Significance
Viruses evolve to evade immune recognition that may otherwise limit transmission. Presentation of virus peptides by human leukocyte antigen (HLA) class I is a necessary step in the recognition of infection by immune cells. Virus adaptation to evade this immune recognition has not been formally tested across the diversity of HLA class I allotypes and virus strains. We analyzed genetic diversity of three human herpesviruses across peptides that bind diverse HLA class I allotypes. We find that adaptation to evade HLA class I recognition may be a general phenomenon shaping human herpesvirus genetic diversity, particularly for those proteins expressed during viral latency. This broad scope, across human and virus diversity, provides a unique comparative perspective of human–herpesvirus coevolution.
Keywords: HLA, herpesvirus, population genetics, EBV, HCMV
Abstract
Herpesviruses are ubiquitous, genetically diverse DNA viruses, with long-term presence in humans associated with infrequent but significant pathology. Human leukocyte antigen (HLA) class I presents intracellularly derived peptide fragments from infected tissue cells to CD8+ T and natural killer cells, thereby directing antiviral immunity. Allotypes of highly polymorphic HLA class I are distinguished by their peptide binding repertoires. Because this HLA class I variation is a major determinant of herpesvirus disease, we examined if sequence diversity of virus proteins reflects evasion of HLA presentation. Using population genomic data from Epstein–Barr virus (EBV), human cytomegalovirus (HCMV), and Varicella–Zoster virus, we tested whether diversity differed between the regions of herpesvirus proteins that can be recognized, or not, by HLA class I. Herpesviruses exhibit lytic and latent infection stages, with the latter better enabling immune evasion. Whereas HLA binding peptides of lytic proteins are conserved, we found that EBV and HCMV proteins expressed during latency have increased peptide sequence diversity. Similarly, latent, but not lytic, herpesvirus proteins have greater population structure in HLA binding than nonbinding peptides. Finally, we found patterns consistent with EBV adaption to the local HLA environment, with less efficient recognition of EBV isolates by high-frequency HLA class I allotypes. Here, the frequency of CD8+ T cell epitopes inversely correlated with the frequency of HLA class I recognition. Previous analyses have shown that pathogen-mediated natural selection maintains exceptional polymorphism in HLA residues that determine peptide recognition. Here, we show that HLA class I peptide recognition impacts diversity of globally widespread pathogens.
Herpesviridae is a family of large, double-stranded DNA viruses, with nine human-infecting members spread across three subfamilies: α (e.g., Varicella–Zoster virus [VZV]), β (e.g., human cytomegalovirus [HCMV]), and γ (e.g., Epstein–Barr virus [EBV]) (1). Although seroprevalence varies considerably across populations, VZV, HCMV, and EBV each infect more than 80% of humans (2–4), usually with mild or no symptoms. In a minority but significant number of cases, herpesvirus infection associates with severe malignancy, such as Burkitt’s lymphoma or nasopharyngeal carcinoma (NPC) (5–7). Coinfection (8, 9) or immunodeficiency (10–12) may also accompany herpesvirus pathogeneses, including postherpetic neuralgia caused by VZV reactivation in older adults and HCMV disease in transplant patients. Such widespread infection with low incidence of morbidity suggests that coevolution across millions of years reduced the fitness cost of infection (13) and that herpesvirus-associated disease represents disruption of an otherwise tolerated balance between host and virus (14).
Herpesviruses exhibit lytic and latent infection stages. Lytic infection is characterized by expression of most of the viral genes and production of infectious virions, thereby spreading the infection within and between hosts. In latency, those genes expressed in the lytic cycle are down-regulated, virion production halts, and a more limited gene expression program directs the establishment and maintenance of the viral genome (15). Through this reduced gene expression, herpesviruses may evade immune detection in the tissues where they become latent. EBV infects naïve tonsillar B cells, where latency proteins force B cell proliferation, survival, and differentiation into the memory B cell pool (16, 17). HCMV has a broad lytic tropism, ultimately establishing latency in early myeloid progenitors and monocytes, where the gene expression signature has been difficult to define but may retain similarity to the late lytic cycle (18–20). VZV infects tonsillar epithelial cells, T cells, and skin, ultimately establishing latency in sensory nerve cells (21, 22).
Critical to the immune-mediated control of viral infections are human leukocyte antigen (HLA) class I molecules. HLA class I (HLA-A, HLA-B, and HLA-C) presents intracellular peptides to circulating CD8+ T and natural killer (NK) cells (23–25) (hereafter referred to as “cytotoxic lymphocytes”), thereby orchestrating immune recognition of viruses. The HLA class I loci are hyperpolymorphic, with allotype frequencies differentially structured across human populations (26, 27). This striking pattern has been attributed to host–pathogen coevolution, which can lead to the maintenance of genetic variation (28, 29). HLA polymorphism localizes to the peptide binding groove, such that specific HLA allotypes can be distinguished by their repertoire of presented peptides (30). The result is that individuals and populations differ in the specific peptides that may be presented by HLA and thus, recognized by cytotoxic lymphocytes (31–33). Suggesting these differences in peptide repertoires are significant in the control of herpesvirus infections, specific HLA class I genetic variants are consistently associated with herpesvirus diseases. Indeed, genome-wide association studies for NPC and shingles (complications of EBV and VZV infection, respectively) identified the most significant genetic predictors to be HLA class I loci (34–38).
EBV, HCMV, and VZV are genetically diverse, with varying degrees of differentiation across human populations (39–50). In this regard, EBV and VZV clades are highly structured by population (46, 48, 50), whereas specific HCMV variants tend to be common across populations (41). As evidenced either by association with pathogenesis (51–53) or by the impact of diversifying natural selection (42, 54, 55), the genetic variants can have functional consequence. Functional variation may be enriched in EBV latency genes, which show much higher diversity and interspecific divergence than those involved exclusively in the lytic cycle (40, 54, 56, 57), likely through sustained challenge by the immune system. Natural selection has also resulted in cosegregation of specific variants in divergent regions of EBV and HCMV, contrasting otherwise substantial genomic recombination. Most notable in this regard are the EBNA-2 and EBNA-3 latency genes, where divergent alleles of each define type 1 vs. type 2 EBV strains (58, 59) and 21 HCMV genes that have multiple nonrecombining variants (41). For EBV, these genetic characteristics are enriched in proteins recognized by CD8+ T cells (40, 60), implicating natural selection driven by HLA class I in the global distribution of EBV genetic diversity.
In this study, we test the hypothesis that herpesviruses adapt to their local HLA class I environment. We focus on EBV, HCMV, and VZV due to the wealth of genomic data available for these viruses spanning diverse human populations. We split EBV, HCMV, and VZV proteins into regions likely bound by HLA class I or not, allowing a comparative analysis of global diversity with respect to HLA-mediated immune recognition. Because EBV latency genes harbor greater genetic diversity than genes expressed during lytic replication, we consider these two classes of genes separately for each of the three virus species. Consistent with previous reports (61–63), we find that HLA binding peptides are derived from conserved regions of herpesvirus lytic proteins. By contrast, HLA binding regions of EBV and HCMV latency proteins have significantly greater rates of diversity (ratio of nonsynonymous polymorphisms per site to synonymous polymorphisms per site [pN/pS]). In addition, we show that unlike lytic cycle proteins, HLA binding regions of latency proteins have greater population structure than nonbinding regions. Consistent with a model of local adaptation, we find patterns consistent with EBV latent protein evasion of HLA allotypes that are at higher frequency in the population from which the isolate is derived. We conclude that herpesvirus protein diversity is altered in predicted HLA class I binding sites, with a pattern consistent with the action of diversifying natural selection during latency mediated by HLA class I.
Results
EBV, HCMV, and VZV Differ in Magnitude of Population Differentiation, Genetic Diversity, and Constraint.
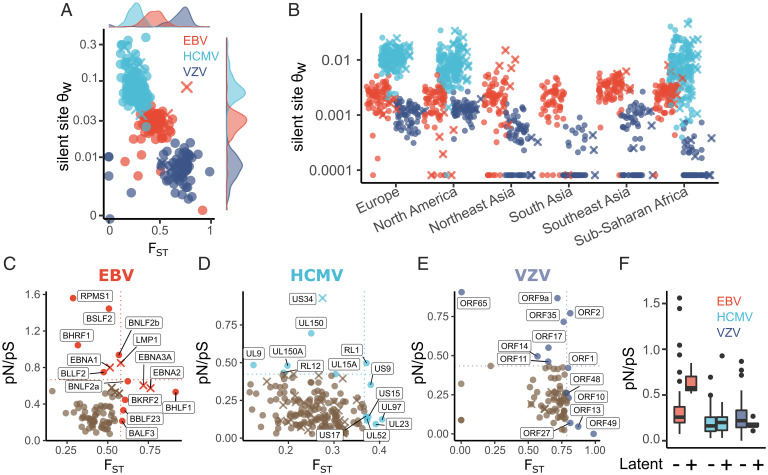
We analyzed genetic diversity of EBV, HCMV, and VZV across 15, 13, and 17 human populations, respectively, representing six major geographic regions (SI Appendix, Fig. S1). For this purpose, we compared the mean pairwise FST for the coding regions of 79 EBV (72 lytic, 7 latent), 170 HCMV (140 lytic, 30 latent), and 77 VZV genes (72 lytic, 5 latent) (Fig. 1A). The posterior estimate for mean FST of EBV genes was 0.43 (95% CI: 0.40 to 0.48). The mean FST of HCMV genes was significantly lower (FST = 0.24; 95% CI: 0.21 to 0.27; P < 0.001) than EBV, and the mean FST of VZV genes was significantly higher (FST = 0.65; 95% CI: 0.61 to 0.68; P < 0.001). We also calculated Watterson’s θ (θW) per silent site for each gene (Fig. 1A). Consistent with previous reports, HCMV genes had the greatest silent site diversity, and VZV had the lowest. Although HCMV sequences were lacking for some populations, we show that this hierarchy in θW is likely consistent within each major geographical region (Fig. 1B).
