Significance
Modeling interactions between short peptides and their receptors is a challenging docking problem due to the peptide flexibility, resulting in a formidable sampling problem of peptide conformation in addition to its orientation. Alternatively, the peptide can be viewed as a piece that complements the receptor monomer structure. Here, we show that the peptide conformation can be determined based on the receptor backbone only and sampled using local structural motifs found in solved protein monomers and interfaces, independent of sequence similarity. This approach outperforms current peptide docking protocols and promotes new directions for peptide interface design.
Keywords: peptide docking, structure matching, surface complementation, protein structure, structural motifs
Abstract
Peptide docking can be perceived as a subproblem of protein–protein docking. However, due to the short length and flexible nature of peptides, many do not adopt one defined conformation prior to binding. Therefore, to tackle a peptide docking problem, not only the relative orientation, but also the bound conformation of the peptide needs to be modeled. Traditional peptide-centered approaches use information about peptide sequences to generate representative conformer ensembles, which can then be rigid-body docked to the receptor. Alternatively, one may look at this problem from the viewpoint of the receptor, namely, that the protein surface defines the peptide-bound conformation. Here, we present PatchMAN (Patch-Motif AligNments), a global peptide-docking approach that uses structural motifs to map the receptor surface with backbone scaffolds extracted from protein structures. On a nonredundant set of protein–peptide complexes, starting from free receptor structures, PatchMAN successfully models and identifies near-native peptide–protein complexes in 58%/84% within 2.5 Å/5 Å interface backbone RMSD, with corresponding sampling in 81%/100% of the cases, outperforming other approaches. PatchMAN leverages the observation that structural units of peptides with their binding pocket can be found not only within interfaces, but also within monomers. We show that the bound peptide conformation is sampled based on the structural context of the receptor only, without taking into account any sequence information. Beyond peptide docking, this approach opens exciting new avenues to study principles of peptide–protein association, and to the design of new peptide binders. PatchMAN is available as a server at https://furmanlab.cs.huji.ac.il/patchman/.
Peptide–protein interactions—namely, interactions mediated by short segments or motifs often located in disordered regions—are very common in the cell, constituting 40% or more of the overall protein interactions (1). Such interactions participate in many important cellular processes, like regulation and cell-signaling (2). Therefore structural characterization of such interactions is crucial for the understanding of many biological pathways and their potential in the development of therapeutic targets and other biotechnological applications (3). However, such interactions are often weaker and more transient than globular protein interactions and therefore more challenging to characterize experimentally, highlighting the need for developing computational tools for modeling their structures.
The intuitive way to look at protein–peptide docking is as a subproblem of protein–protein docking. However, this approach presents several hurdles, since in addition to the problem of finding the relative orientation between the two partners, the peptide conformation is often not known or does not even assume a defined structure before binding the receptor (4). When the binding site is known and a coarse model of a peptide–protein complex is available, it can be further refined to high accuracy by local refinement protocols, such as Rosetta FlexPepDock (5, 6). In the absence of such information, however, global docking has to be performed. To reduce the conformational space needed to sampling both the peptide conformation and its location on the receptor, many currently existing peptide-docking approaches tackle this problem by decoupling the folding and docking steps, generating a peptide conformational ensemble for subsequent docking (7). For example, in the PIPER-FlexPepDock (PFPD) protocol (8), a conformer ensemble is generated using the Rosetta Fragment Picker (9) [similar to the first step in traditional ab initio folding (10)]. This ensemble is then rigid-body–docked using PIPER (11) and further refined by FlexPepDock. This approach is also implemented in the InterPep2 docking protocol (12). MDockPeP2 uses sequence-similar fragments extracted from monomers (13), while in HADDOCK and pepATTRACT, peptide conformations are represented by idealized secondary structure fragments (14, 15), and the CABS-dock protocol uses random peptide conformations for subsequent docking and refinement (16). All these approaches are united by the idea that the peptide, as a separate protein, carries enough information for its separate folding, or at least the determination of a conformer ensemble that represents its conformational preferences. But what if the conformational ensemble of the peptide does not include the conformation that it adopts upon binding? In such a case, the rigid-body step of the docking protocol will not be able to fit the peptide into the binding pocket. An alternative solution for finding the bound conformation of the peptide is template-based modeling. Many protein–protein interactions can be modeled based on a solved structure of a homolog complex (17), and the same can be applied to protein–peptide interactions (18). However, such an approach is restricted to a limited amount of solved protein–peptide complexes.
