Significance
Plasma membranes are composed of a lipid bilayer in which phosphatidylserine (PtdSer) is confined to the inner leaflet by the action of flippase that translocates PtdSer from the outer to inner leaflets. Two P4-ATPases (ATP11A and ATP11C) work as flippase at plasma membranes. Here, we report that the mouse placenta expresses only ATP11A, and Atp11a-deficient mouse embryos die during embryogenesis due to inefficient formation of syncytiotrophoblasts in the placental labyrinth. The flippase-null mutation inactivates human choriocarcinoma BeWo cells to translocate PtdSer into the inner leaflet and undergo cell fusion. These findings highlight the importance of flippase to regulate the distribution of phospholipids for cell fusion, at least in trophoblast fusion.
Keywords: flippase, phosphatidylserine, cell fusion, placenta, trophoblast
Abstract
The P4-ATPases ATP11A and ATP11C function as flippases at the plasma membrane to translocate phosphatidylserine from the outer to the inner leaflet. We herein demonstrated that Atp11a-deficient mouse embryos died at approximately E14.5 with thin-walled heart ventricles. However, the cardiomyocyte- or epiblast-specific Atp11a deletion did not affect mouse development or mortality. ATP11C may have compensated for the function of ATP11A in most of the cell types in the embryo. On the other hand, Atp11a, but not Atp11c, was expressed in the mouse placenta, and the Atp11a-null mutation caused poor development of the labyrinthine layer with an increased number of TUNEL-positive foci. Immunohistochemistry and electron microscopy revealed a disorganized labyrinthine layer with unfused trophoblasts in the Atp11a-null placenta. Human placenta-derived choriocarcinoma BeWo cells expressed the ATP11A and ATP11C genes. A lack of ATP11A and ATP11C eliminated the ability of BeWo cells to flip phosphatidylserine and fuse when treated with forskolin. These results indicate that flippases at the plasma membrane play an important role in the formation of syncytiotrophoblasts in placental development.
Plasma membranes in eukaryotes comprise a lipid bilayer in which phosphatidylserine (PtdSer) is exclusively confined to the inner leaflet (1, 2). PtdSer is reversely or irreversibly exposed on the cell surface and serves as a signal or scaffold for enzymes in various biological processes (3–5). PtdSer irreversibly exposed on the surface of apoptotic cells serves as an “eat me” signal, that on activated platelets binds to blood clotting factors and enhances their protease activities, and that exposed on tumor cells or in their environment suppresses the immune system (3).
ATP11A and ATP11C, members of the P4-type Adenosine triphosphatase (ATPase) family (6, 7), are membrane proteins that carry 10 transmembrane regions and a cytoplasmic ATPase domain (8–10). They are localized to the plasma membrane with a CDC50A chaperone and function as flippases that translocate PtdSer and phosphatidylethanolamine from the outer to inner leaflet. When PtdSer is exposed to the cell surface, these two flippases at the plasma membrane are inactivated by a high concentration of Ca2+ or caspase-mediated cleavage (8, 9). To conform to the rapid exposure of PtdSer on apoptotic cells and activated platelets, another enzyme called scramblase is activated and nonspecifically scrambles phospholipids between the outer and inner leaflets (5).
Although ATP11A and ATP11C are ubiquitously expressed in various cells with a redundant function, a loss-of-function mutation in Atp11c in humans and mice causes multiple phenotypes. Atp11cy/− mice developed B cell lymphopenia, cholestasis, and mild anemia (11–14), while a human patient defective in ATP11C exhibited hemolytic anemia (15). We previously reported that mouse B cell precursors did not express Atp11a at the pre–B cell stage (16). Therefore, Atp11c-deficient pre–B cells do not have flippases, expose PtdSer, and are engulfed alive by surrounding macrophages in bone marrow, which leads to B cell lymphopenia.
In the present study, we demonstrated that Atp11a-deficient mice were embryonic lethal or died at approximately embryonal day (E) 14.5 with improper heart development and anemia. Nevertheless, the cardiomyocyte- or epiblast-specific deletion of Atp11a did not adversely affect mouse development. We found that the mouse placenta, particularly trophoblasts in the labyrinthine placental layer, expressed ATP11A but not ATP11C flippase. The labyrinth is a porous location in the placenta at which nutrients, oxygen, and waste are exchanged between maternal and fetal blood (17). Two tightly adhered layers of syncytiotrophoblasts (SynT-I and SynT-II), generated by the fusion of trophoblasts, separate the maternal and fetal blood compartments in the mouse labyrinth (18). Syncytiotrophoblasts in the Atp11a-deficient placenta were not well developed, possibly due to inefficient cell fusion. Accordingly, the deletion of the ATP11A and ATP11C genes strongly reduced the ability of the human BeWo choriocarcinoma cell line to fuse in response to forskolin. These results indicate that the flipping of PtdSer into the plasma membrane inner leaflet is necessary for efficient cell fusion to establish syncytiotrophoblasts.
Results
Developmental Defects and Embryonic Lethality in Atp11akomp/komp Mice.
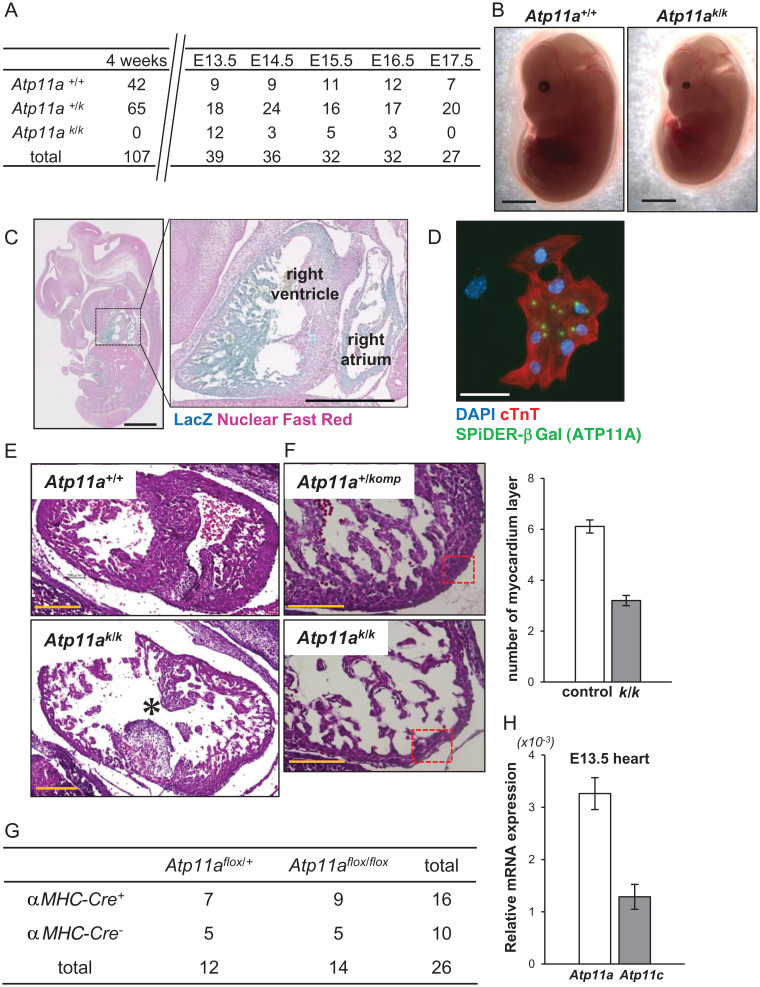
Atp11a+/komp mice harbor the knockout-first allele in the Atp11a gene (SI Appendix, Fig. S1). Atp11a+/komp mice grew normally and had no abnormalities. When they were intercrossed, no Atp11akomp/komp (Atp11ak/k) mice were found among 107 offspring at the age of 4 wk (Fig. 1A). A genotype analysis of embryos indicated that Atp11ak/k mice survived according to Mendelian law until E13.5 but started to die thereafter (Fig. 1A). The surviving E13.5 Atp11ak/k embryos were significantly smaller (by ∼10%) than littermate Atp11a+/komp or Atp11a+/+ embryos and showed anemia (Fig. 1B).
Fig. 1.