Fig. 1.
Genetic diversity in HLA and certain herpesvirus genes is differentiated across populations. (A) Per-gene estimates of synonymous site diversity (θW) plotted against FST. (B) θW measured within major geographical regions (as defined by the Allele Frequencies database) that had more than one sequenced genome from at least two viruses. (C–E) Per-gene estimates of FST and pN/pS for each herpesvirus gene. Genes expressed during latency are marked with an “X” and labeled when their FST or pN/pS values occurred in the top 10% for EBV and VZV or 5% for HCMV. (F) Summary of pN/pS values presented in C–E, split by latent and lytic cycle proteins.
We next analyzed the rates of single-nucleotide mutations within the three viruses using a Poisson mixed model of genic mutation counts. In this analysis, we considered synonymous and nonsynonymous sites separately and whether each gene is expressed during the lytic or latent stage of infection. The model we used is similar in structure to Selection Inference Using a Poisson Random Effects (SnIPRE), proposed as an alternative to McDonald–Kreitman tests but has the divergence parameters removed (Materials and Methods) (64, 65). We observed significant evolutionary constraint (e.g., pN/pS < 1) on nonsynonymous sites (P < 0.001), suggesting widespread purifying selection in all three viruses (Fig. 1 C–F). EBV latency genes exhibit increased synonymous polymorphism relative to genes expressed during lytic replication (P = 0.003). Accounting for synonymous polymorphism rates, EBV latency genes show greater nonsynonymous polymorphism rates than lytic genes (P = 0.045) (Fig. 1F). Genetic diversity is not significantly altered in HCMV or VZV genes that can be expressed during latency, as compared with lytic genes (Fig. 1F). Finally, we observed greater constraint, evidenced by a reduced pN/pS (Materials and Methods), in HCMV genes as compared with EBV (P < 0.001) and VZV (P = 0.007) (Fig. 1F).
Proteins Expressed during Lytic Replication Have Reduced Diversity in Regions Recognized by HLA.
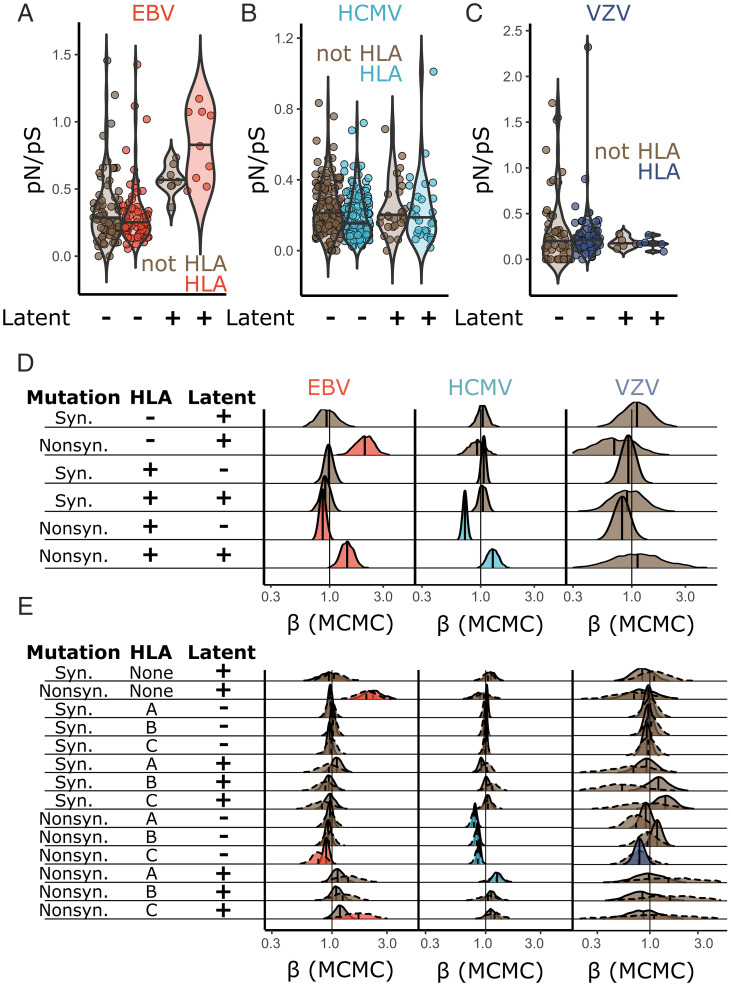
Linkage disequilibrium is maintained between nonsynonymous sites in genes that encode for EBV T cell epitopes, consistent with the action of natural selection imposed by immune recognition (60). We hypothesized that adaptation in response to immune recognition may also be reflected in the rates of protein polymorphism in HLA class I binding peptides. Therefore, we compared regions of herpesvirus proteins predicted to be recognized by HLA with corresponding gene-matched protein regions not recognized by HLA using an SnIPRE-like Poisson mixed model of mutation counts (64, 65).
Using a sliding window, each protein was split into peptide fragments predicted to be recognized or not recognized by HLA-A, -B, and/or -C (SI Appendix, Fig. S2). We included HLA allotypes that were at >10% frequency in at least one population from which a virus genome was derived. HLA-recognized regions were distributed across the length of viral proteins (SI Appendix, Fig. S3). The mutation counts in HLA binding and nonbinding regions were used to calculate pN/pS (Fig. 2 A–C and SI Appendix, Fig. S4) and as input for the SnIPRE-like models (Fig. 2 D and E). In analyzing the lytic cycle genes, there were no differences in synonymous polymorphism rates between regions encoding peptides recognized by HLA or not across any virus or HLA protein (A, B, or C) (Fig. 2 D and E, rows where mutation = “Syn.”). However, in both EBV and HCMV but not VZV, regions of lytic cycle proteins recognized by HLA had significantly reduced rates of nonsynonymous polymorphism, relative to synonymous diversity (EBV: P = 0.018; HCMV: P < 0.001) (Fig. 2D, row 5). These results are consistent with a model whereby HLA recognizes the most conserved regions of viral proteins expressed during the lytic cycle.
Fig. 2.
HLA recognition of latency genes is associated with higher levels of amino acid polymorphism. (A–C) pN/pS was calculated in HLA binding and nonbinding regions of each herpesvirus protein. Analogous data for regions recognized by HLA-A, HLA-B, and HLA-C are presented in SI Appendix, Fig. S4. The significance between observed pN/pS values (A–C) was assessed using a Bayesian SnIPRE-like Poisson mixed model in Markov chain Monte Carlo (MCMC)glmm (SI Appendix, Eq. 1); the output is summarized in D. Each distribution in D is the posterior distribution of the model coefficients (β), where a value of less than one indicates a smaller polymorphism count associated with the corresponding class of mutation. A value of greater than one indicates a greater count associated with the corresponding class. Colored distributions reflect significance at P < 0.05. E presents the same model as D but with regions of each viral protein split into HLA-A, HLA-B, or HLA-C. Solid lines indicate that the data modeled used all peptides recognized by a particular HLA protein (A, B, or C); dotted lines show a model using only peptides recognized uniquely by a single HLA protein (A, B, or C).
Some posterior distributions for VZV parameter estimates had wide CIs (Fig. 2D, row 6). Therefore, we tested if patterns of diversity in regions of viral proteins predicted to bind HLA varied across HLA-A, HLA-B, or HLA-C binding peptides (Fig. 2E). We split regions predicted to encode HLA-presented peptides into three separate classifications relating to presentation by HLA-A, HLA-B, or HLA-C. In these comparisons, two sets of data were considered. In the first, regions of each viral protein were split into regions recognized by each HLA protein (A, B, or C) irrespective of whether they also bind another. For example, a polymorphism in a peptide that binds HLA-A and HLA-C would contribute to the mutation counts of both. In the second, only regions of proteins that uniquely bind one of HLA-A, -B, or -C were considered. The former provides more data, perhaps allowing more accurate parameter estimates, while the latter will not be confounded by any nonadditive effects in the corecognition of a peptide by multiple HLA proteins.
When considering the dataset allowing multiple HLA molecules to recognize a single peptide, we found that regions of lytic cycle proteins predicted to bind HLA-C had significantly lower rates of nonsynonymous polymorphism as compared with their non-HLA binding counterparts (EBV: P = 0.047; HCMV: P < 0.001; VZV: P = 0.018) (Fig. 2E, row 11). Regions of HCMV lytic cycle proteins recognized by HLA-A or HLA-B were similarly more conserved than gene-matched regions that do not bind HLA (HLA-A: P < 0.001; HLA-B: P < 0.001) (Fig. 2E, rows 9 and 10). While pN/pS was not significantly different between HLA-A, HLA-B, or HLA-C binding peptides in EBV and HCMV, VZV peptides recognized by HLA-C had reduced pN/pS as compared with those recognized by HLA-B (P = 0.006) (Fig. 2E, comparing rows 10 and 11). However, reduced pN/pS in HLA-C–presented VZV peptides, alongside the difference between patterns of diversity in HLA-B and HLA-C, was not significant in a model that only considered peptides uniquely bound to HLA-C (Fig. 2E). These analyses suggest that VZV lytic gene regions presented by HLA-C are more conserved, with lower pN/pS values, and that HLA-C may more reliably bind VZV peptides across viral diversity.
Proteins Expressed during Latency Have Increased Diversity in Regions Recognized by HLA Class I.
We next considered polymorphism patterns in viral genes expressed during latency. Similar to lytic proteins, we did not observe altered synonymous polymorphism in HLA peptides from latent proteins (Fig. 2 D and E). However, regions of latent proteins recognized by HLA class I had significantly greater pN/pS relative to lytic proteins (Fig. 2D, row 6) (EBV: P = 0.004; HCMV: P = 0.035). Note that this effect (Fig. 2D, row 6) is measured relative to the rate of nonsynonymous polymorphism observed in non-HLA binding regions of latent proteins (Fig. 2D, row 2). Use of a more conservative set of HCMV latency proteins (Materials and Methods) resulted in a similar trend (all HCMV latency proteins: = 0.29; conservative HCMV latency proteins = 0.33). The latter was not significant (P = 0.12), likely due to reduced power through inclusion of fewer genes.