We present here an approach for blind peptide docking, which we name PatchMAN (Patch-Motif AligNments), that combines a global search with template-based modeling, benefitting from both strategies. We look at peptide docking from the viewpoint of the receptor, building on the assumption that the protein surface carries enough information to determine the peptide-bound conformation. This is based on the previously proposed theory that peptide–protein interactions often mimic structural characteristics that are typical to monomeric folds (19), hinting at a large reservoir of information that can further be used for peptide–protein docking. PatchMAN uses surface patches, defined as bundles of disjoint backbone segments, to search for similar “pockets” that contain a peptide stretch interacting with it in a dataset of protein structures that includes monomers, as well as protein–protein and protein–peptide complexes. The backbone conformation of such peptide stretches is then superimposed back to the receptor protein, and is used as a starting point for local peptide-docking refinement.
PatchMAN shows performance superior to current peptide-docking methods, including our recent implementation of AlphaFold2 (AF2) (20) for peptide docking (21). As such, PatchMAN opens new opportunities to model more complicated protein–peptide-like interactions, in addition to facilitating design of new peptide binders.
Results
General Overview of the PatchMAN Approach: Docking by Globally Mapping the Receptor Surface with Local Motif Templates.
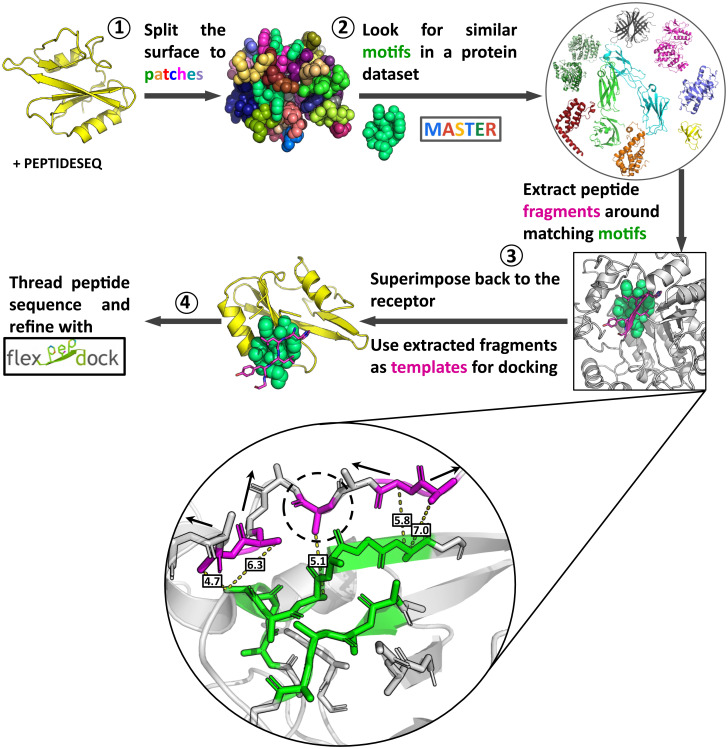
In general, protein–peptide as well as protein–protein interaction modeling can be split into two categories: template-based modeling, in which new interactions are modeled based on solved structures of similar interactions, and free modeling, in which a large number of new rigid-body orientations and internal degrees of freedom are sampled. In PatchMAN we suggest combining the two by generating peptide templates on the whole protein surface, thus sampling the binding sites and “folding” the peptide at the same time. The protocol consists of four consecutive steps (Fig. 1): 1) definition of surface patches on the receptor; 2) identification of structural motif matches in protein structures; 3) generation of the peptide–protein complex template structure by copying the peptide fragment interacting with the matched structural motif onto the receptor; 4) replacing side chains according to the peptide sequence (threading), refinement, and scoring of the model.
Fig. 1.
The PatchMan protocol: flowchart. The input is a receptor PDB file and a peptide sequence. 1) Definition of surface motifs on the receptor: the protein surface is defined based on solvent accessibility, and then split into small structural surface patches. 2) Identification of structural matches in protein structures: matches are detected using MASTER search against a nonredundant dataset of protein structures. 3) Generation of the peptide–protein complex structure: the peptide fragment is determined (see enlargement) and superimposed onto the receptor. 4) Threading, refinement and scoring: the peptide sequence is threaded onto the identified complementing fragment and the structure is refined using the Rosetta FlexPepDock refinement protocol. The generated structures are clustered, and top-scoring cluster representatives are selected as final predictions. Enlargement in step 3: Extracting peptide fragments. Neighboring residues (magenta) around the matching motif (green) are defined as Cβ distance within 8 Å of the motif. Consecutive backbone stretches are then elongated in both directions to the desired peptide length. Arrows indicate stretches that can be elongated. Single residue (indicated with dashed circle) will not be elongated. See Methods for more details.
