Significance
Antagonistic pleiotropy (AP) is a prevailing theory of the evolution of aging; however, it lacks direct experimental evidence at an individual gene level. We performed unbiased translatome analyses of Caenorhabditis elegans recovering from starvation and identified that the trl-1 gene hidden in a pseudogene generates proteinaceous products upon refeeding. Compared with wild-type animals, trl-1 mutants increased brood sizes, shortened the animals’ lifespan, and specifically impaired germline deficiency–induced longevity. The TRL-1 protein undergoes liquid–liquid phase separation, through which TRL-1 granules recruit vitellogenin messenger RNA and inhibit its translation. These results provide evidence that trl-1 regulates the reproduction–longevity tradeoff by optimizing nutrient production for the next generation, thereby supporting the AP theory of aging at the single-gene level.
Keywords: antagonistic pleiotropy, reproduction, longevity, liquid–liquid phase separation
Abstract
The antagonistic pleiotropy theory of aging proposes that genes enhancing fitness in early life limit the lifespan, but the molecular evidence remains underexplored. By profiling translatome changes in Caenorhabditis elegans during starvation recovery, we find that an open reading frame (ORF) trl-1 “hidden” within an annotated pseudogene significantly translates upon refeeding. trl-1 mutant animals increase brood sizes but shorten the lifespan and specifically impair germline deficiency–induced longevity. The loss of trl-1 abnormally up-regulates the translation of vitellogenin that produces copious yolk to provision eggs, whereas vitellogenin overexpression is known to reduce the lifespan. We show that the TRL-1 protein undergoes liquid–liquid phase separation (LLPS), through which TRL-1 granules recruit vitellogenin messenger RNA and inhibit its translation. These results indicate that trl-1 functions as an antagonistic pleiotropic gene to regulate the reproduction–longevity tradeoff by optimizing nutrient production for the next generation.
The antagonistic pleiotropy theory of aging suggests that aging is caused by pleiotropic genes whose functions are beneficial to the organism’s fitness early in life, whereas these functions are detrimental to its fitness later. Natural selection has been compelled to accept the latter as a necessary cost of the former (1–5). Quantitative genetic studies demonstrated tradeoffs between fecundity and lifespan: Fruit flies selected for late-life reproductive success showed prolonged lifespan with a decline in early-life fecundity and mating success compared with lines selected for early-life reproductive success (6, 7). Similar tradeoffs were observed in naturally segregating polymorphisms (8, 9) and studies selecting directly for longevity (10). These studies described the apparent tradeoff between traits, which is consistent with the idea of antagonistic pleiotropy but is also explainable by multiple tightly linked genes bearing opposite fitness effects at different life stages. The identification of antagonistic pleiotropy at an individual gene resolution remains scarce (11).
Experimental evolution studies reveal that mutations can modulate longevity in various model organisms, but evidence on early-life fitness components is available for only a few (4, 5). Caenorhabditis elegans, Drosophila melanogaster, and the mouse can increase their lifespans without fitness costs (4, 12–14). Temporal modulation of the C. elegans insulin/IGF1 pathway allows dissociating its roles in longevity and reproduction (15). Signals from the C. elegans germline and somatic gonad have opposite effects on aging, suggesting an inherent relationship between the reproductive state, but not fecundity per se, and longevity (12). These observations challenged the antagonistic pleiotropy theory as a major explanation for aging.
This study performed unbiased translatome and transcriptome analyses of C. elegans recovering from starvation and identified that the trl-1 gene generates proteinaceous products. We showed that the TRL-1 protein undergoes liquid–liquid phase separation (LLPS) with vitellogenin RNA to regulate the reproduction–lifespan tradeoff under starvation and physiological conditions, which supports the antagonistic pleiotropy theory of aging at the single-gene level.
Results
Translatome Profiling on C. elegans Recovering from Starvation.
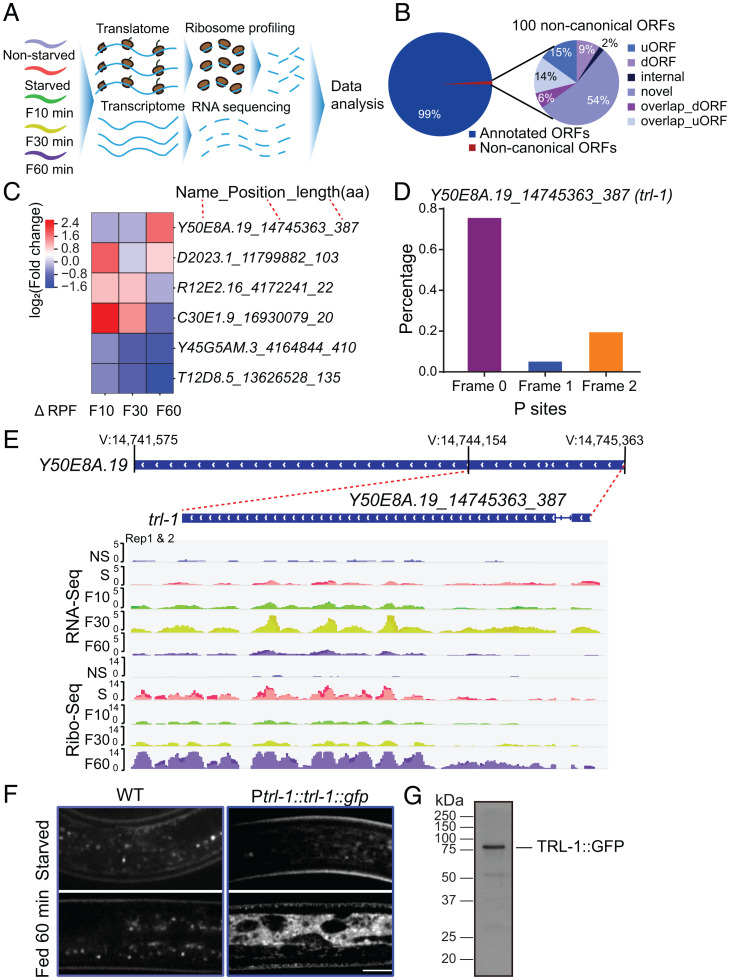
In response to famine, the nematode C. elegans larvae form dauer larvae, which reverse their long-term survival to reproduction after refeeding (16). To monitor translatome changes during the recovery, we starved C. elegans larvae for 6 d and collected them after refeeding with bacterial food for 0, 10, 30, and 60 min. We generated libraries for ribosome profiling (Ribo sequencing [Ribo-Seq]) and RNA sequencing (RNA-Seq) and collected a reference sample from the nonstarved (NS) animals (Fig. 1A). Our RNA-Seq and ribosome profiling measurements show high reproducibility (SI Appendix, Fig. S1A) and are distinct from controls (SI Appendix, Fig. S1B). Sequenced ribosomal footprints map primarily to coding sequences (CDSs) of genes (SI Appendix, Fig. S1C) and display the three-nucleotide codon movement characteristic of actively translating ribosomes (SI Appendix, Fig. S1D).
Fig. 1.
trl-1 translates upon refeeding. (A) Schematic overview of the experimental procedure. (B) Pie chart of the percentage breakdown of protein-coding genes annotated from Ribo-Seq by RiboCode (CPM ≥ 1). The noncanonical ORFs are classified into uORF, dORF, internal ORF, novel ORF, overlap_dORF, and overlap_uORF. (C) Heatmap of the top differentially regulated novel ORFs [log2(fold change), adjusted P ≤ 0.05] associated with ribosomes after refeeding stimulation in C. elegans. (D) RiboCode analysis of ribosome P sites distribution in the expected reading frame for noncanonical ORF Y50E8A.19_14745363_387(trl-1). (E) Patterns of RNA-Seq transcriptional reads (Top) and ribosome profiling translational reads (Bottom) for trl-1 from nonstarved (NS), starved (S), refed 10-min (F10), 30-min (F30), and 60-min (F60) worms. The trl-1 gene structure is in the center, with a thin blue line representing the intron and wide blue rectangles indicating exons. (F) Representative imaging of WT (N2) and TRL-1::GFP dauer under starvation or refed 60-min conditions. TRL-1::GFP stands for the GFP fusion to the C-terminal of TRL-1. (Scale bar, 5 μm.) (G) Western blot analysis of the TRL-1::GFP protein in C. elegans.