In considering HLA-A, HLA-B, and HLA-C separately, we only observed significant evidence for increased pN/pS in HCMV latent cycle peptides bound to HLA-A, as compared with non-HLA binding regions (P = 0.006) (Fig. 2E, row 12). This observation extended to EBV latent-stage peptides uniquely recognized by HLA-C (P = 0.029) (Fig. 2E, row 14). We observed no significant differences in latent gene diversity between peptides bound by HLA-A, HLA-B, or HLA-C (Fig. 2E, comparing posterior distributions from rows 12 to 14).
Thus, protein diversity of EBV and HCMV is increased in HLA binding regions of proteins expressed during latency compared with those expressed during lytic replication. For EBV, this is especially evident in the EBNA-1, EBNA-2, EBNA-3A, and LMP-1 genes (Fig. 2A). These data are consistent with stronger HLA-mediated natural selection pressures on latent-stage EBV and HCMV proteins than corresponding lytic cycle proteins. Underlying this natural selection may be immune detection by CD8+ T or NK cells, which can recognize HLA class I–presented virus peptides. In this model, elimination of latently infected cells by cytotoxic lymphocytes could select for mutations in latent proteins that are less efficiently presented by HLA or recognized by lymphocytes.
Greater Population Structure in Regions of Viral Proteins Predicted to Bind HLA Class I.
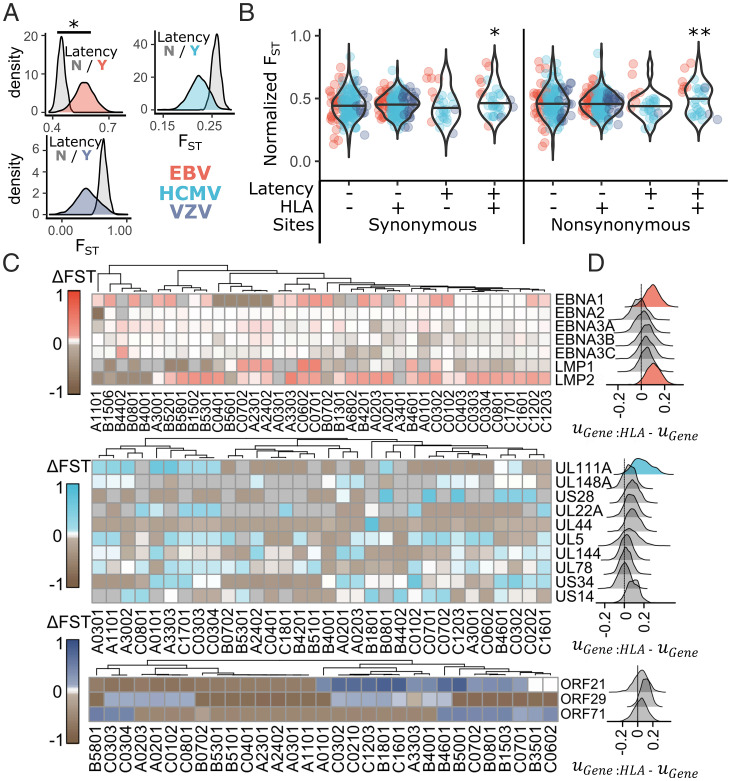
A viral fitness landscape molded by HLA class I–dependent immune recognition (e.g., by CD8+ T and NK cells) may be expected to differ across populations, dependent on the HLA allotype frequencies and associated peptide binding repertoires in each population. A prediction, therefore, is that genetic diversity of regions encoding peptides presented by HLA class I would be more structured across populations than regions less likely to be presented by HLA class I. To test this prediction, we compared FST estimates across latent and lytic viral genes split into regions predicted to encode HLA-bound peptides or not. We performed analyses using FST estimated from either synonymous or nonsynonymous sites. Prior to analysis, we filtered out genes (Materials and Methods) with poor estimates of FST due to limited or no polymorphism, resulting in inclusion of 60.7% of EBV, 81.1% of HCMV, and 38.9% of VZV genes in the synonymous sites model. FST estimated from only nonsynonymous sites resulted in inclusion of 59.4% of EBV, 81.1% of HCMV, and 36.3% of VZV genes (SI Appendix, Fig. S5).
FST estimated from either synonymous or nonsynonymous sites was higher in EBV latency genes (P < 0.001) but not in HCMV or VZV latency genes (Fig. 3A). In lytic cycle genes, recognition by HLA was not associated with differences in FST (EBV: P = 0.35; HCMV: P = 0.57; VZV: P = 0.85). However, in latency genes, regions predicted as encoding HLA binding peptides had significantly greater FST estimates than regions not predicted to be presented when considering all sites (P = 0.021), only nonsynonymous sites (P = 0.008), and only synonymous sites (P = 0.04) (Fig. 3B). FST was estimated to be increased by 0.051 (95% CI: 0.007 to 0.075) in HLA binding peptides of latency genes when using synonymous sites and by 0.067 (95% CI: 0.02 to 0.12) when using only nonsynonymous sites. Using the conservative set of HCMV latency genes yielded similar results, with an estimated 0.073 greater nonsynonymous FST in HLA binding peptides of latency proteins (P = 0.02). Consideration of a model that allowed FST to vary between viruses in HLA binding regions of latency genes was not supported by deviance information criterion (DIC) scores and did not identify significantly different patterns of FST in latency genes in any single virus. We conclude that the altered population genetic patterns in latent HLA binding peptides include increased population structure.
Fig. 3.
Herpesvirus latent peptides that bind HLA have increased population differentiation. Estimates of FST were compared between latent and nonlatent genes and between HLA binding and nonbinding sites in each herpesvirus protein using a linear mixed model to determine differences in FST. (A) The posterior distribution for FST in genes expressed during latency or not is plotted for each virus. (B) Normalized FST, where virus-specific effects on FST were removed from FST estimates for each herpesvirus gene (Materials and Methods), was plotted for HLA binding and nonbinding regions of latent or nonlatent herpesvirus genes using either synonymous or nonsynonymous polymorphism. Significance was assessed by the proportion of MCMC iterations that overlap zero for the posterior distribution of the interaction effect between HLA binding and latent expression ( in SI Appendix, Eq. 2). (C) The difference between FST in HLA binding vs. nonbinding regions of each herpesvirus gene (ΔFST) was calculated for every herpesvirus protein:HLA allotype combination. All EBV and VZV latency proteins included in the model and the 10 HCMV latency proteins with the highest ΔFST values are plotted, with box color corresponding to ΔFST. Allotypes were clustered based on similarity in ΔFST values across latent genes. (D) Plotted is the posterior distribution of the difference between and , which reflects the impact of HLA presentation on population structure of individual genes. Those with the 90% highest posterior density intervals that do not overlap zero are shaded with color. *P < 0.05; **P < 0.01.
HCMV UL111A and EBV LMP-2 Have Increased Population Differentiation in Predicted HLA Binding Peptides.
To visualize differences in FST associated with predicted HLA recognition of latent-stage viral proteins, we calculated ΔFST (FSTHLA − FSTnotHLA) using only nonsynonymous sites separately for regions identified by each HLA allotype for all allotypes included in the analysis (Fig. 3C). For most viral latent-stage proteins, we observed drastically variable ΔFST values across HLA allotypes. Altogether, there was little evidence for single HLA allotypes having a general effect on patterns of FST across viral latency proteins. However, EBV LMP-2, HCMV UL111A, and to a lesser extent, EBV EBNA-1 tended toward positive ΔFST values across HLA allotypes. To test whether these specific latency genes had generally higher FST values in HLA binding sites across allotypes, we compared the posterior distributions of the random effects associated with regions of each gene that are recognized or not by HLA. For each, the estimate (mean of the posterior distribution) for the difference between HLA binding and nonbinding regions was positive (Fig. 3D). HCMV UL111A had the greatest evidence for elevated FST in HLA binding regions (P = 0.025) followed by EBV LMP-2 (P = 0.052) and EBV EBNA-1 (P = 0.16) (Fig. 3D).
Predicted Detection of EBV Latent Proteins by HLA Is a Function of HLA Allele Frequency.
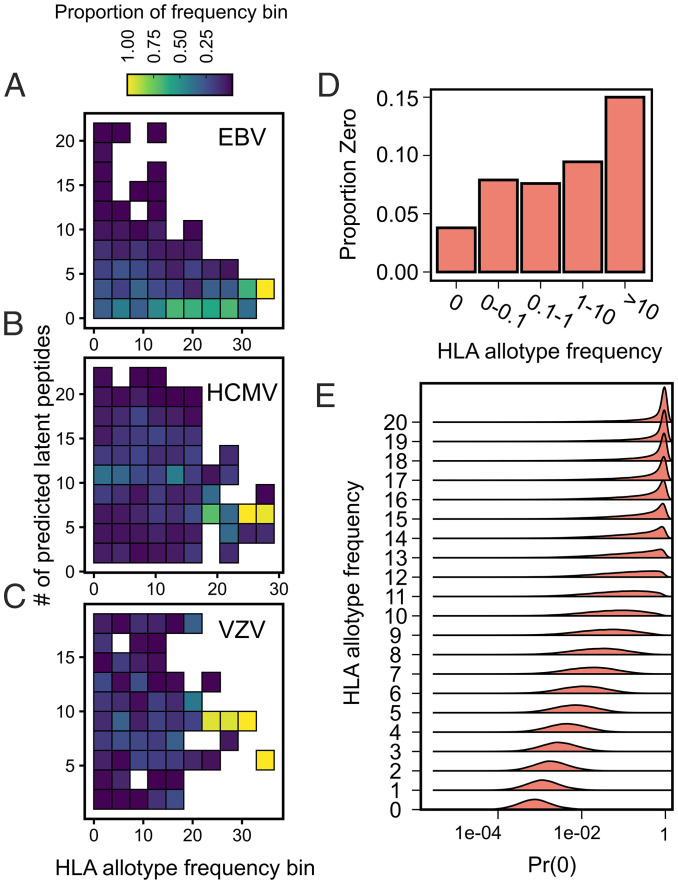
Natural selection on a virus to evade presentation by HLA or detection by HLA-dependent immune processes may have an expected dependence on HLA allotype frequency in that population. Because we observed the strongest evidence for altered population genetic patterns in latency proteins, we tested if the number of latent viral peptides from a particular viral isolate recognized by a particular HLA allotype was correlated with the frequency of the HLA allotype in the population from which the isolate was derived (Fig. 4 A–C). For HCMV and VZV, we used a Poisson mixed model that accounts for multiple observations per HLA allotype and per isolate alongside the correlation in HLA allotype frequencies across populations (Materials and Methods). We used a hurdle Poisson mixed model with a similar structure for EBV because the distribution of the number of predicted latent peptides per EBV isolate was zero inflated [P < 0.001, score test (66)].
