wells and stained with a separate phycoerythrin (PE)-conjugated antibody as indicated in Table 1. Imputation was performed using the InfinityFlow R package following the developer’s instructions (26), except that output files were concatenated in silico and output events were set to 10,000 per well (240,000 events per imputed file). Concatenated cytometry files were analyzed using the CATALYST (27) and Seurat (28) R packages.
For phospho-STAT staining, single-cell suspensions were stained with fixable viability dye as above, then fixed with 1% paraformaldehyde and permeabilized in ice-cold 90% methanol for at least 10 min. Following fixation and permeabilization, cells were stained using surface and intracellular markers together in 1× PBS supplemented with 1% FBS and 1 mM EDTA.
In Vitro Treg Destabilization.
Single-cell suspensions were prepared as above and Treg were isolated by sorting YFP+ CD4 T cells from FoxP3Cre mice. Isolated Treg were cultured in complete RPMI supplemented with 5 ng/mL IL2, anti-CD3/CD28 beads (Miltenyi) according to the manufacturer’s instructions, and 10 μg/mL of the indicated antibody for 24 h in an incubator at 37 °C and 5% CO2.
Tumor Models.
MC38 colon carcinoma (ATCC) was cultured in complete RPMI and passaged when 70% confluent. Cells were expanded for eight passages and then frozen in multiple aliquots so that all experiments were performed with cells from the same passage number. For a given experiment, cells were thawed and expanded for 2 days, then harvested using trypsin and EDTA (Gibco) and washed twice in PBS before use in any experiments.
Flank tumors were induced by subcutaneous injection of 2 × 105 (unless otherwise indicated) MC38 cells in 100 μL 1× PBS. Mice were shorn and cleaned at the injection site to ensure good subcutaneous injection and to allow early detection of small tumors. Tumor growth was monitored every 72 h until palpable tumors were detected, then tumors were measured every 48 h with calipers. Tumor measurements were undertaken by a researcher blind to experimental conditions. Mice were humanely euthanized by CO2 inhalation when any measured diameter exceeded 16 mm or after reaching humane endpoints as defined by the DFCI IACUC. Tumor growth curve plotting and statistical analysis were performed using the R statistical programming language.
Treg RNA Sequencing.
Treg from IL23R-flox and congenic controls were sorted as described above and subjected to low-input RNA sequencing by GeneWiz. The resulting fasta files were aligned to the GRCm38 primary assembly and feature-counted using Rsubread (29). The resulting sample-wise gene expression table was analyzed using edgeR (30) with default parameters. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using pathfindR (31), using mmu KEGG as the input gene set. Manual annotation of Th1 and Treg gene functions among significant DEGs was performed by generating a list of all significant DEGs contributing to immune-relevant KEGG pathways, blinding their differential expression state, and then performing a literature search for their function in Treg or T cells. The differential expression status of all genes was only revealed after the literature searches were completed. The results of this literature search are presented in SI Appendix, Fig. S2.
Antibody Treatment.
MC38 flank tumors were induced as above. Once tumors were palpable on any mouse, mice were treated with three doses of indicated antibody every 48 h beginning when first palpated (100 μg/injection in 100 μL PBS unless otherwise indicated). The antibodies used were anti-IL23R clone 12B2B64 (Biolegend), anti-GITR clone DTA-1 (BioXCell), or Rat IgG2b isotype control (BioXCell). Mice were stratified into one of the four treatment groups randomly by initial tumor size (i.e., mice with the four largest tumors were randomly assigned each to a different antibody treatment, then the next four largest, and so on).
Autoantibody Enzyme-Linked Immunosorbent Assay.
Blood was collected from the saphenous vein of aged mice, according to IACUC-approved procedures. Antinuclear antibodies were measured using an ANA enzyme-linked immunosorbent assay kit (BioVision) and a plate-based spectrophotometer (BioRad). Analysis was performed using the R statistical programming language.
Statistical Analysis
Unless otherwise indicated, statistical analysis was performed using nonparametric, unpaired tests: the Wilcoxon rank-sum test or Kruskal–Wallis one-way test followed by Wilcoxon posthoc tests with Holm correction for multiple comparisons. P values are indicated by asterisks: *P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001. For high-dimensional flow analysis, statistical analysis was performed using the Seurat package with default options, after constructing a single-cell sequencing object using CATALYST, with options set for flow cytometry data instead of the CyToF defaults, as indicated by the developers. For tumor growth curve analysis, statistical comparisons were made on the area under the curve of each tumor curve. Tumor growth is plotted as mean ± SEM, with a locally estimated scatterplot smoothing (loess) regression line showing the imputed continuous growth rate.
Supplementary Material
Acknowledgments
We thank V. Kuchroo and P. Rosentiel for the provision of key mouse models, A. Luoma and and F. Alvarez-Calderon for flow cytometry support, the DFCI Animal Resource Facility for veterinary support, E. Becht for technical support with InfinityFlow, and A. Angel for assistance with manuscript preparation and editing. This work was supported in part by research grants from the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH under award numbers R01AI037562 and R01AI048125 (H.C.); the U.S. Army Medical Research and Materiel Command (W81XWH) (H.C.) and a DFCI Accelerator Award (H.C.); the AAI Intersect Fellowship and Claudia Adams Barr Program in Innovative Basic Cancer Research (A.E.W.); the NRSA Fellowship (NIH/NIAID T32AI074549) (J.M.S.); and the CRI-Irvington Fellowship (H.N.).
Footnotes
Reviewers: T.H., Rheinische Friedrich-Wilhelms-Universitat Bonn; and K.K., Mayo Clinic Minnesota.