In the following, we describe the protocol in more details (see also Methods). For the sampling step, we first identify the surface residues based on surface accessible area. Next, the surface is split into patches consisting of one or more peptide segments, each centered on a surface residue. Those patches are then used to search for similar motifs in a diverse nonredundant database of protein structures (maximum 30% pairwise sequence identity), using MASTER (22). Peptide stretches around every found motif are extracted (Fig. 1, enlargement of step 3). If an interacting fragment is shorter than the required peptide length, it is elongated in both directions so that even patches only partially covering the binding site can lead to generation of a near-native template. The extracted peptide fragments are then superimposed back to the receptor protein using the rotational matrices from the patch-motif alignment. At this point the receptor protein surface is fully mapped with templates for local peptide docking. The peptide sequence is then threaded onto the generated peptide templates. These starting structures are refined using Rosetta FlexPepDock (5). Finally, all models are scored and the best models are selected. Additional and more extensive details on each step, including specific parameters, are described in Methods.
PatchMAN Performance.
For the initial estimation of the method performance, we ran PatchMAN on a nonredundant dataset containing 26 solved protein–peptide complexes previously used to assess performance of PIPER-FlexPepDock (8). It includes two subsets of complexes: one with known binding motifs (here the motifs are eukaryotic linear motifs) (23), and the second for which no motifs have (yet) been reported. For all the complexes, free (unbound) receptor structures are available, and those were used in the present study to reflect a blind, real-world scenario. To prevent bias, all the structures categorized under the same UniProt number as the receptor protein were filtered out from the template set used by MASTER to find matching structural motifs.
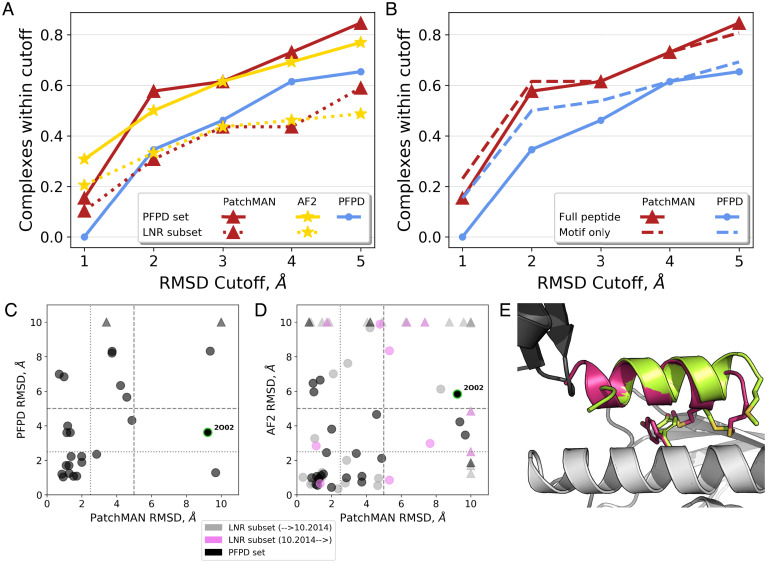
For assessing PatchMAN performance, we used RMSD measured over the peptide interface residues (after aligning the receptor; rmsBB_if calculated by Rosetta FlexPepDock). PatchMAN generates and identifies for 84%/58% of the complexes a near-native model within 5 Å/2.5 Å RMSD, respectively (among the top 10 cluster representatives) (Fig. 2A and Table 1). It outperforms the PFPD blind peptide docking protocol that also uses FlexPepDock in the refinement step, showing performance similar to our recent application of AF2 to peptide docking (21). PatchMAN also outperforms other approaches, including more recently reported methods InterPep2 (12) and MDockPeP2 (13), as well as previously developed protocols: HADDOCK (14, 15), pepATTRACT (14, 15), and CABS-dock (16) (SI Appendix, Fig. S1). A more detailed comparison reveals that while PFPD performs much better for docking the binding motif than for full-length peptides, performance of PatchMAN is not significantly affected when full-length peptides containing flanking regions are modeled (Fig. 2B). In most cases PatchMAN outperforms PFPD (Fig. 2C). We inspected what caused PatchMAN to fail on a few examples: For 1MFG, docking of the motif region without the flanking sequence generated accurate models (SI Appendix, Table S1 for detailed results of the “motif only” runs), but the flanking region was not well modeled. For 1ER8, we observed no sampling at the binding site (SI Appendix, Fig. S2). We found that this is due to the absence of matching motifs for the patches covering the binding site. This can be solved by either further fine-tuning of the patch definition, increasing the template dataset, or loosening the RMSD threshold for the match search.