Fig. 4.
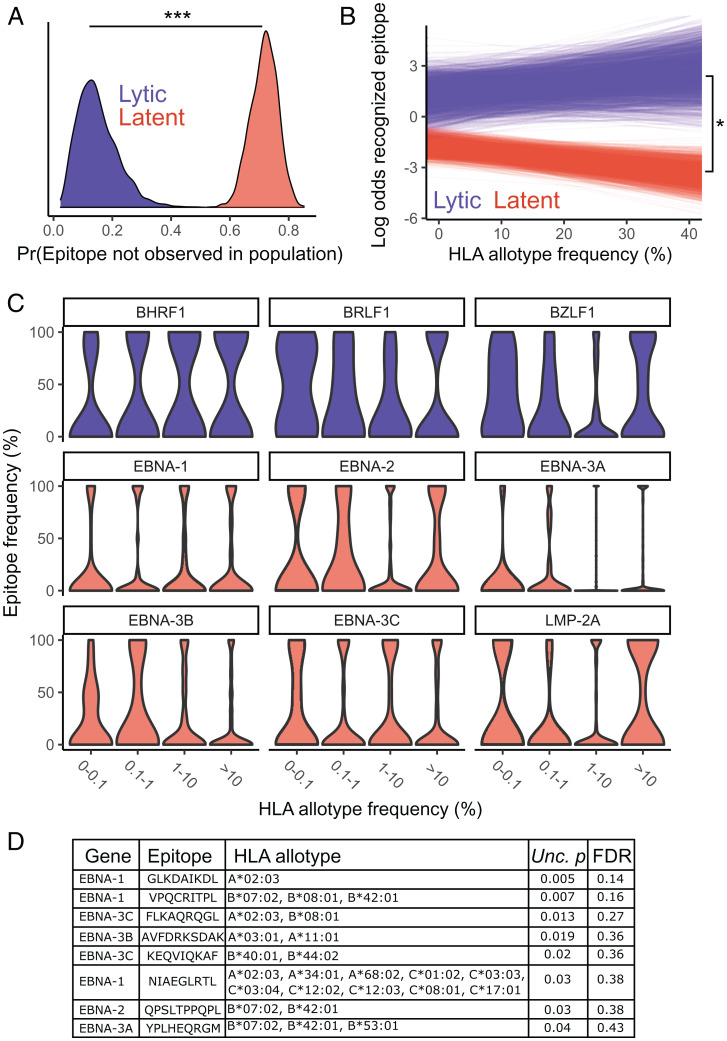
EBV latent proteins encode fewer peptides predicted to bind high-frequency HLA allotypes. The numbers of HLA binding latent peptides per isolate of EBV (A), HCMV (B), and VZV (C) were determined for each HLA allotype and plotted against HLA allotype frequency. Data were jointly binned by allotype frequency and number of predicted peptides and normalized by HLA allotype frequency bin, such that each column adds to 100%. (D) The proportion of allotype:isolate pairs from the bottom row of A, except using only one isolate per population, was plotted against HLA allotype frequency bins. E shows the estimated posterior distributions from the hurdle model for the probability of observing zero HLA binding latent EBV peptides [Pr(0)] across a range of HLA allotype frequencies.
We did not observe a significant relationship between HLA allotype frequency and the number of bound latent peptides for HCMV (βFreq = −0.004 [−0.019 to 0.011]; P = 0.60) or VZV (βFreq = −0.001 [−0.018 to 0.017]; P = 0.87) (Fig. 4 B and C). Similar results were obtained when using the conservative set of HCMV latency proteins (βFreq = −0.017 [−0.075 to 0.029]; P = 0.52). We also did not observe a significant relationship between the number of (nonzero) EBV latent peptides per isolate and HLA allotype frequency (Fig. 4A) (βFreq = 0.01 [−0.03 to 0.04]; P = 0.61). However, the proportion of EBV isolate:HLA pairs that resulted in zero predicted latent peptides increased with HLA allotype frequency (Fig. 4A). As such, the estimated probability of observing zero recognized latent peptides was significantly associated with HLA allotype frequency (P = 0.007) (Fig. 4E). To exclude the influence of variable sampling of isolates across populations, we confirmed this trend in a reduced dataset that contained one isolate per population chosen based on the completeness of genome assemblies (Fig. 4D). In this reduced dataset, we similarly observed a significantly greater failure to recognize latent viral proteins by higher-frequency HLA allotypes (P = 0.02). There were no relationships observed between the number of lytic peptides predicted to be recognized and HLA allotype frequency for any virus.
Because HLA class I binding is not necessarily indicative of CD8+ T cell recognition, we next aimed to confirm the relationship between HLA allotype frequency and EBV latent peptide recognition using verified CD8+ T cell epitopes from the Immune Epitope Database (IEDB). We assembled IEDB CD8+ T cell epitopes (8 to 12 amino acids) from the EBV proteins BHRF1, BRLF1, BZLF1, EBNA-1, EBNA-2, EBNA-3, and LMP-2A and determined their frequency across 18 populations. We matched IEDB epitopes with our peptide binding predictions and retained those that we could assign to at least one HLA allotype. The IEDB epitopes were highly concordant with the netMHCpan4.1 peptide predictions; 172 of 182 IEDB epitopes were contained within the set of predictions at a percentile rank binding affinity of 0.05, the threshold used for analyses in Fig. 4. Our analyses of predicted peptides focused on nonamers, a total of 807 predicted across HLA allotypes. The set of IEDB epitopes includes 113 nonamers, and all of these were identified with netMHCpan4.1 at a 0.05 percentile rank. Therefore, IEDB epitopes are a subset of the predicted peptides with a stricter requirement for experimentally confirmed T cell stimulation.
Using a similar statistical framework as presented in Fig. 4, we modeled epitope frequency across populations using a hurdle binomial model. Relative to lytic cycle epitopes, latent epitopes were both more likely to be missing from a population (P < 0.001) (Fig. 5A) and at a lower baseline frequency (P = 0.001) within populations (Fig. 5B, y intercept), consistent with greater protein diversity and population structure in latent epitopes. We also found that the relationship between the frequencies of epitopes and their corresponding HLA allotypes significantly differed between latent and lytic cycle epitopes (P = 0.02) (Fig. 5B). As expected, latent-stage epitope frequencies were inversely correlated with the HLA allotype frequencies that recognize them, whereas lytic cycle epitopes exhibited a weak positive correlation with HLA allotype frequencies.
Fig. 5.
IEDB epitope frequency is correlated with HLA allotype frequency. (A) Posterior distribution for the probability of not observing a particular latent or lytic epitope in a particular population. (B) The modeled relationship between the log odds of observing a latent or lytic epitope and the highest-frequency HLA allotype that recognizes that epitope in a population. The intercept and slope are plotted across 1,000 sampled iterations of the Markov chain. (C) Epitope frequencies from each EBV protein are plotted by HLA allotype frequency bin. (D) A table of epitopes with the frequencies most significantly negatively correlated with the frequency of HLA recognition alongside the allotypes predicted to recognize them, uncorrected P (Unc. p) values, and false discovery rate (FDR) across the 211 tested latent epitopes. *P < 0.05; ***P < 0.001.
To determine if the relationship between epitope and HLA frequencies varies across EBV latent proteins, we modeled latent proteins individually. Modeling latent proteins individually, rather than as a group, resulted in a lower DIC score for the former (1,277 vs. 1,414), suggesting gene-specific patterns. In comparing individual latent proteins with lytic proteins (Fig. 5C), we observed that the frequency of EBNA-2, EBNA-3A, and EBNA-3B epitopes was inversely correlated with HLA allotype frequency (EBNA-2: P = 0.02; EBNA-3B: P = 0.09; EBNA-3C: P = 0.02). Given that EBNA-2 and EBNA-3 proteins most notably differentiate type 1 and type 2 EBV, these results could suggest a role of CD8+ T cell–mediated natural selection on the establishment, maintenance, or population distribution of EBV type–defining diversity.