Competing interest statement: Author H.C. and reviewer T.H. were listed as coauthors in 2020 (PMC7031256), but this does not represent a financial or personal competing interest, since the contribution of H.C. was limited to the provision of a mutant mouse model in 2013.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200757119/-/DCSupplemental.
Data Availability
Raw and processed RNA-Seq data have been deposited in Gene Expression Omnibus (Accession number GSE197287). All other data pertaining to the paper are presented here. Code for the analysis of flow cytometry data in R is available at GitHub, https://github.com/NKInstinct/flowGate.
All study data are included in the article and/or SI Appendix.
References
- 1.Shimizu J., Yamazaki S., Sakaguchi S., Induction of tumor immunity by removing CD25+CD4+ T cells: A common basis between tumor immunity and autoimmunity. J. Immunol. 163, 5211–5218 (1999). [PubMed] [Google Scholar]
- 2.Sun J.-Y., et al. , Resistance to PD-1/PD-L1 blockade cancer immunotherapy: Mechanisms, predictive factors, and future perspectives. Biomark. Res. 8, 35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakagawa H., et al. , Instability of Helios-deficient Tregs is associated with conversion to a T-effector phenotype and enhanced antitumor immunity. Proc. Natl. Acad. Sci. U.S.A. 113, 6248–6253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yates K., Bi K., Haining W. N., Cantor H., Kim H.-J., Comparative transcriptome analysis reveals distinct genetic modules associated with Helios expression in intratumoral regulatory T cells. Proc. Natl. Acad. Sci. U.S.A. 115, 2162–2167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coe D., et al. , Depletion of regulatory T cells by anti-GITR mAb as a novel mechanism for cancer immunotherapy. Cancer Immunol. Immunother. 59, 1367–1377 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahne A. E., et al. , Dual roles for regulatory T-cell depletion and costimulatory signaling in agonistic GITR targeting for tumor immunotherapy. Cancer Res. 77, 1108–1118 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Amoozgar Z., et al. , Targeting Treg cells with GITR activation alleviates resistance to immunotherapy in murine glioblastomas. Nat. Commun. 12, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zappasodi R., et al. , Rational design of anti-GITR-based combination immunotherapy. Nat. Med. 25, 759–766 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langowski J. L., et al. , IL-23 promotes tumour incidence and growth. Nature 442, 461–465 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., et al. , IL-23 selectively promotes the metastasis of colorectal carcinoma cells with impaired Socs3 expression via the STAT5 pathway. Carcinogenesis 35, 1330–1340 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Caughron B., Yang Y., Young M. R. I., Role of IL-23 signaling in the progression of premalignant oral lesions to cancer. PLoS One 13, e0196034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parham C., et al. , A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 168, 5699–5708 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Bloch Y., et al. , Structural activation of pro-inflammatory human cytokine IL-23 by cognate IL-23 receptor enables recruitment of the shared receptor IL-12Rβ1. Immunity 48, 45–58.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwakura Y., Ishigame H., The IL-23/IL-17 axis in inflammation. J. Clin. Invest. 116, 1218–1222 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duvallet E., Semerano L., Assier E., Falgarone G., Boissier M.-C., Interleukin-23: A key cytokine in inflammatory diseases. Ann. Med. 43, 503–511 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Chan I. H., et al. , Interleukin-23 is sufficient to induce rapid de novo gut tumorigenesis, independent of carcinogens, through activation of innate lymphoid cells. Mucosal Immunol. 7, 842–856 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Liu J., Wang L., Wang T., Wang J., Expression of IL-23R and IL-17 and the pathology and prognosis of urinary bladder carcinoma. Oncol. Lett. 16, 4325–4330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao Y., et al. , Combined effect of IL-12Rβ2 and IL-23R expression on prognosis of patients with laryngeal cancer. Cell. Physiol. Biochem. 50, 1041–1054 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Yan J., et al. , Experimental lung metastases in mice are more effectively inhibited by blockade of IL23R than IL23. Cancer Immunol. Res. 6, 978–987 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Liu T., Soong L., Liu G., König R., Chopra A. K., CD44 expression positively correlates with Foxp3 expression and suppressive function of CD4+ Treg cells. Biol. Direct 4, 40 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H., et al. , Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science 350, 334–339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aden K., et al. , Epithelial IL-23R signaling licenses protective IL-22 responses in intestinal inflammation. Cell Rep. 16, 2208–2218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awasthi A., et al. , Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J. Immunol. 182, 5904–5908 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen E., et al. , Control of germinal center localization and lineage stability of follicular regulatory T cells by the Blimp1 transcription factor. Cell Rep. 29, 1848–1861.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papadopoulos K. P., et al. , Phase 1 study of MK-4166, an anti-human glucocorticoid-induced tumor necrosis factor receptor (GITR) antibody, as monotherapy or with pembrolizumab (pembro) in patients (pts) with advanced solid tumors. J. Clin. Oncol. 37, 9509 (2019). [Google Scholar]
- 26.Becht E. . et al., High-throughput single-cell quantification of hundreds of proteins using conventional flow cytometry and machine learning. Sci. Adv. 7, eabg0505 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowicka M., et al. , CyTOF workflow: Differential discovery in high-throughput high-dimensional cytometry datasets. F1000 Res. 6, 748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao Y., et al. , Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Y., Smyth G. K., Shi W., The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulgen E., Ozisik O., Sezerman O. U., pathfindR: An R package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front. Genet. 10, 858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed RNA-Seq data have been deposited in Gene Expression Omnibus (Accession number GSE197287). All other data pertaining to the paper are presented here. Code for the analysis of flow cytometry data in R is available at GitHub, https://github.com/NKInstinct/flowGate.
All study data are included in the article and/or SI Appendix.