Developmental heart defect in Atp11a-null embryos. (A) Atp11a+/komp mice were intercrossed, and the genotype of littermates at the age of 4 wk or the indicated embryonal day was identified. (B) The appearance of E13.5 Atp11a+/+ and Atp11ak/k littermate embryos. Atp11ak/k embryos were smaller and paler than wild-type embryos. Scale bar, 2 mm. (C) E13.5 Atp11ak/k embryos were stained with X-gal (blue) and counterstained with nuclear fast red (pink). Scale bar, 1 mm. Right: Enlarged area of the dotted box. Scale bar, 500 μm. (D) Cardiomyocytes prepared from E13.5 Atp11a+/komp hearts were stained with SPiDER-βGal (green), anti-cTnT Ab (red), and DAPI (blue). Scale bar, 50 μm. (E and F) Sections of E13.5 Atp11a+/+ or Atp11a+/komp and Atp11ak/k littermate embryos were stained with H&E. The ventricular septum (E) and right ventricular apex (F) are shown. Scale bar, 200 μm (E), 100 μm (F). The asterisk in E points to the ventricular septal defect in the Atp11ak/k embryo. The myocardial layer is thinner in Atp11ak/k embryos (F). The number of myocardial layers in the boxed area was quantified for four to six sections per mouse (n = 3), and the average values were plotted with SD (bar) on the Right. (G) Atp11aflox/flox and αMhc-Cre+-Atp11aflox/+ mice were crossed, and the genotype of obtained pups at the age of 4 wk was identified. The number of mice with the indicated genotype is shown. (H) mRNAs for the Atp11a and Atp11c genes in E13.5 wild-type mouse fetal hearts (n = 3) were quantified by real-time RT-PCR, and their relative levels against Gapdh mRNA were plotted with SD (bar).
The Atp11akomp allele carries the LacZ gene in intron 6 of the Atp11a gene (SI Appendix, Fig. S1). When E13.5 Atp11ak/k embryos were subjected to the in situ 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal) staining of whole-mount embryos, intense staining was observed in the choroid plexus, heart, dorsal root ganglion, mesentery, and tongue (Fig. 1C). Strong β-galactosidase (βGal) activity was detected in the ventricles and atrium regions. Cells in the E13.5 heart were dissected and stained for βGal activity using the spirobased immobilisable diethylrhodol-βGal (SPiDER) technique (19). As shown in Fig. 1D, cardiac troponin T (cTnT)–positive cardiac myocytes showed positive signals, indicating that the Atp11a gene was explicitly expressed in cardiac myocytes.
A histological analysis showed a defect at the ventricular septum of the heart from the E13.5 Atp11ak/k embryo (Fig. 1E). In addition, the heart myocardium of E13.5 mutant mice was thinner than that of the littermates due to a reduced number of myocardium layers at the ventricular apex (Fig. 1F). These results suggested that developmental disorders in Atp11ak/k mice were caused by the lack of ATP11A in cardiac myocytes. Therefore, we generated the Atp11a allele flanked by loxP (Atp11aflox/flox mice) (SI Appendix, Fig. S1) and crossed these mice with mice carrying the Cre gene under the promoter of the α-Myosin heavy-chain gene (αMhc-Cre) that specifically expresses Cre in cardiac myocytes (20). Surprisingly, the Atp11aflox/flox mice carrying αMhc-Cre were born and developed normally (Fig. 1G). Genotyping with DNA from the heart indicated that the Atp11a allele was mutated as expected and designated as Atp11aΔ/Δ. These results showed that abnormal cardiac development in Atp11ak/k mice was not due to the Atp11a deficiency in cardiac myocytes. We previously reported that among 14 members of the P4-ATPase family, ATP11A and ATP11C localized to the plasma membrane were ubiquitously expressed and exhibited redundant activity (8). A real-time RT-PCR analysis indicated that Atp11c messenger ribonucleic acid (mRNA) was present at ∼30% of Atp11a mRNA in the heart (Fig. 1H), suggesting that flippase activity provided by ATP11C was sufficient to maintain homeostasis in the heart.
Lethal Effect of the Abnormal Placenta in Atp11a-Null Mice.
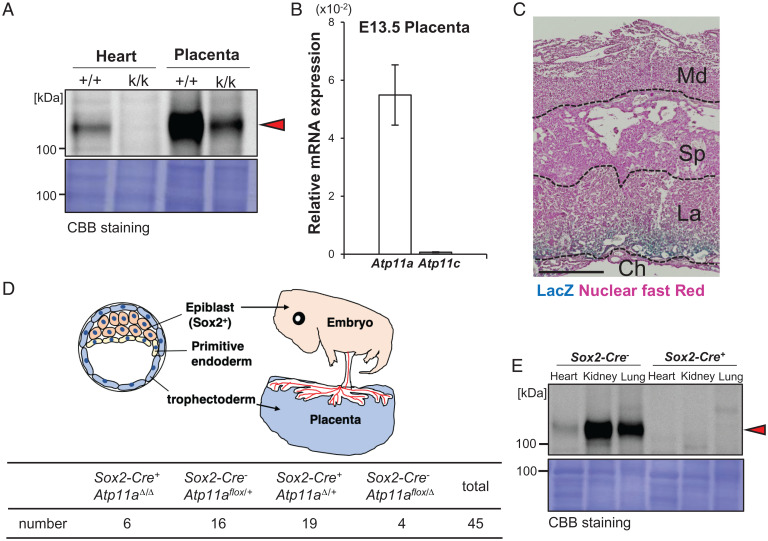
Perez-Garcia et al. (21) reported that among embryonic lethal knockout mice, 35% of those dying by E9.5 to E14.5 had placental abnormalities, which were often accompanied by the abnormal development of the heart or brain (21–23). We attributed the lethality of Atp11a-deficient mice to a defect in the placenta. Western blotting with a monoclonal antibody (mAb; clone 4-C11) against mouse ATP11A showed an ∼130-kDa band in the wild-type heart but not in the Atp11ak/k heart (Fig. 2A), which confirmed the specificity of the mAb. The intensity of 130-kDa ATP11A in lysates from the wild-type mouse placenta was approximately 10-fold stronger than in those from the heart. A markedly reduced but still significant level of the ATP11A band was observed in the Atp11ak/k placenta, potentially from maternal (Atp11a+/komp)–derived decidual cells. A real-time RT-PCR analysis indicated that the level of Atp11a mRNA normalized to that of glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA was more than 10-fold higher in the placenta (Fig. 2B) than in the heart (Fig. 1H). In contrast, Atp11c mRNA expression was negligible in the E13.5 placenta (Fig. 2B). X-gal staining of the whole placenta revealed that cells in the labyrinthine layer, but not in the spongiotrophoblast layer, expressed the βGal gene under the Atp11a gene promoter (Fig. 2C).
Fig. 2.
Normal development of epiblast-specific Atp11a-deficient mice. (A) Tissue homogenates (8.5 μg of protein) from the heart and placenta of E13.5 wild-type (+/+) and Atp11ak/k (k/k) embryos were separated by 7.5% SDS-PAGE, transferred to a PVDF membrane, and analyzed by Western blotting with anti-ATP11A mAb. The molecular mass of standard proteins is shown in kilodaltons. The red arrowhead shows ATP11A. Bottom: Membrane was stained with Coomassie Brilliant Blue (CBB). (B) mRNAs for Atp11a and Atp11c in the E13.5 wild-type mouse placenta (n = 3) were quantified by real-time RT-PCR, and their relative levels against Gapdh mRNA were plotted with SD (bars). (C) Male Atp11a+/komp mice were crossed with wild-type female mice. On day 13 of pregnancy, the placenta was dissected and stained by X-gal (blue) and nuclear fast red (pink). Md, maternal decidua; Sp, spongiotrophoblast layer; La, labyrinthine layer; Ch, chorion. Scale bar, 500 μm. (D) Sox2-Cre+Atp11aΔ/+ mice were crossed with Atp11aflox/flox mice, and the genotypes of 45 pups at the age of 4 wk were identified. The number of mice carrying the indicated genotype is shown. Upper: Schematic for blastocyst development. The blastocysts consist of epiblasts, primitive endoderm, and trophectoderm. Epiblasts differentiate into the embryo, the primitive endoderm into the yolk sac, and the trophectoderm into the placenta. (E) Tissue lysates (6 μg of protein) from the heart, kidney, and lung of Atp11aflox/flox and Atp11aΔ/Δ mice at the age of 9 to 16 wk were separated by 7.5% SDS-PAGE and analyzed by Western blotting with anti-ATP11A mAb. The molecular mass of the standard proteins is shown in kilodaltons. The red arrowhead shows the band ATP11A. Lower: Membrane stained with CBB.
