Significance
A potential outbreak of swine acute diarrhea syndrome coronavirus (SADS-CoV) in the human population could be devastating. Using genomewide CRISPR knockout screening, we identified the placenta-associated 8 protein (PLAC8) as an essential host factor for SADS-CoV infection, uncovering a novel antiviral target for CoV infection. The PLAC8-related pathway may also have implications for other CoV infections. Given the ability of SADS-CoV to infect human primary cultures without adaptation, our findings lay the foundation for pandemic preparedness for the potential emergence of SADS-CoVs in response to the One Health Initiative.
Keywords: PLAC8, coronavirus, CRISPR, swine acute diarrhea syndrome coronavirus
Abstract
Zoonotic transmission of coronaviruses poses an ongoing threat to human populations. Endemic outbreaks of swine acute diarrhea syndrome coronavirus (SADS-CoV) have caused severe economic losses in the pig industry and have the potential to cause human outbreaks. Currently, there are no vaccines or specific antivirals against SADS-CoV, and our limited understanding of SADS-CoV host entry factors could hinder prompt responses to a potential human outbreak. Using a genomewide CRISPR knockout screen, we identified placenta-associated 8 protein (PLAC8) as an essential host factor for SADS-CoV infection. Knockout of PLAC8 abolished SADS-CoV infection, which was restored by complementing PLAC8 from multiple species, including human, rhesus macaques, mouse, pig, pangolin, and bat, suggesting a conserved infection pathway and susceptibility of SADS-CoV among mammals. Mechanistically, PLAC8 knockout does not affect viral entry; rather, knockout cells displayed a delay and reduction in viral subgenomic RNA expression. In a swine primary intestinal epithelial culture (IEC) infection model, differentiated cultures have high levels of PLAC8 expression and support SADS-CoV replication. In contrast, expanding IECs have low levels of PLAC8 expression and are resistant to SADS-CoV infection. PLAC8 expression patterns translate in vivo; the immunohistochemistry of swine ileal tissue revealed high levels of PLAC8 protein in neonatal compared to adult tissue, mirroring the known SADS-CoV pathogenesis in neonatal piglets. Overall, PLAC8 is an essential factor for SADS-CoV infection and may serve as a promising target for antiviral development for potential pandemic SADS-CoV.
Coronaviruses (CoVs) infect a wide range of animals and are global public health, as well as economic, threats (1). In the last two decades, there have been three highly pathogenic CoV outbreaks in the human population—severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) (2), Middle East respiratory syndrome coronavirus (MERS-CoV) (3), and the ongoing SARS-CoV-2 pandemic (4, 5)—as well as several emerging CoVs in swine, such as porcine epidemic diarrhea virus and porcine delta-CoV (6). SARS-CoV-1 and MERS-CoV originated via zoonotic transmission from bats and camels, respectively. Although still the subject of ongoing study, SARS-CoV-2 likely emerged from natural zoonotic transmission events from an unknown source, most probably bats (7). Under the global One Health Initiative, companion and farm animals are a major focus of concern for future zoonosis events. In particular, modern industrial farming techniques promote an environment with high animal population densities and close human contact, potentiating the next zoonotic transmission or even pandemic.
Swine acute diarrhea syndrome coronavirus (SADS-CoV), also known as swine enteric alpha-CoV and porcine enteric alpha-CoV, is a highly pathogenic alpha-CoV, which caused two large outbreaks in Guangdong, China, in 2017 and 2019 (8–10). Fortunately, workers from the outbreak farms were not seroconverted, suggesting limited pig-to-human transmission. Infected piglets die within 2 to 6 d after the onset of symptoms, which include severe diarrhea and vomiting and, consequently, dehydration and weight loss. Mortality rates approach 90% in piglets less than 5 d old, but death rates drop dramatically to ∼5% in piglets greater than 8 d of age (8, 9). Necropsies suggest that infected piglets suffer from intestinal lesions and villus atrophy, as well as a reduced villus height to crypt depth ratio in the jejunum and ileum. Viral antigen is found in the epithelium of the small intestine (9).
SADS-CoV has 95% sequence identity to the bat CoV HKU2 identified in 2004, and retrospective studies on bat samples suggest that SADS-related CoVs (SADSr-CoVs) have been circulating in Rhinolophus affinis and Rhinolophus sinica bats since 2013 (8, 11). Recently, a related rat CoV, Lucheng Rn rat CoV, was identified, suggesting possible zoonotic transmission between different animals in nature and highlighting the danger of, and a potential path for, uncontrolled transmission to humans (11). Although SADS-CoV is an alpha-CoV, phylogenetic analysis shows that SADS-CoV and SADSr-CoV spike proteins form a distinct group, alpha-CoV-1, which is more closely related to beta-CoVs than the contemporary alpha-CoV-2 strains, which include the human CoVs (hCoVs) NL63 and 229E (8, 11). This unique feature of spike protein has been confirmed by structural studies, which show that although the SADS-CoV N-terminal domain (NTD) resembles the subtype I NTD from alpha-CoVs, the C-terminal domain is closely related to beta-CoVs, with the signature one-layer beta strand fold in the core domain (12, 13). Taken together, the data suggest that SADS-CoV and HKU2 may have arisen from an ancestral recombination event between alpha- and beta-CoVs, which warns of potential recombination events with other hCoVs. In agreement with the separate phylogenetic grouping of the SADS Spike protein, SADS-CoV Spike displays distinct antigenicity and cannot be neutralized by antisera raised against the circulating human alpha-CoV hCoV-NL63, forewarning the lack of preexisting immune protection in the human population (14). Functional analyses have also shown that SADS-CoV spike does not use any known CoV receptors, including aminopeptidase N, angiotensin coverting enzyme 2, or dipeptidyl-peptidase 4, for viral entry (14, 15).
In experimental infections, SADS-CoV is capable of infecting a wide variety of cell lines from multiple species, highlighting its preemergent potential for cross-species transmission (8, 15, 16). More importantly, SADS-CoV is able to infect a variety of primary human lung and intestinal cells, implying the ability to infect humans in vivo (14). Currently, the only potential countermeasure against SADS-CoV is the nucleoside analog remdesivir, which inhibits viral replication (14). Given the importance of SADS-CoV in the livestock industry, its potential to infect humans, and the lack of knowledge about and antivirals against SADS-CoV, an outbreak of SADS-CoV or any related viruses could be devastating. In this study, we performed a genomewide CRISPR-Cas9 knockout (GeCKO) screen and identified PLAC8 as the single most essential host factor for SADS-CoV infection and a possible drug target for antiviral development.
Results
GeCKO Screen Identifies Essential Host Factors for SADS-CoV Infection.
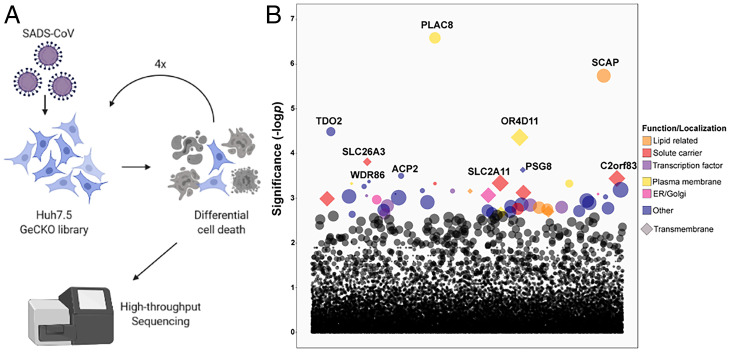
To identify host factors essential for SADS-CoV infection, we established a GeCKO library on human hepatoma-derived Huh7.5 cells using the previously developed Brunello CRISPR library (17). Deep sequencing confirmed our CRISPR library contains roughly 75,000 unique guide RNAs (97% library coverage) with an average of 175x coverage per guide RNA. The Huh7.5 library was infected with a high multiplicity of infection (MOI) of SADS-CoV, and surviving cells were passaged and reinfected with SADS-CoV a total of four times (Fig. 1A). Guide RNAs that were enriched in the cell population after the fourth round were identified by deep sequencing (Fig. 1B and SI Appendix, Table S1). We generated polyclonal knockout (KO) Huh7.5 cell lines for 5 of the top 10 hits (with two guide RNAs each), including placenta-associated 8 protein (PLAC8), sterol regulatory element-binding protein cleavage-activating protein (SCAP), olfactory receptor 4D11 (OR4D11), and two members of the solute carrier family, SLC26A3 and SLC2A11. Comparison of our top 10 target genes with published literature on CoV host factors revealed overlapping hits (SCAP, PLAC8, SLC26A3, PSG8, and ACP2) with other CRISPR screens based on Huh7.5 cells but no overlap with genes identified using screens against other CoVs on different cell types (SI Appendix, Table S2). In particular, SCAP, which has been shown to be involved in cholesterol pathways for SARS-CoV-2 infection, has been enriched in multiple studies (18). Although PLAC8 was not enriched in any SARS-CoV-2 screen, it was identified in a hCoV-NL63 screen and another SADS-CoV screen (19, 20), arguing for the potential importance of PLAC8 in other alpha-CoV infections. The KO cell lines were infected with wild-type (WT) SADS-CoV and SADS-CoV-RFP (red fluorescent protein) reporter virus. While KO of OR4D11, SLC26A3, or SLC2A11 showed little qualitative difference in RFP expression or quantitative change in viral growth kinetics, KO of PLAC8 and SCAP reduced the viral titer of WT SADS-CoV at 72 hours postinfection (hpi) by 350- and 15-fold, respectively (SI Appendix, Fig. S1 A and B). Strikingly, PLAC8 KO almost completely abolished infection and was therefore selected for further investigation. To ensure homogeneity of the cells, we selected a clonal PLAC8 KO cell line for better characterization (SI Appendix, Fig. S2).
Fig. 1.