Ribo-Seq facilitates the detection of pervasive but uncharacterized translation outside of canonical CDSs (17), which prompts us to identify noncanonical open reading frames (ORFs) that translate upon refeeding. Using the established bioinformatic pipelines (18) (Materials and Methods), we identified 8,850 annotated CDSs (99% of the identified CDSs) and uncovered 100 distinct, noncanonical CDSs (Fig. 1B and Dataset S1). Among the noncanonical CDSs, 54 were CDSs on transcripts lacking previous protein-coding annotations (“novel,” i.e., pseudogene or long noncoding RNAs [lncRNAs]), and 29 CDSs were upstream CDSs (i.e., uORFs or start overlaps: CDSs that overlap annotated start codons in a different reading frame), and 15 were downstream ORFs (dORFs or stop overlaps: CDSs that overlap annotated stop codons in a different reading frame) (Fig. 1B and Dataset S1).
trl-1 Translates upon Refeeding.
By searching for protein-coding noncanonical ORFs whose translation markedly increased after bacterial food stimulation (Fig. 1C and Dataset S2), we identified an ORF (trl-1) that is “hidden” within a previously annotated pseudogene Y50E8A.19. The average ribosome density of the trl-1 CDS resembles footprints from annotated coding regions with strong three-nucleotide periodicities (Fig. 1D), a hallmark of active translation. The previous RNA-Seq of nonstarved animals detected little Y50E8A.19 transcription (19), confirmed by our work (Fig. 1E and SI Appendix, Fig. S1E). We revealed a dynamic change of its transcription in starved animals responding to food supplies: trl-1 up-regulates its transcription 10 min after refeeding, reaches the maximum level at 30 min, and down-regulates at 60 min (Fig. 1E and SI Appendix, Fig. S1E). The translation of trl-1 messenger RNA (mRNA) is detectable at starvation and reaches the peak at 60 min, leading to the maximum translational efficiency at 60 min after refeeding (Fig. 1E and SI Appendix, Fig. S1E). We noticed that TRL-1 translation was low during the 10 to 30 min of refeeding but increased at 60 min. Intriguingly, ribosome biogenesis-related genes were translationally up-regulated immediately after refeeding (SI Appendix, Fig. S1F and Dataset S3, Gene Ontology [GO] term). We suspect that animals starved for 6 d probably had a low level of ribosome proteins, and that refed animals must first generate a sufficient amount of ribosomes before translating other proteins, thereby resulting in a high trl-1 translation 60 min after refeeding.
To validate Ribo-Seq results, we generated a green fluorescence protein (GFP)–based translational reporter under the control of the trl-1 promoter. In the fed or starved animals, the TRL-1::GFP signal is barely detectable (Fig. 1F); neither did we visualize GFP at 10 or 30 min after refeeding. By contrast, TRL-1::GFP animals recovering 60 min with food supplies produced the GFP fluorescence (Fig. 1F), reflecting its high translation level, speeds of transcription, translation, and GFP fluorophore maturation. To verify the TRL-1 protein production, we affinity purified GFP-tagged TRL-1 fusion protein from the refeeding animals and detected the protein at the expected molecular weight (Fig. 1G). By subjecting the purified TRL-1::GFP protein to mass spectrometry, we found that lytic protease-digested peptides mapped uniquely to the predicted TRL-1 ORF-encoded protein (SI Appendix, Fig. S2A and Dataset S4, 60% total protein coverage). Using a heat-shock promoter to overexpress trl-1, we found that its overexpression caused embryonic lethality or larval arrest from two independent transgenic lines (SI Appendix, Fig. S2 B and C), indicating that TRL-1 overproduction is deleterious to C. elegans. These results show that trl-1 translates into protein upon refeeding and that the animals control an appropriate TRL-1 protein level.
trl-1 Deletions Increase Brood Sizes by Up-Regulating Vitellogenin Translation.
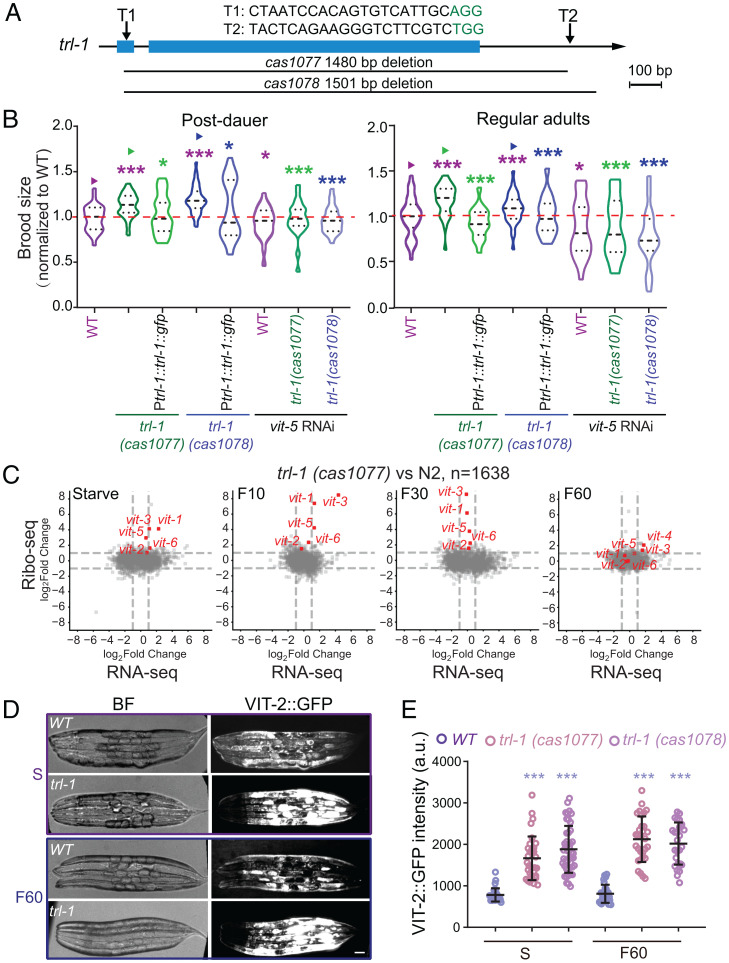
To dissect the function of trl-1, we implemented the CRISPR-Cas9–based gene deletion strategy to remove its coding region, generating two putative null alleles of trl-1 (Fig. 2A). Both trl-1 alleles are indistinguishable from the wild type (WT) in animal morphology or growth rates under the optimal culture conditions, consistent with low transcriptional and translational levels of trl-1. We have not detected any apparent defects in animal survival or efficiency to recovery from starvation in trl-1 mutants. By measuring brood sizes from the postdauer adults, L1 diapause or adult diapause adults, we found that trl-1 deletions caused an ∼15% increase of brood sizes in both alleles compared to that from WT animals under the same culture conditions (Fig. 2B and SI Appendix, Fig. S2D). The expression of the Ptrl-1::trl-1::gfp transgene rescued the increased brood size phenotype in trl-1 mutant animals (Fig. 2B). These results indicate that trl-1 restricts the brood size in starved animals after refeeding and provide evidence that the TRL-1::GFP translational reporter is functional. The reduced brood size by gene deletion is expected; however, the null genetic mutations that increase brood sizes have not been well documented (11).