Fig. 2.
Highly accurate modeling of peptide–protein complexes with PatchMAN. (A) Comparison of performance of PatchMAN to PFPD and the AF2 implementation for peptide docking, on the PFPD benchmark (n = 26 complexes; solid lines) and on the LNR subset test set (n = 39; dotted lines). The y axis shows the cumulative success, namely, the percentage of complexes modeled within the RMSD threshold indicated in the x axis. The top-performing model is considered for each complex (i.e., the best RMSD among the top 10 cluster representatives; peptide interface residue backbone RMSD values are reported). (B) Modeling only the motif sequence (dashed lines, extracted from the full peptide sequence) significantly improves performance of PFPD but only slightly affects PatchMAN performance. (C and D) Detailed comparison of PatchMAN performance to PFPD (C) and AF2 (D). PatchMAN and AF2 complement each other, successfully modeling all the complexes from the PFPD dataset (black markers) within the 5 Å cutoff, except for the 2O02 complex for which only PFPD produced a model within 5 Å (indicated with green outline). For 7 of 39 complexes from the LNR subset (gray and pink markers, solved before and after the date of the MASTER dataset compilation, respectively) neither PatchMAN nor AF2 succeeded to produce a near-native model. Of note, new complexes from the LNR subset show similar performance to the general performance. Triangles indicate structures for which the best RMSD model was larger than 10 Å. (E) Modeling of the interaction of a UBA5-derived peptide bound to UFC1, a structure solved after the development of PatchMAN [PDB ID code 7NW1 (24)]: The modeled peptide (green) is very similar to the native structure (magenta), with the most significant differences found at the peptide termini. The two monomers of the crystal structure are shown in grey and black.
Table 1.
Summary of performance for the representative, nonredundant benchmark (PFPD, from ref. 8)
| Complex PDB ID | Unbound receptor | Best model RMSD* (Å) | Best sampled RMSD (Å) | Peptide length |
|---|---|---|---|---|
| 1AWR | 2ALF | 0.9 | 0.8 | 6 |
| 1CZY | 1CA4 | 2.8 | 2.1 | 7 |
| 1EG4 | 1EG3 | 19.6 | 3.9 | 13 |
| 1ELW | 1A17 | 2.0 | 1.2 | 8 |
| 1ER8 | 4APE | 9.7 | 3.7 | 8 |
| 1JD5 | 1JD4 | 3.7 | 2.5 | 8 |
| 1JWG | 1JWF | 1.4 | 1.3 | 5 |
| 1MFG | 2H3L | 9.4 | 1.8 | 9 |
| 1NTV | 1P3R | 1.2 | 1.2 | 10 |
| 1NVR | 2QHN | 0.7 | 0.5 | 5 |
| 1NX1 | 1ALV | 1.6 | 1.2 | 11 |
| 1OU8 | 1OU9 | 4.2 | 2.6 | 8 |
| 1RXZ | 1RWZ | 1.1 | 1.1 | 11 |
| 1SSH | 1OOT | 1.3 | 0.8 | 11 |
| 1U00 | 2V7Y | 2.0 | 2.0 | 9 |
| 1X2R | 1X2J | 0.9 | 0.9 | 9 |
| 2A3I | 2AA2 | 1.1 | 1.0 | 12 |
| 2B9H | 2B9F | 3.4 | 2.2 | 12 |
| 2C3I | 2J2I | 3.7 | 2.1 | 8 |
| 2CCH | 1H1R | 4.9 | 3.0 | 12 |
| 2DS8 | 2DS7 | 1.3 | 1.3 | 6 |
| 2FMF | 1JBE | 4.6 | 2.4 | 13 |
| 2H9M | 2H14 | 1.3 | 1.3 | 5 |
| 2HPL | 2HPJ | 1.7 | 1.7 | 5 |
| 2O02 | 2BQ0 | 9.2 | 3.8 | 14 |
| 3D1E | 3D1G | 1.0 | 1.0 | 6 |
*In this table, and in the SI Appendix, Tables, we refer to the top-RMSD model among the 10 top-scoring cluster representatives.
To assess robustness of performance, we applied PatchMAN to an additional, nonredundant set of 39 peptide–protein complexes (a subset with available free receptor structures of the Large-Nonredundant [LNR] dataset described in our study of AF2 implementation for peptide docking (21), Methods). In this dataset, PatchMAN showed performance very similar to that of AF2, slightly falling behind in the highest-resolution modeling bin (10% vs. 21% of cases were modeled with RMSD better than 1 Å), but outperforming AF2 for less strict cutoffs, producing models within 5 Å cutoff in 62% of the cases versus 49% for AF2 (Fig. 2 A and D and SI Appendix, Table S2).