GeCKO library screening to identify critical genes involved in SADS-CoV infection. (A) Schematic of the CRISPR KO screening procedure on Huh7.5 cells. (B) Guide RNAs enriched after four rounds of SADS-CoV infection determined by high-throughput sequencing.
Transcriptome Analysis Reveals Multiple Pathways Affected by PLAC8 KO and SADS Infection.
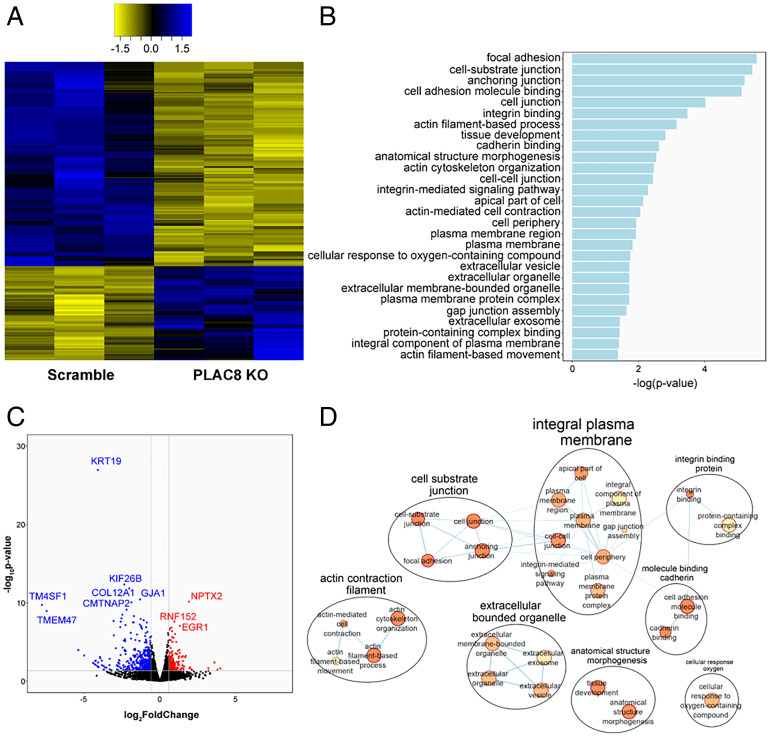
Prior to the study herein, PLAC8 was not known to be involved in viral pathogenesis. PLAC8 is a small cysteine-rich protein highly expressed in placenta, lung, and intestine (21, 22). Reports of PLAC8 cellular localization vary depending on cell type, from cytosolic, to lysosomal (23, 24), to the apical domain of differentiated human intestinal epithelium (25) and/or the inner leaflet of the plasma membrane (24), although cell surface localization has not been demonstrated. PLAC8 is reported to be involved in multiple complex cellular pathways, including autophagy (24–27), epithelial-mesenchymal transition (EMT), cell motility, innate immune functions (22, 28), mitochondrial metabolism, and DNA repair (29). In particular, autophagy is known to be important in CoV infections. To understand the global cellular changes in PLAC8 KO cells, we performed transcriptomic analysis. We identified 200 differentially expressed genes (DEGs) between Huh7.5 Scramble and PLAC8 KO cell lines, the majority of which were down-regulated in PLAC8 KO cells (Fig. 2A). Top down-regulated genes included keratin-19 (KRT19), kinesin family member 26B (KIF26B), and collagen type XII alpha 1 chain (COL12A1), while top up-regulated genes included neuronal pentraxin-2 (NPTX2), ring finger protein 152 (RNF152), and early growth response 1 (EGR1) (Fig. 2B). Interestingly, PRSS23, a trypsin family member serine protease, is down-regulated upon PLAC8 KO, as is F2RL1, a trypsin receptor reported to interact with SARS-CoV-2 ORF9c (30). Glycoproteins such as HSPG2, PARM1, and MUC13 are also down-regulated in PLAC8 KO cells. Notably, none of the top DEGs in PLAC8 KO cells are known pro- or antiviral factors, suggesting a nonimmune role of PLAC8 in SADS-CoV infection. Gene ontology (GO) analysis identified significantly affected pathways, including plasma membrane components, cell junctions, and actin organization (Fig. 2 C and D), correlating with known roles of PLAC8 in autophagy and EMT.
Fig. 2.
Transcriptomic analysis of PLAC8 KO Huh7.5 cells. (A) Heatmap of the normalized counts of significant DEGs between Huh7.5 Scramble and PLAC8 KO cells. Yellow indicates relatively lower expression, and blue indicates relatively higher expression. Data are shown from three replicate samples from each experimental condition. (B) Bar plot of the significantly enriched GO pathways between Huh7.5 Scramble and PLAC8 KO as determined by g:Profiler. (C) Volcano plot of DEGs between Scramble and PLAC8 KO Huh7.5 cells. Red denotes significantly up-regulated genes, and blue denotes significantly down-regulated genes. (D) Cytoscape EnrichmentMap of significantly enriched GO pathways as determined by g:Profiler.
Additionally, we performed transcriptomics on Huh7.5 Scramble cells infected with SADS-CoV and identified many DEGs due to viral infection. Top down-regulated genes included collagen type II alpha 1 chain (COL2A1), inhibitor of DNA binding 3 (ID3), and histone H2B type 1-O (HIST1H2BO), while top up-regulated genes included 5.8S ribosomal RNA N4 (RNA5-8SN4), decidual protein induced by progesterone 1 (DEPP1), and CCAAT enhancer binding protein delta (CEBPD) (SI Appendix, Fig. S3A). GO analysis identified several pathways affected by SADS-CoV infection, including cell signaling/development, complement activation, blood coagulation/hemostasis, programmed cell death/apoptosis, extracellular matrix organization, and metabolic processes (SI Appendix, Fig. S3B). We compared DEGs in PLAC8 KO versus Scramble, Scramble versus SADS-infected cells, and enriched genes identified by the GeCKO screen. Notably, several genes were shared between groups, including 38 genes between PLAC8 KO and SADS-infected cells (SI Appendix, Fig. S3C). Interestingly, one of the three shared genes found in all experiments is OLFML3, an extracellular matrix glycoprotein involved in cell adhesion, which could be a candidate for further study.
Development of SADS-CoV-nLuc for Quantitative Measurement.
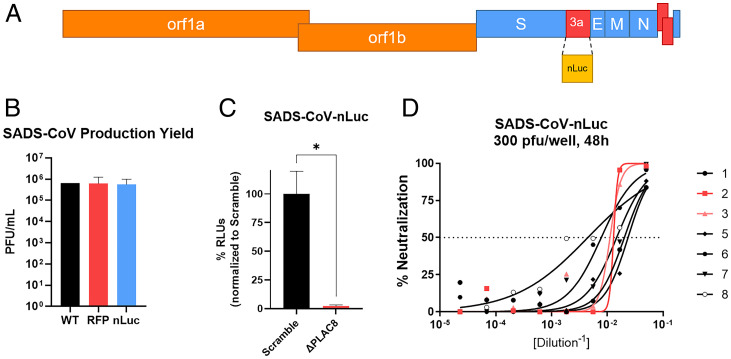
To quantitatively measure SADS-CoV infection in an efficient manner, we developed a recombinant nano-Luciferase (nLuc) reporter SADS-CoV (SADS-CoV-nLuc) via reverse genetics by replacing ORF3a with nLuc (Fig. 3A). Replacement of ORF3a does not affect viral growth in vitro, with viral titers reaching 105.5, similar to WT and RFP viruses (Fig. 3B). The infectivity of SADS-CoV-nLuc recapitulates WT and RFP viruses, as SADS-CoV-nLuc infection is reduced by 95% in PLAC8 KO cells compared to Scramble cells (Fig. 3C). SADS-CoV-nLuc also shows a dose-dependent reduction in relative light units (RLUs) in the presence of anti-SADS-CoV Spike mouse serum, demonstrating a large dynamic range of infectivity and its usage for serological studies (Fig. 3D).
Fig. 3.
Development of SADS-CoV-nLuc reporter virus via reverse genetics. (A) The design of SADS-CoV-nLuc virus with ORF3a replaced by nLuc. (B) Relative production yield of SADS-CoV infectious clone (WT), SADS-CoV-RFP (RFP), and SADS-CoV-nLuc (nLuc). (C) RLU/infectivity of SADS-CoV-nLuc (MOI: 0.0075) at 48 hpi in Scramble and PLAC8 KO Huh7.5 cells. (D) nLuc-based neutralization assay of SADS-CoV-nLuc against SADS-CoV Spike mouse antisera from eight mice. Error bars represent ± 1 SD, *P < 0.05 by Student's t test.
PLAC8 KO Cells Are Resistant to SADS-CoV Infection.