Fig. 2.
trl-1 deletions increase brood sizes by up-regulating vitellogenin translation. (A) Schematic design of the CRISPR-Cas9–assisted knockout for trl-1 in C. elegans. T1 and T2 represent the single-guide RNA (sgRNA) targeting the trl-1. cas1077 and cas1078 are the two independent deletion alleles of trl-1. (Scale bar, 100 bp.) (B) Normalized brood sizes of WT (N2), trl-1(cas1077), trl-1(cas1078), vit-5 RNAi, and trl-1 overexpression animals in postdauer stages (Left) and regular adult stages (Right) (mean ± SD). n ≥ 17, statistical significances were calculated by Student’s t test, *P < 0.05, and ***P < 0.001. For each worm, the brood size was normalized to the mean value of matched WT controls. (C) Scatterplots of the gene expression fold changes of RNA-Seq and Ribo-Seq in trl-1(cas1077) worms under starved (S), refed 10-min (F10), 30-min (F30), and 60-min (F60) conditions. Red dots show vitellogenin genes. Gray dots show differentially expressed genes in the two independent trl-1 deletion alleles, n = 1,638. (D) Brightfield (BF) and fluorescence images of the VIT-2::GFP knockin show WT (N2) or trl-1 mutants on the starved or refed 60-min (F60) conditions. (Scale bar, 5 μm.) (E) Quantifications of the VIT-2::GFP fluorescence intensity (mean ± SD) in WT and trl-1 mutant animals (n = 35 to 50 animals). Statistical significances were calculated by Student’s t test, ***P < 0.001.
How can trl-1 deletions cause an increase in brood size? By performing RNA-Seq and Ribo-Seq analyses of trl-1 mutant animals recovering from starvation, we revealed a global change of 1,638 genes either in their transcriptional or translational levels at any time point (Dataset S5). The top hits for the translationally up-regulated genes are the vitellogenin gene family (Fig. 2C and SI Appendix, Fig. S2E). In the C. elegans genome, six vitellogenin genes encode yolk proteins that are produced and secreted from the intestine to the germline, providing lipoprotein nutrient resources for the development of the next generation (20). Compared to WT animals upon refeeding, the loss of trl-1 caused an abnormal translational up-regulation among vitellogenin genes (Fig. 2C and SI Appendix, Fig. S2E). To validate Ribo-Seq results, we genetically crossed the vit-2::gfp knockin (KI) into trl-1 deletion strains. VIT-2::GFP signals increased in starved trl-1 mutants recovering from feeding compared with WT animals (Fig. 2 D and E). In line with the translation regulation of vitellogenin, TRL-1::GFP protein distributes in the cytosol of C. elegans intestinal cells where vitellogenin is produced (Fig. 1F). Previous studies established that the increased vitellogenin level leads to increased brood sizes, whereas reducing vitellogenin decreases brood sizes (21, 22). We propose that the ectopic up-regulation of vitellogenin translation might be responsible for increasing brood sizes in trl-1 mutants.
We carried out RNA interference (RNAi) against the vit-5 gene to test this. Protein sequences of the six vitellogenin are highly conserved (SI Appendix, Fig. S2F) (20), and the vit-5 RNAi treatment was previously shown to disrupt other vitellogenin genes (22). We confirmed that RNAi of vit-5 markedly reduced the GFP fluorescence in the VIT-2::GFP KI animals (SI Appendix, Fig. S2G). We found that RNAi of vit-5 restored the abnormally increased brood size in trl-1 mutants to the WT range (Fig. 2B), which indicates that TRL-1 restricts brood sizes by modulating vitellogenin generation.
We measured the maximal brood size of trl-1 hermaphrodites mated with WT males. Compared to the maximal brood size of WT hermaphrodites mating with WT males, we detected an ∼30% larger brood size from trl-1 hermaphrodites mated with WT males (SI Appendix, Fig. S2D). This result suggests that the increased numbers of progeny from trl-1 mutant hermaphrodites is not simply the result of an increase in sperm production compared to the WT, but reflects increased reproductive capacity of the oocyte pool, in agreement with the notion that the trl-1 gene controls the supply of vitellogenin produced from the intestine.
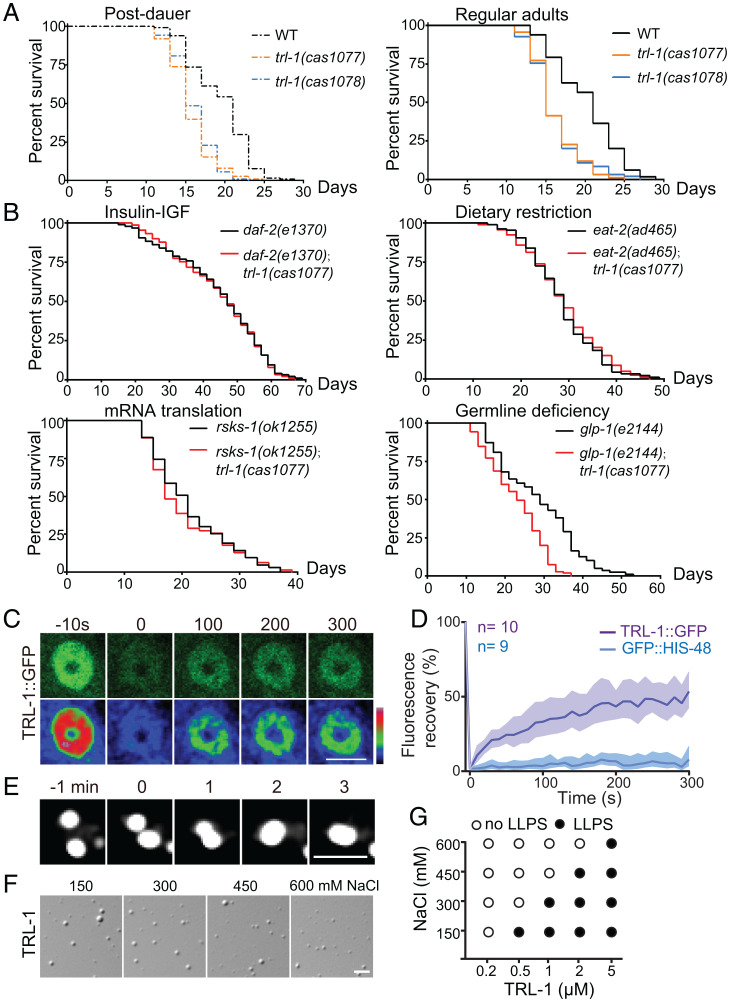
trl-1 Deletions Shorten Lifespan and Impair Germline Deficiency–Induced Longevity.
We wondered whether trl-1 regulates lifespan. Overproduction of vitellogenin has been shown to inhibit germline deficiency–induced longevity (23), whereas RNAi of vitellogenin promoted lifespan (21, 24). We predicted that the increased translation of vitellogenin in trl-1 mutant animals might reduce the lifespan. Indeed, we found a 15% or 20% reduction of lifespan in trl-1(cas1077) or trl-1(cas1078) L1 diapause– or postdauer–stage animals after refeeding, respectively (Fig. 3A and SI Appendix, Figs. S3A and S4A). Multiple genetic pathways, including insulin-IGF1 (daf-2), germline deficiency (glp-1), mRNA translation (rsks-1), and dietary restriction (eat-2) pathways, regulate longevity in C. elegans (25). We introduced trl-1(cas1077) into daf-2(e1370), glp-1(e2144), rsks-1(ok1255), and eat-2(ad465) mutant animals. By measuring the lifespan in the double mutant animals with ample food, we found that trl-1 mutation specifically shortened the lifespan of germline-deficiency glp-1 animals but had negligible effects on other longevity mutants (Fig. 3B and SI Appendix, Fig. S3A), which is consistent with the observation that trl-1 restricts vitellogenin production. By performing intestine-specific overexpression of trl-1, we showed that overexpression of trl-1 at the intestine caused an 18% reduction of brood size compared with control (SI Appendix, Fig. S4B). By contrast, the intestine-specific RNAi of trl-1 caused a 14% increase in brood size (SI Appendix, Fig. S4B). The intestine-specific overexpression of trl-1 led to a 5% increase in lifespan, whereas intestine-specific RNAi of trl-1 caused a 15% reduction in lifespan (SI Appendix, Figs. S3A and S4C). Using the heat-shock promoter-driven inducible overexpression of trl-1 at the L4 stage, we detected a 60% brood size reduction but a 9% lifespan increase (SI Appendix, Figs. S3A and S4 B and D). Together, these results indicate that trl-1 functions in the intestine to regulate the tradeoff between reproduction and longevity.
Fig. 3.
