Significance
We successfully identified the duper allele as a null mutation of Cryptochrome 1 in Syrian hamsters. Here, we have shown the use of fast homozygosity mapping as an effective approach to identify causal mutations in mammals, despite lacking chromosomal genome information. In the course of this work, we improved the draft Syrian hamster genome and generated datasets necessary to exploit Syrian hamsters as a modern genetic research model. The unique physiological features of Syrian hamsters make them a desirable model to investigate human diseases, including circadian disorders, cancer, heart function, metabolism, and infectious diseases (e.g., severe acute respiratory syndrome coronavirus 2).
Keywords: fast homozygosity mappings, Cry1-null, short circadian period length, Syrian hamster genome
Abstract
The duper mutation is a recessive mutation that shortens the period length of the circadian rhythm in Syrian hamsters. These animals show a large phase shift when responding to light pulses. Limited genetic resources for the Syrian hamster (Mesocricetus auratus) presented a major obstacle to cloning duper. This caused the duper mutation to remain unknown for over a decade. In this study, we did a de novo genome assembly of Syrian hamsters with long-read sequencing data from two different platforms, Pacific Biosciences and Oxford Nanopore Technologies. Using two distinct ecotypes and a fast homozygosity mapping strategy, we identified duper as an early nonsense allele of Cryptochrome 1 (Cry1) leading to a short, unstable protein. CRY1 is known as a highly conserved component of the repressive limb of the core circadian clock. The genome assembly and other genomic datasets generated in this study will facilitate the use of the Syrian hamster in biomedical research.
Biological clocks coordinate the timing of biochemical, physiological, and behavioral processes. In mammals, the circadian clock is regulated by a transcriptional-translational negative feedback loop (TTFL) (1). This TTFL drives circadian oscillations of many other clock-regulated genes and results in a gene network that oscillates with a 24-h cycle. Disruption of circadian rhythms is associated with many diseases, including metabolic syndrome, susceptibility to infections, and cancers (2).
tau was one of the first mammalian period length mutants identified (3). This allele shortens the free running period of locomotor activity under constant dark conditions (τ) to ∼22 h in heterozygote and 20 h in homozygote (“supershort”) animals. tau was identified as a gain of function mutation in a key clock protein kinase, casein kinase 1 epsilon (CKIε), which results in hyperphosphorylation and accelerated degradation of the PER2 protein (4–6). As a consequence, the repressive phase of the TTFL is abbreviated, shortening the period length of locomotor activity.
In prior work, a spontaneous mutation (duper) was identified on the homozygous tau background (7). These animals had a τ of approximately 18 h and a greatly expanded range of entrainment. To isolate duper from the tau locus, these animals were backcrossed to the Lakeview (LVG) ecotype. The free running period of locomotor activity rhythms for homozygous duper animals was 22.8 h. duper heterozygotes had a normal τ, suggesting duper is a recessive mutation (7). In contrast to tau mutants, duper hamsters maintain a high amplitude phase response curve (8, 9).
The limited genetic resources available for Mesocricetus auratus (Syrian or Golden hamsters) presented a major obstacle to cloning duper. This caused the duper mutation to remain unknown for over a decade. In 2000, the tau mutation was identified using genetically directed representational difference analysis (GDRDA) (4, 10). GDRDA identifies the polymorphic DNA markers that are tightly linked to a monogenic trait through comparison of “tester” and “driver” pools. GDRDA is a powerful method because it does not need prior knowledge of the chromosomal location of that trait or the availability of genetic maps for the organism. However, GDRDA is a complex and arduous task, especially for a recessive mutation (11).
Fast homozygosity mapping (FHM), a mapping-by-sequencing approach, has been widely applied to screen for recessive mutations (including complex traits) in zebrafish and plants using whole-genome sequencing (12–14). To apply FHM, individuals with the mutation are first crossed to an individual with a distant genetic background. The principle behind FHM is that a fraction of the genome of offspring with the phenotype would be expected to be homozygous, and linked to the phenotypic parental genetic background. Pooling DNA from a large cohort of phenotypic offspring will help to further segregate the mutations and localize the causal variants.
A good genome assembly for the species is needed for the FHM approach. Efforts to build a reference genome for the Syrian hamster started recently (15, 16) because this species is an excellent model to study many types of cancers, viruses, and heart disease. The contiguity of the genome assembly is important to map the allele using FHM. We used Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) long-read sequencing to reassemble the Syrian hamster genome and improve its contiguity. Using the newly assembled reference genome, we successfully applied FHM to identify a ∼10.6 Mbp genomic region linked to duper. Further analysis revealed that duper is a Cryptochrome 1 (CRY1) null mutation of Syrian hamsters, caused by a frameshift deletion in exon 4 that leads to an early stop codon. CRY1 is known as a highly conserved component of the core circadian clock (17). Our study identified duper as a CRY1-null Syrian hamster. Tools and datasets generated through the course of this work will help to facilitate the use of this species as a modern genetic animal model to study circadian, metabolic, infectious, and cardiovascular diseases.
Results
duper Allele Isolation and Characterization.