Finally, we evaluated PatchMAN performance also on a truly blind case whose structure was published only after the development of the PatchMAN protocol: the recently solved structure of UFC1 bound to a UBA5-derived peptide (PDB ID code 7NW1) (24). PatchMAN produced a model with 2.5 Å RMSD from the native structure in its top 10 predictions, using an unbound receptor for the simulation (Fig. 2E). The most significant differences between the models and the crystal structure are located at the peptide termini. Discrepancy at the N terminus is explained by the fact that a monomer structure was used for simulation, while in the solved structure there is a crystal contact between the monomers, forcing the N terminus of the peptide to turn aside. The two methionines in the peptide that were shown to be critical for the interaction (24) adopt a different conformation in the PatchMAN model. This is a result of side chain flipping of receptor residue Tyr36, opening a pocket at the interface that is filled by peptide residue Met401. Consequently, the critical interaction created by Met401 in the solved structure is compensated by Met404 in the model, leading to the aforementioned change in the C terminus of the peptide.
The Receptor Surface Can Be Mapped by Local Structural Motif Matches.
Our results demonstrate that even with a relatively small nonredundant set of proteins (Methods), we can model the conformation of the peptide bound to its receptor (Fig. 2). We show that in all cases, PatchMAN samples peptide conformations within ∼5 Å RMSD from the native complex structure (Table 1 and SI Appendix, Table S2). This is one of the many possible conformations generated that cover the whole receptor surface (SI Appendix, Fig. S3, and the energy landscapes presented throughout this paper and in SI Appendix, Fig. S2). This implies that even within a limited dataset of proteins, there are many motifs that are similar to receptor surface patches, and include complementing peptide stretches fitting into these surface pockets. Increasing the size of the database for the template search can help introduce more diversification of the sampling step. More diverse motifs will help in finding less trivial matches, thus introducing more intrinsic flexibility to the receptor and aiding in solving more complicated cases.
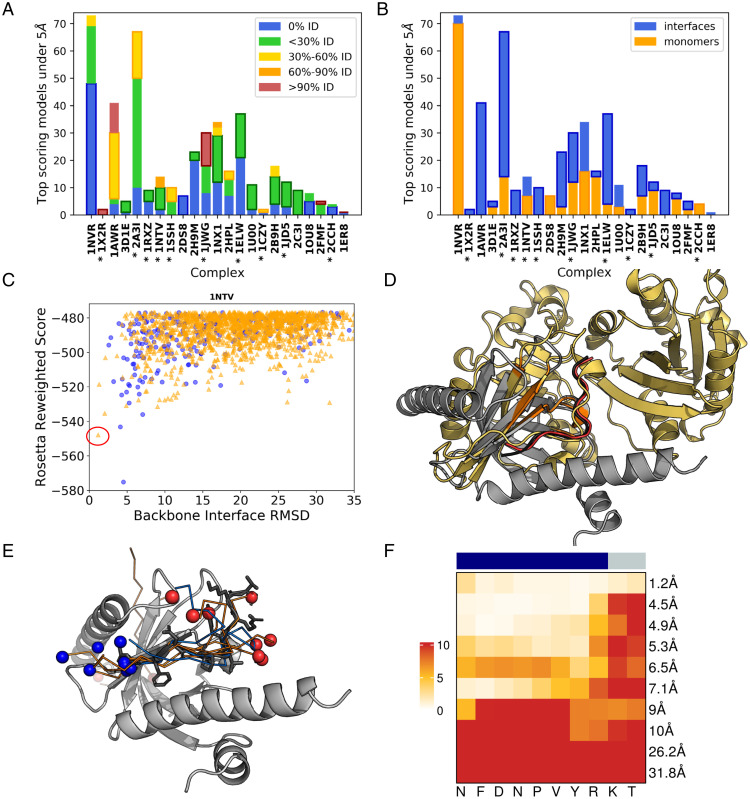
We analyzed the sequence similarity between the peptide templates and the docked peptide that led to generation of the near-native models (top 1% best scoring models within 5 Å RMSD) (Fig. 3A). We found that sequence identity of the peptide templates is predominantly below 30%, with many templates showing no sequence identity to the native peptide. For most cases (14 of 22), the best model is derived from a template with less than 30% sequence identity, while only in three cases were peptides derived from templates with more than 60% sequence identity. These results indicate that peptide-bound conformation can be sampled based on the receptor surface conformation only, without regard to the peptide sequence.