We then deleted the Atp11a gene in an epiblast-specific manner by crossing Atp11aflox/flox mice with Sex determining region Y-box 2 (Sox2)-Cre transgenic mice that expressed Cre in epiblasts (24–26). As shown in Fig. 2D, Atp11a-deficient mice with the Sox2-Cre+Atp11aΔ/Δ genotype were born at a lower number than expected (Fig. 2D) but grew normally. A Western blot analysis indicated that the ATP11A protein was not present in the heart, kidneys, or lungs of Sox2-Cre+Atp11aΔ/Δ mice (Fig. 2E). Histological analysis of the heart from the mice at 16 mo of age showed no apparent abnormality (SI Appendix, Fig. S2). These results showed that ATP11A was dispensable for mouse embryos and adulthood but was indispensable for placental development.
Abnormal Labyrinthine Layer in the Atp11a-Null Placenta.
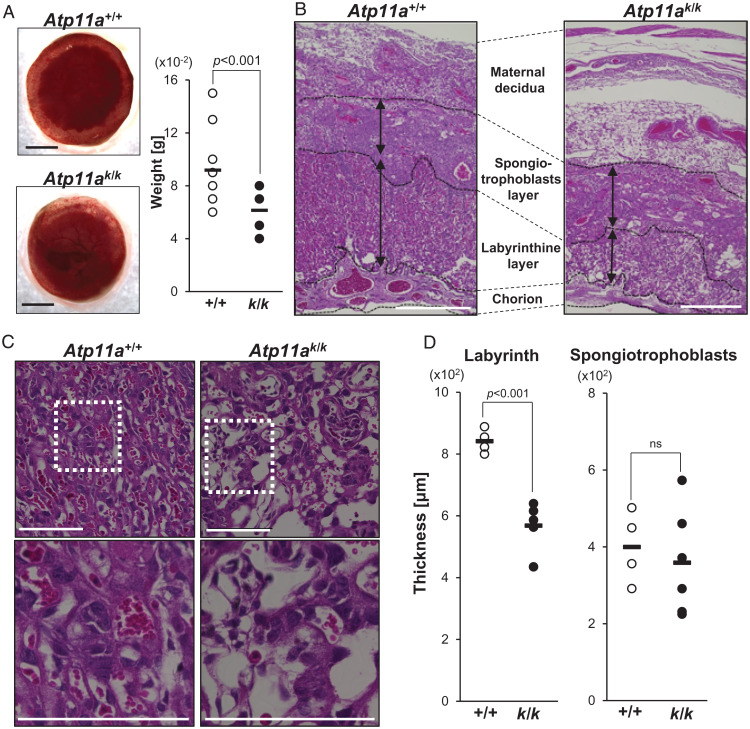
When Atp11a+/komp mice were intercrossed, the E13.5 placentas of Atp11ak/k embryos were smaller and thinner than those of their littermates of the Atp11a+/+ or Atp11a+/komp genotype (Fig. 3A). The weight of placentas was ∼70% of those for the control littermates. The mouse placenta comprises the maternal decidua and fetal parts that consist of the spongiotrophoblast and labyrinthine cell layers and chorion (Fig. 3B). A histological analysis of the placenta in E13.5 Atp11ak/k mice showed that their labyrinthine layer was 30% thinner than that in Atp11a+/+ mice (Fig. 3D). In contrast, the size of the spongiotrophoblast layer was similar to that of the control littermates.
Fig. 3.
The abnormal labyrinthine layer of the Atp11a-deficient placenta. Atp11a+/komp mice were intercrossed, and placentas were removed at E13.5 and genotyped. (A) Left: Representative appearance of the Atp11a+/+ (+/+) and Atp11ak/k (k/k) placenta. Scale bar, 1 mm. Right: Weights of the placentas from control (Atp11a+/+ or Atp11a+/komp) (n = 22) and Atp11ak/k (n = 7) mice were plotted with the average (bars). P values were calculated by Student’s t test. (B and C) Histological analysis of the placenta (B) and labyrinthine layer (C). Paraffin sections from the Atp11a+/+ or Atp11ak/k placenta were stained with H&E and observed by microscopy. Bottom: Higher-magnification images of the boxed regions. Scale bar, 500 μm (B), 200 μm (C). (D) The lengths of the Sp and La in the center of the placenta indicated by black arrows in B were measured in three sections per placenta dissected from different pregnant mice (n = 4 for Atp11a+/+ and n = 6 for Atp11ak/k). The thickness of each placenta was plotted with the average value (bars). P values were calculated by Student’s t test. ns, not significant.
A close examination of the wild-type labyrinth showed juxtaposed maternal blood lacunae and fetal blood vessels separated by syncytial and mononuclear trophoblastic cell layers (Fig. 3C). Fetal blood vessels lined by endothelial cells contained primitive nucleated erythrocytes that were easily distinguished from maternal sinusoids carrying enucleated erythrocytes. In comparison with the wild-type placenta labyrinth, the Atp11ak/k placenta labyrinth was less packed and only a few fetal blood vessels containing nucleated erythrocytes were present (Fig. 3C). Maternal sinusoids were also not surrounded well by syncytiotrophoblasts. A similar abnormality was observed in the labyrinth of E11.5 Atp11ak/k placenta (SI Appendix, Fig. S3).
Abnormal Syncytiotrophoblasts in the Labyrinthine Layer of the Atp11a-Null Placenta.
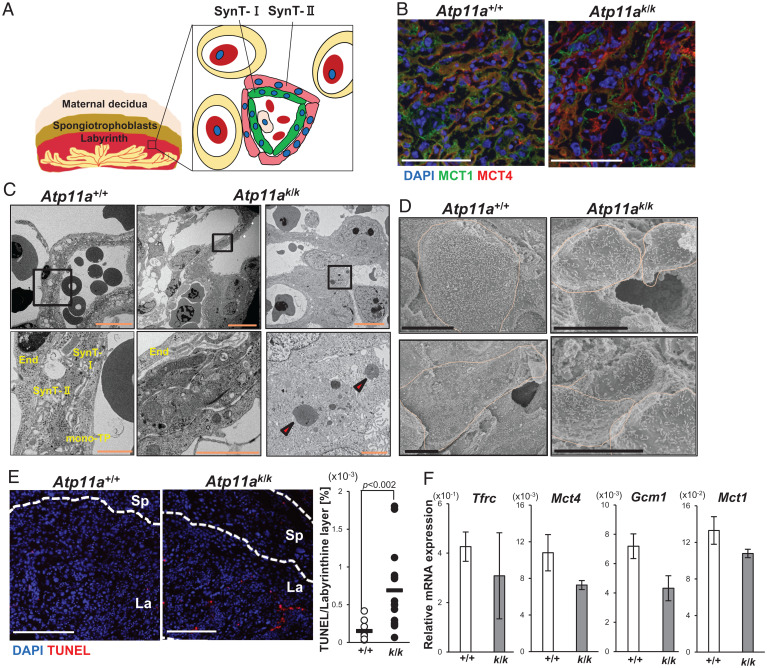
In the mouse placenta, the SynT-I and SynT-II layers surrounded maternal sinusoids in the labyrinthine layer (Fig. 4A). Immunostaining with anti-monocarboxylate transporter (MCT)1 and anti-MCT4, which are explicitly expressed in SynT-I and SynT-II, respectively (27), showed that SynT-I and SynT-II were closely associated in the wild-type placental labyrinth. On the other hand, the cell layers expressing MCT1 or MCT4 were not well connected in the Atp11ak/k placenta (Fig. 4B), suggesting that syncytiotrophoblasts were not appropriately developed in the Atp11ak/k placenta.
Fig. 4.