TRL-1 regulates lifespan and germline deficiency–induced longevity and undergoes liquid–liquid phase separation. (A) The lifespan of trl-1 mutant worms compared with WT (N2) under the postdauer stage (Left) and the regular adult stage (Right) with ample food. Postdauer-stage worms were refed with food, and the adulthood day 1 is as t = 1 in lifespan analyses. n = 94 to 115 animals; survival plots are generated by nonparametric methods. (B) Lifespan of trl-1 double mutants compared with the corresponding single mutants [rsks-1(ok1255), daf-2(e1370), eat-2(ad465), and glp-1(e2144)] growing with ample food. The adulthood day 1 is as t = 1 in lifespan analyses. n = 62 to 104 animals; survival plots are generated by nonparametric methods. The results are consistent in three replicates. (C) Recovery of the TRL-1::GFP fluorescence in C. elegans after FRAP. (Scale bar, 5 μm.) (D) Quantifications of FRAP data. Data are mean ± 95% CI from independent animals, n = 9 to 10 animals. (E) Droplet fusion of TRL-1::GFP puncta in C. elegans. (Scale bar, 2 μm.) (F) Droplet formation of 5 μM TRL-1 protein in vitro. (Scale bar, 5 μm.) (G) Formation of TRL-1 droplets in buffers containing different NaCl concentrations.
We extended our studies to the trl-1 null animals that grew in the presence of plentiful food. trl-1 deletions increased brood sizes but decreased their lifespans (Figs. 2B and 3A and SI Appendix, Fig. S3A), revealing similar roles of TRL-1 under physiological conditions. We did not detect any significant difference in the peak egg–laying time between WT and trl-1 mutants after refeeding (SI Appendix, Fig. S4 E, Left). However, when trl-1 mutants grew with ample food, they increased brood sizes but showed a delayed peak egg–laying time (SI Appendix, Fig. S4 E, Right), indicating that increased brood size is not always advantageous. Together, these results demonstrate that trl-1 is an antagonistic pleiotropic gene that restricts the brood size in early life and maintains the regular lifespan later, thereby regulating the tradeoff between reproduction and lifespan (trl).
TRL-1 Protein Undergoes Liquid–Liquid Phase Separation.
We investigated the mechanism through which TRL-1 regulates vitellogenin production. The N terminals of TRL-1 exhibit a significantly high content of intrinsically disordered regions (IDRs) enriched with the polar amino acids glycine (G) and serine (S) residues (SI Appendix, Fig. S5A), which implies a tendency toward LLPS (26–28). We showed that GFP-tagged TRL-1 formed punctate structures in C. elegans or HEK293T cells (SI Appendix, Figs. S2B and S5B). Fluorescence recovery after photobleaching (FRAP) assays revealed a quick recovery of the GFP fluorescence from TRL-1::GFP puncta (Fig. 3 C and D, SI Appendix, Fig. S5 C–E, and Movies S1–S3, GFP-tagged histone H2B as a negative control). Two encountering TRL-1 puncta fused and coalesced into a larger sphere (Fig. 3E, SI Appendix, Fig. S5F, and Movies S4 and S5). These results indicate that TRL-1 puncta are dynamic and possess liquid-like properties in living cells.
Next, we determine whether the TRL-1 protein is the scaffold or a client of LLPS by in vitro biochemical reconstitution. Using differential interference contrast (DIC) microscopy, we found that the purified TRL-1 protein quickly formed spherical droplets in a protein concentration– and salt concentration–dependent manners (Fig. 3 F and G and SI Appendix, Fig. S5G). The fluorescent dye Cy3-labeled TRL-1 droplets formed and grew larger within 30 s (SI Appendix, Fig. S5 H and I and Movie S6). The fluorescent signal of the Cy3-labeled TRL-1 droplets showed a 50% recovery within 300 s after photobleaching inside the droplets (SI Appendix, Fig. S6 A and B and Movie S7). These results indicate that the TRL-1 protein forms droplets with liquid-like properties in test tubes.
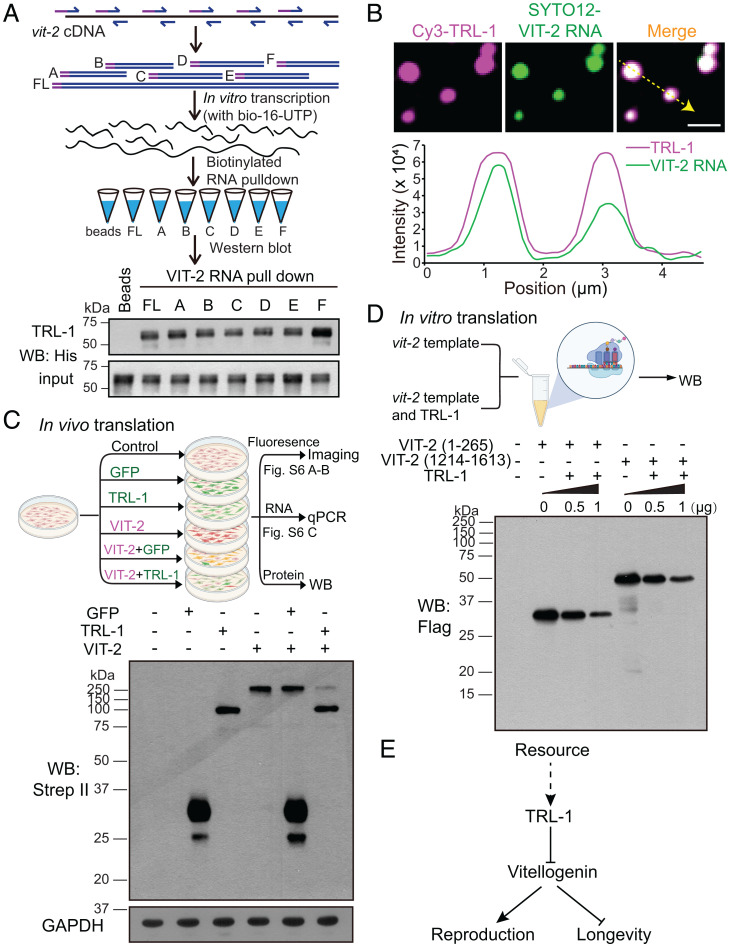
TRL-1 Interacts with Vitellogenin RNA.
The C terminus of TRL-1 protein contains 31% positively charged residuals (SI Appendix, Fig. S6C), raising the possibility that TRL-1 may bind to RNA molecules through electrostatic interactions. To determine whether TRL-1 directly interacts with vitellogenin RNA, we used biotin to label the full length or fragments of the vit-2 RNA and performed an in vitro RNA–protein binding assay (Fig. 4A). Western blotting analyses of His-tagged TRL-1 protein showed that the full length or fragments of vit-2 RNA pulled down TRL-1, but the empty bead control did not (Fig. 4A). We found that RNA fragments of the actin (act-1) and tubulin (tbb-4) genes also pull down TRL-1 protein (SI Appendix, Fig. S6D). By mixing the Cy3-labeled TRL-1 and SYTO12-labeled C. elegans total RNA, we detected the colocalization of the total worm RNA with TRL-1 droplets (SI Appendix, Fig. S6E). Considering that TRL-1 C terminus is positively charged and that all the tested RNA molecules bind to TRL-1, we suggest that TRL-1 may interact with RNA molecules through electrostatic interactions rather than a specific sequence or tertiary structure.
Fig. 4.
TRL-1 binds to vitellogenin RNA and inhibits its translation. (A) Schematic design of the biotin-labeled vit-2 RNA pull-down assays: Fragments of vit-2 RNA or its full length were labeled with biotin-16-UTP by in vitro transcription with T7 polymerase, then capturing and enriching TRL-1 using Dynabeads MyOne Streptavidin C1 beads (Top). Western blotting analysis of the interactions between the full-length and fragmented vit-2 RNA molecules with the TRL-1 protein (Bottom). (B) Representative imaging of the colocalization of TRL-1 proteins (magenta) and vit-2 RNA (green) in droplets and quantification of TRL-1 and vit-2 RNA fluorescence intensity along the yellow line. Magenta line: Cy3-TRL-1; green line: vit-2 RNA. (Scale bar, 2 μm.) (C) Diagram of vit-2, trl-1, and gfp expression in HEK293T cells followed by fluorescence analysis, qPCR, and Western blotting (Top). Western blotting shows the reduction of VIT-2 production in the presence of TRL-1 (Bottom). (D) Diagram of the in vitro translation assay using the human HeLa cell lysate (Top). Western blotting of the VIT-2 translation levels in the presence of TRL-1 at different concentrations (Bottom). (E) A proposed model summarizes that TRL-1 regulates the reproduction–lifespan tradeoff by modulating the translation of vitellogenin.