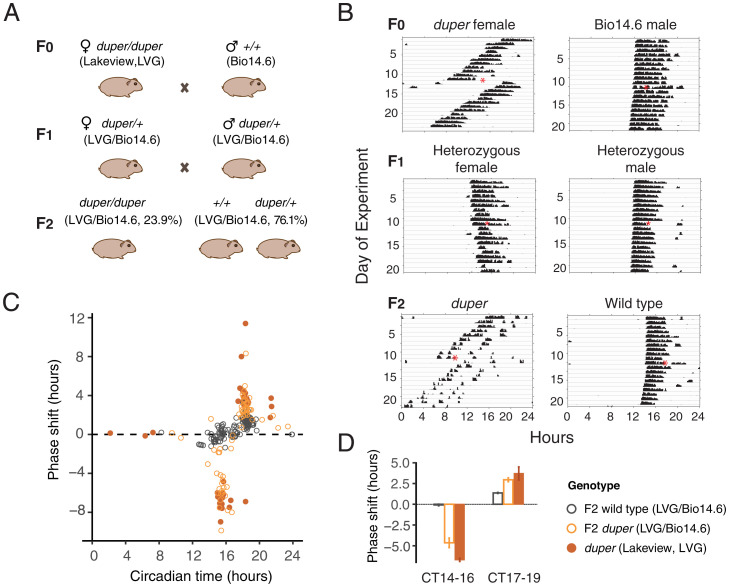
duper arose on the homozygous tau background. A female duper hamster (LVG ecotype) was crossed with a wild-type male hamster (Bio14.6 ecotype) to move the duper allele to a different genetic background (Fig. 1A). The paternal line arose as a spontaneous mutation and was maintained for over 50 y by BioBreeders (Fitchburg, MA) as a separate lineage for studies of cardiomyopathy and muscular dystrophy resulting from deletion of d-sarcoglycan (18). Similar to previous observations (7), the duper phenotype transmitted as a recessive mutation in this cross. The τ of the F1 heterozygotes was ∼24 h, similar to the period of wild-type hamsters (Fig. 1B). We inbred the F1 heterozygotes and generated F2 animals (n = 197). F2 hamsters were phenotypically separated using the period length of their free-running locomotor activity. In total, 47(23.9%) hamsters showed the short τ (duper) phenotype, with a mean period of 22.73 ± 0.1 h (mean ± SEM). The remaining F2 hamsters had a mean period of 24.04 ± 0.01 h.
Fig. 1.
Isolation and characterization of duper allele in Syrian hamsters. (A) Mating scheme for fast homozygosity mappings. (B) Actograms of duper and wild type hamsters. Red asterisks indicate when a 15′ light pulse was given. (C) Phase response curve established the response to 15′ light pulses. F2 wild-type (LVG/Bio14.6), F2 duper (LVG/Bio14.6), and duper (LVG) are shown as gray open circles, orange open circles, and filled orange points, respectively. (D) Bar graph show phase delays and advances in response to 15′ light pulse presented at CT14-16 and CT17-19, respectively (mean ± SEM).
To map the phase response of the F2 hamsters, animals maintained in DD were exposed to a 15-min (15′) light pulse in the early or late subjective night. Hamsters with the duper mutation showed a mean phase delay of 4.64 ± 0.57 h when given a light pulse at CT15.29 ± 0.08 h. When these hamsters were given a 15′ light pulse at CT18.27 ± 0.06 h, they showed a mean phase advance of 2.95 ± 0.24 h. In contrast, wild-type F2 hamsters given light pulses at CT15.17 ± 0.08 h showed a phase delay of 0.08 ± 0.10 h. Light pulses at CT18.36 ± 0.05 h elicited advances of 1.36 ± 0.07 h (Fig. 1 C and D). Thus, responses of the F2 duper hamsters to 15′ light pulses were comparable to those of the F0 duper hamsters of the LVG background. The duper phenotype transmits without apparent modification on the Bio14.6 background.
Reassembly of the Syrian Hamster Reference Genome.
FHM identifies genetic regions that are homozygous and conserved in the F2 offspring that match to the phenotypic parental genetic background. The contiguity of the assembly is important to successfully map the region, because longer scaffolds of the reference genome allow us to identify genomic regions linked to the phenotype with higher confidence. The first reference genome of the Syrian hamster (MesAur1.0) was published in 2013 using short sequencing reads. This genome assembly has 21,483 scaffolds and 237,699 contigs, with N50 of 12,753,307 and 22,512, respectively. This assembly is fragmented, with many gaps and missing information at the low complexity region. To improve the structure of the reference genome, we used PacBio (Sequel System) and ONT (GridION) to generate long-reads and then reassembled the reference genome of the Syrian hamster (Table 1). Our reference genome greatly reduced the total contigs and scaffolds to 3,056, with a N50 of 26,783,296. Benchmarking Universal Single-Copy Orthologs (BUSCO) assessments with the 9,226 mammalian markers (19), the estimated completeness of this assembly was 88.2%.
Table 1.
Statistics of the de novo genome assembly
| Parameters | Measurements |
|---|---|
| Contigs and scaffolds | 3,056 |
| Total length (bp) | 2,500,764,195 |
| N50/N90 (bp) | 26,783,296/2,215,234 |
| GC% | 41.8% |
| BUSCO | 88.2% |
Fast Homozygosity Mapping Identifies the duper Allele in Syrian Hamsters.
Genomic DNA was isolated from a duper (LVG) animal, a wild-type (Bio14.6) animal, and a pool of 45 phenotypically selected F2 homozygous duper mutants (LVG/Bio14.6). Whole-genome sequencing was performed using paired-end Illumina sequencing. Read alignment and variant calling were done following GATK best practices (20). Variants with at least 5× total coverage were used for further analysis.