The observed significant relationship between the frequencies of specific HLA allotypes and EBV epitopes is almost certainly driven by a subset of allotype:epitope combinations. To identify IEDB epitopes that are most likely to have responded to HLA-mediated selection, we modeled epitope frequency with only the frequency of HLA recognition () as a covariate using simple linear models. After multiple testing, no individual epitope was significantly associated with HLA allotype frequency. This finding suggests that the significant relationship between HLA allotype and viral epitope frequencies (Figs. 4A and 5B) is driven by smaller effects across epitope:HLA combinations, rather than a few epitopes that have coevolved extremely closely with specific allotypes. The 10 epitopes exhibiting the strongest negative relationship (P < 0.05, before multiple correction) with frequency of HLA recognition are shown in Fig. 5D. The relationships of some of these peptides with HLA allotype frequency are unlikely to be driven by immune recognition. For example, VPQCRITPL has only two other possible peptides across EBNA-1 diversity, and each is predicted to be recognized by the same three HLA allotypes. However, others may represent candidate loci for HLA evasion. The most frequency-dependent peptide, GLKDAIKDL, was correlated with A*02:03 frequency. These positions of EBNA-1 are extremely variable, with eight possible peptide sequences identified across EBV isolates and each peptide recognized by one to four HLA allotypes (A*02:01, A*02:03, B*15:06, or B*46:01). Similarly, FLKAQRQGL is one of four EBNA-3C peptides at this position, where the other three nonamers are predicted to be presented by C*06:02, C*07:01, and C*07:02. As such, FLKAQRQGL may evade HLA-C recognition, allowing susceptibility to presentation by specific HLA-A and HLA-B allotypes (Fig. 5D).
Discussion
We observed greater population differentiation and rates of protein polymorphism in the residues of herpesvirus latency proteins that are presented by HLA class I. Complex demographic and selective forces shape herpesvirus diversity within and between hosts, which could confound efforts to localize the causes of natural selection. Bottlenecks have been documented through the progression of infection of each virus, promoting genetic drift and reducing diversity. For example, only one to three VZV virions establish the skin infection that underlies a single lesion, and interhost divergence of HCMV can be comparable with intrahost divergence between tissue compartments due to bottlenecks following superinfection (67–70). Thus, the viral genome sequenced from a particular individual will depend on the sampled tissue site. However, demography may be assumed to similarly influence all sites in a linked region, and comparison of interspersed sites within a gene may avoid the misinterpretation of demographic forces as selective ones. Our analyses specifically compare HLA-targeted regions with nontargeted ones, and we consider it unlikely that bottlenecks, or other demographic forces, could discriminate between these regions under neutrality.
Altered patterns of genetic diversity in HLA class I–presented peptides, most apparent in EBV and HCMV, are generally consistent with the action of diversifying, population-specific natural selection pressures from local HLA allotypes with variable peptide binding repertoires. These altered patterns of diversity were only apparent in genes expressed during latency. Notably, some latent-expressed proteins are also expressed during the lytic stage, and our modeling cannot distinguish natural selection during latency from natural selection on latent proteins during the lytic cycle. However, the latter would require stronger natural selection on latent proteins expressed during the lytic cycle as compared with other lytic proteins. Because this scenario lacks biological precedence or a clear mechanism, we favor the hypothesis that altered diversity of latent proteins reflects natural selection during the establishment or maintenance of latency.
EBV latency proteins are established in having greater polymorphism than most lytic proteins (40, 54, 71). Our analyses support a generally increased pN/pS in EBV latency genes, with significantly greater values in the regions that bind HLA class I. Therefore, while our results suggest that HLA class I promotes the maintenance of EBV latent protein diversity, it does not alone explain the observed increased diversity, and other diversifying natural selection forces likely target EBV latent proteins. For example, HLA class II also coordinates immune detection of EBV and may be especially important given EBV infects B cells.
Latently expressed HCMV proteins have not previously been considered separately in population genetic analyses, perhaps because the exact nature of HCMV latency is still debated (18, 20, 72). We find that HCMV genes expressed in latency do not have generally higher rates of polymorphism but that pN/pS is specifically increased in regions encoding for peptides presented by HLA class I. Our results were similar when considering a conservative set of HCMV latency proteins or a larger set recently characterized by transcriptome sequencing of HCMV latently infected hematopoietic progenitor cells (18).
In contrast to EBV and HCMV, we did not observe patterns consistent with HLA class I–mediated natural selection in VZV latent proteins. This difference could be caused by true biological differences in the immune responses to these viruses or by a reduced power to detect natural selection in VZV. Population genetic patterns may be a reflection of virus-specific immune responses; both EBV and HCMV are robustly targeted by CD8+ T cells, with large expansions in primary infection, while the HLA class II–dependent CD4+ T cells are primarily studied in the VZV immune response (6, 33, 73, 74). However, VZV has a lower recombination rate and lower levels of polymorphism than EBV and HCMV, with global VZV diversity reflecting a recent lineage replacement (43, 47). Thus, there could have been insufficient time for VZV to accumulate a detectable number of mutations in HLA binding peptides, or linked selection could create correlated patterns of diversity in the interspersed HLA binding and nonbinding regions of each protein. Further, VZV is the only herpesvirus with an available vaccine, which could alter the fitness landscape imposed by immune recognition.
Altered patterns of variation in HLA class I–presented peptides of EBV and HCMV are likely due to detection by cytotoxic CD8+ T cells or NK cells. During early EBV infection, CD8+ T cell responses against highly immunogenic lytic antigens abound, followed by a dominant response to EBNA-3 epitopes in the transition to latency (6). Subdominant T cell responses toward latent proteins EBNA-1, EBNA-2, and LMP-2 also occur and can be HLA restricted (75–77). HCMV-reactive CD8+ T cells continue to expand after primary infection, a phenomenon termed “memory expansion” (78). Cytotoxic CD4+ and CD8+ T cells also target HCMV latency-associated proteins (79, 80), which overlap considerably with the genes expressed during early lytic infection (18). The importance of CD8+ T cells in the control of EBV and HCMV is consistent with the altered patterns of polymorphism in HLA class I binding peptides of these two viruses. In addition to CD8+ T cells, NK cells can also underlie peptide-specific immune recognition. NK cells canonically target cells with reduced HLA class I expression; however, more recent evidence suggests that activating killer cell immunoglobulin-like receptors (KIRs) can recognize peptide–HLA complexes as ligands to eliminate infected cells (23–25). While certain EBV peptides can alter HLA–KIR binding (81–83), the prevalence and determinants of peptide-specific KIR–HLA binding are not well described, precluding a clear suggestion of whether NK cells are likely to markedly impact herpesvirus evolution through HLA recognition.
HLA-mediated natural selection on herpesviruses could conceivably progress through two distinct but not mutually exclusive modes. In the first, herpesviruses could adapt to their individual host over the course of their infection, resulting in evasion of the specific HLA allotypes present in that host. Indeed, EBV and HCMV genomes sampled early in infection are genetically diverse, suggesting superinfection by a population of strains that could then be targeted by natural selection (70, 71). Intrahost selective sweeps are evident during HCMV infection, and EBV diversity converges toward a reference strain from the sampled population (71, 84, 85). Thus, it is likely that most virus genome sequences included in this study have been targeted by some degree of within-host natural selection. This may be especially true for EBV genomes sampled from tumors, which can evade detection by HLA through mutations in latent genes. For example, in HLA-A*02+ NPC patients, antigenic mutations arise in LMP-1 of tumor-associated isolates that were not present in peripheral blood (40, 86, 87). Especially strong natural selection on EBV+ tumors to evade immune detection could conceivably drive the relationship between HLA allotype frequency and HLA recognition of EBV latency proteins (Fig. 3). However, polymorphism in EBV latent epitopes restricted to high-frequency allotypes has been discovered outside of EBV+ tumors, suggesting that intrahost evolution alone is unlikely to drive these patterns. An example is populations with high HLA-A*11 frequency, such as Papua New Guinea and southern China, where there are high-frequency mutations with evidence of recent positive selection in EBV EBNA-3B epitopes that are immunodominant in other populations with lower HLA-A*11 frequencies (88–90). Our results suggest that this may be a more general occurrence across HLA allotypes.
The second possible mode of herpesvirus evolution in response to HLA is a more persistent coevolution between humans and their herpesviruses, which could lead to concurrent population differentiation, ultimately resulting in virus strains adapted to the average local HLA landscape. Correlations between the phylogenies of primates and lymphocryptoviruses (91), cytomegaloviruses (92), and varicella viruses (45) suggest that EBV, HCMV, and VZV likely cospeciated with humans. Their prolonged relationship could offer a sufficient timescale for codifferentiation to occur, especially given the possibility of intrahost evolution. A prediction would be differentiation of population-wide epitope frequencies, which could be reflected in the higher FST values observed in predicted HLA binding latent peptides.
We observed patterns consistent with HLA class I evasion specifically in latent proteins but not lytic proteins. Rather, we observed significantly reduced protein polymorphism (pN/pS) (Fig. 1) in HLA binding regions of EBV, HCMV, and VZV lytic proteins. These observations are consistent with other analyses that show that HLA binding peptides tend to have greater constraint across pathogen and human proteomes (61–63). Thus, HLA class I proteins may be better suited to recognize certain highly conserved amino acid residues or sequences (e.g., functional protein domains). Presentation of peptides derived from conserved protein domains could then offer a pathogen with a trade-off between immune recognition and optimal domain function. Further, if uncontrolled lytic replication is detrimental to herpesvirus fitness, an immune-mediated self-domestication borne of lytic epitope conservation may be beneficial to both host and virus. An assured rapid recognition of lytic proteins could increase the importance of establishing latency, from which reactivation and transmission could occur. A transmission strategy that requires latency could drive strong natural selection in latent proteins to evade immunity, even in the presence of a trade-off with optimal protein function.
The diversity of EBV latency genes has been attributed to more direct or sustained exposure to patrolling immune cells. EBV latency genes are expressed at different points during the establishment of latency, but once latency is established, there is exclusive and only sporadic expression of EBNA-1 (17). However, dysregulation of other EBNA and LMP genes has been described in EBV+ cancers, such as Hodgkin’s lymphoma and NPC (6). Thus, a particularly strong natural selection for immune evasion during the establishment of latency or following tumorigenesis could drive the patterns of diversity observed in latent EBV proteins. Alternatively, HCMV latency proteins may have sustained contact with the adaptive immune system, where continual reactivation of latency-associated genes could underlie memory expansion of CD8+ T cells, ultimately selecting for escape variants.