We examined whether TRL-1 droplets recruit vit-2 RNA in vitro. By mixing the Cy3-labeled TRL-1 and SYTO12-labeled vit-2 RNA, we detected the colocalization of vit-2 RNA with TRL-1 droplets (Fig. 4B). The addition of vit-2 RNA increased the TRL-1 droplet sizes in an RNA concentration–dependent manner, despite the high RNA level (up to 100 ng/μL) reducing TRL-1 droplet sizes (SI Appendix, Fig. S6 F and G). FRAP analyses showed that the TRL-1 plus vit-2 RNA (TRL-1–RNA) droplet recovery was slower than TRL-1 alone (SI Appendix, Fig. S6 A and B and Movie S8), indicating that the TRL-1–RNA droplets are less dynamic. After mixing TRL-1 with vit-2 RNA for 2 h, TRL-1–RNA granules changed from sphere to irregular shape and became even less mobile (SI Appendix, Fig. S6 A, B, H, and I and Movie S9), which suggests that TRL-1–RNA droplets mature over time to form kinetically trapped and stable structures, explaining why the TRL-1 level must be low and transient in C. elegans. Together, these biochemical results indicate that TRL-1 droplets recruit vitellogenin RNA.
TRL-1 Inhibits Vitellogenin RNA Translation.
We investigated the potential functional impact of which TRL-1 droplets sequestered vitellogenin RNA. The loss of TRL-1 increases vitellogenin production but does not significantly impact vitellogenin transcription (Fig. 2 C–E and SI Appendix, Fig. S2E), suggesting that TRL-1 may inhibit vitellogenin translation. To test this hypothesis, we transfected a VIT-2::Scarlet reporter with GFP or TRL-1::GFP in HEK293T cells (Fig. 4C). By measuring the red VIT-2::Scarlet fluorescence, we found a significant reduction of VIT-2 protein signal in the presence of TRL-1 (SI Appendix, Fig. S7 A and B). In support of the fluorescence data, Western blots showed that the VIT-2 protein level was decreased in trl-1::gfp and vit-2 cotransfected cells compared with that in gfp and vit-2 cotransfected cells (Fig. 4C). Quantitative PCR experiments of the total, cytoplasmic, and nuclear RNAs showed that vit-2 transcription levels were indistinguishable in differently transfected HEK293T cells (SI Appendix, Fig. S7C), consistent with the notion that TRL-1 restricts VIT-2 generation at the translational level. Using an in vitro translation (IVT) system (29), we further examined whether TRL-1 inhibits vitellogenin RNA translation (Fig. 4D). Considering that TRL-1 binds to vitellogenin RNA fragments (Fig. 4A), we translated the N terminus (1 to 265 amino acids [aa]) or C terminus (1,214 to 1,613 aa) of VIT-2 (Fig. 4D). The addition of purified TRL-1 protein reduced the translation of the N- or C-terminal region of VIT-2 in a dose-dependent manner (Fig. 4D), but the same amount of an irrelevant ciliary protein IFT-20 did not affect VIT-2 translation (SI Appendix, Fig. S7D). These results provide evidence that TRL-1 inhibits vitellogenin RNA translation.
Discussion
This study identifies a noncanonical ORF trl-1 as an antagonistic pleiotropic gene that regulates the reproduction–longevity tradeoff. We show that trl-1 null animals up-regulate vitellogenin translation, which benefits reproduction but at the cost of lifespan (Fig. 4E). Other molecules undergoing a similar translational change to TRL-1 may also be involved in the reproduction–lifespan tradeoff or starvation recovery (Dataset S3). Quantitative traits, such as brood sizes and lifespan, are challenging to be scored on a large scale, making forward genetic screens impractical to identify mutants defective in brood sizes or lifespan, even in a simple model organism like C. elegans. This work isolated candidate regulators by profiling translatome changes in animals recovering from starvation, establishing an alternative platform to dissect the molecular regulation of both traits.
TRL-1 controls this tradeoff in animals growing with ample food (Figs. 2B and 3A), despite the low trl-1 expression level. We suspect that TRL-1 granules may recruit other RNA molecules via electrostatic interactions and block their translation; therefore, animals must produce a low level of TRL-1 beyond the GFP reporter’s detection limit. Indeed, inducible overexpression of TRL-1 caused embryonic lethality and larval arrest (SI Appendix, Fig. S2C), perhaps resulting from a general effect of TRL-1 on protein translation.
LLPS regulates various biological activities, ranging from chromatin organization, autophagic degradation, assembly of signaling clusters, and cytoskeletal networks to asymmetric segregations of cell fate determinants (26–28). To our knowledge, the contribution of LLPS to understanding the reproduction–lifespan tradeoff has not been previously documented. The formation of protein–RNA granules via LLPS appears to be a common mechanism to inhibit RNA translation. The cytoplasmic stress granules assemble from pools of mRNA molecules that stall at the translation initiation stage (30, 31). Similarly, processing bodies recruit mRNAs and translational repressors (30, 31). TRL-1–vitellogenin mRNA droplets resemble these ribonucleoprotein granules, sequestering vitellogenin RNA from the translation machinery and down-regulating its translation. The addition of purified TRL-1 protein into the in vitro translation system is sufficient to reduce vitellogenin mRNA translation, demonstrating the role of TRL-1 in translational regulation. TRL-1 granules may contain other inhibitors to limit mRNA translation in live cells.
Our results shed light on the molecular regulation of the reproduction–lifespan tradeoff in other species. Vitellogenin has been known as an evolutionary ancestor of the human apoB protein, the principal component of the low-density lipoprotein (LDL) (20, 32). In addition to the well-established effects of apoB on lipid-mediated atherogenesis, a recent study reported that the high human apoB gene expression levels correlate with a reduction of lifespan by months to years (33). Sequence homology analyses have not identified the trl-1 homologs in mammals; however, functional modules rather than protein sequences are more likely to be preserved. TRL-1 has two operating units: Its N-terminal IDR region facilitates phase separation, and its positively charged domain enables RNA recruitment. By searching for both modules in a single protein, we found that the human genome encodes 44 candidates that contain an IDR domain and a positively charged region (Dataset S6). Future study will determine whether some of these candidates regulate the reproduction–lifespan tradeoff in higher organisms and address whether LLPS-dependent translational regulation is involved.
Materials and Methods
C. elegans Strains, Genetics, and DNA Manipulations.
SI Appendix, Table S1 summarizes the strains used in this study. C. elegans strains were raised with Escherichia coli strain OP50 on nematode growth medium (NGM) plates at 20 °C. Worms at mixed stages were collected to perform Ribo-Seq, RNA-Seq, and mass spectrometry. For transgenesis, 1 to 20 ng/μL DNA was injected into C. elegans germlines. All the plasmids and primers are listed in SI Appendix, Table S2. All the animal experiments were performed following governmental and institutional guidelines.
Cell Culture and Transfection.
HEK293T cells were maintained at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium (Corning, 10-013-CV) containing 10% fetal bovine serum. pCMV-GFP, pCMV-VIT-2-Scarlet or pCMV-TRL-1-GFP, pCMV-VIT-2-Scarlet were cotransfected into the cell using Lipofectamine 3000 (Invitrogen, L3000008) following the manufacturer’s instructions. Cells were incubated at 37 °C for 24 h and subsequently imaged by spinning disk confocal microscopy (Zeiss) or harvested for Western blot and RT-qPCR.
Ribosome Profiling and RNA-Seq Libraries Processing.