FHM used unique homozygous single nucleotide polymorphisms (SNPs) from either parent as markers to identify genomic regions in the F2 phenotypic offspring that linked to the parental genetic background. We identified 215,699 and 801,403 unique homozygous SNPs in wild type (Bio14.6) and duper (LVG), respectively (Table 2). The newly assembled reference genome was generated from a Syrian hamster from the LVG ecotype. Its genetic distance is nearer to the duper (LVG) background in comparison to wild type (Bio14.6). Therefore, the number of SNPs called in wild type (Bio14.6) is 3-fold more than duper. The curated markers have a higher density of Bio14.6 SNPs such that it will dominate the homozygosity score calculation.
Table 2.
Number of unique homozygous SNPs in the two Syrian hamster ecotypes
| Ecotypes | No. of unique homozygous SNPs |
|---|---|
| Lakeview (F0 duper/duper) | 215,699 |
| Bio14.6 (F0 +/+) | 801,403 |
We modified the scoring approach to reduce the impact of imbalanced SNPs from the parental background. Instead, we scanned through all SNPs (both homozygous and heterozygous) from F2 duper pools with coverage larger or equal to 5 (n = 1,232,905) and assigned different weights to these SNPs. Detailed scoring criteria is described in the Material and Methods. We calculated the average homozygosity score in a rolling 100 SNPs window. Regions with homozygosity score >0.9 and <0.1 suggested the region is highly conserved in the F2 duper pools and were linked to LVG and Bio14.6, respectively (Fig. 2A).
Fig. 2.
Identification of genomic regions linked to duper. (A) Schematic diagram representing the principle underlying homozygosity mapping. (B) Fast homozygosity mapping identified two regions that have high homozygosity scores and was linked to the duper LVG ecotype (brown and light brown). (C) Both LVG-linked homozygous regions mapped to 10qC1-C2 in the mouse genome (mm10). Cry1 (red) is the only known clock gene of the TTFL loop found in this region.
Using this approach, we identified two highly conserved homozygous regions (∼4.8 and 5.8 Mbp, respectively) in the F2 duper pools inherited from the LVG background (Fig. 2B). The Syrian hamster reference genome still lacks chromosomal-level information. For that reason, we used the Basic Local Alignment Search Tool (BLAST) to map these two regions to the mouse genome. Interestingly, both regions mapped near each other on mouse chromosome 10 (build mm10) at 10qC1-C2 (Fig. 2C). Given these findings, we reasoned that the duper locus resides in this ∼10.6 Mbp genomic region.
We restricted our search for candidates to unique duper variants (both SNPs and INDELs). These candidates represent variants found in F0 and F2 duper but not in our mapping ecotype, Bio14.6. We annotated these variants with SnpEff (21) and filtered out intergenic, intronic, and synonymous variants (Fig. 3A). This identified 15 variants to have high- or moderate-putative impact. We manually screened these variants and filtered out those from low confidence genomic regions. After applying these filters, four candidate variants remained (Fig. 3A). Among these candidate variants, Cry1 is the only gene known to encode a core clock protein (1). A 1-bp frameshift deletion in exon 4 of Cry1 (c.578delC; p.P193fs) was predicted to cause an early stop codon in the CRY1 protein (Fig. 3B). In duper, the 1-bp deletion was validated using Sanger sequencing. This mutation, associated with the duper phenotype, was not found in the wild-type hamsters (Fig. 3C).
Fig. 3.
Identification of the duper allele. (A) Filtering process to identify candidate variants of duper alleles. One high-putative impact variant (Cry1) and 3 moderate-putative impact variants remained after the filtering process. (B) The p.Pro193fs mutation in Cry1 was predicted to cause an early stop codon in the CRY1 protein in duper. (C) Sanger sequencing validation of the 1bp deletion in duper.
CRY1 Protein Is Not Expressed in duper Hamsters.
The duper allele is predicted to cause an early stop codon in Cry1 in Syrian hamsters. We performed immunocytochemistry using an antibody directed at the N-terminal region of CRY1 to determine whether protein is expressed in brain regions of Syrian hamsters. We observed a profound reduction of CRY1 staining in duper relative to wild-type hamsters at ZT12 in both suprachiasmatic nucleus (SCN) and piriform cortex (Fig. 4A). Next, we performed Western blot analysis using an N-terminal CRY1 antibody on whole cell lysates of liver tissues collected at ZT3, ZT7, ZT11, ZT15, ZT19, and ZT23. The predicted truncated protein is 22.5 kDa, while the full size CRY1 is 66.3 kDa. We detected full size CRY1 proteins in wild-type animals, but detected neither the full size nor the truncated CRY1 protein in duper animals (Fig. 4B). Taken together, these data suggest that duper is indeed a CRY1-null in hamsters.
Fig. 4.
Validation of CRY1-null in the duper hamsters. (A) Representative immunocytochemistry images of duper and wild-type hamster brain sections from ZT12 containing SCN and piriform cortex. (Scale bar: 100 μm.) Bar plot indicates the quantifications of CRY1 signal intensity in SCN and piriform cortex, respectively. (B) Western blotting was performed on total protein extracts with N-terminal CRY1 antibody in wild-type and duper hamsters from liver tissues collected at ZT3, ZT7, ZT11, ZT15, ZT19, and ZT23. Normalized levels of the full size CRY1 protein (66.3 kDa) are shown in the plot. Significance of the rhythmicity test was performed using CircaCompare and represented with solid dark gray line (P = 0.075) and dotted orange line (P = 0.148).