Overall, our analyses provide reasonable evidence to conclude that HLA class I peptide presentation has shaped global patterns of diversity in EBV and HCMV latency–associated genes. HLA class I genetic variation is associated with HCMV reactivation in transplant patients (93, 94) and pathogenesis of EBV malignancies (34–36, 95, 96). Our observations provide historical context for these associations and could aid in future development of inclusive therapies and vaccines.
Materials and Methods
Herpesvirus Genomes.
We obtained EBV, HCMV, and VZV genomes (strains) from the National Center for Biotechnology Information (NCBI) Virus database. We filtered out duplicates, those that did not contain geographical information, and laboratory-derived transgenic strains and confirmed the geographical metadata. In the cases where a single virus strain had multiple genome sequences, we kept the one that had the higher number of genes annotated. We kept 691 EBV (52, 97–104), 258 HCMV (41, 42, 105–112), and 163 VZV (43, 44, 68, 69, 113–118) strains from 25, 16, and 18 populations, respectively, for further analysis (Dataset S1). SI Appendix, SI Methods discusses the bioinformatic processing of virus genomes.
For analyses, we classified LMP and EBNA proteins as EBV “latency proteins”; US28, UL144, UL138, UL111A, and genes expressed with Fragments Per Kilobase of transcript per Million mapped reads > 12,500 in Cheng et al. (18) as HCMV latency proteins; and ORF21, ORF29, ORF71, ORF70, and ORF66 products as VZV latency proteins (119, 120). HCMV analyses were also repeated with a more conservative set of latency proteins: US28, UL144, UL138, and UL111A. Latent and lytic classifications were assumed to be mutually exclusive. While some genes may be expressed in latent and lytic stages, our modeling approach allows for comparison of genes that can be expressed during latency with those with no known latent-stage expression.
Prediction of Herpesvirus Peptides Bound by HLA.
We next predicted herpesvirus-derived peptides expected to bind HLA class I allotypes. We predicted binding peptides for any allotype that occurs at high frequency (>10%) in at least one of the populations from which the herpesvirus strains were isolated. We compiled HLA allotype frequency data from the Allele Frequencies Database (121) (Dataset S2), in each case using two field–resolution (denoting a unique polypeptide sequence, also known as allotype) HLA genotypes. Herpesvirus strains that were isolated from populations without high-resolution HLA typing data were excluded. For populations with complex ancestries and for which HLA genotypes were available across ancestries (the United States and Brazil), we used average weighted frequencies derived from the expected proportions of those ancestries (Dataset S2).
We used netMHCpan4.1 to predict the herpesvirus peptides bound to the set of HLA allotypes that reached >10% frequency in a population (122). For each virus (EBV, HCMV, VZV), we used a sliding window to consider all unique peptide nonamers derived from the annotated coding sequence (CDS) of the set of sequenced strains as input to predict those that bound HLA allotypes of interest. We only considered “strong binding” peptides, defined as less than a 0.5% percentile rank binding affinity. Percentile rank is a preferable filter to absolute binding affinity, as predictions of the latter can vary considerably across HLA allotypes (123, 124). Comprehensive epitope mapping and netMHCpan benchmarking suggest that this threshold should account for 75 to 90% of T cell epitopes and response magnitude (125, 126). We mapped strong binding peptides back to the herpesvirus multiple sequence alignment, thereby splitting each protein alignment into residues predicted to bind HLA class I and those not expected to bind HLA class I. The latter includes nested classes of residues bound by HLA-A, HLA-B, and HLA-C as well as the peptides bound by specific allotypes. To avoid spuriously high rates of protein polymorphism in predicted HLA binding sites, we only used the consensus sequence to classify regions as HLA binding or nonbinding, rather than all variants across isolates.
Calculation of Population Genetic Statistics.
pN/pS and FST were calculated independently for each entire CDS and for various subsets of residues defined by HLA-binding properties. These residue subsets included CDS regions not bound by HLA class I, bound by HLA class I, bound by any HLA-A allotype, bound by any HLA-B allotype, bound by any HLA-C allotype, or bound by an individual HLA allotype. Columns of the multiple sequence alignment were subset and used as input to calculate pN/pS and FST. We calculated pN/pS by simple counting of the observed number of segregating amino acid and synonymous polymorphisms and dividing each by the respective number of nonsynonymous and synonymous sites estimated with the YN00 model implemented in PAML (127). As such, intraspecies pN/pS is similar to the interspecies statistic ratio of divergence at nonsynonymous and synonymous sites, except without fitting a substitution model. We calculated the average pairwise nucleotide FST (128) as implemented in PopGenome (v2.7.5) (129). SI Appendix, SI Methods has a description of statistical analyses. The code and data used for all analyses are provided in Figshare (DOI: 10.6084/m9.figshare.19119743).
Analysis of IEDB Epitopes.
A set of 11 EBV genes was selected for analysis. The selection included most protein-coding latency genes (EBNA-1, EBNA-2, EBNA-3A, EBNA-3B, EBNA-3C, LMP-2A), which show the strongest geographical stratification, evidence for persistent positive selection (40, 54, 98), and signals of association with diseases (130). The subset was complemented by five additional genes with critical roles for the EBV’s lytic phase (BDLF4, BGLF4, BHRF1, BRLF1, BZLF1) that have suggested associations with a wide range of EBV-related diseases (131–134). The analyses included the totality of the 7– to 12–amino acid epitopes falling within these selected EBV genes described in the IEDB.
For each EBV gene, all available protein sequences with geographical information were downloaded from GenBank, including the translation of the sequences generated in Telford et al. (50). Lists of the accession identifications of all sequences and epitopes are available in Dataset S1. Each protein sequence was aligned to the NC_007605 reference sequences using T-coffee, with default settings. The sequences showing poor alignments (more than 10% of gaps or X’s in the nonrepeating parts of the proteins) were excluded from the subsequent analyses. A “callable” region was defined as the parts of the epitopes that were covered by high-quality sequences in all our dataset (i.e., excluding repetitive regions and regions with originally low coverage). The callable region covered more than 98% of the epitope list. The remaining complete sequences were selected for each epitope and used to calculate population frequencies of each epitope haplotypic variant. SI Appendix, SI Methods has a description of the statistical analysis.
Supplementary Material
Acknowledgments
This work was supported AQ15 by NIH Grants R56 AI151549 (to P.J.N) and R01 AI158410 (to P.J.N). W.H.P is supported by NIH Grant F32 AI161790. A.N. is supported by Ministerio de Ciencia e Innovación, Spain Grant PGC2018-101927-B-I00, Ministerio de Economía y Competitividad/Fondo Europeo de Desarrollo Regional, Union Europea and by “Unidad de Excelencia María de Maeztu” funded by MINECO Grant MDM-2014-0370. G.S. is supported by Instituto de Salud Carlos III (Spain) Grant MS20/00064 and cofunded by European Social Fund Grant PID2019-104700GA-I00 funded by Agencia Estatal de Investigación, Spain and NIH Grant R01HG010898-01. We thank Oscar MacLean for helpful comments and suggestions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123248119/-/DCSupplemental.
Data Availability
Population genetic data tables, R code, and Python code have been deposited in Figshare (DOI: 10.6084/m9.figshare.19119743) (135).