C. elegans starved for 6 d and refed at different time points were flash frozen in liquid nitrogen and stored at −80 °C. Approximately 107 worms were collected for deep sequence. Libraries were performed as the previously published protocol (34) described with a few modifications. The sample pellets were crushed into powder in a mortar filled with liquid nitrogen and then lysed in 400 μL lysis buffer containing 20 mM Tris⋅HCl (pH 7.4) (Invitrogen, AM9850G), 150 mM NaCl (Invitrogen, AM9760G), 5 mM MgCl2 (Invitrogen, AM9530G), 1 mMdithiothreitol (Sigma-Aldrich, 43816), 100 μg/mL cycloheximide (Sigma-Aldrich, C4859), 1% Triton X-100 (Sigma-Aldrich, T8787), and 25 U/mL Turbo DNase I (Invitrogen, AM2238). A total of 200 μL of the clarified supernatant was used to purify the ribosome protected fragments (RPFs), and 100 μL of the supernatant was used for the total RNA-Seq in parallel.
For the RPFs, 15 U RNase I (Epicentre, N6901K) was added to 200 μL of the lysate, and the lysate was incubated with gentle mixing at room temperature for 45 min. To stop nuclease digestion, we added 10 μL of SUPERase In RNase Inhibitor (Life Technologies, AM2696) to the lysate and chilled on ice. RPFs were purified by Sephacryl S400 spin column chromatography (GE Healthcare, 27514001) and extracted with TRIzol (Invitrogen, 15596018) following the manufacturer’s protocol. The rRNA was depleted using the RiboZero kit (Illumina, MRZPL1224). The 25- to 34-nt fragments were excised from the 15% urea gel and eluted in 400 μL RNA gel extraction buffer (300 mM ammonium acetate [Invitrogen, AM9070G], 0.25% vol/vol of 10% sodium dodecyl sulfate (SDS) [Invitrogen, AM9823], 1 mM ethylene diamine tetraacetic acid (EDTA) [Invitrogen, AM9260G], followed by isopropanol [Sigma-Aldrich, I9030] precipitation). The purified RPFs were then treated with T4 polynucleotide kinase (NEB, M0201S) to dephosphorylate and further treated with T4 Rnl2(tr) K227Q (NEB, M0351S) to ligate with a preadenylated DNA linker. The RNA–DNA hybrid was used as a template for reverse transcription. The 145-nt fragments were excised from the 15% urea gel and eluted in 400 μL DNA gel extraction buffer (300 mM sodium chloride [Invitrogen, AM9760G], 10 mM Tris [Invitrogen, pH 8.0, AM9855G], 1 mM EDTA [Invitrogen, AM9260G], followed by isopropanol (Sigma-Aldrich, I9030) precipitation, followed by circularization with CircLigase II [Epicentre, CL9021K]). Deep sequencing libraries were generated from the cDNA circles with different indexes and sequenced on an Illumina HiSeq platform.
Total RNA from the corresponding samples was isolated with TRIzol (Invitrogen, 15596018) from 100 μL clarified lysate, and the rRNA was depleted using the RiboZero kit (Illumina, MRZPL1224). The rRNA-depleted total RNA was fragmented using polynucleotide kinase buffer (NEB, M0201L) at 95 °C for 20 min. Deep sequencing libraries were generated from the fragments and sequenced on an Illumina HiSeq platform.
RNA-Seq and Ribo-Seq Data Analyses.
Ribosome profiling data were processed according to a previously published protocol (34). In short, linker sequences were trimmed from the 3′ end of each sequencing read, and low-quality reads were discarded with CASAVA 1.8. Bowtie was used to remove rRNA and other contaminants present in the libraries. The unaligned reads were aligned to the C. elegans reference genome (ce11) with Tophat v2.1.1, allowing for two mismatches per read, and unique alignments were quantified by HTSeq v0.9.1. Transcriptome and translatome changes were calculated using the Bioconductor package DESeq2 with adjusted P ≤ 0.05. Translational efficiency was calculated by normalizing Ribo-Seq counts per million (CPM) to RNA-Seq CPM as reported (34). The ribosome P-site triplet periodicity and noncanonical ORFs were analyzed by RiboCode (18). Noncanonical ORFs were identified within the Ribo-Seq data using start codons ATG, CTG, TTG, and GTG. The active translation of noncanonical ORFs was measured by the three-nucleotide periodicity of the ribosome P site. A combined analysis P < 0.05 was used to determine protein-coding potential.
Mass Spectrometry.
Mass spectrometry was processed as described previously (35) with a few modifications. In brief, unsynchronized transgenic worms raised on 80 to 90, 90-mm NGM plates were harvested and flash frozen in liquid nitrogen and stored at −80 °C. Lysates were made from 3-mL pellets in 2× lysis buffer (50 mM Tris⋅HCl, pH 7.4, 1 M NaCl, 1% Nonidet P-40, 10% glycerol, 5 mM Na3VO4, 1× mixture of protease inhibitors [Complete, EDTA free], and 40 mM NaF), and two volumes of 0.5-mm diameter glass beads using FastPrep-24 (MP Biomedicals) vortexed until the pellets were homogenized. The supernatants were incubated with GFP-Trap agarose beads (Chromoteck, gta-20) at 4 °C for 1 h on a shaker. The beads were washed with 1× wash buffer (25 mM Tris⋅HCl, pH 7.4, 500 mM NaCl, 0.5% Nonidet P-40, 5% glycerol, 1 mM Na3VO4, 1× mixture of protease inhibitors [Complete, EDTA free], and 10 mM NaF) three times and then washed with 10 mM NH4HCO3. Samples were dried using a vacuum drier and digested with lytic protease at 37 °C overnight. The resultant peptides were desalted with ZipTip pipette tips (Merck Millipore) and dried again using a vacuum drier. For liquid chromatograph mass spectrometer/mass spectrometer analysis, peptides were separated by an EASY-nLCII integrated nano-high-performance liquid chromatography system (Proxeon) to a Thermo Scientific Q Exactive mass spectrometer at a flow rate of 0.250 mL/min. Xcalibur 2.1.2 software was used to operate the Q Exactive mass spectrometer in data-dependent acquisition mode. Proteome Discoverer (version PD1.4) was used to search against the C. elegans proteome database and the noncanonical ORF TRL-1.
Western Blot Analysis.
HEK293T cells were harvested using phosphate buffer saline (PBS), and cell pellets were resuspended with 1× loading buffer (4% SDS, 20% glycerol, 10% β-mercaptoethanol, 0.02% bromophenol blue, 125 mM Tris⋅HCl pH 6.8) and boiled for 5 min at 100 °C for Western blotting. For RNA–protein interaction analysis, the streptavidin beads were boiled for 5 min at 100 °C in 1× SDS loading buffer, and the protein that was pulled down was analyzed by Western blotting. For the in vitro translation analysis, the reaction mixtures were boiled for 5 min at 100 °C in 5× SDS loading buffer for further analyses. Proteins were separated by 10 to 15% SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Bio-Rad). The membrane was blocked by 5% skimmed milk and incubated overnight with the primary antibodies at 4 °C. After washing with tris buffered saline containing 1‰ Tween 20 three times, the membrane was incubated with the secondary antibodies for 1 h on a shaker. Protein bands were visualized by chemiluminescence using the SuperSignal West Dura (Thermo Fisher Scientific, 34076). The following antibodies were used: Mouse anti-flag (Sigma, F1804), mouse anti-strep II (Abcam, ab18224), mouse anti-glyceraldehyde-3-phosphate dehydrogenase (Abcam, ab8245), rabbit anti-GFP (Abcam, ab290), mouse anti-his (Abcam, ab18184), mouse anti-HA (Abclonal, AE008), goat anti-rabbit IgG (Bioeasytech, BE0101-100), and goat anti-mouse IgG (Bioeasytech, BE0102-100).
Brood Size Assay.