Commercially available hamster-specific antibodies are very limited. In addition to CRY1, we were able to detect three other core clock proteins (PER1, PER3, and CRY2) in hamsters using these antibodies (SI Appendix, Fig. S1 and Table S1). While total protein levels do not show a significant difference, we observed an ∼6 h phase advance in PER3 abundance in the liver of duper animals (CircaCompare, P = 0.004). No significant changes were observed between duper and wild type in the time course of expression of PER1 or CRY2 in the liver.
We also compared genes expressed in three tissues (cortex, SCN, and liver, RNA sequencing [RNA-seq]) between a pool of four duper and four wild-type hamsters collected at single time point (CT15). As expected, expression levels of Cry1 were significantly reduced in duper animals (SI Appendix, Fig. S2). In accordance with Western blot results, Per3 expression was reduced in the liver at CT15. However, similar changes were not observed in the SCN and cortex, suggesting the changes of PER3 might be liver specific. No significant changes were observed for other core clock genes at the transcriptional level at CT15.
Additionally, we identified 39 genes differentially expressed in the three tested tissues in duper (SI Appendix, Fig. S3 and Dataset S1). Several of these genes are not well-characterized in the Syrian hamster genome. We selected the 15 well-annotated differentially expressed genes for pathway enrichment analysis. Five of these genes, including Nat2, Cryl1, Alas2, Aldh1a3, and Gapdh, are enriched in metabolic pathways (P = 0.09). Two of them, Irf7 and Isg15, are enriched in RIG-I-like receptor signaling pathways (P = 0.09). Further experiments with time-series tissue collections and more technical replicates would be helpful to elaborate transcriptomic changes of duper, a CRY1-null hamster model.
Discussion
We identified the underlying mutation of duper as a frameshift deletion in exon 4 of Cry1 that generates a premature stop codon, resulting in the absence of CRY1 protein. Cry1 plays a prominent role in regulating the period length of locomotor activity in mice. Studies in CRY1-null mice found shortening of τ similar to duper (22). A large-scale N-ethyl-N-nitrosourea mutagenesis screen in mice identified a mutant termed part-time with a short period circadian locomotor activity phenotype. This mutation was mapped and found to be a nonsense mutation at the 3′-exonic region of exon 2 in Cry1 (23). In humans, the c.1657 + 3A > C allele results in skipping of exon 11 in CRY1 (CRY1 Δ11) and segregates with delayed sleep-wake phase disorder. In contrast to the loss-of-function CRY1-null, CRY1 Δ11 represents a gain-of-function mutation. Exon 11 plays an important role in the interaction between the CRY1 tail and its photolyase homology region. CRY1 Δ11 results in enhanced interactions with CLOCK:BMAL1 proteins and increased inhibition of transcription for the CLOCK:BMAL1 targeted genes (24, 25)
Other than shortening τ, we also observed a dramatic shortening of the latency to re-entrain upon 8 h or 12 h shifts of the light:dark (L:D) cycle in duper hamsters compared to wild-type. Similar responses are observed in Cry1-null mice upon 6 h shifts of the L:D cycle (26). Additionally, duper hamsters show a high amplitude phase response curve, with shifts of up to 12 circadian hours in response to a 15′ light pulse (Fig. 1C) (8). Such a marked increase in amplitude of the PRC has not been reported in CRY1-null mice (27).
Notable evolutionary differences exist in light entrainment. Drosophila cryptochrome is a flavin adenine dinucleotide (FAD)-dependent circadian photoreceptor, whereas mammalian cryptochromes (CRY1 and CRY2) are integral clock components that repress CLOCK/BMAL1-dependent transcription (28). Interestingly, while cryptochromes play a role in light entrainment in flies, in mammals, it functions as the critical repressive component of the TTFL. Recent studies suggest that the FAD pocket of mammalian CRY2, but not CRY1, retains some function and possibly works in a light-dependent manner (29). A mutation in the CRY2 FAD binding domain, p.A260T, was segregated with familial advanced sleep-wake phase disorder. This mutation promotes FBXL3 binding and therefore increased degradation of CRY2. The humanized p.A260T CRY2 transgenic mice show a subtle shortened period and altered sensitivity to photic entrainment (30). This suggests CRYs play an important role in both the period of the circadian clock and light-induced phase shifting in humans. duper may provide a unique model for light entrainment studies.
duper was segregated from a spontaneous mutation that arose on the homozygous tau hamster background. Interestingly, the combination of duper and tau mutations in hamsters (termed “super duper”) showed significant shortening of τ to ∼18 h, which is the shortest circadian period of locomotor activity so far recorded in mammals (9). The interaction between mutations affecting PER and CRY was described in studies of mice carrying both the tau and afterhours mutations. tau mutations lead to gain of function CKIε that hyperphosphorylates PER proteins and destabilizes them, leading to a shortened repressive phase. Afterhours is caused by a mutation in FBXL3 that stabilizes CRY proteins and lengthens the period. The circadian effects of Fbxl3Afh and CKIεtau were independent and additive (31). This contrasts with the phenotype of the super duper hamsters, in which the combination of the CKIε gain-of-function with the effective loss of CRY1 results in an epistatic interaction: the shortening of period in duper (~6 h) is more than additive of CKIε (~4 h) and CRY1-null (~1 h). We hypothesize that super duper hamsters have hyperphosphorylated PER2 that accelerates nuclear entry of PER:CRY2 complexes to initiate the early repressive phase. In the absence of CRY1, the late repressive phase may be truncated or abbreviated in duper animals. Since CRY proteins prevent ubiquitylation of PER2 (32), PER may be further destabilized in the super duper hamsters. Further exploration of the mechanism by which tau and duper mutations interact, not only to shorten period but also to amplify phase resetting, will provide further insights into interactions of components of the TTFL.