Abnormal syncytiotrophoblasts in the Atp11a-deficient placenta. (A) A schematic representation of syncytiotrophoblasts in the mouse placental labyrinthine layer. Two layers of syncytiotrophoblasts, SynT-I (green) and SynT-II (pink), separate fetal and maternal blood. SynT-I and SynT-II express MCT1 and MCT4, respectively. Nuclei are shown in the blue circle. Red blood cells are in red. (B) Cryosections of the E13.5 Atp11a+/+ and Atp11ak/k placenta were stained with anti-MCT1 (green), anti-MCT4 (red), and DAPI (blue). Scale bar, 100 μm. (C and D) Electron microscopy of the labyrinthine layer of the placenta. Sections of the placenta in E13.5 Atp11a+/+ and Atp11ak/k mice were analyzed by transmission electron microscopy (C) or scanning electron microscopy (D). The boxed area (Upper) was enlarged in the Bottom in (C). The arrowheads indicate degenerated nuclei. The cell boundary was surrounded by an orange line in D. Scale bar, 100 μm (C, Upper), 20 μm (C, Lower), 10 μm (D). End, endothelial cell; mono-TP, mononuclear trophoblasts. (E) Cryosections from the E13.5 Atp11a+/+ and Atp11ak/k placenta were stained with TUNEL. Red, TUNEL; blue, DAPI. Scale bar, 250 μm. Right: Total TUNEL-positive area was quantified with three sections per placenta, and the ratio against the DAPI-positive area in the labyrinthine layer was plotted (n = 3 for Atp11a+/+ or Atp11a+/k and n = 6 for Atp11ak/k). P values calculated by Student’s t test are shown. (F) RNA from the E13.5 Atp11a+/+ (+/+) and Atp11ak/k (k/k) placenta (n = 3 for each) was subjected to real-time RT-PCR to assess the mRNA levels of the indicated genes . The average values were plotted with SD (bars).
To further characterize the defect in the Atp11ak/k placenta, we observed the E13.5 placenta under a transmission electron microscope. As shown in Fig. 4C, the wild-type labyrinth showed the layer of mononuclear sinusoidal trophoblast giant cells, the SynT-I cell layer, and the vacuole-containing SynT-II cell layer (17, 28). On the other hand, although endothelial cells and vacuole-containing cells were identified in the labyrinth of the Atp11ak/k placenta, the SynT-I and SynT-II layers were poorly organized (Fig. 4C). Scanning electron micrography of the wild-type labyrinthine layer from the maternal blood side showed giant cells that were expected to be syncytiotrophoblasts (Fig. 4D). On the other hand, the majority of cells were small in the Atp11ak/k labyrinth, while few giant cells were present, suggesting the inefficient fusion of trophoblasts. Transmission electron microscopy showed many trophoblasts carrying degenerated nuclei in the Atp11ak/k labyrinth (Fig. 4C). Accordingly, the number of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-positive cells in the Atp11ak/k placental labyrinthine layer was about fivefold higher than in the Atp11a+/+ placental labyrinthine layer (Fig. 4E). In addition, a real-time RT-PCR analysis showed that the mRNA levels of Mct1, transferrin receptor protein (Tfrc), Mct4, and chorion-specific transcription factor (Gcm1) genes, which are expressed in syncytiotrophoblasts (29, 30), were lower in the E13.5 Atp11ak/k placenta than in the wild-type placenta (Fig. 4F). These results suggested that Atp11a-deficient trophoblasts incapable of forming syncytiotrophoblasts died.
Inefficient Cell Fusion of ATP11A−/−ATP11C−/− BeWo Cells.
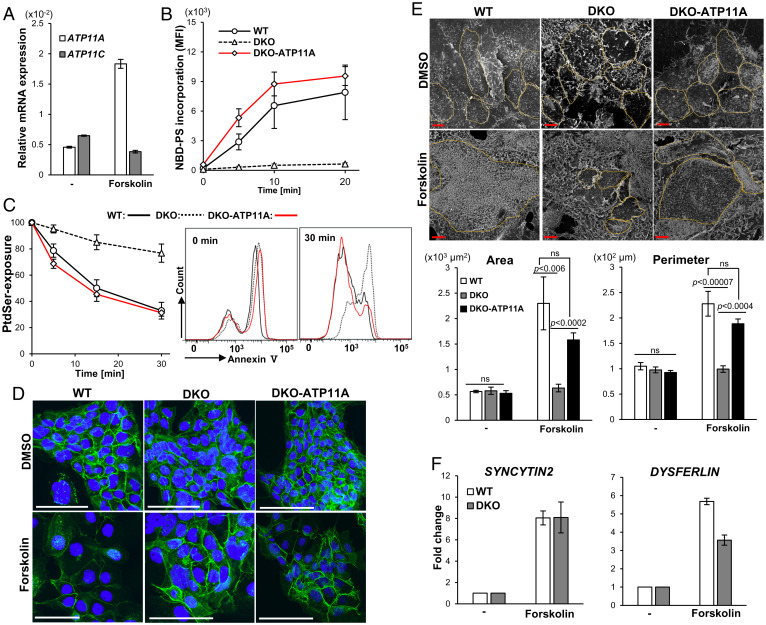
To examine the effects of flippase on the formation of syncytiotrophoblasts, we investigated human BeWo cells, which undergo cell fusion into syncytiotrophoblast-like cells when treated with forskolin (31). Real-time RT-PCR showed that BeWo cells expressed the ATP11A and ATP11C genes (Fig. 5A), and the expression level of ATP11A mRNA increased following the treatment with 20 μM forskolin for 48 h. We established ATP11A−/−ATP11C−/− (DKO) BeWo cells by the CRISPR-Cas9 system (32) ( SI Appendix, Fig. S4). As shown in Fig. 5B, wild-type BeWo cells incorporated nitrobenzoxadiazole (NBD)-labeled PtdSer (NBD-PS); however, this activity was completely lost in DKO-BeWo cells. The transformation of DKO-BeWo with human ATP11A rescued DKO-BeWo to flip or translocate NBD-PS into cells. To further confirm the contribution of ATP11A to the establishment of an asymmetrical distribution of PtdSer, cells were treated with 5 μM ionomycin, a Ca2+ ionophore, to activate Ca2+-dependent scramblases, such as TMEM16F (33). This treatment resulted in the exposure of PtdSer in most populations of wild-type, DKO, and DKO-ATP11A BeWo cells (Fig. 5C). When the ionomycin was removed, PtdSer on the surface of wild-type BeWo cells gradually disappeared at 25 °C, indicating that PtdSer was translocated into the inner leaflet of the plasma membrane. This internalization of PtdSer was severely retarded in DKO-BeWo cells, and more than 80% of the cell population continued to expose PtdSer after a 30-min incubation. On the other hand, DKO-expressing ATP11A regained the ability to internalize PtdSer. These results indicated that ATP11A and ATP11C flippases at the plasma membrane were essential for rapidly establishing the asymmetrical distribution of PtdSer, similar to mouse WR19L lymphoma cells (16).
Fig. 5.
Inefficient cell fusion of human BeWo cells lacking ATP11A and ATP11C. (A) Human BeWo cells were treated for 48 h with 20 μM forskolin. In forskolin-treated or untreated cells, mRNAs for ATP11A and ATP11C were quantified by real-time RT-PCR, and their levels are shown relative to that of β-ACTIN (ACTB). The experiment was performed three times, and average values are shown with SD (bars). (B) Wild-type (WT) BeWo, DKO-BeWo, and DKO-ATP11A cells were incubated at 15 °C with NBD-PS for the indicated period, treated with fatty acid–free BSA, and subjected to flow cytometry. Experiments were performed three times, and the average value of mean fluorescence intensity of incorporated NBD-PS in the Sytox blue–negative population was plotted with SD (bar). (C) Reestablishment of the asymmetrical distribution of PtdSer. WT-BeWo, DKO, and DKO-ATP11A cells were treated at 15 °C for 5 min with 5 μM ionomycin and incubated at 25 °C for the indicated time in the culture medium containing EGTA and BAPTA-AM. The exposure of PtdSer was then analyzed by the binding of Cy5-annexin V. Experiments were performed three times, and the average percentage of the annexin V–positive population in PI-negative cells was plotted with SD. (D–F) Forskolin-induced cell fusion. WT-BeWo, DKO, and DKO-ATP11A cells were treated at 37 °C for 48 h with 20 μM forskolin. (D) Cells were stained with anti–E-cadherin Ab (green) and DAPI (blue). Scale bar, 100 μm. (E) Cells were observed by scanning electron microscopy. Scale bar, 10 μm. The size of the cellular area and perimeter length were assessed for at least six randomly selected cells. Lower: Mean value ± SEM (bar). Statistical analyses were performed using Student’s t test, and P values are shown. ns stands for “not significant”. ns, not significant. (F) mRNA levels for SYNCYTIN 2 and DYSFERLIN were assessed by real-time RT-PCR and normalized to ACTB mRNA. Experiments were performed three times, and the average values were presented with SD (bars).