Fig. 3.
Peptide templates leading to high-resolution models show no sequence similarity and can be extracted from monomers. (A and B) Detailed results for the different complexes (shown are the 1% top-scoring models within 5 Å RMSD; the category that includes the best RMSD model is marked with a bold outline). (A) Most of the top-scoring near-native models are modeled using templates with low sequence identity. (B) The source of low-RMSD templates comes from monomers (orange) as well as interfaces (blue). Complexes are sorted in increasing order of the best RMSD. Interactions with known binding motifs are marked with an asterisk. (C–F) Details of the prediction for 1NTV (43). (C) Energy landscape. Models generated based on templates originating from monomers and interfaces are indicated in orange triangles and blue circles, respectively. The model shown in D is marked with a red circle on the energy landscape. SI Appendix, Fig. S2 shows more energy landscapes. (D) Structure of the interaction, together with the template that was used for modeling [PDB ID code 2A5D, chain A: Cholera enterotoxin (44)]. The free receptor structure (1P3B) (45) is shown in gray, the native peptide in black, top-scoring model in red and the monomer from which the template was extracted in gold. The matching motif is colored in orange. (E) The top 10 cluster representative models converge to the binding site. In ribbon models (coloring scheme is the same as in C, depending on whether the templates originate from monomers/interfaces), gray sticks represent native peptide; N and C termini in blue and red spheres, accordingly. (F) Heatmap of backbone per residue RMSD for the models shown in E. The upper bar indicates the motif and the flanking region with blue and gray color, respectively. For each model, the per-residue heatmap is followed by the value of the overall RMSD.
Many Templates for Near-Native Models Can Be Extracted from Monomer Structures.
Analysis of the templates revealed that although most of the templates originate from monomers (an average of 77%), there is a great diversity in the source of the templates that leads to the final near-native complex, depending on the type of interaction (Fig. 3B and SI Appendix, Fig. S2). In many cases, successful templates were extracted from both interfaces and monomers, where the interfaces include both peptide–protein complexes as well as protein–protein complexes. However, for several cases (1NTV, 2DS8, 2CCH, 1OU8) the only near-native complexes originated from monomers. For example, for the PatchMAN prediction of the 1NTV complex (Disabled-1 [Dab1] PTB domain-ApoER2 peptide complex) (Fig. 3 C–F), the top-scoring structure (RMSD = 1.2 Å) (Fig. 3C) was generated using a template extracted from the structure of the monomer of Cholera enterotoxin (Fig. 3D). This is an example of nontrivial template extraction that requires finding a “nonperfect match”: in this case the patch consists of four disjoint segments with patch-motif alignment RMSD of 1.4 Å. This is in contrast to cases where homologous complexes of a peptide–protein interaction are available as templates (as, for example, for 1X2R). These results demonstrate that protein monomers can indeed serve as models for peptide conformations and should be utilized in peptide-protein docking.
Inspection of the extracted templates at the 1NTV binding site reveals that while they show considerable variability (SI Appendix, Fig. S3C), the best-scoring models selected after clustering converge toward the near-native peptide conformation, and do not include conformations of different secondary structure or opposite orientation (Fig. 3E). At the same time we do see a local diversity of the models in the binding site, in particular outside the motif region (Fig. 3F).
PatchMAN Overcomes Conformational Changes Induced by Ligand Binding: The FERM Domain Example.
One of the big challenges in protein docking, and in peptide docking specifically, is that the binding pocket can undergo conformational changes upon binding of the ligand. Given that at the sampling stage we use only the receptor surface information, it is crucial that the representation of the surface will be robust to such changes. This challenge is addressed in PatchMAN in two ways. 1) Backbone-based search: the surface patches that we use for screening of matching motifs are represented as bundles of backbone segments, thus allowing for flexibility at the side-chain level with surface rotamers. 2) Diversification of matches: for each surface patch we use matching motifs with very low RMSD for finding easy templates (e.g., homologous structures), but also more distant motifs with RMSD ∼1.5 Å to capture cases of possible backbone conformational changes.