References
- 1.Arvin A., et al. , Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis (Cambridge University Press, 2007). [PubMed] [Google Scholar]
- 2.Vyse A. J., Gay N. J., Hesketh L. M., Morgan-Capner P., Miller E., Seroprevalence of antibody to varicella zoster virus in England and Wales in children and young adults. Epidemiol. Infect. 132, 1129–1134 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowd J. B., Palermo T., Brite J., McDade T. W., Aiello A., Seroprevalence of Epstein-Barr virus infection in U.S. children ages 6-19, 2003-2010. PLoS One 8, e64921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ibrahim S., et al. , Sociodemographic factors associated with IgG and IgM seroprevalence for human cytomegalovirus infection in adult populations of Pakistan: A seroprevalence survey. BMC Public Health 16, 1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azevedo L. S., et al. , Cytomegalovirus infection in transplant recipients. Clinics (São Paulo) 70, 515–523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor G. S., Long H. M., Brooks J. M., Rickinson A. B., Hislop A. D., The immunology of Epstein-Barr virus-induced disease. Annu. Rev. Immunol. 33, 787–821 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Opstelten W., et al. , Herpes zoster and postherpetic neuralgia: Incidence and risk indicators using a general practice research database. Fam. Pract. 19, 471–475 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Stewart J. A., Reef S. E., Pellett P. E., Corey L., Whitley R. J., Herpesvirus infections in persons infected with human immunodeficiency virus. Clin. Infect. Dis. 21 (suppl. 1), S114–S120 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Rochford R., Cannon M. J., Moormann A. M., Endemic Burkitt’s lymphoma: A polymicrobial disease? Nat. Rev. Microbiol. 3, 182–187 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Damania B., Münz C., Immunodeficiencies that predispose to pathologies by human oncogenic γ-herpesviruses. FEMS Microbiol. Rev. 43, 181–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ansari R., et al. , Primary and acquired immunodeficiencies associated with severe Varicella-Zoster virus infections. Clin. Infect. Dis. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steininger C., Clinical relevance of cytomegalovirus infection in patients with disorders of the immune system. Clin. Microbiol. Infect. 13, 953–963 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Azab W., Dayaram A., Greenwood A. D., Osterrieder N., How host specific are herpesviruses? Lessons from herpesviruses infecting wild and endangered mammals. Annu. Rev. Virol. 5, 53–68 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Cruz-Muñoz M. E., Fuentes-Pananá E. M., Beta and gamma human herpesviruses: Agonistic and antagonistic interactions with the host immune system. Front. Microbiol. 8, 2521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen J. I., Herpesvirus latency. J. Clin. Invest. 130, 3361–3369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorley-Lawson D. A., EBV persistence: Introducing the virus. Curr. Top. Microbiol. Immunol. 390, 151–209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Münz C., Latency and lytic replication in Epstein-Barr virus-associated oncogenesis. Nat. Rev. Microbiol. 17, 691–700 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Cheng S., et al. , Transcriptome-wide characterization of human cytomegalovirus in natural infection and experimental latency. Proc. Natl. Acad. Sci. U.S.A. 114, E10586–E10595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shnayder M., et al. , Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. MBio 9, e00013–e00018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz M., Stern-Ginossar N., The transcriptome of latent human cytomegalovirus. J. Virol. 93, e00047–e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arvin A. M., et al. , Varicella-zoster virus T cell tropism and the pathogenesis of skin infection. Curr. Top. Microbiol. Immunol. 342, 189–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zerboni L., Sen N., Oliver S. L., Arvin A. M., Molecular mechanisms of varicella zoster virus pathogenesis. Nat. Rev. Microbiol. 12, 197–210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naiyer M. M., et al. , KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA-C. Sci. Immunol. 2, eaal5296 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Das J., Khakoo S. I., NK cells: Tuned by peptide? Immunol. Rev. 267, 214–227 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Guethlein L. A., Norman P. J., Hilton H. G., Parham P., Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol. Rev. 267, 259–282 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Z., et al. , Adaptive admixture of HLA class I allotypes enhanced genetically determined strength of natural killer cells in east Asians. Mol. Biol. Evol. 38, 2582–2596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandt D. Y. C., César J., Goudet J., Meyer D., The effect of balancing selection on population differentiation: A study with HLA genes. G3 (Bethesda) 8, 2805–2815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abi-Rached L., et al. , The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334, 89–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radwan J., Babik W., Kaufman J., Lenz T. L., Winternitz J., Advances in the Evolutionary Understanding of MHC Polymorphism (Elsevier Ltd., 2020), vol. 36, pp. 298–311. [DOI] [PubMed] [Google Scholar]
- 30.van Deutekom H. W. M., Keşmir C., Zooming into the binding groove of HLA molecules: Which positions and which substitutions change peptide binding most? Immunogenetics 67, 425–436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyun S.-J., et al. , Comprehensive analysis of cytomegalovirus pp65 antigen-specific CD8+ T cell responses according to human leukocyte antigen class I allotypes and intraindividual dominance. Front. Immunol. 8, 1591–1591 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barquera R., et al. , Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA 96, 277–298 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forrest C., Hislop A. D., Rickinson A. B., Zuo J., Proteome-wide analysis of CD8+ T cell responses to EBV reveals differences between primary and persistent infection. PLoS Pathog. 14, e1007110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang M., et al. , The principal genetic determinants for nasopharyngeal carcinoma in China involve the HLA class I antigen recognition groove. PLoS Genet. 8, e1003103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tse K. P., et al. , Genome-wide association study reveals multiple nasopharyngeal carcinoma-associated loci within the HLA region at chromosome 6p21.3. Am. J. Hum. Genet. 85, 194–203 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bei J. X., et al. , A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat. Genet. 42, 599–603 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Tian C., et al. , Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat. Commun. 8, 599–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crosslin D. R., et al. , Genetic variation in the HLA region is associated with susceptibility to herpes zoster. Genes Immun. 16, 1–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiara M., et al. , Geographic population structure in Epstein-Barr virus revealed by comparative genomics. Genome Biol. Evol. 8, 3284–3291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palser A. L., et al. , Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J. Virol. 89, 5222–5237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lassalle F., et al. , Islands of linkage in an ocean of pervasive recombination reveals two-speed evolution of human cytomegalovirus genomes. Virus Evol. 2, vew017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sijmons S., et al. , High-throughput analysis of human cytomegalovirus genome diversity highlights the widespread occurrence of gene-disrupting mutations and pervasive recombination. J. Virol. 89, 7673–7695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zell R., et al. , Sequencing of 21 varicella-zoster virus genomes reveals two novel genotypes and evidence of recombination. J. Virol. 86, 1608–1622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters G. A., et al. , A full-genome phylogenetic analysis of varicella-zoster virus reveals a novel origin of replication-based genotyping scheme and evidence of recombination between major circulating clades. J. Virol. 80, 9850–9860 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grose C., Pangaea and the out-of-Africa model of Varicella-Zoster virus evolution and phylogeography. J. Virol. 86, 9558–9565 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagenaar T. R., Chow V. T., Buranathai C., Thawatsupha P., Grose C., The out of Africa model of varicella-zoster virus evolution: Single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 21, 1072–1081 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Pontremoli C., Forni D., Clerici M., Cagliani R., Sironi M., Possible European origin of circulating Varicella Zoster virus strains. J. Infect. Dis. 221, 1286–1294 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Barrett-Muir W., et al. , Genetic variation of varicella-zoster virus: Evidence for geographical separation of strains. J. Med. Virol. 70 (suppl. 1), S42–S47 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Blazquez A. C., et al. , Comprehensive evolutionary analysis of complete Epstein-Barr virus genomes from Argentina and other geographies. Viruses 13, 1172 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Telford M., et al. , Expanding the geographic characterisation of Epstein-Barr virus variation through gene-based approaches. Microorganisms 8, 1686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaymaz Y., et al. , Epstein-Barr virus genomes reveal population structure and type 1 association with endemic Burkitt lymphoma. J. Virol. 94, e02007–e02019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu M., et al. , Genome sequencing analysis identifies Epstein-Barr virus subtypes associated with high risk of nasopharyngeal carcinoma. Nat. Genet. 51, 1131–1136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arcangeletti M.-C., et al. , Combined genetic variants of human cytomegalovirus envelope glycoproteins as congenital infection markers. Virol. J. 12, 202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santpere G., et al. , Genome-wide analysis of wild-type Epstein-Barr virus genomes derived from healthy individuals of the 1,000 Genomes Project. Genome Biol. Evol. 6, 846–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mozzi A., et al. , Past and ongoing adaptation of human cytomegalovirus to its host. PLoS Pathog. 16, e1008476 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang F., Rivailler P., Rao P., Cho Y., Simian homologues of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 489–497 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGeoch D. J., Gatherer D., Lineage structures in the genome sequences of three Epstein-Barr virus strains. Virology 359, 1–5 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Sample J., et al. , Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 64, 4084–4092 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen J. I., Wang F., Mannick J., Kieff E., Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. U.S.A. 86, 9558–9562 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wegner F., Lassalle F., Depledge D. P., Balloux F., Breuer J., Co-evolution of sites under immune selection shapes Epstein-Barr Virus population structure. Mol. Biol. Evol. 36, 2512–2521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hertz T., et al. , Mapping the landscape of host-pathogen coevolution: HLA class I binding and its relationship with evolutionary conservation in human and viral proteins. J. Virol. 85, 1310–1321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Comas I., et al. , Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 42, 498–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forni D., et al. , Antigenic variation of SARS-CoV-2 in response to immune pressure. Mol. Ecol. 30, 3548–3559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Palmer W. H., Hadfield J. D., Obbard D. J., RNA-interference pathways display high rates of adaptive protein evolution in multiple invertebrates. Genetics 208, 1585–1599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eilertson K. E., Booth J. G., Bustamante C. D., SnIPRE: Selection inference using a Poisson random effects model. PLOS Comput. Biol. 8, e1002806 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van den Broek J., A score test for zero inflation in a Poisson distribution. Biometrics 51, 738–743 (1995). [PubMed] [Google Scholar]