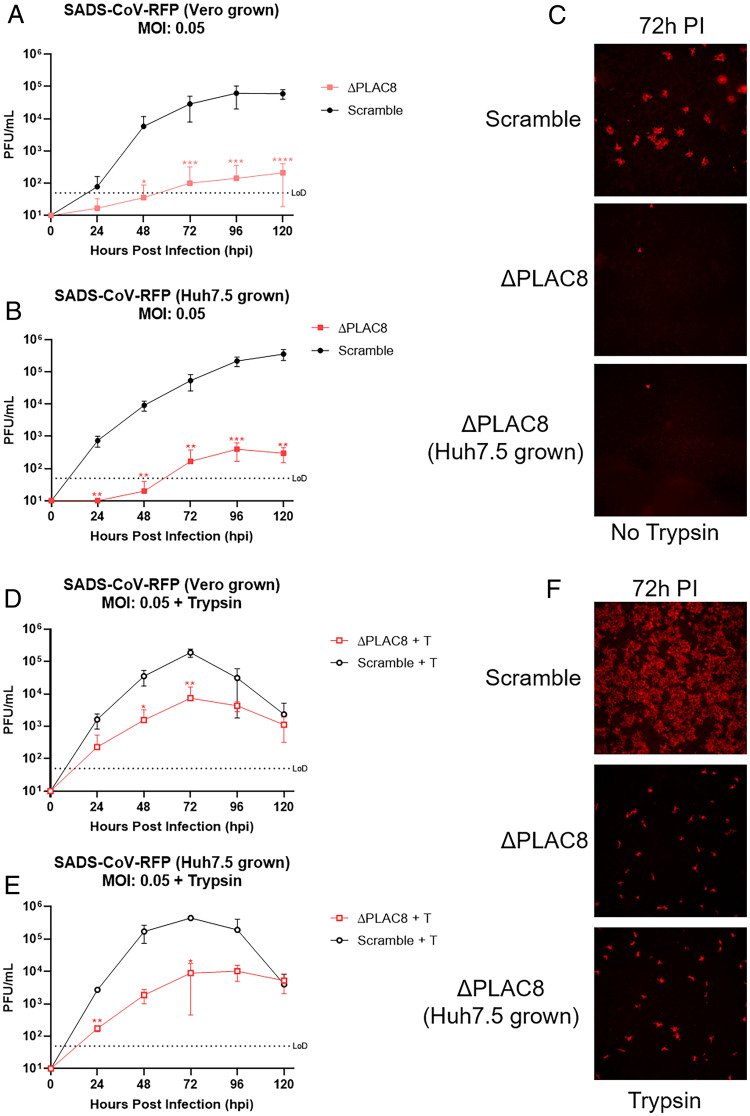
We next performed viral growth assays using clonal PLAC8 KO cells. As expected, our results were comparable to those seen with the polyclonal PLAC8 KO cells, showing no detectable virus until 72 hpi and a 3-log reduction of viral titer at 120 hpi compared to Scramble cells (Fig. 4A). The origin of virus did not affect the results, as SADS-CoV grown in both Huh7.5 cells and Vero cells require PLAC8 for infection. (Fig. 4B). Detection of RFP after SADS-CoV-RFP infection was used as a surrogate for early stages of infection, as it is encoded by a subgenomic mRNA (Fig. 4C). While a robust RFP signal is detected in Scramble cells, PLAC8 KO cells showed little to no RFP signal, supporting an early block in virus replication (Fig. 4C).
Fig. 4.
Growth kinetics of SADS-CoV-RFP. Infections were performed at a MOI of 0.05. (A) Growth kinetics of Vero-produced virus examined every 24 h for 120 hpi. (B) Growth kinetics of Huh7.5-produced virus examined every 24 h for 120 hpi. (C) Representative fluorescence images (4x) of SADS-CoV-RFP infection at 72 hpi. Growth kinetics of the virus were examined every 24 h for 120 hpi for virus produced in Vero cells (D) or Huh7.5 cells (E) in the presence of trypsin (1 µg/mL). (F) Representative fluorescence images (4x) of SADS-CoV-RFP infection in the presence of trypsin at 72 hpi. Error bars represent ± 1 SD, *P < 0.05, **P < 0.01, ***P < 0.001 by Student's t test.
PLAC8 Acts Downstream and Partially Independent of Trypsin-Mediated Entry Pathway.
Previous reports have demonstrated that SADS-CoV replication is enhanced by trypsin. To test whether PLAC8 functions in trypsin-mediated entry pathways, we performed growth kinetic analyses of SADS-CoV on Scramble and PLAC8 KO Huh7.5 cells in the presence of trypsin. As expected, addition of trypsin increased the infection kinetics of SADS-CoV in Scramble cells, reaching peak titer at 72 hpi. Although SADS-CoV shows better growth in PLAC8 KO cells in the presence of trypsin, it maintains a 1.5-log reduction of peak titer (103.5) compared to Scramble cells (Fig. 4 D and E). Such low infectivity even in the presence of trypsin suggests that the role of PLAC8 on viral infection is downstream and partially independent of the trypsin-mediated entry pathway. Similarly to untreated cells, no difference was observed between viruses produced in Vero cells versus Huh7.5 cells.
Ectopic Expression of PLAC8 Restores SADS-CoV Infection.
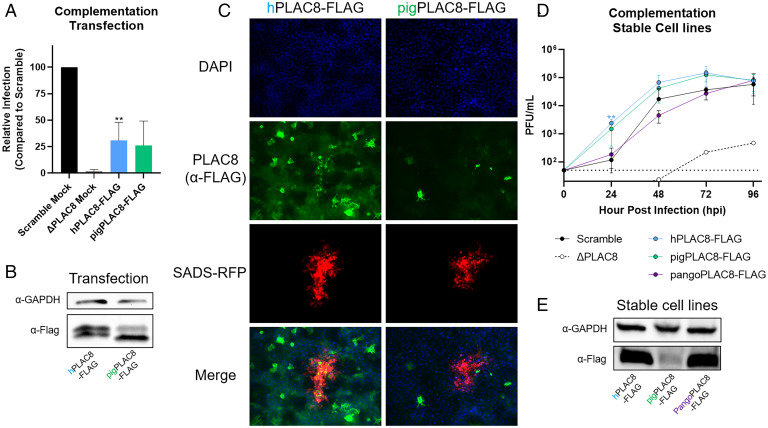
To confirm that PLAC8 is necessary and sufficient for SADS-CoV infection, and to determine the species specificity and zoonotic potential of SADS-CoV infection with regard to PLAC8, we tested PLAC8 proteins (authentic sequence, no tag) from human (hPLAC8), rhesus monkey (rhPLAC8), mouse (mPLAC8), pig (pigPLAC8), pangolin (pangoPLAC8), and bat (bPLAC8), in descending order of percent identity compared to hPLAC8 (SI Appendix, Fig. S4 A and B). Although bPLAC8 had the lowest sequence identity to hPLAC8, the bat protein displayed the highest level of rescue, at 36%, compared to Scramble cells. mPLAC8 and rhPLAC8 proteins showed similar levels of rescue as hPLAC8 at 22 and 15%, respectively. Finally, pig PLAC8 (pigPLAC8) and pangoPLAC8 had relatively low levels of rescue (SI Appendix, Fig. S4C). Western blotting with a PLAC8 antibody was unable to detect PLAC8 from other species (SI Appendix, Fig. S4D). We therefore measured mRNA expression and found that bPLAC8 expressed robustly compared to hPLAC8 and rhPLAC8 and mPLAC8 were moderately expressed; however, pigPLAC8 and pangoPLAC8 had very low expression levels (SI Appendix, Fig. S4E). We therefore generated C-terminal 3xFLAG-tagged human PLAC8 (hPLAC8-FLAG) and codon-optimized pigPLAC8-FLAG. With codon optimization, ectopic expression by transient transfection of both hPLAC8-FLAG and pigPLAC8-FLAG rescued the ability of SADS-CoV-nLuc to infect and restore luciferase activity to ∼29 and 25% of the infected Scramble cells, respectively (Fig. 5A). Western blotting confirmed equivalent expression of both PLAC8s after transient transfection (Fig. 5B). Following SADS-CoV-RFP infection, the RFP signal colocalized with the PLAC8-FLAG signal, suggesting a cell-autonomous (cis-acting) mechanism (Fig. 5C).
Fig. 5.
Complementation with ectopic or stable PLAC8 expression rescues SADS-CoV infection. (A) PLAC8 KO cells were transiently transfected with human and pigPLAC8-FLAG (codon optimized). Subsequently, transfected cells were infected with SADS-CoV-nLuc (MOI: 0.015) for 48 hpi. RLUs from the infected cells were used as surrogates for viral infection. (B) Western blot images of hPLAC8 and pigPLAC8-FLAG expression on Huh7.5 cells after transient transfection using anti-FLAG antibody. (C) Representative immunofluorescence images of hPLAC8- and pigPLAC8-FLAG–transfected cells infected with SADS-CoV-RFP and stained with anti-FLAG antibody (green), SADS-CoV-RFP (red), and DAPI (blue). (D) SADS-CoV-RFP growth kinetics on Huh7.5 ΔPLAC8 cells stably expressing hPLAC8, codon-optimized pigPLAC8, and pangoPLAC8-FLAG. (E) Western blot images of hPLAC8, pigPLAC8, and pangoPLAC8-FLAG expression in stable cell lines using anti-FLAG antibody. Error bars represent ± 1 SD, *P < 0.05, **P < 0.01, ***P < 0.001 by Student's t test.
Stable Expression of PLAC8 in PLAC8 KO Cells Enhances SADS-CoV Replication.
To minimize variations due to transient transfection, we established stable cell lines overexpressing codon-optimized hPLAC8-FLAG, pigPLAC8-FLAG, or pangoPLAC8-FLAG with a C-terminal 3xFLAG tag in PLAC8 KO Huh7.5 cells. pangoPLAC8 was chosen due to its low levels of both expression and rescue in the transient complementation experiments. We found that all the PLAC8 overexpression cell lines support SADS-CoV replication (Fig. 5D). Interestingly, overexpression of hPLAC8-FLAG and pigPLAC8-FLAG enhanced SADS-CoV infection kinetics, with greater replication at early timepoints compared to Scramble cells, while pangoPLAC8-FLAG only rescued to the level of Scramble cells (Fig. 5D). High levels of PLAC8 expression were observed by Western blot for hPLAC8-FLAG and pangoPLAC8-FLAG, with a lower level of expression of pigPLAC8-FLAG in the stable cell lines (Fig. 5E). Staining revealed that overexpressed PLAC8s display punctate localization in the perinuclear/endoplasmic reticulum (ER) region (SI Appendix, Fig. S4F). pigPLAC8 also shows pronounced plasma membrane localization (SI Appendix, Fig. S4F).
PLAC8 Is Not Required for Cellular Entry of SADS-CoV.