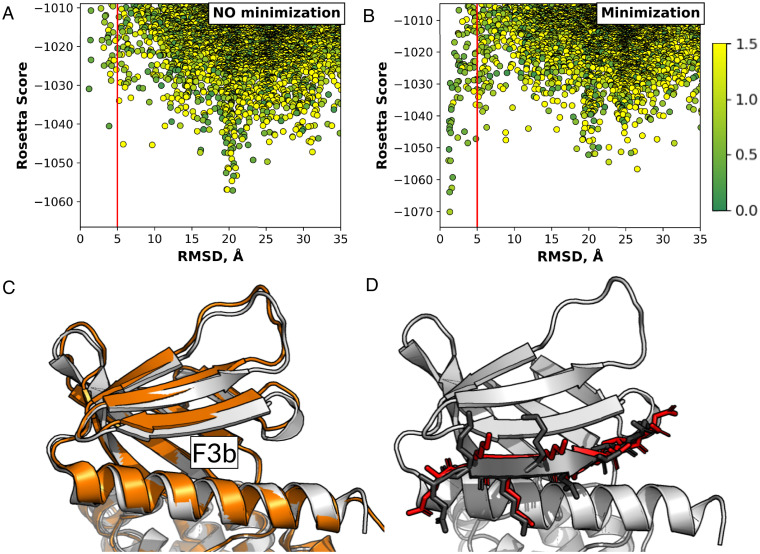
One example of a protein that undergoes such conformational change is the Moesin FERM domain (Fig. 4A). Prior to binding, the F3b binding pocket is closed and inaccessible to the peptide (25, 26). However, in case a binding site is known, the pocket could be opened by positioning a peptide into the binding site, with subsequent refinement of the structure, as implemented previously, for example, in CAPRI target T121 (27). To test the ability of PatchMAN to deal with such conformational changes we docked a CD44-derived peptide, known to bind the F3b pocket of the FERM domain, starting from the structure with the closed pocket. We compared a simulation without receptor backbone flexibility to a simulation in which receptor backbone minimization was added in the FlexPepDock refinement step of the protocol, to allow for opening of the inaccessible binding site (Fig. 4). We found that PatchMAN samples near-native fragments on the free FERM domain (with the closed F3b pocket), but cannot identify it as top-scoring on the rigid receptor structure (Fig. 4 A and C). However, it easily identified the near-native complex structure when backbone minimization was allowed (Fig. 4 B and D). PFPD failed to place the peptide at the closed binding site at the rigid-body docking step, requesting an initial opening of the pocket for successful docking (28). This case demonstrates that PatchMAN is able to identify cryptic binding pockets, with its sampling approach that takes into account motifs with structural variability. Moreover, PatchMan can open such pockets by positioning the peptide and moving the receptor backbone around it during the subsequent refinement step. Thus, this example illustrates the ability of PatchMAN to accommodate for conformational changes of the receptor upon peptide binding.
Fig. 4.
Successful modeling of a peptide into a closed binding pocket using PatchMAN, shown on the example of a CD44-derived peptide binding to the Moesin FERM domain F3b binding site. (A) PatchMAN simulation without receptor backbone minimization samples the correct binding pocket but misses it at the scoring stage. (B) PatchMAN simulation including receptor backbone minimization identifies a clear funnel around the native structure. (A and B) The red line indicates the 5 Å RMSD cutoff. The dots are colored according to a green-yellow scale that reflects source-target patch RMSD in ångstroms. (C) The moesin FERM domain structure, showing the unbound closed [gray, PDB ID code 1EF1 (25)] and the bound open F3b binding pocket [orange, PDB ID code 6TXS (26)] structures. A shift in the β-sheet at the F3b binding site is induced by peptide binding. (D) Comparison of model (red) to crystal structure (black) (for the crystal structure, only the peptide is shown).
The extent to which PatchMAN can account for conformational changes in the receptor may depend among others on the RMSD cutoff applied to select matching templates (set currently to 1.5 Å backbone RMSD) (Methods). We checked the effect of changing the matching RMSD cutoff on several examples of complexes that undergo conformational changes upon binding. Surprisingly, when analyzing the origin of templates from which PatchMAN derived the near-native models in the FERM domain example, we found that the source-target patch RMSD was only 0.9 Å for the top scoring model, and 0.4 Å for the second best (Fig. 4B). This means that changing the cutoff to as low as 0.5 Å will not significantly affect the quality of prediction (SI Appendix, Fig. S4). Of note, the near-native energy funnel also includes a model that originated from a 1.5 Å match; however, this model scored much worse than the models derived from similar templates, highlighting the challenge in refinement of structures originating from an initially distinct template (Fig. 4B). However, increasing the matching cutoff from 1.5 to 2.5 Å for examples for which PatchMAN failed, and which involve conformational changes upon binding, showed only insignificant improvement (SI Appendix, Table S3). Only for one of them was PatchMAN able to identify a model within 5 Å RMSD. This model indeed originated from a patch match with source-target RMSD of ∼2.5 Å [Protein Data Bank (29), PDB ID code 5JIU ()] (SI Appendix, Fig. S5). Further studies are therefore needed to map out the features that govern PatchMAN performance in cases that involve receptor conformational changes upon binding.