Beyond its importance as a repressor of the TTFL in circadian clocks, CRY1 has been found to regulate metabolic oscillations (33, 34) and participate in DNA damage response (35) and cancer (36). Surprisingly, we found no publicly available CRY1-null transcriptomics datasets in any animal models to date. Using our RNA-seq data, we sought to explore the possible functional consequences of CRY1-null and circadian alteration in hamsters at CT15.
RNA expression profiling showed alterations of the RIG-I-like receptor signaling pathway in duper, which may link the CRY1-null duper animals to the viral sensing response. Both the enriched genes, Irf7 and Isg15, are not rhythmic in any tissues in mice (37), suggesting that the observed changes are not likely due to phase shifts in the animals. Uba7, the gene encoding enzyme UBE1L that activates ISG15 protein conjugation (38), was also up-regulated in duper animals. We cross-checked changes of these three genes with a published CRY1 knockdown RNA-seq dataset (GSE144961) (36). Similar to duper animals, these three genes are up-regulated in CRY1 knockdown in human cells, albeit with more subtle changes (SI Appendix, Fig. S4). The interplay between the circadian clock and viral infection has been previously reported (39). ARNTL-null mice were found to be more susceptible to viral infections, however, all these studies explored responses to viral infections in arrhythmic animals (40–42). Higher viral replication is observed at ZT0 in comparison to ZT10 (40). This aligns with the rhythmicity of CRY1 protein levels, where CRY1 peaks at ZT0 and reaches a nadir at ZT10. Whether Cry1 inhibits (directly or indirectly) the RIG-I-like receptor signaling pathway, in particular IRF/ISG transcription, remains to be explored.
A significant decrease in Nat2 in the SCN and cortex is observed in our transcriptomics data. Although AANAT is thought to be the rate limiting enzyme for melatonin synthesis in the pineal gland (43), changes in Nat2 expression may contribute to changes in circadian and seasonal responses in duper animals.
Our pilot RNA-seq datasets do not conclusively represent CRY1-null transcriptomics profiles in hamsters. First, we only tested a single time point (CT15). Second, we only tested three tissues without replicates. We did stringent analysis and found 15 well-annotated genes altered in all three tissues analyzed (cortex, SCN, and liver), 8 of which are enriched in two specific pathways: RIG-I-like receptor signaling and metabolic pathways. This observation suggests the need for future exploration of duper, a CRY1-null model. Increasing temporal resolution and replicates for each tissue will help to better characterize the transcriptomic profiles of the animals.
The utility of Syrian hamsters has declined since the early 2000s, largely due to the lack of genetic and molecular resources available. In the past few years, long-read sequencing has facilitated the effort to build high quality genome assemblies for other mammals (44). In this study, we used long-read sequencing from PacBio and ONT to improve the scaffolding and contiguity of the Syrian hamster genome. Using this assembly and the prior knowledge of the mouse genome, we successfully applied FHM to identify the duper locus. In the course of this work, we also sequenced cDNA collected from three hamster ecotypes, including the LVG, Bio14.6, and hamsters descended from a few generations wild caught (SI Appendix, Fig. S5) (45). Tools and data generated in this study expand the genetic resources available for Syrian hamsters.
The unique physiological features of the Syrian hamster make it a desirable model to investigate human diseases, including cancer, heart function, metabolism, and infectious diseases (e.g., severe acute respiratory syndrome coronavirus 2) (46, 47). Furthermore, hamsters offer a superior model for study of seasonal and photoperiodic responses (48). Future work to complete and annotate the reference genome for Syrian hamsters will enable its exploitation as a modern genetic model for biomedical research.
Materials and Methods
Animal Models.
All Syrian hamsters (M. auratus) were born and raised in a 14:10 L:D cycle and allowed ad libitum access to food and water throughout the study. Wild-type hamsters of the Lakeview (LVG) ecotype were obtained from Charles River Laboratories (Wilmington, MA) or bred in our laboratory from that stock. duper hamsters used in these studies were derived from the original super duper animals crossed to LVG as previously described (7). They descended from animals that were confirmed by restriction digest mapping of their genomic DNA (4, 7) to lack the tau mutation and in which τ was ∼23 h. The Bio14.6 hamsters were obtained from BioBreeders (Farmington, MA). All experiments were approved by the IACUC of the University of Massachusetts Amherst (Protocol 2013–0076).
Light Pulse and Phase Response Curve.
Circadian phenotype was assessed by maintenance in tub cages containing running wheels; locomotor activity rhythms were assessed using Actimetrics software as previously described (8). The phenotype of the parental (F0) Bio14.6 and duper hamsters and of their F1 offspring was evaluated by quantification of free running period over the course of at least 10 d in DD and response to 15′ light pulses administered in early to midsubjective night. In order to confirm the phase shifting phenotype of the F2 animals, activity rhythms of 79 hamsters were assessed in DD and phase shifts were quantified after 15′ light pulses at approximately CT15.5 and CT18. To determine whether the duper phenotype was transmitted on the Bio14.6 background, we constructed a phase response curve to compare shifts elicited in dupers on wild type (LVG) background with the F2 animals. In all cases, the phase shift was determined by linear regression of activity onsets upon establishment of steady state in the 10 d following the light pulse.
Long-Read Sequencing, De Novo Genome Assembly, and Annotation.