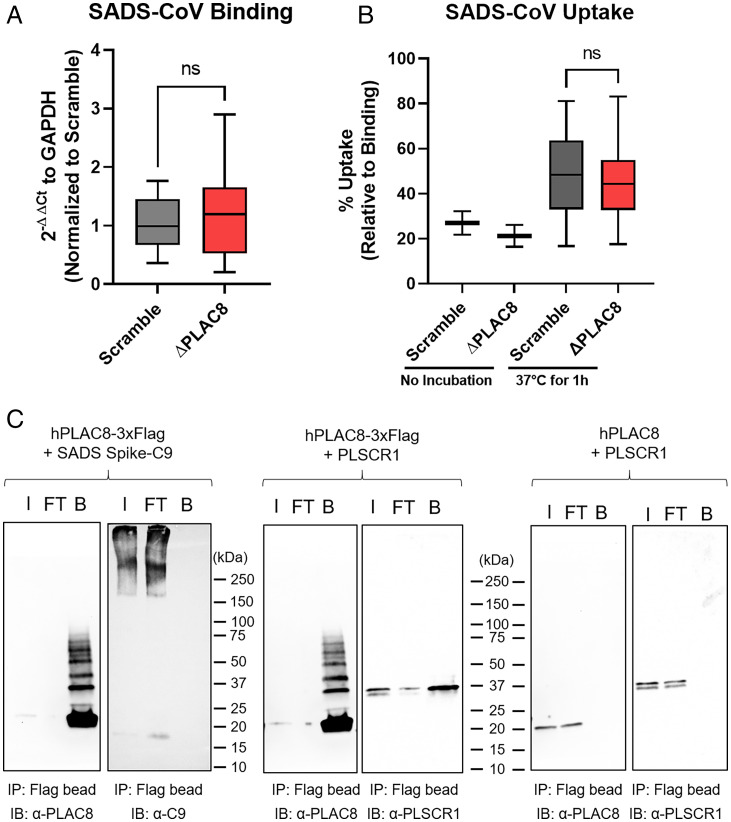
To test whether PLAC8 is the surface receptor for SADS-CoV, we performed a surface binding assay with SADS-CoV on Scramble and PLAC8 KO cells at 4 °C. No statistically significant difference in bound viral genomes was detected by qPCR between the two cell lines, suggesting that PLAC8 does not affect surface binding of SADS-CoV (Fig. 6A). After 1 h incubation at 37 °C and subsequent removal of noninternalized viral particles by extensive trypsin and acid washes, viral uptake was quantified and revealed that 40 to 60% of bound virions were endocytosed in both Scramble and PLAC8 KO cells; therefore, no significant defect in the immediate/early viral entry events was observed (Fig. 6B). We next tested for direct physical interaction between PLAC8 and SADS-CoV by performing an immunoprecipitation in lysate from 293T cells overexpressing SADS-CoV Spike and hPLAC8-FLAG. FLAG-tagged PLAC8 was unable to pull down SADS-CoV Spike even under overexpressing conditions, while PLSCR1, a known PLAC8 interactor, was successfully recovered with this approach (Fig. 6C). To confirm that PLAC8 does not interact with other surface molecules on SADS-CoV, we carried out immunoprecipitation of PLAC8 with intact SADS-CoV virions and were unable to detect any physical interaction between hPLAC8-FLAG and the viral particles (SI Appendix, Fig. S5). Taken together, these results reveal that PLAC8 is neither a receptor nor an attachment factor for SADS-CoV.
Fig. 6.
PLAC8 does not affect binding and internalization of SADS-CoV. Binding (A) and uptake (B) assay of SADS-CoV on Scramble and PLAC8 KO cells. For binding, cells were incubated with virus (MOI: 0.5) at 4 °C for 1 h and washed three times with HBSS. For uptake, after the wash from the binding assay, cells were incubated at 37 °C for 1 h. Surface virions were removed by 1) trypsin wash, 2) pH 2.5 acid wash, and 3) HBSS wash. Total RNA was isolated, and viral gRNA was determined by qRT-PCR. Data were plotted in a box-and-whisker plot, with the boxes representing 95% CI and whiskers representing maximum and minimum values from four independent experiments with three replicates. (C) Immunoprecipitation of PLAC8 with SADS-CoV spike protein. hPLAC8-FLAG and SADS-CoV spike with a C-terminal C9 tag were cotransfected in 293T cells for 48 h. Magnetic beads against FLAG tag were used for IP and blotted with α-PLAC8 or α-C9 antibodies. PLSCR1, a known factor that interacts with PLAC8, was used as the positive control, and PLAC8 without a FLAG tag was used as the negative control. I, input; FT, flow through; B, bound; ns, not significant; IP, immunoprecipitation; IB, immunoblot.
PLAC8 Is Likely Involved in Viral Trafficking prior to Transcription and Translation.
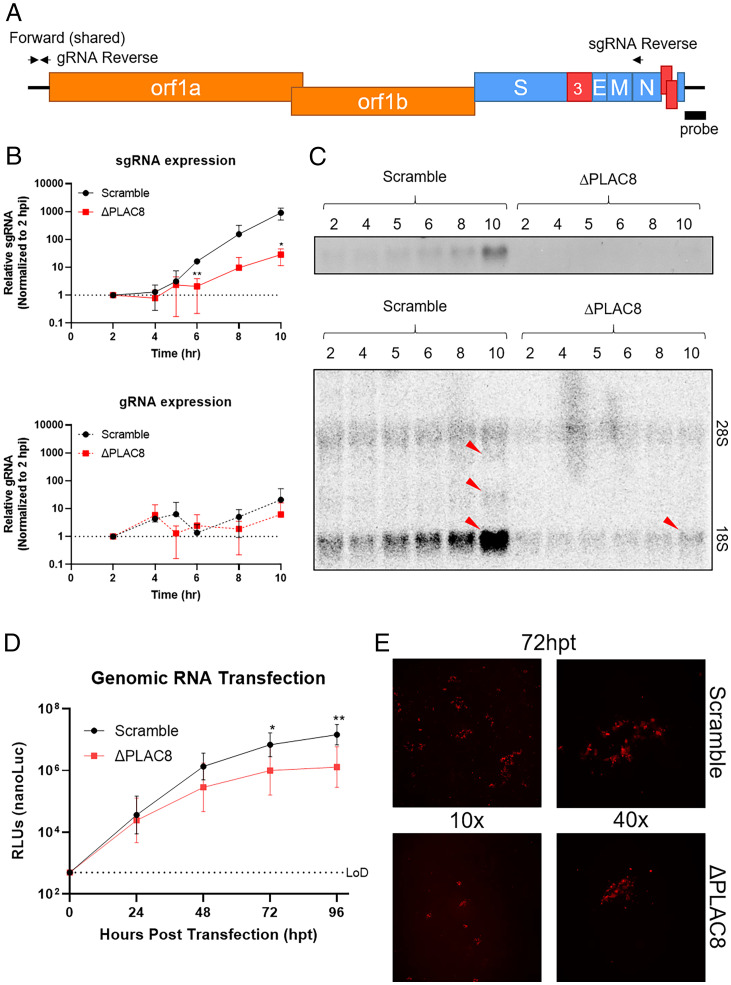
The lack of RFP and luciferase expression from our replication-competent reporter viruses supports the hypothesis that PLAC8 is critical for an early step or steps in the SADS-CoV life cycle, likely prior to protein translation. We performed a single-round growth curve to examine early viral replication events, including pretranscription trafficking, subgenomic RNA (sgRNA) expression, protein translation, and genomic RNA (gRNA) synthesis. As expected, low levels of viral gRNA can be detected and begin to increase between 6 and 8 hpi, indicating the SADS-CoV replication cycle is between 10 and 12 h in Huh7.5 cells (Fig. 7B). We observed that viral sgRNA expression was delayed and reduced by 40-fold in PLAC8 KO cells, suggesting a blockage at or before sgRNA synthesis (Fig. 7B). In addition to detection of a single sgRNA by qPCR, we performed Northern blotting to visualize all viral transcripts over the time course. The predominant RNA detected is the smallest sgRNA (the species detected by qPCR). In Scramble cells, this sgRNA clearly increases over time, whereas sgRNA expression is near the background in KO cells (Fig. 7 C, Top). A longer exposure of the blot revealed low amounts of two other sgRNAs in Scramble cells by 10 hpi (Fig. 7 C, Bottom).
Fig. 7.
PLAC8 reduces viral sgRNA expression during infection and does not affect translation. (A) Schematic of SADS-CoV viral genome, qPCR primer sets, and Northern probe to determine gRNA and sgRNA expression. (B) Kinetics of gRNA and sgRNA expression of SADS-CoV-RFP from 2 to 10 hpi on Scramble and PLAC8 KO cells. (C) Northern blot image of viral RNA from B using radiolabeled probes against the SADS-CoV-RFP C terminus. Red arrows indicate sgRNA bands. (D) Kinetics of nLuc protein expression after viral gRNA transfection into Scramble and PLAC8 KO cells over 4 d. (E) Representative fluorescence images (10 and 40x) of the RFP signal detected in Scramble (Top) and PLAC8 KO (Bottom) cells at 72 hpt. Error bars represent ± 1 SD, *P < 0.05, **P < 0.01, ***P < 0.001 by Student's t test.