Wild-type, DKO, and DKO-ATP11A BeWo cells at ∼10% confluency were then treated for 48 h with 20 μM forskolin or its solvent dimethyl sulfoxide (DMSO) (0.05%). Staining with anti–E-cadherin antibody (Ab) indicated that a high level of E-cadherin was present at the site of cell-to-cell contact in aggregated BeWo cells in control DMSO-treated cells (Fig. 5D). As reported by Coutifaris et al. (34), the treatment of wild-type BeWo cells with forskolin strongly reduced the intensity of E-cadherin or even removed it at several locations, suggesting that these cells underwent cell fusion. On the other hand, the expression of E-cadherin in DKO-BeWo cells remained after the forskolin treatment, while it disappeared in forskolin-treated DKO-BeWo-ATP11A cells. Observations by scanning electron micrography indicated that the area size of wild-type BeWo cells increased 4.1-fold from 565 to 2,299 μm2 after the treatment with forskolin for 48 h, while the perimeter length increased 2.2-fold from 105 to 228 μm (Fig. 5E). Similar increases in the area size and perimeter length were observed in forskolin-treated DKO-ATP11A cells but not DKO-BeWo cells. Omata et al. (35) previously reported that the forskolin-induced fusion of BeWo cells was accompanied by the expression of a number of genes related to cell differentiation or cell fusion. Real-time RT-PCR indicated that the expression of the SYNCYTIN2 gene that can be up-regulated by forskolin treatment before cell fusion increased to a similar extent between wild-type and DKO-BeWo cells after the forskolin treatment (Fig. 5F). The up-regulation of the DYSFERIN gene that occurs after cell fusion (35) was less pronounced in DKO-BeWo cells, supporting a role of the plasma membrane flippase, ATP11A or ATP11C, in the fusion of BeWo cells.
Discussion
The mouse P4-ATPase family comprises 14 members, with ATP11A, ATP11B, and ATP11C belonging to class 6 of the family (6). We herein demonstrated that a global deficiency of Atp11a caused embryonic lethality and mutant mice died after E14.5, possibly due to heart failure. As previously reported in embryonic lethal mutant mice with heart failure (21–23, 36), the placenta was abnormal or poorly developed in Atp11a-null mice. Organogenesis of the mouse heart, accompanied by the proliferation of cardiac myocytes to thicken the ventricular wall, occurs between E8.5 and E14.5 (37, 38). Since the proliferation of cardiac myocytes is strongly affected by blood flow and hemodynamics (39), a decrease in blood flow due to placental defects may cause abnormal heart development in the embryo.
In contrast to the global Atp11a deficiency, the epiblast-specific Atp11a-null mutation had no apparent effect on mouse development. The mice live healthy at least at the age of 16 mo. ATP11A, ATP11B, and ATP11C have 58 to 64% identities on the amino acid sequence and translocate PtdSer when they are reconstituted in liposomes (40). ATP11A and ATP11B, but not ATP11C, are highly expressed in placental trophoblasts (Expression Atlas: https://www.ebi.ac.uk/gxa/home). In contrast, ATP11A and ATP11C are ubiquitously expressed in other tissues (8). ATP11A and ATP11C are present at the plasma membrane, while ATP11B is localized intracellularly or in early or recycling endosomes (8, 41). The severe phenotype of the Atp11a-null placenta indicated that ATP11B, a flippase at endosomes, was unable to compensate for the null mutation of the plasma membrane flippase (ATP11A), whereas ATP11C at the plasma membranes in the embryonal cells can compensate for the Atp11a-null mutation for the development of embryos.
In various biological processes, live cells transiently expose PtdSer to the cell surface (4). Differentiating human trophoblasts expose PtdSer on the cell surface (42), and the Ab against PtdSer inhibits the forskolin-induced fusion of BeWo cells (43). In addition, a high titer of anti-PtdSer Ab in the serum is often associated with recurrent early or late abortions due to the inefficient development of the placenta (44). Based on these findings, Chernomordik’s group proposed that PtdSer functions as a “fuse me” signal (45, 46). Accordingly, Zhang et al. (47) recently claimed that TMEM16F, a Ca2+-dependent phospholipid scramblase (33), was responsible for the exposure of PtdSer during the fusion of trophoblasts. However, the normal development of Tmem16f-null embryos (48) may reject its involvement in this process.
The development of syncytiotrophoblasts was inefficient in the Atp11a-null placenta, which is similar to the phenotype reported in mice deficient in Syncytin-A (28), supporting the role of ATP11A in the efficient fusion of trophoblasts. The function of plasma membrane flippases (ATP11A and ATP11C) is to swiftly internalize PtdSer exposed on the cell surface (16). PtdSer irreversibly exposed on the cell surface due to the lack of flippases functions as an eat-me signal not only for apoptotic cells but also for live cells (16). We previously demonstrated that B-cell lymphopenia in Atp11c-deficient mice was due to the engulfment by macrophages of precursor B cells that failed to internalize the exposed PtdSer due to the lack of plasma membrane flippases (16). On the other hand, the lack of the PtdSer-dependent engulfment system (MerTK and Axl tyrosine kinases) did not rescue the embryonic lethality of Atp11a-null mice (SI Appendix, Table S1). It is likely that the prolonged exposure of PtdSer prevented cell fusion or that PtdSer exposed on the cell surface had to return to the inner leaflet of the plasma membrane to complete cell fusion. PtdSer has been shown to change the surface charge of membranes and regulates protein localization (49). In trophoblasts undergoing fusion, intracellular annexin V localizes via PtdSer between mononuclear trophoblasts and syncytiotrophoblasts to form an adherent junction composed of E-cadherin, β-catenin, and α-catenin (50). A low concentration of PtdSer at the inner leaflet of plasma membranes due to the lack of the flippase (ATP11A) may have resulted in an inefficient adherent junction. A large number of dying trophoblasts in the labyrinthine layer of the Atp11a-null placenta suggests that unfused differentiated trophoblasts cannot survive, which is consistent with previous findings on anti-phospholipid–antibody-treated trophoblasts (51).
The involvement of PtdSer in cell fusion has been suggested not only for trophoblasts but also for myoblasts, osteoclasts, and egg/sperm fusion (45). Two groups recently established mouse myoblasts that lack CDC50A, a shared subunit of multiple P4-ATPases, or ATP11A, and reported contradictory findings (52, 53). One group showed that multiple P4-ATPases were required for the efficient fusion of C2C12 cells (53), while the other claimed that a deficiency in Cdc50a or Atp11a caused the uncontrolled fusion of myoblasts in vitro and in vivo (52). The present results on trophoblasts are consistent with the former findings; however, we cannot rule out the possibility that myoblasts and trophoblasts use different mechanisms for fusion. The exposure of PtdSer has been proposed to serve as a fuse-me signal in the fusion process of various cells (45). The present study showed that the distribution of PtdSer needs to be tightly regulated for efficient fusion, at least for the formation of syncytiotrophoblasts. In order for PtdSer to serve as an eat-me signal, it binds to a specific receptor or receptors on macrophages and activates their engulfment system (54). Further studies are needed to identify which scramblase family members (TMEM16, XKR, or others) are responsible for exposing PtdSer in the fusion process and also to elucidate the molecular mechanisms by which PtdSer is recognized in order to activate the fusion process.
Materials and Methods
Mice.
C57BL/6N mice and MRL-lpr/lpr mice were obtained from Japan SLC. Atp11a+/komp mice (Atp11atm1a(KOMP)/Wtsi) were from the Mutant Mouse Resource and Research Centers. SI Appendix, Fig. S1 shows the structure of the Atp11komp allele. Atp11aflox/+ mice in which two loxP elements flanked exons 7 and 8 of the Atp11a gene were prepared by crossing Atp11a+/komp mice with mice carrying the CAG-FLPe gene [C57BL/6-Tg(CAG-flpe)36Ito/ItoRbrc] (Riken). αMhc-Cre mice [B6.FVB-Tg(Myh6-cre)2182Mds/J] carrying the Cre gene under the promoter of αMhc were from the Jackson Laboratory. Sox2-Cre mice (B6N.Cg-Edil3Tg(Sox2-cre)1Amc/J) in which Cre is under the control of the SRY-box containing gene 2 promoter were from the Jackson Laboratory through R. Nishinakamura (Kumamoto University). All mice were housed in a specific pathogen-free facility at the Research Institute for Microbial Diseases, Osaka University. The Ethics Review Committee at Osaka University approved all mouse studies.