DNA was extracted from whole blood obtained from a female wild-type LVG outbred golden Syrian hamster (M. auratus). Genomic DNA libraries were prepared for sequencing on two different long-read platforms, Oxford Nanopore Technologies (GridION; LSK109) and Pacific Biosciences (Sequel System; SMRTbell Express Template Prep Kit 2.0). Prior to sequencing, the Oxford Nanopore library was processed with the Short Read Eliminator XL Kit (Circulomics) following manufacturer’s directions. One single-molecule real-time (SMRT) cell and four MinION flow cells generated the entirety of data used in the assembly of the Syrian Hamster genome. Oxford Nanopore adapters and reads containing evidence of internal adapters were removed with Porechop v0.2.4 (https://github.com/rrwick/Porechop). The remaining reads were then filtered with Filtlong v0.2.0 (https://github.com/rrwick/Filtlong), requiring a minimum read QV of 9 and length of 5 Kb. Flye v2.7 (49) was used to generate a de novo assembly of the filtered nanopore reads requiring a minimum of 8 Kb overlap among reads. Error correction was performed independently with gcpp v2.0.2 (https://github.com/PacificBiosciences/gcpp) using pbmm2 v1.1.0 alignments (https://github.com/PacificBiosciences/pbmm2) of Pacific Biosciences subread data to the draft assembly. The resulting assembly of error-corrected contigs was assessed for completeness using lineage-specific single-copy orthologs (mammalia_odb10) with BUSCO v4.0.6 (19). Liftoff v1.6.1 (50) was used to annotate the draft assembly with BCM_Maur_2.0 (GCF_017639785.1) serving as the reference annotation source. All portions of the workflow were performed by the University of Wisconsin–Madison Biotechnology Center.
Whole-Genome Sequencing, Genome Alignment, and Variants Calling.
Genome DNA was prepared from F0 male (Bio14.6), F0 female duper (Lakeview) hamster and a pool of phenotypically identified F2 duper offspring (n = 45). Libraries from these three samples were pooled and split equally into three lanes of the flow cells. Whole-genome sequencing was run with 2 × 150 bp sequencing on the Illumina 4000 platform. Sequenced data were aligned to the newly assembled Syrian hamster genome assembly mentioned above, using Burrows-Wheeler Aligner (51) (v0.7.17) with the “bwa mem” command. BAMs from the same samples were merged using samtools (52) (v1.9.0) merge. Duplicate reads were removed and base quality were recalibrated as suggested in the best-practice using GATK (20) (v4.1.2). HaplotypeCaller (53) from GATK were used to call variants in each sample for each individual contig separately. All SNPs/INDELs from the same samples were merged to a vcf file using MergeVcf and filtered with parameter suggested by the best practice guidance of GATK.
Fast Homozygosity Mapping and Variants Filtration.
We modified the fast homozygosity mapping strategy as described in Voz et al. (12). We scanned through all SNPs from F2 duper pools with coverage ≥5 (n = 1,232,905) and calculated the average homozygosity score in a rolling 100 SNPs window, using scoring criteria listed below.
-
I.
For all homozygous SNPs from the duper pools, skipped if it is a homozygous SNP in both the parental samples. Assigned a score of 1 for unique homozygous SNP in the Lakeview sample, a score of 0 for unique homozygous SNP in the Bio14.6 sample, and a score of 0.5 otherwise.
-
II.
For all heterozygous SNPs from the duper pools, we assigned a score of 0.8 for unique homozygous SNP in the Lakeview sample, a score of 0.2 for unique homozygous SNP in the Bio14.6 sample, and a score of 0.5 otherwise.
Windows with homozygosity scores ≥0.9, ≤0.1 were merged to indicate strong linkage to Lakeview or Bio14.6 alleles, respectively.
We restricted the search for candidate mutations in the LVG linked region. SNPs predicted to cause a HIGH or MODERATE putative impact estimated by SnpEff (21) were being screened manually using Integrative Genomics Viewer (54) to exclude potential false-positive variants. Given that this was a drafted assembly, some transcripts, especially those with lower complexity or higher repetitive regions, were mapped with lower confidence. We filtered variant(s) with more than 1 variant found within 50 bp up- or downstream, and variants found in exonic regions with no complete reading frames (e.g., multiple stop codons found in all possible reading frames).
Sanger Sequencing Validation.
To confirm the candidate mutations in Cry1, cDNA was prepared from total RNA extracted from cortex and hypothalamus of wild-type and duper hamsters. We prepared cDNA with 500 ng of RNA input using the High Capacity RT kit (Applied Biosystems) manufacturer’s protocol. cDNA was diluted 1:10, and 2 μL were used as template per reaction for the Phusion High-Fidelity DNA Polymerase (NEB) PCR amplification following the manufacturer’s protocol with an annealing temperature of 60 °C. The forward (GAGATGCCAGCAGAGACCA) and reverse (GGCTCGCAAGCAGGGAAT) primers produce a predicted amplicon of 253 bp, which was confirmed by gel electrophoresis. Sanger sequencing was performed at the DNACore (Cincinnati Children’s Hospital) following an enzymatic digest with ExoSAP-IT (Applied Biosystems). Two individual reactions were sequenced with both forward and reverse primers. Sequencher (GeneCodes) was used for sequence alignment and analysis.
Immunocytochemistry.