To further narrow down the affected pathway, we transfected purified viral gRNA from both RFP and nLuc reporter viruses into Scramble and PLAC8 KO Huh7.5 cells. Luciferase activity and RFP expression activity were monitored for 96 h posttransfection (hpt) and increased over time in both Scramble and PLAC8 KO cells. Luciferase activities were identical at 24 hpt, revealing that transfected viral gRNA was equally capable of transcription and translation in KO cells (Fig. 7D). After the initial round of replication, progeny virions begin a normal viral infection cycle. As expected, after 24 hpt, Scramble cells begin to produce more RLU signal compared to KO cells. Additionally, the number of infected cells by RFP expression was lower in PLAC8 KO cells than in Scramble cells at 72 hpt; however, the intensity of the RFP signal remained the same in infected cells, indicating these cells are capable of translation (Fig. 7E). We quantified the amount of progeny virus from the supernatant of transfected cells. Progeny SADS-CoV-nLuc virions were detected at 48 hpt in Scramble cells, revealing viral replication after gRNA transfection. Although much slower, progeny viruses were also detected at 96 hpt in PLAC8 KO cells (SI Appendix, Fig. S6A). To ensure PLAC8 KO cells do not produce defective progeny virus particles, we transferred supernatant from transfected cells to fresh Scramble cells. Signs of infection were observed at 96 hpt, indicating that transfected PLAC8 KO cell supernatants contained infectious viral particles (SI Appendix, Fig. S6B). These results indicate that PLAC8 KO cells are capable of viral transcription and translation. Taken together with the previous results showing that PLAC8 does not play a role in viral entry, this indicates that PLAC8 is important for either viral trafficking or double-membrane vesicle (DMV) formation after entry.
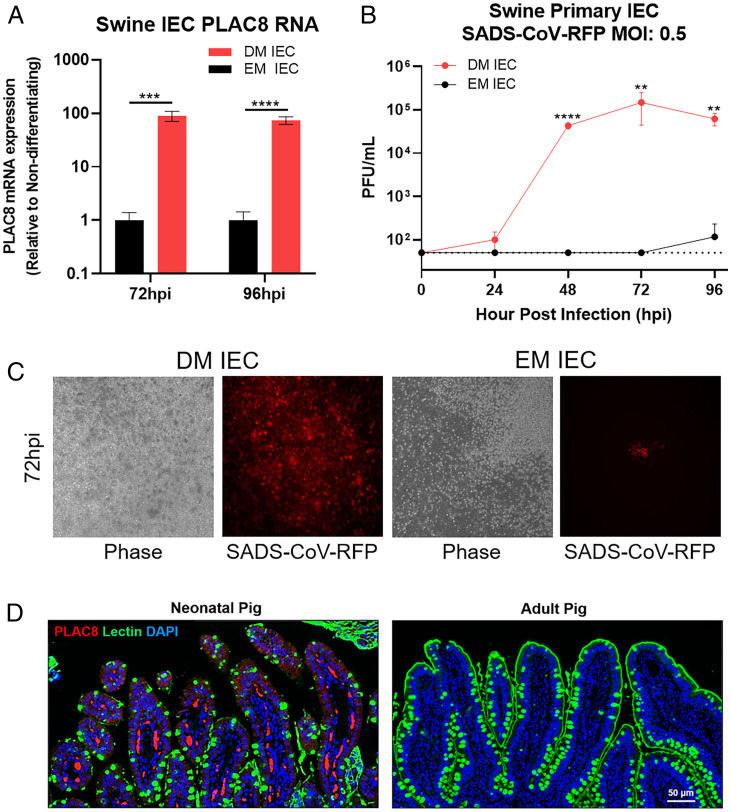
PLAC8 Expression Correlates with SADS-CoV Infectivity in Swine Primary Intestinal Epithelial Cell (IEC).
To investigate the relevance of PLAC8 in SADS-CoV infection in swine cells, we extracted IEC from pig jejunum and established primary cultures for SADS-CoV infection. Primary IEC were maintained either in expansion medium (EM) to preserve the stem- or crypt-like state or in differentiation medium (DM) to adopt a villi-like phenotype. We measured PLAC8 mRNA expression in both EM and DM IECs and observed an 87-fold increased expression in DM compared to EM IECs (Fig. 8C). Remarkably, IECs in DM were able to support SADS-CoV growth and reach peak titer (105) at 72 hpi. In contrast, IECs in EM were completely resistant to SADS-CoV infection (Fig. 8A). RFP expression correlated with viral titer and showed extensive expression of RFP in DM IEC but no expression in EM IEC (Fig. 8B). PLAC8 expression level correlated with SADS-CoV infectivity in swine primary IEC, further supporting the importance of PLAC8 in SADS-CoV infection.
Fig. 8.
SADS-CoV infection in primary swine IECs. (A) PLAC8 mRNA expression of SADS-CoV-RFP–infected IECs in DM or EM at 72 and 96 hpi. (B) Viral growth of SADS-CoV-RFP on primary swine IEC in DM or EM. (C) Representative images of SADS-CoV-RFP–infected IECs in DM and EM at 72 hpi. (D) PLAC8 and glycocalyx integrity in neonatal and adult pig ileum. Neonatal and adult swine ileal sections of naïve pigs were stained with PLAC8 (red), UEA lectin (green), and DAPI (blue). Scale bar is 50 µm. Error bars represent ± 1 SD, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Student's t test.
PLAC8 Is Expressed in Neonatal Swine Ileum.
Neonatal piglets (<5 d old) infected with SADS-CoV have a 90% mortality rate; however, as the swine gastrointestinal tract matures, mortality rates drop dramatically to <5% in adult animals. We examined PLAC8 expression in in vivo conditions using a WT, noninfected pig model. We obtained ileal tissues from neonatal piglets (1 to 2 d old) and adult pigs (>10 mo old) and performed immunostaining with an anti-PLAC8 antibody to examine changes in protein expression and Ulex europaeus agglutinin (UEA) lectin to visualize the glycocalyx coating the enterocytes. In the neonatal ileum, PLAC8 expression was detected with a moderate signal in the enterocytes and a robust signal in the capillaries/lacteal vessels of the villi. In contrast, expression of PLAC8 was not detectable in adult tissue, consistent with the notion that older animals are less affected by SADS-CoV infection (Fig. 8D). Furthermore, lectin stain revealed the presence of a thin and irregular layer lining the neonatal pig ileum, while the adult tissue was coated with a thick and continuous glycocalyx, which may provide a physical barrier against pathogens like SADS-CoV (Fig. 8D).
Discussion
SADS-CoV is an important, emerging veterinary pathogen with the potential to transmit to humans. However, little is known about the fundamental virus–host interaction networks of this pathogen. Therefore, we performed a GeCKO screen and identified PLAC8 as the top host factor for SADS-CoV infection in Huh7.5 cells. Consistent with earlier reports (14, 15), none of the known CoV entry receptors were identified in our CRISPR screen, supporting the hypothesis that SADS-CoV uses an unknown receptor to initiate binding and entry.
PLAC8 plays a critical role in an early stage of SADS-CoV infection during viral trafficking, leading to a delay and reduction in viral sgRNA expression. We did not detect a direct physical interaction between SADS-CoV and PLAC8 by immunoprecipitation. Although we cannot exclude the possibility of indirect binding to other known CoV receptors, such as neuropilin-1 (31, 32), CD147 (33, 34), or L-SIGN (35), the wide tropism of SADS-CoV argues that it either uses a highly conserved generic entry factor, such as sugars or a paralogous gene family, or uses multiple entry pathways. In agreement with the broad tropism of SADS-CoV, our studies demonstrate that PLAC8 from multiple species can support SADS-CoV infection, suggesting PLAC8 is involved in an evolutionarily conserved pathway. While noting that PLAC8 is the top host factor for SADS-CoV infection and may be involved in an uncharacterized endocytic pathway, future studies would benefit from mapping the protein interaction network of PLAC8 with other host proteins and cellular pathways.
PLAC8, also known as onzin, is a small (12.5 kDa) cysteine-rich protein with no previously known function in viral infection. A recent study identified ZDHHC17, an S-acyltransferase that places S-palmitoylation, as a critical factor for SADS-CoV infection (20). PLAC8 is predicted to contain four S-palmitoylation sites and to be a substrate for ZDHHC17 (36). S-palmitoylation is a reversible posttranslational modification formed by attaching a palmitoyl chain to a cysteine residue via a thioester bond. S-palmitoylation influences protein localization, trafficking, and stability and hence is critical for cellular functions (37–40). Further studies of how palmitoylation of PLAC8 affects its ability to support SADS-CoV infection would be of great interest.
Our transcriptome analysis on PLAC8 KO Huh7.5 cells sheds light on the cellular pathway involvement of PLAC8 in SADS-CoV infection. Based on our analysis, PLAC8 KO affects aspects of cellular structure and motility, including the plasma membrane, cell-cell junctions, actin, integrins, and cadherins. Cytoskeletal/structural proteins including actin filaments, microtubules, and intermediate filaments are known to contribute to multiple stages of human and animal coronaviral life cycles, including entry, trafficking, replication, and egress (41, 42). Cytoskeletal components have been identified as differentially expressed and/or virion associated for infectious bronchitis virus (IBV) and transmissible gastroenteritis virus (TGEV) (43, 44).
PLAC8 is also known to be involved in autophagy and EMT pathways, which are important for CoV infection. For example, autophagy and autophagy-related proteins have been shown to be involved in CoV replication (45–47). Studies have shown that CoV-induced DMVs originate from the omegasome, a derivative of the ER (48, 49); however, instead of ER proteins, it is decorated with the autophagosome markers LC3, ATG12, and ATG8 (50). Mouse hepatitis virus, IBV, TGEV, and porcine alpha-CoV infections induce autophagy and require a key regulator of phagophoric extension, ATG5, for viral replication (49–52) although this requirement is cell type dependent (53–55). Nevertheless, the findings showing LC3 is present on DMVs suggest that autophagy-related proteins play an important role in CoV replication (56). Recent CRISPR screens on SARS-CoV-2 and flaviviruses both identified TMEM41B, a cellular factor involved in autophagy, which also plays an important role in viral replication complex formation (19, 57).
PLAC8 is heavily involved in EMT, a pathway that is important in cancer metastasis, wound repair, and organ development (58). The vast difference in mortality rates between <5-d-old piglets and adult pigs coincides with rigorous intestinal development/EMT in newborn piglets (59–61). The field observations and molecular understanding of PLAC8 are in agreement with the robust PLAC8 expression in the ileum of neonatal piglets but not in adult pigs. In our infection model, differentiating primary swine IEC showed robust infection by SADS-CoV correlating with high levels of PLAC8 expression. Our results unequivocally highlight the importance of PLAC8 in SADS-CoV infection in nature. Given the unique involvement of EMT and SADS-CoV infection, future studies to understand the interplay between CoV infection and the EMT pathway are warranted.