Mouse pregnancy was checked by vaginal plugs and confirmed by weight gain (55). The morning when a vaginal plug was observed was noted as E0.5. In some cases, embryos were generated by in vitro fertilization as previously described (56). Genotyping was performed with DNA prepared from tail snips according to the protocol in the Jackson Laboratory (https://www.jax.org/jax-mice-and-services/customer-support/technical-support/genotyping-resources/dna-isolation-protocols) using the primers described in SI Appendix, Table S2.
Cell Lines, Plasmids, Ab, and Reagents.
Human HEK293T (American Type Culture Collection CRL-3216) and BeWo (Riken RCB3678) cells were maintained in Dulbecco's modified eagle medium (DMEM) containing 10% fetal calf serum (FCS) (Gibco) and in Ham’s F-12 medium containing 15% FCS, respectively. Mouse cardiac myocytes were prepared as previously described (57). In brief, the hearts of E13.5 embryos were washed with phosphate-buffered saline (PBS), placed in 300 μL of enzyme solution [350 μg/mL pancreatin (Nacalai Tesque) in 136.9 mM NaCl, 11.1 mM D-glucose, 2.6 mM KCl, 41.7 mM NaH2PO4, and 11.9 mM NaHCO3], and incubated at 37 °C for 10 min. The solution excluding the undigested heart was transferred to a new tube, mixed with 300 μL of DMEM containing 10% FCS, and spun at 1,000 rpm for 10 min. Precipitated myocytes were suspended in DMEM-10% FCS. Fresh enzyme solution was concurrently added to the heart and incubated at 37 °C for 10 min. This procedure (the treatment with enzyme solution and collection by centrifugation) was repeated five to eight times, and all myocytes were combined.
The plasmids pMAL-p2x and pGEX-5x-1 were purchased from New England Biolabs and GE Healthcare, respectively. pX459v2 was from Addgene. pCMV-VSV-G and pCAG-HIVgp were from Riken. The pLVSIN-EF-1α-Pur vector was from Takara Bio.
A horseradish peroxidase (HRP)–labeled mouse anti-FLAG mAb (clone M2) and chicken anti-rat Mct1 Ab were purchased from Sigma-Aldrich. HRP-goat anti-mouse Immunoglobulin (Ig)G2a heavy-chain Ab, rabbit anti-human cTnT mAb (clone EPR3696), Alexa Fluor 568–donkey anti-rat IgG (Heavy and Light chains, H+L) preadsorbed Ab, and Alexa Fluor 568–donkey anti-mouse IgG (H+L) Ab were from Abcam. Rat anti-mouse E-cadherin mAb (clone ECCD-2), Alexa Fluor 488–goat anti-rat IgG (H+L) Ab, Alexa Fluor 488–goat anti-chicken IgY (H+L) Ab, and Alexa Fluor 568–goat anti-rabbit IgG (H+L) Ab were from Invitrogen. HRP-goat anti-mouse IgG Ab was purchased from Dako Agilent. Mouse anti-human Mct4 mAb (clone D-1) was from Santa Cruz.
NBD-PS was purchased from Avanti Polar Lipids. X-gal was from Fujifilm Wako Pure Chemical Co. SPiDER-βGal, DAPI, and O,O’-Bis(2-aminophenyl)ethyleneglycol-N,N,N',N'-tetraacetic acid, tetraacetoxymethyl ester (BAPTA-AM) were from Dojindo Molecular Technologies. Forskolin and ionomycin were from Sigma-Aldrich and Merck, respectively. Cy5-labeled annexin V was purchased from BioVision. Accutase was from Nacalai Tesque.
mAb against Mouse ATP11A.
A hybridoma (clone 4-C11) producing mAb against mouse ATP11A was established by immunizing MRL-lpr/lpr mice as previously described (58) with a recombinant mouse ATP11A protein. In brief, two regions of mouse ATP11A (amino acids 427 to 513 and 408 to 892) were fused to glutathione S-transferase (GST) and maltose-binding protein (MBP), respectively, produced in Escherichia coli BL21 and were purified using glutathione Sepharose (GE Healthcare) and amylose resin (New England Biolabs), respectively.
Female MRL-lpr/lpr mice at the age of 4 wk were injected subcutaneously twice at a 2-wk interval with 50 μg of GST-ATP11A mixed with TiterMax Gold (TiterMax). Mice were further immunized by intraperitoneal injections of 50 μg of the protein in PBS at a 3-d interval. After six injections, the Ab serum titer plateaued. Lymphocytes were prepared from the popliteal and inguinal lymph nodes 1 d after the last injection and fused with NSOBcl-2 myeloma (59). Hybridomas were cultured at 37 °C in DMEM containing 10% FCS, 10% NCTC-109 (Gibco), 1% nonessential amino acids, 5.5 U/mL human IL-6, 100 μM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine, and screened by enzyme-linked immunosorbent assay (ELISA) using MBP-fused ATP11A. A positive hybridoma (clone 4-C11) was cultured in ASF medium 104N (Ajinomoto), and the Ab was purified by (NH4)2SO4 precipitation, followed by dialysis against PBS. The Ig subclass of the 4-C11 mAb, identified by the IsoStrip Mouse Monoclonal Antibody Isotyping kit (Roche Diagnostics), was IgG2a. In ELISA to assess the Ab titer, 96-well plates were coated with 50 ng of MBP-ATP11A. Samples were serially diluted with PBS containing 1% bovine serum albumin (BSA) and 0.05% Tween 20, added to the wells, and incubated at 4 °C for 12 to 16 h. The assay was developed with HRP-goat anti-mouse IgG, and peroxidase activity was detected with a peroxidase-detecting kit (Sumiron, Sumitomo) using o-phenylenediamine as a substrate. Absorbance at 492 nm was measured using a microplate reader (Infinite M200, Tecan).
Real-Time RT-PCR and Western Blotting.
RNA was isolated from tissues and cells as previously described (60) and reverse transcribed using a High Capacity RNA-to-cDNA kit (Applied Biosystems). Real-time PCR was performed using the LightCycler system (Roche Diagnostics) with the primers described in SI Appendix, Table S2. Regarding Western blotting, mouse tissues were suspended in radio-immunoprecipitation assay (RIPA) buffer [50 mM Hepes-NaOH (pH 7.6), 150 mM NaCl, 1 mM ethylene glycol tetraacetic acid (EGTA), 1.5 mM MgCl2, 10% glycerol, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), and 0.5% sodium deoxycholate] containing 1% protease inhibitor mixture (Nacalai Tesque), broken by sonication at 4 °C, and centrifuged at 20,000 × g for 30 min. The supernatants (1.0 to 8.5 μg of protein) were separated by 7.5% SDS-polyacrylamide gel electrophoresis (PAGE) (Extra PAGE One Precast Gel, Nacalai Tesque) and transferred to polyvinylidene difluoride (PVDF) membranes (Merck). After incubating at room temperature for 1 h in buffer A (PBS containing 0.05% Tween 20 and 1 to 5% skim milk), the membrane was incubated at 4 °C for 12 to 16 h with 0.2 μg/mL 4-C11 mAb in buffer A, followed by incubation for 1 h with 1 μg/mL HRP-goat anti-mouse IgG2a heavy-chain Ab. HRP activity was visualized using Western LightningPlus-ECL (PerkinElmer) and detected by LAS4000 (GE Healthcare).
Histological Analyses and TUNEL Staining.
In the histological analysis, tissues were fixed at room temperature for 48 h in PBS-4% paraformaldehyde (PFA), soaked overnight in 70% ethanol, dehydrated, embedded in paraffin, sliced into 4- to 6-μm-thick sections, and mounted on glass slides. Samples were then stained with hematoxylin and eosin (H&E) and observed under a fluorescence microscope. Regarding immunohistological staining, placentas were mounted in optimal cutting temperature (OCT) compound (Sakura Finetek), frozen at −80 °C, and sectioned at a thickness of 5 μm. After fixing with 4.0% PFA for 20 min, cells were permeabilized by treatment with 0.1% Triton X-100 at room temperature for 5 min. Sections were incubated for 60 min with 2 μg/mL chicken anti-rat Mct1 Ab and 5 μg/mL mouse anti-human Mct4 mAb, followed by incubation with Alexa Fluor 488–goat anti-chicken IgY Ab and Alexa Fluor 568–donkey anti-mouse IgG (H+L) Ab. Samples were mounted on FluorSave and observed by confocal fluorescence microscopy (IX81, Olympus).