Adult male duper and wild-type hamsters (n = 3) were anesthetized with pentobarbital (80 mg/kg) and transcardially perfused with cold phosphate-buffered saline (100 mL) and 4% paraformaldehyde (300 mL) at ZT12. Brains were postfixed overnight, infiltrated with 20% sucrose in phosphate buffer, and frozen sectioned at 40 um. Sections were denatured in sodium citrate for 30 min at 92 °C. After buffer rinses and blocking in normal serum, sections were incubated with an antibody directed at the N-terminal of CRY1 (Novus; NBP1-69080; 1:100) overnight followed by a 3-h incubation in Cy3-conjugated donkey anti-rabbit (1:350). Before mounting, sections were incubated in Neurotrace FluroNissl 500/525 for 30 min (ThermoFisher, N21480, 1:300). Immunostaining was assessed using a Zeiss 700 confocal microscope as previously described (55).
Western Blotting.
Liver tissue (30 mg) was processed in RIPA buffer with protease and phosphatase inhibitors (SIGMAFAST Protease Inhibitor Mixture Tablets, EMD Millipore Phosphatase Inhibitor Mixture Set 1 and 2). A dounce homogenizer was used to homogenize the tissues in the RIPA buffer. Homogenized lysates were centrifuged at 15,000 × g for 20 min and the clear phase supernatant was collected as the whole cell lysate. We performed a BCA (bicinchoninic acid) assay (Sigma) to determine protein concentrations in the lysates. We mixed the lysates with NuPAGE LDS Sample Buffer (Thermo Fisher) and boiled them at 95 °C for 10 min. SDS-PAGE was used to separate the proteins based on their molecular weight. We used the Trans-Turbo Transfer System (Bio-Rad) to transfer the proteins from SDS-PAGE to the PVDF membrane. After blocking the membranes with 5% bovine serum albumin, we used CRY1-N-terminal binding antibody provided by the Lamia Lab to detect CRY1. PER1 (ab136451), PER3 (ab177482), and CRY2 (ab93802) antibodies were purchased from Abcam. The ChemiDoc MP (Bio-Rad) is used to detect chemiluminescence signals from the blots and ImageLab analysis software (Bio-Rad) was used to determine the band intensities. Stain-Free Imaging Technology by Bio-Rad is used for normalization purposes. Briefly, the trihalo compound in the gel generates fluorescence signals by binding to tryptophan residues. Thus, the total protein amount in the membrane is detected and used to normalize bands in each lane. Relative protein expressions were tested for circadian expression differences using CircaCompare (56).
Supplementary Material
Acknowledgments
We thank Dr. Katja Lamia for the N-terminal CRY1 antibody. We thank Dr. Christopher Bradfield and Dr. Joshua Hyman for guidance and sequencing core support. We also thank Dr. Nicholas Lahens and Jacqueline Growe for technical support and Dr. Marc Ruben and XingLiang Liu for helpful discussions. This work is supported by National Heart, Lung and Blood Institute (grant 5R01HL138551-02 to E.L.B and J.B.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123560119/-/DCSupplemental.
Data Availability
Genome sequencing data have been uploaded to Sequence Read Archive (accession no. PRJNA754138). Transcriptomics data have been uploaded to Gene Expression Omnibus (accession no. GSE184205). Syrian hamster genome is associated with BioSample (accession no. SAMN26084744). Analysis code for the fast homozygosity mapping is available at GitHub, https://github.com/yyenglee/FHM-duper.
References
- 1.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rijo-Ferreira F., Takahashi J. S., Genomics of circadian rhythms in health and disease. Genome Med. 11, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ralph M. R., Menaker M., A mutation of the circadian system in golden hamsters. Science 241, 1225–1227 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Lowrey P. L., et al. , Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288, 483–492 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Virshup D. M., Eide E. J., Forger D. B., Gallego M., Harnish E. V., Reversible protein phosphorylation regulates circadian rhythms. Cold Spring Harb. Symp. Quant. Biol. 72, 413–420 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Meng Q.-J., et al. , Setting clock speed in mammals: The CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58, 78–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monecke S., Brewer J. M., Krug S., Bittman E. L., Duper: A mutation that shortens hamster circadian period. J. Biol. Rhythms 26, 283–292 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Krug S., McKinley Brewer J., Bois A. S., Bittman E. L., Effects of the duper mutation on circadian responses to light. J. Biol. Rhythms 26, 293–304 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Bittman E. L., Does the precision of a biological clock depend upon its period? Effects of the duper and tau mutations in Syrian hamsters. PLoS One 7, e36119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lisitsyn N. A., et al. , Direct isolation of polymorphic markers linked to a trait by genetically directed representational difference analysis. Nat. Genet. 6, 57–63 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Sheffield V. C., Nishimura D. Y., Stone E. M., Novel approaches to linkage mapping. Curr. Opin. Genet. Dev. 5, 335–341 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Voz M. L., et al. , Fast homozygosity mapping and identification of a zebrafish ENU-induced mutation by whole-genome sequencing. PLoS One 7, e34671 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolman M. A., et al. , A genome-wide screen identifies PAPP-AA-mediated IGFR signaling as a novel regulator of habituation learning. Neuron 85, 1200–1211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jander G., et al. , Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129, 440–450 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tchitchek N., et al. , Sequencing, annotation and analysis of the Syrian hamster (Mesocricetus auratus) transcriptome. PLoS One 9, e112617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alan Harris R., et al. , Construction of a new chromosome-scale, long-read reference genome assembly of the Syrian hamster, Mesocricetus auratus. bioRxiv [Preprint] (2021). 10.1101/2021.07.05.45107 (Accessed 10 August 2021). [DOI] [PMC free article] [PubMed]
- 17.Sancar A., Cryptochrome: The second photoactive pigment in the eye and its role in circadian photoreception. Annu. Rev. Biochem. 69, 31–67 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Homburger F., Baker J. R., Nixon C. W., Whitney R., Primary, generalized polymyopathy and cardiac necrosis in an inbred line of Syrian hamsters. Pharmacology 6, 339–345 (1962). [DOI] [PubMed] [Google Scholar]