Lastly, multiple CoV CRISPR screens also identified similar pathways, such as autophagy and cholesterol metabolism (including SCAP, also identified here), supporting a general similarity of cellular pathways among CoV infections (18, 19, 62–65). Cytoskeletal pathways, solute carrier family members, and EMT were only identified in our SADS-CoV screen but not other CoVs, indicating a unique feature of SADS-CoV infection. Given the strong requirement for PLAC8 in SADS-CoV infection, the identified PLAC8 gene could be a prime target for antiviral development to combat future emerging CoVs.
Materials and Methods
Huh7.5 GeCKO Library and Huh7.5 CRISPR KO Cell Line Generation.
The Huh7.5 CRISPR KO library was generated as described previously (17). In brief, the CRISPR viral guide RNA library was produced from 293T cells cotransfected with pCMV-VSV-G (Addgene 8454), psPAX2 (Addgene 12260), and the human guide RNA library Brunello in lentiCRISPRv2 (Addgene 73179). Huh7.5 cells were transduced by the lentiviral library and selected with puromycin (2.5 µg/mL). Individual CRISPR KO cell lines were generated as described above, except that guide RNAs were cloned individually into lentiCRISPRv2 (Addgene 98290). Polyclonal cells were sequenced via Sanger sequencing to confirm deletion of the target genes and were subsequently clonal selected via limited dilution. Clonal CRISPR KO cells were sequence confirmed via Sanger sequencing.
CRISPR Screen of SADS-CoV.
Two T-75 flasks of low-passage Huh7.5 CRISPR library were infected with SADS-CoV-WT at high MOI for 5 to 7 d in 5% CO2 at 37 °C. Surviving cells were passaged to a clean flask and allowed to continue growing in growth media until reaching 60 to 70% confluency before reinfection, for a total of four cycles. Genomic DNA before selection and from each round of infection was collected from the cell pellet.
High-Throughput Sequencing and Transcriptomics.
For GeCKO library sequencing, genomic DNA from unselected and selected library cells was isolated using the Qiagen Blood & Cell Culture Kit. Amplicons were prepared using the Illumina Truseq system and Q5 Hot Start High Fidelity DNA polymerase (New England Biolabs [NEB]). Primers for first-round PCR were specific for the gRNA sequence with overhangs for the Illumina adapters. After 18 cycles of amplification, PCR products were purified and used as templates for second-round PCR using the standard Illumina P5 and P7 primers. Second-round PCR products were purified and analyzed by BioAnalyzer (Agilent) and a Qubit 4 fluorometer (Invitrogen) and then diluted to 4 nM and pooled for sequencing. Sequencing was carried out on a MiSeq system using a 300-cycle v2 MiSeq Reagent Kit (Illumina), following the manufacturer’s protocol. At least 35 million reads were obtained from unselected samples, and at least 300,000 reads were obtained from selected samples.
For transcriptomics, samples were prepared in triplicate from Huh7.5 Scramble cells, Huh7.5 PLAC8 KO cells, and Huh7.5 Scramble cells 24 hpi with SADS-CoV. Total cellular RNA was isolated using TRIzol reagent following the manufacturer’s protocol. Libraries were prepared using the Illumina Stranded Total RNA Prep kit (Illumina) and sequenced by the University of North Carolina at Chapel Hill (UNC) High-Throughput Sequencing Core Facility on a NovaSeq SP using a 200-cycle v1.5 kit.
Sequencing Analysis.
For GeCKO library analysis, reads were trimmed to guide RNA sequences using a custom perl script. Guide enrichment analysis was performed to compare selected samples to the unselected library using MAGeCK software as written with the --norm-method total command. The results of this analysis were plotted in R version 4.0.3 using a custom script.
For transcriptome analysis, sequences were first trimmed using BBDuK version 38.87 with the parameters ktrim = r k = 23 hdist = 1 minlen = 50. Trimmed reads were aligned to the human genome (hg19) using STAR version 2.7.7a with the parameters --peOverlapNbasesMin 30 --outSAMtype BAM SortedByCoordinate. BAM files were indexed using Samtools version 1.12. Features were counted using featureCounts as part of Subread version 2.0.1 with the parameters -T 4 -s 2 -d 25 -p -P -B -M -O. Differential expression analysis was performed using DESeq2 version 1.30.1 under the Variable design. Heatmaps and volcano plots were created using custom R scripts. A list of significant DEGs was ordered by increased P value and used as input for g:Profiler (https://biit.cs.ut.ee/gprofiler/) with ordered query checked and using GO annotations. The output from g:Profiler was input into Cytoscape version 3.8.2 and the EnrichmentMap tool was used to create a network.
SADS-CoV-nLuc Reverse Genetics.
SADS-CoV-nLuc reporter viruses were constructed using a seven-plasmid cloning strategy as described previously (14). The genome was divided into seven fragments and subcloned into seven separate plasmids. A T7 promoter was introduced into the 5′ end, and ORF3a was replaced with nLuc. Unique type IIS restriction endonuclease cleavage sites are introduced into the 5′ and 3′ ends of each fragment. Plasmid DNA was digested with the corresponding enzymes, gel purified, and ligated together with T4 DNA ligase (NEB). Ligation products were purified by chloroform extraction and used as a template for in vitro transcription with T7 RNA polymerase (Thermo Fisher Scientific). RNA was electroporated into Vero81 cells along with mRNA transcript of SADS-CoV N protein. Virus-containing supernatant was harvested 3 to 4 d postelectroporation and subsequently passaged.
SADS-CoV Growth Kinetics.
Five hundred thousand cells were seeded in a six-well plate 1 d prior to infection. Cells were infected with SADS-CoV or SADS-CoV-RFP at 0.05 MOI, assuming 1 × 106 cells at infection. Cells were prewashed once with phosphate buffered saline (PBS) and incubated with 300 µL of inoculum at 37 °C for 1 h with rocking every 15 min. Viral inoculum was removed, and cells were washed three times with PBS and replenished with 3 mL of infection media. Then, 300 µL of viral supernatant was collected and replaced with fresh media at 0, 24, 48, 72, 96 and 120 hpi. Fluorescence images were also taken at each time point. Titers were determined by a standard plaque assay. In brief, Vero81 cells were seeded at 2.5 × 105 cells/well in a six-well plate. Cells were washed twice with PBS, and 200 µL of serial diluted viral supernatant in Dulbecco's modified Eagle medium (DMEM) + 5% tryptose phosphate broth (Sigma) + 5 µg/mL trypsin was added to each well and incubated for 1 h at 37 °C. Next, 2.5 mL of overlay consisting of DMEM + 0.8% SeaKem Agarose (Lonza) + 5% tryptose phosphate broth + penicillin-streptomycin + 5 µg/mL trypsin was added to each well and incubated for 72 h at 37 °C. Cells were stained with neutral red for 2 h, and plaques were counted manually.
nLuc-Based Infection Assay.
Cells were seeded at 2 × 104 cells/well on a black 96-well plate 1 d before infection. The medium was removed and cells were infected with 100 µL of virus inoculum (600 plaque forming unit [pfu]/well, MOI: ∼0.012) diluted in infection media. The plates were incubated at 37 °C for 48 h before nLuc reading.
PLAC8 Complementation Assays.
PLAC8 KO cells were seeded on a 12-well plate. At 60 to 70% confluency, cells were transfected with 1.2 µg of PLAC8 plasmids using Lipofectamine 3000 (Invitrogen). The transfection medium was replaced with normal growth media after 6 h and incubated overnight. Transfected cells were split into a black 96-well plate with ∼2 × 104 to 5 × 104 cells/well or an 8-well chamber slide with ∼5 × 104 to 10 × 104 cells/well and incubated overnight. At 80 to 100% confluency, the medium was removed and cells were infected with SADS-CoV-nLuc at 600 pfu/well, ∼0.01 MOI (96-well plate), or SADS-CoV-RFP at 5 × 104 pfu/well, ∼0.5 MOI (8-well chamber slides), for 48 h at 37 °C.
nLuc Assay, Immunostaining, and Western Blotting.
For the nLuc assay, cells were equilibrated at room temperature (RT) for 10 min. Then, 25 µL of Nano-Glo Luciferase Substrate (Promega, N3100) was added to each well and incubated at RT for 5 min. Luciferase readings were detected via Glo-Max Explorer (Promega). For immunostaining, cells were fixed in 10% formalin in PBS and permeabilized with PBS + 0.3% Triton X-100 (Sigma). Monoclonal mouse anti-FLAG (Sigma F3165, 1:100) was used as the primary antibody, and goat anti-mouse Alexa 488 was used as the secondary antibody. RFP was the endogenous signal from the SADS-CoV-RFP viruses. Slides were mounted with ProLong gold reagent with DAPI (Invitrogen, P36931) and images were taken by a Keyence BZ-X810 fluorescence microscope. For Western blotting, cell were lysed in 1% Triton X-100, 100 mM Tris, 2M NaCl, and 100 mM ethylenediaminetetraacetic acid (EDTA). Cell lysates were run in sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto polyvinylidene fluoride membrane. PLAC8 bands were detected using polyclonal rabbit anti-PLAC8 (Thermo Fisher Scientific, PA5-59189, 1:500) and goat anti-rabbit horseradish peroxidase (HRP), or mouse anti-FLAG (Sigma, F3165, 1:2,000) and goat anti-mouse HRP.
SADS-CoV Binding, Uptake, and One-Step Growth Assay.