In TUNEL staining, placentas were fixed at room temperature for 48 h in PBS-4% PFA, dehydrated, embedded in paraffin, and sliced into 6-μm-thick sections. TUNEL staining was performed using the In Situ Cell Death Detection kit, tetramethylrhodamine (TMR) red (Roche Diagnostics). Sections were counterstained with 1 μg/mL DAPI, mounted on FluorSave, and observed by fluorescence microscopy (BioRevo BZ-9000, Keyence). TUNEL-positive areas were quantitated using ImageJ software (https://imagej.nih.gov/ij/).
X-Gal and SPiDER-βGal Staining.
Tissues were fixed at 4 °C for 1 h in PBS containing 4% PFA and washed with X-gal buffer (PBS containing 2 mM MgCl2, 0.02% Nonidet P-40, and 0.01% sodium deoxycholate). After pricking several places with a needle, tissues were immersed in 2 mg/mL X-gal in X-gal buffer containing 5 mM potassium ferricyanide and incubated in the dark at 37 °C for 24 to 48 h. Tissues were fixed at room temperature for 48 h in 4% PFA, soaked overnight in 70% ethanol, and dehydrated. Samples were then embedded in paraffin using CT-Pro20 (Japan Genetics) and sliced into 12-μm-thick sections using a microtome (RM2245, Leica). After being deparaffinized and rehydrated, samples were counterstained with 0.1% nuclear fast red (Sigma) in 5% (wt/vol) aluminum sulfate and observed on a fluorescence microscope.
In SPiDER-βGal staining (19), mouse cardiomyocytes cultured on fibronectin-coated glass coverslips were fixed at room temperature for 10 min in PBS-0.2% PFA and incubated at 37 °C for 1 to 2 h in PBS containing 3 to 5 μM SPiDER-βGal and 0.1% Triton X-100. Samples were further incubated at room temperature for 60 min in PBS-2% BSA and stained with 2 μg/mL rabbit anti-human cTnT Ab, followed by incubation with 1 μg/mL Alexa Fluor 568–goat anti-rabbit IgG Ab. Samples were stained with 1 μg/mL DAPI, mounted with FluorSave, and observed under a confocal fluorescence microscope (IX83, Olympus).
Electron Microscopy.
On day 13 postmating, pregnant mice were anesthetized by an intraperitoneal injection of three anesthetics [medetomidine hydrochloride (Zenoaq), midazolam (Astellas Pharma), and butorphanol (Meiji Seika Pharma)] and subjected to cardiac perfusion with 200 to 300 mL of 0.1 M sodium phosphate buffer (pH 7.4) containing 2% glutaraldehyde and 2% PFA. The placenta was removed, fixed at 4 °C overnight, and left at room temperature for 3 d in 0.1 M sodium phosphate buffer (pH 7.4). Samples were postfixed with 1% OsO4 at 4 °C for 2 h and dehydrated. Samples were incubated twice in propylene oxide (PO) for 20 min, in a 3:1 mixture of PO and epoxide for 1 h, in a 1:3 mixture of PO and epoxide for 1 h, and in epoxide overnight. After incubating in epoxide at 60 °C for 3 d, ultrathin sections (thickness of 70 to 80 nm) were prepared with an Ultracut UCT ultramicrotome (Leica), stained with uranyl acetate and lead citrate, and observed with a transmission electron microscope (H-7650, Hitachi High-Technologies) or scanning electron microscope (Helios NanoLab 660, Field Electron and Ion Company).
Gene Editing and Transformation of Cell Lines.
The ATP11A and ATP11C genes in BeWo cells were knocked out using the CRISPR-Cas9 system (9, 32). Complementary oligonucleotides carrying the single-guide (sg)RNA target sequence for human ATP11A and ATP11C genes (SI Appendix, Table S2) were annealed and ligated into pX459v2. Plasmid DNA was introduced into BeWo cells using FuGENE®︎HD (Promega). Single clones were isolated by limiting dilutions and genotyped by sequencing the target region of chromosomal DNA. Human ATP11A cDNA (NM_032189.4) was previously described (8). The DNA fragment for Flag-tagged ATP11A was inserted into pLVSIN-EF-1α Neo and introduced into HEK293T cells with pCAG-HIVgp and pCMV-VSV-G-RSV-rev using Fugene6 (Promega). After culturing for 2 d, the virus in the supernatant was collected with the Lenti-X Concentrator (Takara Bio) and used to infect DKO-BeWo cells. Transformants were selected with 250 μg/mL G418.
Flippase Assay.
Flippase activity for endogenous PtdSer or exogenous PtdSer analog was assessed as previously described (16). In brief, 5.0 × 104 BeWo cells in a 12-well plate were cultured overnight, washed with Hanks' balanced salt solution (HBSS) (−) without phenol red (buffer B), and incubated at 15 °C with 1 μM NBD-PS in 200 μL of buffer B. Cells were incubated at 4 °C for 1 min in buffer B containing 5 mg/mL fatty acid–free BSA, treated at room temperature for 3.5 min with Accutase, mixed with 200 μL of buffer B containing 5 mg/mL fatty acid–free BSA and 500 nM Sytox blue (Thermo Fisher Scientific), and analyzed by fluorescence-activated cell sorting (FACS) Canto II (BD Biosciences). Data were assessed by FlowJo software (BD Biosciences). To evaluate flippase activity for endogenous PtdSer, BeWo cells were cultured as above, detached by a treatment with Accutase, and collected by centrifugation at 200 × g at 4 °C for 2 min. Cells were then treated at 15 °C for 5 min with 5 μM ionomycin in 800 μL of annexin V buffer [10 mM Hepes-NaOH (pH 7.6), 140 mM NaCl, 2.5 mM CaCl2] to expose PtdSer. Cells were collected by centrifugation, resuspended in 600 μL of DMEM containing 15% FCS, 5 mM EGTA, and 20 μM BAPTA-AM, and incubated at 25 °C. Cells were then suspended in 400 μL of annexin V buffer containing 1,000-fold diluted Cy5-labeled annexin V and 5 μg/mL propidium iodide (PI; Merck) and analyzed by FACS Canto II.
Cell Fusion with BeWo Cells.
The fusion of BeWo cells was performed according to Omata et al. (35). In brief, 1 × 105 BeWo cells in a six-well plate were treated for 48 h with 20 μM forskolin with a medium change at 24 h. Cell fusion was evaluated by immunohistochemical staining followed by morphological observations and by scanning electron micrography. Cells were fixed with 4% PFA, permeabilized with 0.1% Triton X-100, and stained at room temperature for 1 h with 2 μg/mL rat anti-mouse E-cadherin mAb, followed by incubation with 1 μg/mL Alexa Fluor 488–goat anti-rat IgG (H+L) Ab. Samples were counterstained with 5 μg/mL DAPI, mounted by FluorSave, and observed under a confocal fluorescence microscope.
Regarding scanning electron microscopy, cells were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4), postfixed with 2% OsO4, and dehydrated with a graded series of ethanol. After treatment with t-butyl alcohol, samples were freeze-dried with a freeze dryer (VFD-30, Vacuum Device) and mounted on aluminum stubs with carbon paste. Dried specimens were coated with osmium with a Neoc-Pro osmium coater (Meiwafosis) and observed under the field-emission scanning electron microscope (JSM-IT800, JEOL). To estimate cellular areas and perimeters, cell boundaries were traced and analyzed using ImageJ software. Data were expressed as the area (in squared micrometers) and perimeter (in micrometers) of the syncytium surrounded by the cell boundary.
Statistical Analysis.
Data obtained from each experiment were expressed as the mean ± SEM or the mean ± SD. Statistical analyses were performed using Student’s t test.
Supplementary Material
Acknowledgments
We thank M. Kamada for secretarial assistance. This work was supported in part by grants-in-aid from the Japan Society for the Promotion of Science (21H04770) to S.N.