- 19.Seppey M., Manni M., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245 (2019). [DOI] [PubMed] [Google Scholar]
- 20.McKenna A., et al. , The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Horst G. T., et al. , Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Siepka S. M., Yoo S.-H., Park J., Lee C., Takahashi J. S., Genetics and neurobiology of circadian clocks in mammals. Cold Spring Harb. Symp. Quant. Biol. 72, 251–259 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patke A., et al. , Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell 169, 203–215.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parico G. C. G., et al. , The human CRY1 tail controls circadian timing by regulating its association with CLOCK:BMAL1. Proc. Natl. Acad. Sci. U.S.A. 117, 27971–27979 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sisson C., Bahiru M. S., Manoogian E. N. C., Bittman E. L., The duper mutation reveals previously unsuspected functions of Cryptochrome 1. Proc. Natl. Acad. Sci. U.S.A. 119, e2121883119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spoelstra K., Albrecht U., van der Horst G. T. J., Brauer V., Daan S., Phase responses to light pulses in mice lacking functional per or cry genes. J. Biol. Rhythms 19, 518–529 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Czarna A., et al. , Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153, 1394–1405 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Vanderstraeten J., Gailly P., Malkemper E. P., Light entrainment of retinal biorhythms: Cryptochrome 2 as candidate photoreceptor in mammals. Cell. Mol. Life Sci. 77, 875–884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano A., et al. , A Cryptochrome 2 mutation yields advanced sleep phase in humans. eLife 5, e16695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maywood E. S., et al. , Tuning the period of the mammalian circadian clock: Additive and independent effects of CK1εTau and Fbxl3Afh mutations on mouse circadian behavior and molecular pacemaking. J. Neurosci. 31, 1539–1544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yagita K., et al. , Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 21, 1301–1314 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnaiah S. Y., et al. , Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab. 25, 961–974.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang H., Lee G., Kim J. B., SREBP1c-CRY1 axis suppresses hepatic gluconeogenesis upon insulin. Cell Cycle 16, 139–140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papp S. J., et al. , DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. eLife 4, e04883 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shafi A. A., et al. , The circadian cryptochrome, CRY1, is a pro-tumorigenic factor that rhythmically modulates DNA repair. Nat. Commun. 12, 401 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., Hogenesch J. B., A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 111, 16219–16224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perng Y.-C., Lenschow D. J., ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 16, 423–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borrmann H., McKeating J. A., Zhuang X., The circadian clock and viral infections. J. Biol. Rhythms 36, 9–22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar R. S., et al. , Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 113, 10085–10090 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majumdar T., Dhar J., Patel S., Kondratov R., Barik S., Circadian transcription factor BMAL1 regulates innate immunity against select RNA viruses. Innate Immun. 23, 147–154 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Issah Y., et al. , Loss of circadian protection against influenza infection in adult mice exposed to hyperoxia as neonates. eLife 10, e61241 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Backlund P. S., et al. , Daily rhythm in plasma N-acetyltryptamine. J. Biol. Rhythms 32, 195–211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dijk E. L., Jaszczyszyn Y., Naquin D., Thermes C., The third revolution in sequencing technology. Trends Genet. 34, 666–681 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Weinert D., Fritzsche P., Gattermann R., Activity rhythms of wild and laboratory golden hamsters (Mesocricetus auratus) under entrained and free-running conditions. Chronobiol. Int. 18, 921–932 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Valentine H., Daugherity E. K., Singh B., Maurer K. J., “The experimental use of Syrian hamsters” in The Laboratory Rabbit, Guinea Pig, Hamster, and Other Rodents, Suckow M., Stevens K., Wilson R., Eds. (Elsevier, 2012), p. 875. [Google Scholar]
- 47.Imai M., et al. , Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U.S.A. 117, 16587–16595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daan S., Pittendrigh C. S., A functional analysis of circadian pacemakers in nocturnal rodents. J. Comp. Physiol. 106, 253–266 (1976). [Google Scholar]
- 49.Kolmogorov M., Yuan J., Lin Y., Pevzner P. A., Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Shumate A., Salzberg S. L., Liftoff: Accurate mapping of gene annotations. Bioinformatics 37, 1639–1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poplin R., et al. , Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv [Preprint] (2018). 10.1101/201178 (Accessed 2 September 2021). [DOI]
- 54.Robinson J. T., et al. , Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manoogian E. N. C., Kumar A., Obed D., Bergan J., Bittman E. L., Suprachiasmatic function in a circadian period mutant: Duper alters light-induced activation of vasoactive intestinal peptide cells and PERIOD1 immunostaining. Eur. J. Neurosci. 48, 3319–3334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parsons R., Parsons R., Garner N., Oster H., Rawashdeh O., CircaCompare: A method to estimate and statistically support differences in mesor, amplitude and phase, between circadian rhythms. Bioinformatics 36, 1208–1212 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequencing data have been uploaded to Sequence Read Archive (accession no. PRJNA754138). Transcriptomics data have been uploaded to Gene Expression Omnibus (accession no. GSE184205). Syrian hamster genome is associated with BioSample (accession no. SAMN26084744). Analysis code for the fast homozygosity mapping is available at GitHub, https://github.com/yyenglee/FHM-duper.