Synchronized L4 worms or dauer-stage worms were transferred singly to NGM plates with E. coli OP50 bacteria cultured with Luria-Bertani broth. L4-stage worms were combined with well-fed males (five males to one L4 hermaphrodite) for mating broods. After 24 h of mating, males were removed to minimize mating-induced trauma. Worms were repeatedly transferred to a freshly seeded NGM plate every 24 h until reproduction ceased. We counted the number of progeny at their L3/L4 stages. Our measured WT brood size is consistent with the previous results (36). Total brood sizes were analyzed using ANOVA statistics running a Student’s t test.
Lifespan Assay.
Lifespan assays were carried out at 20 °C in the presence of floxuridine (20 μg/mL) to prevent progeny production. We used adulthood day 1 as t = 1 for lifespan analyses. At least 60 worms were used for each experiment, and animals were transferred to newly seeded NGM plates every week. Animals were marked as dead when they did not move, pump, or respond to stimuli. Survival plots and mean life spans were generated by nonparametric methods. P values were calculated using the Log-rank (Mantel–Cox) method.
Entry of L1 Diapause and Adult Diapause.
Larva diapause was processed as described previously (37). To complete synchronization, we harvested eggs from bleaching and placed them on NGM plates without OP50. For L1 diapause, the hatched L1-stage animals were left on NGM plates without OP50 for 6 d. For adult diapause, the hatched L1-stage animals were then transferred to NGM plates plus OP50. The mid-L4–stage animals were collected and washed six times using M9 buffer. Animals were then plated onto NGM plates with no food at a density of ∼104 worms per plate for 6 d. Only those adults containing one or two viable embryos in utero that did not hatch and cause bagging were considered in adult reproductive diapause and were selected for further analysis.
RNAi.
RNAi knockdown of vitellogenins in C. elegans was achieved by feeding worms with E. coli HT115 expressing vit-5 dsRNA or a control plasmid (L4440). To obtain synchronous worms, 20 to 30 adult VIT-2::GFP knockin animals laid eggs for ∼1 h on NGM plates supplemented with 100 μg/mL ampicillin and 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG). Animals were exposed to vit-5 RNAi from hatching until grown to day 1 at 20 °C, and images were captured for GFP fluorescence analysis. For intestine-specific RNAi, DCL907[rde-1(mkc36) V; kbIS7(Pnhx-2::rde-1+rol-6)] animals were fed with E. coli HT115 expression trl-1 dsRNA or control plasmid (L4440). Animals were exposed to trl-1 RNAi from hatching at 20 °C for brood size and lifespan analysis.
Protein Expression and Purification.
trl-1 was cloned into the pETduet vector to produce His6-tag–fused recombinant protein. The fused protein was expressed in E. coli Rosetta induced by 1 mM IPTG for 16 h at 16 °C. To purify His-tagged TRL-1, we resuspended the E. coli sediments in binding buffer (20 mM Hepes pH 7.5, 500 mM NaCl, and 10 mM imidazole), lysed with a high-pressure homogenizer, and sedimented at 18,000 rpm, 30 min at 4 °C. The supernatants were incubated with Ni-NTA agarose beads (QIAGEN) and washed with extensive binding buffers. The proteins were then eluted with His6 elution buffer (20 mM Hepes pH 7.5, 500 mM NaCl, and 500 mM imidazole) and further purified with a HiPrep 26/60 Sephacryl S-200 HR column (GE Healthcare) on an AKTA purifier (GE Healthcare). The proteins were finally eluted with a buffer containing 20 mM Hepes pH 7.5, 500 mM NaCl, concentrated by centrifugal filtrations (Millipore), and then stored in aliquots at −80 °C.
In Vitro Phase Separation Assays.
TRL-1 proteins dissolved in a buffer containing 20 mM Hepes pH 7.5, 500 mM NaCl were diluted to desired molarity with different concentrations of NaCl. For imaging, droplets were observed on a glass slide for DIC or fluorescence imaging (Zeiss LSM 900) with a 63 × 1.40 oil-immersion objective lens at room temperature and analyzed by ImageJ. All the buffers were RNase-free and supplemented with RNase inhibitor for phase separation with RNA. TRL-1 proteins and vit-2 RNA were mixed at the indicated concentrations (including 2 μM TRL-1 labeled with Cy3) in 20 mM Hepes pH 7.5, 150 mM NaCl.
Protein Fluorescence Labeling.
Sulfo-cyanine3 maleimide (Lumiprobe) was dissolved in dimethyl sulfoxide and incubated with TRL-1 proteins for 1 h at room temperature (fluorophore-to-protein molar ratio was 1:100). Buffer containing 20 mM Hepes pH 7.5, 500 mM NaCl was added to quench the reaction. The fluorophores and other small molecules were removed from the reaction through a desalting column (Thermo Fisher, 89882).
FRAP Assays.
Cy3-labeled TRL-1 proteins were used to represent liquid droplets in vitro. In the FRAP assay in C. elegans and HEK293T, TRL-1::GFP proteins were used to label the liquid droplets. FRAP experiments were performed on a confocal microscope (LSM 900, Zeiss) with a 63 × 1.40 oil-immersion objective lens at room temperature. Either a 488-nm (for in vivo droplets) or a 561-nm (for in vitro droplets) laser was used to photobleach defined regions, and the fluorescence intensities in these regions were collected every 10 s and normalized to the initial intensity before bleaching. Image intensity was measured by mean region of interest and further analyzed by Prism GraphPad.
Biotin-Labeled RNA Pull-Down Assay.
According to the manufacturer’s instructions, the fragments and full length of vit-2 RNA molecules and the fragments of act-1 RNA and tbb-4 RNA molecules were labeled with biotin-16-uridine triphosphate (UTP) by in vitro transcription with T7 polymerase (Lucigen, ASB71110). The Dynabeads MyOne Streptavidin C1 beads (Invitrogen, 65002) and the reaction tubes were blocked by 5% bovine serum albumin at 4 °C for 1 h. The RNA fragments and purified TRL-1 proteins in the binding buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride, RNase inhibitor) were incubated with Dynabeads MyOne Streptavidin C1 beads for 3 h at 4 °C on a shaker. After being washed three times with wash buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 0.5 mM PMSF, RNase inhibitor) at 4 °C for 5 min each, the beads were boiled for 5 min at 100 °C in 1× SDS loading buffer, and the protein pulled down was analyzed by Western blotting.
Isolation of Total, Cytoplasmic, and Nuclear RNA.
HEK293T cells were harvested using PBS and divided into two parts. Total RNA was isolated using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Cytoplasmic and nuclear RNA were isolated using Cytoplasmic and Nuclear RNA Purification Kit (Norgen Biotek, 21000). Briefly, cell pellets were lysed by 200 μL lysis buffer J with vortexing, and the lysates were centrifuged at 14,000 rpm for 10 min at 4 °C. The supernatants were enriched by the cytoplasmic RNA, and the pellets were enriched by the nuclear RNA. The pellets were resuspended in buffer SK from the Cytoplasmic and Nuclear RNA Purification Kit with vortex. The supernatant and pellet fractions were further bound to the columns, and the columns were washed three times before eluting cytoplasmic RNA and nuclear RNA.
RT-qPCR.
The first-strand cDNA was synthesized using Takara PrimeScript RT reagent kit with gDNA Eraser (Takara, RR047A) and diluted 1:10 in nuclease-free water as a template for qPCR. For total and cytoplasmic RNA, 1 μg RNA was used as the template for reverse transcription. For nuclear RNA, 75 ng RNA was used as the template. RT-qPCR was carried out with PowerUp SYBR Green Master Mix (ABI, A25742) and performed in four replicates using the Bio-Rad CFX96 Real-Time System. The vit-2 expression values relative to the housekeeping genes GAPDH were calculated by the Ct differences (ΔCt), and the fold change of the gene expression relative to the control sample was calculated by the difference between the ΔCt values (ΔΔC). The experiments were repeated three times on independent RNA preparations. The gene-specific primers were shown as follows: VIT-2-F: 5′-TACGTCAAGGCTGACAAGAAGATCC-3′, VIT-2-R: 5′-TGGGAGAATGTCCTCGTTTTCCA-3′, GFP-F: 5′-TGAGCAAGGGCGAGGAGCTG-3′, GFP-R: 5′-TGAACTTCAGGGTCAGCTTG-3′, GAPDH-F: 5′-GTCTCCTCTGACTTCAACAGCG-3′, GAPDH-R: 5′-ACCACCCTGTTGCTGTAGCCAA-3′.