Footnotes
Reviewers: R.B., Rutgers The State University of New Jersey; and Y.H., Tokyo Daigaku.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200582119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Harayama T., Riezman H., Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 19, 281–296 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Leventis P. A., Grinstein S., The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407–427 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Birge R. B., et al. , Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 23, 962–978 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevers E. M., Williamson P. L., Getting to the outer leaflet: Physiology of phosphatidylserine exposure at the plasma membrane. Physiol. Rev. 96, 605–645 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Nagata S., Suzuki J., Segawa K., Fujii T., Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 23, 952–961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen J. P., et al. , P4-ATPases as phospholipid flippases—Structure, function, and enigmas. Front. Physiol. 7, 275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.López-Marqués R. L., Theorin L., Palmgren M. G., Pomorski T. G., P4-ATPases: Lipid flippases in cell membranes. Pflugers Arch. 466, 1227–1240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segawa K., Kurata S., Nagata S., Human type IV P-type ATPases that work as plasma membrane phospholipid flippases and their regulation by caspase and calcium. J. Biol. Chem. 291, 762–772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segawa K., et al. , Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344, 1164–1168 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Nakanishi H., et al. , Crystal structure of a human plasma membrane phospholipid flippase. J. Biol. Chem. 295, 10180–10194 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siggs O. M., et al. , The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nat. Immunol. 12, 434–440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siggs O. M., Schnabl B., Webb B., Beutler B., X-linked cholestasis in mouse due to mutations of the P4-ATPase ATP11C. Proc. Natl. Acad. Sci. U.S.A. 108, 7890–7895 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yabas M., et al. , Mice deficient in the putative phospholipid flippase ATP11C exhibit altered erythrocyte shape, anemia, and reduced erythrocyte life span. J. Biol. Chem. 289, 19531–19537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yabas M., et al. , ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat. Immunol. 12, 441–449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arashiki N., et al. , ATP11C is a major flippase in human erythrocytes and its defect causes congenital hemolytic anemia. Haematologica 101, 559–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segawa K., et al. , Phospholipid flippases enable precursor B cells to flee engulfment by macrophages. Proc. Natl. Acad. Sci. U.S.A. 115, 12212–12217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takata K., Fujikura K., Shin B.-C., Ultrastructure of the rodent placental labyrinth: A site of barrier and transport. J. Reprod. Dev. 43, 13–24 (1997). [Google Scholar]
- 18.Hu D., Cross J. C., Development and function of trophoblast giant cells in the rodent placenta. Int. J. Dev. Biol. 54, 341–354 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Doura T., et al. , Detection of LacZ-positive cells in living tissue with single-cell resolution. Angew. Chem. Int. Ed. Engl. 55, 9620–9624 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Agah R., et al. , Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Invest. 100, 169–179 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Garcia V., et al. , Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature 555, 463–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weninger W. J., et al. , Phenotyping structural abnormalities in mouse embryos using high-resolution episcopic microscopy. Dis. Model. Mech. 7, 1143–1152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karp N. A., Heller R., Yaacoby S., White J. K., Benjamini Y., Improving the identification of phenotypic abnormalities and sexual dimorphism in mice when studying rare event categorical characteristics. Genetics 205, 491–501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood H. B., Episkopou V., Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 86, 197–201 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Zappone M. V., et al. , Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development 127, 2367–2382 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Hayashi S., Lewis P., Pevny L., McMahon A. P., Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119 (suppl. 1), S97–S101 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Nagai A., Takebe K., Nio-Kobayashi J., Takahashi-Iwanaga H., Iwanaga T., Cellular expression of the monocarboxylate transporter (MCT) family in the placenta of mice. Placenta 31, 126–133 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Dupressoir A., et al. , Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. U.S.A. 106, 12127–12132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueno M., et al. , c-Met-dependent multipotent labyrinth trophoblast progenitors establish placental exchange interface. Dev. Cell 27, 373–386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh B., Blelloch R., Single nuclei RNA-seq of mouse placental labyrinth development. eLife 9, e60266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wice B., Menton D., Geuze H., Schwartz A. L., Modulators of cyclic AMP metabolism induce syncytiotrophoblast formation in vitro. Exp. Cell Res. 186, 306–316 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki J., Umeda M., Sims P. J., Nagata S., Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Coutifaris C., et al. , E-cadherin expression during the differentiation of human trophoblasts. Development 113, 767–777 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Omata W., Ackerman W. E. IV, Vandre D. D., Robinson J. M., Trophoblast cell fusion and differentiation are mediated by both the protein kinase C and a pathways. PLoS One 8, e81003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maruyama E. O., et al. , Extraembryonic but not embryonic SUMO-specific protease 2 is required for heart development. Sci. Rep. 6, 20999 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gessert S., Kühl M., The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ. Res. 107, 186–199 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Foglia M. J., Poss K. D., Building and re-building the heart by cardiomyocyte proliferation. Development 143, 729–740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thornburg K. L., O’Tierney P. F., Louey S., Review: The placenta is a programming agent for cardiovascular disease. Placenta 31 (suppl.), S54–S59 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., et al. , Proteomic analysis and functional characterization of P4-ATPase phospholipid flippases from murine tissues. Sci. Rep. 8, 10795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takatsu H., et al. , ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J. Biol. Chem. 286, 38159–38167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katsuragawa H., et al. , Monoclonal antiphosphatidylserine antibody reactivity against human first-trimester placental trophoblasts. Am. J. Obstet. Gynecol. 172, 1592–1597 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Das M., et al. , Phosphatidylserine efflux and intercellular fusion in a BeWo model of human villous cytotrophoblast. Placenta 25, 396–407 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Žigon P., et al. , Anti-phosphatidylserine/prothrombin antibodies are associated with adverse pregnancy outcomes. J. Immunol. Res. 2015, 975704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitlock J. M., Chernomordik L. V., Flagging fusion: Phosphatidylserine signaling in cell-cell fusion. J. Biol. Chem. 296, 100411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brukman N. G., Uygur B., Podbilewicz B., Chernomordik L. V., How cells fuse. J. Cell Biol. 218, 1436–1451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., et al. , TMEM16F phospholipid scramblase mediates trophoblast fusion and placental development. Sci. Adv. 6, eaba0310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang H., et al. , TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell 151, 111–122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeung T., et al. , Membrane phosphatidylserine regulates surface charge and protein localization. Science 319, 210–213 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Degrelle S. A., Gerbaud P., Leconte L., Ferreira F., Pidoux G., Annexin-A5 organized in 2D-network at the plasmalemma eases human trophoblast fusion. Sci. Rep. 7, 42173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viall C. A., Chamley L. W., Histopathology in the placentae of women with antiphospholipid antibodies: A systematic review of the literature. Autoimmun. Rev. 14, 446–471 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Tsuchiya M., et al. , Cell surface flip-flop of phosphatidylserine is critical for PIEZO1-mediated myotube formation. Nat. Commun. 9, 2049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grifell-Junyent M., et al. , CDC50A is required for aminophospholipid transport and cell fusion in mouse C2C12 myoblasts. J. Cell Sci. 135, jcs258649 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagata S., Apoptosis and clearance of apoptotic cells. Annu. Rev. Immunol. 36, 489–517 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Heyne G. W., et al. , A simple and reliable method for early pregnancy detection in inbred mice. J. Am. Assoc. Lab. Anim. Sci. 54, 368–371 (2015). [PMC free article] [PubMed] [Google Scholar]
- 56.Tokuhiro K., Ikawa M., Benham A. M., Okabe M., Protein disulfide isomerase homolog PDILT is required for quality control of sperm membrane protein ADAM3 and male fertility [corrected]. Proc. Natl. Acad. Sci. U.S.A. 109, 3850–3855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodgers L. S., Schnurr D. C., Broka D., Camenisch T. D., An improved protocol for the isolation and cultivation of embryonic mouse myocytes. Cytotechnology 59, 93–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tabata N., et al. , Establishment of monoclonal anti-retroviral gp70 autoantibodies from MRL/lpr lupus mice and induction of glomerular gp70 deposition and pathology by transfer into non-autoimmune mice. J. Virol. 74, 4116–4126 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ray S., Diamond B., Generation of a fusion partner to sample the repertoire of splenic B cells destined for apoptosis. Proc. Natl. Acad. Sci. U.S.A. 91, 5548–5551 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chomczynski P., Sacchi N., Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 (1987). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.