Cells were seeded at 2 × 105 cells/well in a 48-well plate 1 d before infection. Cells were prechilled on ice, and the medium was removed and infected with SADS-CoV-nLuc or RFP viruses at a high MOI (0.5 to 1) for 1 h at 4 °C. After 1 h, cells were washed with 1x Hank's balanced salt solution (HBSS) twice to remove unbound virions. Then, 200 µL of TRIzol was used to collect total RNA from the binding samples. For uptake assays, after the 2x HBSS wash from binding assay, prewarmed infection medium was added to each well and incubated at 37 °C in a 5% CO2 incubator for 1 h. To remove noninternalized virus, cells were washed with trypsin wash, acid wash with potassium phosphate buffer pH 2.5, and 1x HBSS wash. Cell pellets were resuspended in 200 µL of TRIzol for total RNA extraction. For one-step growth assay, after the 2x HBSS washes from the binding assay, prewarmed infection medium was added to each well and incubated at 37 °C in a 5% CO2 incubator. The medium was removed and TRIzol was added at 2, 4, 5, 6, 8, and 10 hpi to collect total RNA.
RNA Extraction and qRT-PCR.
RNA collected in TRIzol reagent was extracted following the manufacturer’s protocol. Up to 5 μg of RNA was treated with DNase using the Turbo DNA-free kit (Invitrogen). Equal nanogram amounts were used to synthesize complementary DNA (cDNA) using the High-Capacity RNA-to-cDNA kit (Applied Biosystems). Equal volumes of cDNA were used to run qPCR using SYBR green master mix (Roche).
Northern Blotting.
RNA was resuspended in denaturing buffer (Sigma R1386), incubated at 65 °C for 7 min, and cooled on ice. Samples were separated on a 1.2% denaturing formaldehyde-agarose gel in a 1X 3-morpholinopropane-1-sulfonic acid running buffer and subsequently transferred to a Hybond N+ membrane (GE Healthcare). A radiolabeled probe was generated using the Prime-It II Random Primer Labeling Kit (Agilent Technologies). DNA templates for probe labeling were generated by PCR from a plasmid template. The radiolabeled probe was purified using Illustra MicroSpin G-50 columns (GE Healthcare). The probe was then hybridized to the membrane in ULTRAhyb buffer (Invitrogen) at 42 °C for 4 h. After washing, blots were visualized by exposure to film or to a PhosphorImager screen followed by scanning and using Amersham Typhoon (GE Healthcare).
Immunoprecipitation.
Confluent 10-cm plates of 293T cells were transfected with equal microgram amounts of the indicated plasmids using polyethylenimine (PEI) Max. At 2 to 3 d posttransfection, cells were lysed in 5 mL of Nonidet P-40 lysis buffer (50 mM Tris HCl pH 7.4, 150 mM NaCl, and 0.2% Nonidet P-40) + a protease inhibitor mixture on ice for 30 min. Lysates were spun at 16,000 x g for 5 min to remove cellular debris. The clarified lysates were combined with 12.5 μL of FLAG beads and incubated with rocking at 4 °C for 2 h. The beads were washed 3x in lysis buffer, and then bound protein was eluted in 100 μL of lysis buffer + 5 μL of 3x FLAG peptide at 4 °C with rocking for 2 h. Input, unbound, and eluted samples were prepared with a final concentration of a 1x Laemmli sample buffer and 0.1 M dithiothreitol (DTT). Samples were then analyzed by Western blot as described above. For SADS spike C9 protein, monoclonal mouse anti-C9 (GeneTex, GTX25417) and goat anti-mouse HRP were used as primary and secondary antibodies. For PLSCR1, monoclonal mouse anti-PLSCR1 (Millipore, MABS482, 1:1,000) and goat anti-mouse HRP were used as primary and secondary antibodies.
For live virus immunoprecipitations, confluent 10-cm plates of 293T cells were transfected with the indicated PLAC8 plasmids using PEI Max. At 3 d posttransfection, cells were treated for lysis and immunoprecipitation as above. Following incubation of beads and lysate, beads were washed followed by transport into the biosafety level 3 laboratory space. Beads were combined with 1 mL of viral supernatant and incubated for a further 2 h at 4 °C, with manual mixing every 15 min. Beads were washed 3x as before and then resuspended with 100 μL of Nonidet P-40 lysis buffer with a final concentration of a 1x Laemmli sample buffer and 0.1 M DTT. Samples were heated at 65 °C for 30 min and then analyzed by Western blot as described above.
Viral gRNA Extraction and Transfection.
SADS-CoV-RFP and SADS-CoV-nLuc at 3 × 106 pfu each were mixed, concentrated, and washed twice with PBS using 100,000 molecular weight cut-off centrifuge filters (Prometheus, 84-568). Viral gRNA was extracted using TRIzol-LS (Thermo Fisher Scientific) following the manufacturer’s protocol. Cells at 50% confluency were transfected with 6 × 105 pfu equivalents + 1.5 µg of carrier transfer RNA via a TransIT-mRNA Kit (Mirus Bio), following the manufacturer’s protocol. Media and immunofluorescence images were collected every day for 4 d after transfection. Luciferase activities and titers were determined as described above.
SADS-CoV-nLuc Neutralization Assays.
Mouse α-SADS spike protein sera generated from virus replicon particle vaccination were serially diluted in infection media and mixed at a 1:1 ratio with 300 pfu/well of SADS-CoV-nLuc and incubated at 37 °C for 1 h. The virus and antibody mixture was then added to Huh7.5 Scramble cells and incubated at 37 °C for 48 h. A nLuc assay was used to determine relative infectivity. The percentage of inhibition was calculated by the following equation: 100*(1 − [RLUsample/RLUmock-treatment]).
Pig Tissue Collection and Staining.
All animal work with adult pigs and neonatal piglets was approved by the Institutional Animal Care and Use Committee at the North Carolina State University (NCSU) Veterinary School under protocol 19-019B according to guidelines outlined by the Association for the Assessment and Accreditation of Laboratory Animal Care and the US Department of Agriculture. Following euthanasia of neonatal (1 to 2 d old) and adult (>10 mo old) naïve Yorkshire crossbred pigs with pentobarbital (85 to 100 mg/kg, intravenously), the ileum was identified and collected. Tissues were rinsed with 1X PBS and opened longitudinally along the antimesenteric border. Tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned (∼5- to 8-µm thickness). Following deparaffinization, sections were processed for immunolabeling. Unstained sections were incubated with heated 1X citrate buffer in deionized water for 10 min and blocked for 1 h in 3% bovine serum albumin (BSA) solution. Slides were incubated with a rabbit polyclonal anti-PLAC8 (Thermo Fisher Scientific, PA5-67482, 1:100) and biotinylated UEA lectin (vector ZE0627) overnight at 4 °C, followed by secondary antibodies anti-rat Alexa Fluor 594 and antistreptavidin Alexa 488 (Invitrogen R37119 and S11223), with incubation for 1 h at RT. Slides were incubated with DAPI (Invitrogen 2031179) for 5 min at RT and mounted using FluorSave reagent (Millipore Sigma 345789). Fluorescence was detected with a VS120 virtual scanning microscope (Olympus Microscopy, Shinjuku, Japan).
Swine Primary IEC Cultures.
Intestinal crypts were extracted and isolated from jejunum tissue of Yorkshire crossbred pigs obtained from NCSU as described in Gonzalez et al. (66). Briefly, tissue sections were washed with ice-cold PBS, placed in 50-mL conical tubes containing 30 mM EDTA, 10 mM Y-27632, and 1 mM dithiothreitol in PBS on ice on a rocker at 60 rpm for 30 min with vigorous shaking every 5 min, and then incubated in a PBS solution containing 30 mM EDTA and 10 mM Y-27632 for 10 min at 37 °C, followed by vigorous shaking to remove the villi or fully differentiated enterocytes. Subsequently, villi-free tissue was washed for 5 min in ice-cold PBS containing 1X antibiotic-antimycotic and gently shaken to detach crypts, the intestinal structure containing stem cells. Isolated crypts were expanded in cell culture on neutralized rat tail type I collagen (Corning 356236) hydrogels in 12-well plates using the monolayer culture system, as described in Wang et al. (67). Briefly, freshly isolated jejunum epithelial cells were seeded at 250,000 cells/well and maintained with EM, promoting expansion and preventing differentiation to maintain cells in a stem- or crypt-like state. In addition to base media, EM contained the following compounds: ROCK inhibitor (Y-27632, ApexBio A3008-200), p38 MAPK inhibitor (SB202190, Selleckchem, S1077), gastrin (Anaspec, AS-64149) for mitosis induction, PGE2 (Caymen Chemicals, 14010) for stem cell proliferation, and B27 (Thermo Fisher Scientific, 12587010) for stem cell growth. One day after seeding, half of the cells were switched to DM, which induced cell differentiation and allowed enterocytes to adopt a villi-like phenotype. In addition to base media, DM contained murine epidermal growth factor (PeproTech, 315-09) to stimulate epithelial cell growth and differentiation. Undifferentiated and differentiated cells were infected 48 h after seeding on collagen hydrogels with MOI = 0.5, and apical media and cells were collected at 72 and 96 hpi for titers and mRNA extraction, respectively. Swine IEC cultures were lysed in TRIzol reagent (Invitrogen, Carlsbad, CA) and processed for total mRNA extraction, followed by cDNA reverse transcription and qPCR as described in Morrison et al. (68). In brief, total RNA was purified via the Direct-Zol RNA miniprep kit (Zymo Research, Irvine, CA). RNA was reverse transcribed to cDNA using iScript Reverse Transcription Supermix for qRT-PCR (Bio-Rad Laboratories Inc., 1708841), and qPCR was performed on a QuantStudio6 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA). Expression of pigPLAC8 was detected using a TaqMan PLAC8 qPCR probe (assay ID: Ss06903341_m1) with SsOAdvanced Universal Probes Supermix (172581). Pig GAPDH was used as a housekeeping gene for normalization, and qPCR was performed using a PrimePCR GAPDH probe (12001961, assay ID: qSscCEP0038854) with SsOAdvanced Universal Probes Supermix.