In Vitro Translation Assay.
IVT assays were performed using a 1-Step Human Coupled IVT Kit (Thermo Scientific, 8882) with a few modifications. Briefly, VIT-2 (1 to 265 aa) and VIT-2 (1,214 to 1,613 aa) were cloned into the Thermo Scientific pT7CFE1 Vector to produce Flag-tag–fused recombinant proteins. Purified TRL-1 proteins or an irrelevant ciliary protein IFT20 (as a negative control) was added to 25 μL HeLa lysate IVT mixtures containing the vit-2 gene for 180 min at 30 °C. After incubation at 30 °C, the reaction mixtures were boiled for 5 min at 100 °C in 5× SDS loading buffer for Western blotting analysis. In control reactions, no template was added to the IVT mixture.
Search for Homologs.
Homologs were obtained by searching with two functional modules in the human genome: The IDR and the positively charged domain. IDR regions were predicted using the Predictor of Natural Disordered Regions (PONDR, http://www.pondr.com). The candidates are listed in Dataset S6.
Quantification and Statistical Analysis.
For each group, quantification was represented by the mean value ± SD (Fig. 2 B and E and SI Appendix, Figs. S2D, S4B, S6G, and S7 B and C) and the mean ± 95% confidence interval (CI) (Fig. 3D and SI Appendix, Figs. S5D and S6B). Two-tailed Student’s t test analysis (Fig. 2 B and E and SI Appendix, Figs. S2D, S4B, S6G, and S7 B and C) and Log-rank (Mantel–Cox) method (SI Appendix, Fig. S3A) were used to examine significant differences between groups as indicated in the figure legends. Statistical significance was determined when the P value is lower than 0.05 and N.S., not significant (SI Appendix, Figs. S4B and S7 B and C) . N represents the number of animals used for the brood size assay (Fig. 2B and SI Appendix, Figs. S2D and S4 B and E), the number of animals used for lifespan analyses (Fig. 3 A and B and SI Appendix, Figs. S3A and S4 A, C, and D), the number of animals or cells used for FRAP assay (Fig. 3D and SI Appendix, Fig. S5D), the number of animals or cells used for fluorescence intensity analysis (Fig. 2E and SI Appendix, Fig. S7B), the number of droplets used for diameter analysis (SI Appendix, Fig. S6G), or the number of droplets used for FRAP assay (SI Appendix, Fig. S6B).
Supplementary Material
Acknowledgments
We thank Prof. Xiang Y. for supporting TRL-1 protein purification and Profs. W. Qian, W. Wen, P. Li, and X. Yang for their comments on this project. We thank the Caenorhabditis Genetics Center (CGC) for providing some strains and the Protein Chemistry Facility at the Center for Biomedical Analysis of Tsinghua University for sample analyses. This work was supported by the National Basic Research Program of China (973 Program; Grants 2019YFA0508401, 2017YFA0503501, and 2017YFA0102900) and the National Natural Science Foundation of China (Grants 31991191, 31730052, 31671444, 31871352, and 31971092).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120311119/-/DCSupplemental.
Data Availability
The Ribo-Seq and RNA-Seq data were deposited in Gene Expression Omnibus (accession no. GSE184209). All other study data are included in the article and/or supporting information.
References
- 1.Williams G. C., Pleiotropy, natural-selection, and the evolution of senescence. Evolution 11, 398–411 (1957). [Google Scholar]
- 2.Hamilton W. D., Moulding of senescence by natural selection. J. Theor. Biol. 12, 12–45 (1966). [DOI] [PubMed] [Google Scholar]
- 3.Wallace D. C., Inevitability of growing old. J. Chron. Dis. 20, 475–486 (1967). [DOI] [PubMed] [Google Scholar]
- 4.Austad S. N., Hoffman J. M., Is antagonistic pleiotropy ubiquitous in aging biology? Evol. Med. Public Health 2018, 287–294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campisi J., et al. , From discoveries in ageing research to therapeutics for healthy ageing. Nature 571, 183–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sgrò C. M., Partridge L., A delayed wave of death from reproduction in Drosophila. Science 286, 2521–2524 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Rose M. R., Laboratory evolution of postponed senescence in Drosophila melanogaster. Evolution 38, 1004–1010 (1984). [DOI] [PubMed] [Google Scholar]
- 8.Paaby A. B., Bergland A. O., Behrman E. L., Schmidt P. S., A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution 68, 3395–3409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paaby A. B., Schmidt P. S., Functional significance of allelic variation at methuselah, an aging gene in Drosophila. PLoS One 3, e1987 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nusbaum T. J., Rose M. R., The effects of nutritional manipulation and laboratory selection on lifespan in Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci. 54, B192–B198 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Leroi A. M., et al. , What evidence is there for the existence of individual genes with antagonistic pleiotropic effects? Mech. Ageing Dev. 126, 421–429 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Hsin H., Kenyon C., Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399, 362–366 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Hwangbo D. S., Gershman B., Tu M. P., Palmer M., Tatar M., Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429, 562–566 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Blüher M., Kahn B. B., Kahn C. R., Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299, 572–574 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Dillin A., Crawford D. K., Kenyon C., Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298, 830–834 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Baugh L. R., Hu P. J., Starvation responses throughout the Caenorhabditis elegans life cycle. Genetics 216, 837–878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingolia N. T., Hussmann J. A., Weissman J. S., Ribosome profiling: Global views of translation. Cold Spring Harb. Perspect. Biol. 11, a032698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao Z., et al. , De novo annotation and characterization of the translatome with ribosome profiling data. Nucleic Acids Res. 46, e61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J. S., et al. , FMRFamide-like peptides expand the behavioral repertoire of a densely connected nervous system. Proc. Natl. Acad. Sci. U.S.A. 114, E10726–E10735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez M. F., Lehner B., Vitellogenins - Yolk gene function and regulation in Caenorhabditis elegans. Front. Physiol. 10, 1067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ezcurra M., et al. , C-elegans eats its own intestine to make yolk leading to multiple senescent pathologies. Curr. Biol. 28, 2544–2556.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez M. F., Francesconi M., Hidalgo-Carcedo C., Lehner B., Maternal age generates phenotypic variation in Caenorhabditis elegans. Nature 552, 106–109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seah N. E., et al. , Autophagy-mediated longevity is modulated by lipoprotein biogenesis. Autophagy 12, 261–272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy C. T., et al. , Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Kenyon C. J., The genetics of ageing. Nature 464, 504–512 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., et al. , Liquid-liquid phase separation in biology: Mechanisms, physiological functions and human diseases. Sci. China Life Sci. 63, 953–985 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Mikami S., Masutani M., Sonenberg N., Yokoyama S., Imataka H., An efficient mammalian cell-free translation system supplemented with translation factors. Protein Expr. Purif. 46, 348–357 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Protter D. S. W., Parker R., Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roden C., Gladfelter A. S., RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 183–195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker M. E., Is vitellogenin an ancestor of apolipoprotein B-100 of human low-density lipoprotein and human lipoprotein lipase? Biochem. J. 255, 1057–1060 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson T. G., et al. , Effects of apolipoprotein B on lifespan and risks of major diseases including type 2 diabetes: A mendelian randomisation analysis using outcomes in first-degree relatives. Lancet Healthy Longev. 2, e317–e326 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingolia N. T., Brar G. A., Rouskin S., McGeachy A. M., Weissman J. S., The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi P., Li W. J., Dong M. Q., Ou G., Dynein-driven retrograde intraflagellar transport is triphasic in C. elegans sensory cilia. Curr. Biol. 27, 1448–1461.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Cong W., et al. , Evaluation of the influence of fullerenol on aging and stress resistance using Caenorhabditis elegans. Biomaterials 42, 78–86 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Angelo G., Van Gilst M. R., Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science 326, 954–958 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Ribo-Seq and RNA-Seq data were deposited in Gene Expression Omnibus (accession no. GSE184209). All other study data are included in the article and/or supporting information.