- 67.Renzette N., et al. , Rapid intrahost evolution of human cytomegalovirus is shaped by demography and positive selection. PLoS Genet. 9, e1003735 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Depledge D. P., et al. , Deep sequencing of viral genomes provides insight into the evolution and pathogenesis of varicella zoster virus and its vaccine in humans. Mol. Biol. Evol. 31, 397–409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Depledge D. P., et al. , High viral diversity and mixed infections in cerebral spinal fluid from cases of Varicella Zoster virus encephalitis. J. Infect. Dis. 218, 1592–1601 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cudini J., et al. , Human cytomegalovirus haplotype reconstruction reveals high diversity due to superinfection and evidence of within-host recombination. Proc. Natl. Acad. Sci. U.S.A. 116, 5693–5698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weiss E. R., et al. , Early Epstein-Barr virus genomic diversity and convergence toward the B95.8 genome in primary infection. J. Virol. 92, e01466-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forte E., Zhang Z., Thorp E. B., Hummel M., Cytomegalovirus latency and reactivation: An intricate interplay with the host immune response. Front. Cell. Infect. Microbiol. 10, 130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laing K. J., Ouwendijk W. J. D., Koelle D. M., Verjans G. M. G. M., Immunobiology of Varicella-Zoster virus infection. J. Infect. Dis. 218 (suppl. 2), S68–S74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sylwester A. W., et al. , Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202, 673–685 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khanna R., Burrows S. R., Nicholls J., Poulsen L. M., Identification of cytotoxic T cell epitopes within Epstein-Barr virus (EBV) oncogene latent membrane protein 1 (LMP1): Evidence for HLA A2 supertype-restricted immune recognition of EBV-infected cells by LMP1-specific cytotoxic T lymphocytes. Eur. J. Immunol. 28, 451–458 (1998). [DOI] [PubMed] [Google Scholar]
- 76.Lee S. P., et al. , HLA A2.1-restricted cytotoxic T cells recognizing a range of Epstein-Barr virus isolates through a defined epitope in latent membrane protein LMP2. J. Virol. 67, 7428–7435 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blake N., et al. , Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity 7, 791–802 (1997). [DOI] [PubMed] [Google Scholar]
- 78.Klenerman P., Oxenius A., T cell responses to cytomegalovirus. Nat. Rev. Immunol. 16, 367–377 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Tey S.-K., Goodrum F., Khanna R., CD8+ T-cell recognition of human cytomegalovirus latency-associated determinant pUL138. J. Gen. Virol. 91, 2040–2048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lim E. Y., Jackson S. E., Wills M. R., The CD4+ T cell response to human cytomegalovirus in healthy and immunocompromised people. Front. Cell. Infect. Microbiol. 10, 202–202 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hansasuta P., et al. , Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 34, 1673–1679 (2004). [DOI] [PubMed] [Google Scholar]
- 82.Stewart C. A., et al. , Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. U.S.A. 102, 13224–13229 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stewart-Jones G. B., et al. , Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur. J. Immunol. 35, 341–351 (2005). [DOI] [PubMed] [Google Scholar]
- 84.Renzette N., Bhattacharjee B., Jensen J. D., Gibson L., Kowalik T. F., Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog. 7, e1001344 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Renzette N., Kowalik T. F., Jensen J. D., On the relative roles of background selection and genetic hitchhiking in shaping human cytomegalovirus genetic diversity. Mol. Ecol. 25, 403–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Edwards R. H., Sitki-Green D., Moore D. T., Raab-Traub N., Potential selection of LMP1 variants in nasopharyngeal carcinoma. J. Virol. 78, 868–881 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin J. C., et al. , Amino acid changes in functional domains of latent membrane protein 1 of Epstein-Barr virus in nasopharyngeal carcinoma of southern China and Taiwan: Prevalence of an HLA A2-restricted ‘epitope-loss variant’. J. Gen. Virol. 85, 2023–2034 (2004). [DOI] [PubMed] [Google Scholar]
- 88.de Campos-Lima P. O., et al. , HLA-A11 epitope loss isolates of Epstein-Barr virus from a highly A11+ population. Science 260, 98–100 (1993). [DOI] [PubMed] [Google Scholar]
- 89.Midgley R. S., Bell A. I., McGeoch D. J., Rickinson A. B., Latent gene sequencing reveals familial relationships among Chinese Epstein-Barr virus strains and evidence for positive selection of A11 epitope changes. J. Virol. 77, 11517–11530 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Midgley R. S., et al. , HLA-A11-restricted epitope polymorphism among Epstein-Barr virus strains in the highly HLA-A11-positive Chinese population: Incidence and immunogenicity of variant epitope sequences. J. Virol. 77, 11507–11516 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ehlers B., et al. , Lymphocryptovirus phylogeny and the origins of Epstein-Barr virus. J. Gen. Virol. 91, 630–642 (2010). [DOI] [PubMed] [Google Scholar]
- 92.Murthy S., et al. , Cytomegalovirus distribution and evolution in hominines. Virus Evol. 5, vez015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fernández-Ruiz M., et al. , Influence of Age and HLA Alleles on the CMV-Specific Cell-Mediated Immunity among CMV-Seropositive Kidney Transplant Candidates (Blackwell Publishing Ltd., 2015), vol. 15, pp. 2525–2526. [DOI] [PubMed] [Google Scholar]
- 94.Hassan J., et al. , Cytomegalovirus infection in Ireland: Seroprevalence, HLA class I alleles, and implications. Medicine (Baltimore) 95, e2735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hjalgrim H., et al. , HLA-A alleles and infectious mononucleosis suggest a critical role for cytotoxic T-cell response in EBV-related Hodgkin lymphoma. Proc. Natl. Acad. Sci. U.S.A. 107, 6400–6405 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Niens M., et al. , HLA-A*02 is associated with a reduced risk and HLA-A*01 with an increased risk of developing EBV+ Hodgkin lymphoma. Blood 110, 3310–3315 (2007). [DOI] [PubMed] [Google Scholar]
- 97.Borozan I., Zapatka M., Frappier L., Ferretti V., Analysis of Epstein-Barr virus genomes and expression profiles in gastric adenocarcinoma. J. Virol. 92, e01239-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Correia S., et al. , Sequence variation of Epstein-Barr virus: Viral types, geography, codon usage, and diseases. J. Virol. 92, e01132-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Correia S., et al. , Natural variation of Epstein-Barr virus genes, proteins, and primary microRNA. J. Virol. 91, e00375-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Y., et al. , Genome-wide analysis of Epstein-Barr virus (EBV) isolated from EBV-associated gastric carcinoma (EBVaGC). Oncotarget 7, 4903–4914 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lei H., et al. , Epstein-Barr virus from Burkitt Lymphoma biopsies from Africa and South America share novel LMP-1 promoter and gene variations. Sci. Rep. 5, 16706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hui K. F., et al. , High risk Epstein-Barr virus variants characterized by distinct polymorphisms in the EBER locus are strongly associated with nasopharyngeal carcinoma. Int. J. Cancer 144, 3031–3042 (2019). [DOI] [PubMed] [Google Scholar]
- 103.Tu C., et al. , Identification of genomic alterations in nasopharyngeal carcinoma and nasopharyngeal carcinoma-derived Epstein-Barr virus by whole-genome sequencing. Carcinogenesis 39, 1517–1528 (2018). [DOI] [PubMed] [Google Scholar]
- 104.Peng R. J., et al. , Genomic and transcriptomic landscapes of Epstein-Barr virus in extranodal natural killer T-cell lymphoma. Leukemia 33, 1451–1462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cunningham C., et al. , Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J. Gen. Virol. 91, 605–615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dargan D. J., et al. , Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J. Gen. Virol. 91, 1535–1546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ourahmane A., et al. , Inclusion of antibodies to cell culture media preserves the integrity of genes encoding RL13 and the pentameric complex components during fibroblast passage of human cytomegalovirus. Viruses 11, 221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jung G. S., et al. , Full genome sequencing and analysis of human cytomegalovirus strain JHC isolated from a Korean patient. Virus Res. 156, 113–120 (2011). [DOI] [PubMed] [Google Scholar]
- 109.Dolan A., et al. , Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85, 1301–1312 (2004). [DOI] [PubMed] [Google Scholar]
- 110.Sinzger C., et al. , Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89, 359–368 (2008). [DOI] [PubMed] [Google Scholar]
- 111.Davison A. J., et al. , The human cytomegalovirus genome revisited: Comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84, 17–28 (2003). [DOI] [PubMed] [Google Scholar]
- 112.Murrell I., et al. , Impact of sequence variation in the UL128 locus on production of human cytomegalovirus in fibroblast and epithelial cells. J. Virol. 87, 10489–10500 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Norberg P., et al. , Recombination of globally circulating Varicella-Zoster Virus. J. Virol. 89, 7133–7146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jeon J. S., et al. , Analysis of single nucleotide polymorphism among Varicella-Zoster Virus and identification of vaccine-specific sites. Virology 496, 277–286 (2016). [DOI] [PubMed] [Google Scholar]
- 115.Jensen N. J., et al. , Analysis of the reiteration regions (R1 to R5) of varicella-zoster virus. Virology 546, 38–50 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Loparev V. N., et al. , Distribution of varicella-zoster virus (VZV) wild-type genotypes in northern and southern Europe: Evidence for high conservation of circulating genotypes. Virology 383, 216–225 (2009). [DOI] [PubMed] [Google Scholar]
- 117.Garcés-Ayala F., et al. , Full-genome sequence of a novel Varicella-Zoster virus clade isolated in Mexico. Genome Announc. 3, e00752-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim M. H., et al. , Characterization and phylogenetic analysis of Varicella-zoster virus strains isolated from Korean patients. J. Microbiol. 55, 665–672 (2017). [DOI] [PubMed] [Google Scholar]
- 119.Cohen J. I., Cox E., Pesnicak L., Srinivas S., Krogmann T., The varicella-zoster virus open reading frame 63 latency-associated protein is critical for establishment of latency. J. Virol. 78, 11833–11840 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Eshleman E., Shahzad A., Cohrs R. J., Varicella zoster virus latency. Future Virol. 6, 341–355 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gonzalez-Galarza F. F., et al. , Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 48, D783–D788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M., NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 48, W449–W454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Paul S., et al. , HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J. Immunol. 191, 5831–5839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao W., Sher X., Systematically benchmarking peptide-MHC binding predictors: From synthetic to naturally processed epitopes. PLOS Comput. Biol. 14, e1006457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Croft N. P., et al. , Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. U.S.A. 116, 3112–3117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Paul S., et al. , Benchmarking predictions of MHC class I restricted T cell epitopes in a comprehensively studied model system. PLOS Comput. Biol. 16, e1007757 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 128.Hudson R. R., Slatkin M., Maddison W. P., Estimation of levels of gene flow from DNA sequence data. Genetics 132, 583–589 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pfeifer B., Wittelsbürger U., Ramos-Onsins S. E., Lercher M. J., PopGenome: An efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 31, 1929–1936 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Farrell P. J., Epstein-Barr virus and cancer. Annu. Rev. Pathol. 14, 29–53 (2019). [DOI] [PubMed] [Google Scholar]
- 131.Ji K. M., Li C. L., Meng G., Han A. D., Wu X. L., New BZLF1 sequence variations in EBV-associated undifferentiated nasopharyngeal carcinoma in southern China. Arch. Virol. 153, 1949–1953 (2008). [DOI] [PubMed] [Google Scholar]
- 132.Jin Y., Xie Z., Lu G., Yang S., Shen K., Characterization of variants in the promoter of BZLF1 gene of EBV in nonmalignant EBV-associated diseases in Chinese children. Virol. J. 7, 92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jing Y. Z., Wang Y., Jia Y. P., Luo B., Polymorphisms of Epstein-Barr virus BHRF1 gene, a homologue of bcl-2. Chin. J. Cancer 29, 1000–1005 (2010). [DOI] [PubMed] [Google Scholar]
- 134.Bristol J. A., et al. , A cancer-associated Epstein-Barr virus BZLF1 promoter variant enhances lytic infection. PLoS Pathog. 14, e1007179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Palmer W. H., HLA class I binding of herpesvirus proteins. Figshare. 10.6084/m9.figshare.19119743. Deposited 21 March 2022. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Population genetic data tables, R code, and Python code have been deposited in Figshare (DOI: 10.6084/m9.figshare.19119743) (135).