Statistical Analysis.
Statistical analysis was carried out using GraphPad Prism version 9.0 and Student’s t test. Significance symbols are defined as follows: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data are graphed as mean ± SD.
Supplementary Material
Acknowledgments
We thank members of the entire Baric laboratory for helpful discussions, Amanda Ziegler for supplying the pig intestines, and Shlok Joshi for technical assistance. This project received support from NIH grants AI110700, AI151797, and AI116484 to R.S.B.; K01 OD019911-01A1 to L.M.G.; and P30 DK065988-17 to C.E. S.T.M. received grants from the Center for Gastrointestinal Biology and Disease (P30 DK034897) and Cystic Fibrosis Center (P30DK065988 and R01DK109559). L.V.T. is the recipient of the Pfizer NCBiotech Distinguished Postdoctoral Fellowship in Gene Therapy. This research was also supported by funding from Vertex Research Innovation awarded to C.E. and from the Chan Zuckerberg Initiative awarded to R.S.B. We gratefully acknowledge technical support from the UNC High-Throughput Sequencing Facility. This facility is supported by the University Cancer Research Fund, a Comprehensive Cancer Center Core Support grant (P30-CA016086), and a UNC Center for Mental Health and Susceptibility grant (P30-ES010126).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118126119/-/DCSupplemental.
Data Availability
High-throughput sequencing data from the CRISPR screen have been deposited in the Sequence Read Archive (BioProject PRJNA826466) (69). RNAseq data have been deposited in the Gene Expression Omnibus (Accession GSE200982) (70). All other study data are included in the article and/or SI Appendix.
References
- 1.Gallagher T., Perlman S., Broad reception for coronavirus. Nature 495, 176–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., et al. , Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D. M. E., Fouchier R. A. M., Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Wu F., et al. , A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., et al. , A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q., Vlasova A. N., Kenney S. P., Saif L. J., Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 34, 39–49 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F., The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P., et al. , Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556, 255–258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan Y., et al. , Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 211, 15–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong L., et al. , A new bat-HKU2–like coronavirus in swine, China, 2017. Emerg. Infect. Dis. 23, 1607–1609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y. L., Yu J. Q., Huang Y. W., Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): An update three years after its discovery. Virus Res. 285, 198024 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J., Qiao S., Guo R., Wang X., Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution. Nat. Commun. 11, 3070 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan H., et al. , Cryo-electron microscopy structure of the swine acute diarrhea syndrome coronavirus spike glycoprotein provides insights into evolution of unique coronavirus spike proteins. J. Virol. 94, e01301-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards C. E., et al. , Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. U.S.A. 117, 26915–26925 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y.-L., et al. , Broad cross-species infection of cultured cells by bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J. Virol. 93, e01448-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Y., et al. , Broad cell tropism of SADS-CoV in vitro implies its potential cross-species infection risk. Virol. Sin. 36, 559–563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R., et al. , Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell 184, 106–119.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider W. M., et al. , Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell 184, 120–132.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y., et al. , Identification of ZDHHC17 as a potential drug target for swine acute diarrhea syndrome coronavirus infection. mBio 12, e0234221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galaviz-Hernandez C., et al. , Plac8 and Plac9, novel placental-enriched genes identified through microarray analysis. Gene 309, 81–89 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Ledford J. G., Kovarova M., Koller B. H., Impaired host defense in mice lacking ONZIN. J. Immunol. 178, 5132–5143 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Kinsey C., et al. , Plac8 links oncogenic mutations to regulation of autophagy and is critical to pancreatic cancer progression. Cell Rep. 7, 1143–1155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaistha B. P., et al. , PLAC8 localizes to the inner plasma membrane of pancreatic cancer cells and regulates cell growth and disease progression through critical cell-cycle regulatory pathways. Cancer Res. 76, 96–107 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Li C., et al. , Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer. J. Clin. Invest. 124, 2172–2187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang M. L., et al. , Plac8-mediated autophagy regulates nasopharyngeal carcinoma cell function via AKT/mTOR pathway. J. Cell. Mol. Med. 24, 7778–7788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., et al. PLAC8 promotes adriamycin resistance via blocking autophagy in breast cancer. J. Cell. Mol. Med. 25, 6948–6962 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson R. M., Kerr M. S., Slaven J. E., Plac8-dependent and inducible NO synthase-dependent mechanisms clear Chlamydia muridarum infections from the genital tract. J. Immunol. 188, 1896–1904 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daghino S., et al. , Yeast expression of mammalian Onzin and fungal FCR1 suggests ancestral functions of PLAC8 proteins in mitochondrial metabolism and DNA repair. Sci. Rep. 9, 6629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon D. E., et al. , A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantuti-Castelvetri L., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daly J. L., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370, 861–865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K., et al. , CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 5, 283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z., et al. , Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J. Infect. Dis. 191, 755–760 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amraei R., et al. , CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. ACS Cent. Sci. 7, 1156–1165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemonidis K., MacLeod R., Baillie G. S., Chamberlain L. H., Peptide array-based screening reveals a large number of proteins interacting with the ankyrin-repeat domain of the zDHHC17 S-acyltransferase. J. Biol. Chem. 292, 17190–17202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobocińska J., Roszczenko-Jasińska P., Ciesielska A., Kwiatkowska K., Protein palmitoylation and its role in bacterial and viral infections. Front. Immunol. 8, 2003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blaskovic S., Blanc M., van der Goot F. G., What does S-palmitoylation do to membrane proteins? FEBS J. 280, 2766–2774 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Mitchell D. A., Vasudevan A., Linder M. E., Deschenes R. J., Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 47, 1118–1127 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Linder M. E., Deschenes R. J., Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Wen Z., Zhang Y., Lin Z., Shi K., Jiu Y., Cytoskeleton-a crucial key in host cell for coronavirus infection. J. Mol. Cell Biol. 12, 968–979 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z., et al. , Porcine hemagglutinating encephalomyelitis virus enters neuro-2a cells via clathrin-mediated endocytosis in a Rab5-, cholesterol-, and pH-dependent manner. J. Virol. 91, 1–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dent S. D., et al. , The proteome of the infectious bronchitis virus Beau-R virion. J. Gen. Virol. 96, 3499–3506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X., et al. , Identification of cellular proteome using two-dimensional difference gel electrophoresis in ST cells infected with transmissible gastroenteritis coronavirus. Proteome Sci. 11, 31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller K., et al. , Coronavirus interactions with the cellular autophagy machinery. Autophagy 16, 2131–2139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maier H. J., Britton P., Involvement of autophagy in coronavirus replication. Viruses 4, 3440–3451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Haan C. A. M., Reggiori F., Are nidoviruses hijacking the autophagy machinery? Autophagy 4, 276–279 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Axe E. L., et al. , Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cottam E. M., et al. , Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 7, 1335–1347 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prentice E., Jerome W. G., Yoshimori T., Mizushima N., Denison M. R., Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279, 10136–10141 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo X., et al. , Porcine epidemic diarrhea virus induces autophagy to benefit its replication. Viruses 9, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu L., Mou C., Yang X., Lin J., Yang Q., Mitophagy in TGEV infection counteracts oxidative stress and apoptosis. Oncotarget 7, 27122–27141 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Z., et al. , Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3, 581–585 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Ko S., et al. , Rapamycin-induced autophagy restricts porcine epidemic diarrhea virus infectivity in porcine intestinal epithelial cells. Antiviral Res. 146, 86–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo L., et al. , Autophagy negatively regulates transmissible gastroenteritis virus replication. Sci. Rep. 6, 23864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reggiori F., et al. , Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7, 500–508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffmann H. H., et al. , TMEM41B is a pan-flavivirus host factor. Cell 184, 133–148.e20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalluri R., Weinberg R. A., The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Everaert N., et al. , A review on early gut maturation and colonization in pigs, including biological and dietary factors affecting gut homeostasis. Anim. Feed Sci. Technol. 233, 89–103 (2017). [Google Scholar]
- 60.McCance R. A., The effect of age on the weights and lengths of pigs’ intestines. J. Anat. 117, 475–479 (1974). [PMC free article] [PubMed] [Google Scholar]
- 61.Choudhury R., et al. , Impact of early-life feeding on local intestinal microbiota and digestive system development in piglets. Sci. Rep. 11, 4213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baggen J., et al. , Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 53, 435–444 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Daniloski Z., et al. , Identification of required host factors for SARS-CoV-2 infection in human cells. Cell 184, 92–105.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu Y., et al. , A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 12, 961 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei J., et al. , Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 184, 76–91.e13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonzalez L. M., et al. , Preservation of reserve intestinal epithelial stem cells following severe ischemic injury. Am. J. Physiol. Gastrointest. Liver Physiol. 316, G482–G494 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y., et al. , Self-renewing monolayer of primary colonic or rectal epithelial cells. Cell. Mol. Gastroenterol. Hepatol. 4, 165–182.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morrison C. B., et al. , Treatment of cystic fibrosis airway cells with CFTR modulators reverses aberrant mucus properties via hydration. Eur. Respir. J. 59, 2100185 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.R. M. Meganck, L. V. Tse, R. S. Baric, Genome-wide CRISPR knockout screen identified PLAC8 as an essential factor for SADS-CoVs infection. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/PRJNA826466. Deposited 13 April 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R. M. Meganck, L. V. Tse, R. S. Baric, Genome-wide CRISPR knockout screen identified PLAC8 as an essential factor for SADS-CoVs infection. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200982. Deposited 18 April 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing data from the CRISPR screen have been deposited in the Sequence Read Archive (BioProject PRJNA826466) (69). RNAseq data have been deposited in the Gene Expression Omnibus (Accession GSE200982) (70). All other study data are included in the article and/or SI Appendix.