Discussion
Peptide–protein docking poses particular challenges due to the flexible nature of the peptide partner. The sampling space is vast and complex, as it involves both peptide-internal degrees of freedom as well as rigid-body orientation, and often also receptor flexibility. Many different approaches were developed to tackle this sampling challenge, usually by breaking it into several smaller, independent sampling steps. However, the biological process of peptide binding is likely to be less modular. It can be seen as a subproblem of monomer folding, in which the peptide complements the receptor structure in a way structurally similar to what is observed within other monomers. Here we present a template-based approach that builds upon these biological observations and aims to bridge the gaps between the sampling steps.
PatchMAN leverages information on local structural motifs to search for complementary fragments of the protein surface. These local motifs can be derived from interfaces, but also from completely nonrelated, monomer structures, as demonstrated in this work (Fig. 3B and SI Appendix, Fig. S2). The definition of the structural patches is crucial for success in finding complementing peptides and requires thorough optimization. As was shown previously in Verschueren et al. (31), matching single fragments (e.g., using pairs of fragments, one from the receptor and the other from the peptide) instead of patches composed of multiple segments (as in PatchMAN) can be useful in some cases, but is not enough to generate a robust sampling strategy. We show that the PatchMAN patch definition is coarse enough to be flexible for conformational changes and thus able to identify near-native templates even from divergent structures, but still specific enough to keep the hit number tractable. We demonstrate that the sampling at the binding site is specific to the native peptide-like conformation, while at the same time diverse, resulting in multiple starting points to local peptide docking, increasing the chances to model a native-like conformation at the refinement step (Fig. 3 E and F and SI Appendix, Fig. S3).
It is important to note that fast screening of such arbitrary structural motifs is challenging. Here we use MASTER (22), a fast and exhaustive RMSD-based search tool that uses the Kabsch algorithm (32) for fast alignment of backbone fragments, managing the growing complexity of multiple segment motif alignments by on-the-fly filtering of nonpromising matches. This approach does not include any heuristics, finding all existing alignments within the cutoff RMSD in a matter of seconds, and allowing for fast and efficient sampling.
PatchMAN also demonstrates lower sensitivity to parameters that limit the performance of other methods, such as the peptide length and modeling flanking regions of the peptide. As shown in Fig. 2B, PatchMAN performs equally well on full peptides and on peptide motifs, compared to PFPD performance, which decreases when adding the flanking regions. Those findings suggest that using PatchMAN docking can be further improved by connecting templates on the protein surface, to model more complex interactions involving long intrinsically disordered proteins wrapping around a structured partner, a problem only addressed by a few studies to date (33).
In PatchMAN, instead of using the sequence of the peptide as the key to modeling its backbone conformation, the focus shifts toward the receptor context. The receptor dictates the ensemble of possible peptide structures, making the sampling strategy invariant to peptide sequence. As shown here, most of the selected fragments share very low sequence identity to the docked peptide (Fig. 3A). PatchMAN opens an avenue for improved peptide design based on these principles. For a targeted receptor pocket, peptide conformations could be extracted and modeled with new sequences. Additionally, peptide backbones could be pieced together to design peptides that interact with multiple adjacent pockets of the receptor.
Moving the focus to the receptor surface also allows for improved modeling by including intrinsic local flexibility (Fig. 4). For each receptor surface patch, PatchMAN assembles an ensemble of similar motifs from different structures. Hence, even if the surface patch on the receptor is in a “closed” conformation, it can be identified by finding a similar pocket in an “open” conformation. Such a pocket can then be opened by superimposing an extracted template followed by short structural refinement. We believe that enriching the hit pool with matches from receptor homologous structures will further improve PatchMAN performance, and specifically may be helpful for cases of conformational changes.
Preliminary examination showed no evidence that calibration of the RMSD cutoff at the matching step affects PatchMAN performance (SI Appendix, Fig. S5 and Table S3). This could be thanks to the ability of PatchMAN to identify hits to patches that often cover only part of a binding pocket, but locate the peptide at an accurate position, allowing the opening of the pocket in the following refinement step that includes receptor flexibility (Fig. 4 B and D). Thus, one standard cutoff can fit a wide range of conformational changes. However, PatchMAN still needs to overcome challenges posed by cases for which the closed pocket clashes significantly with the backbone of the bound peptide (as for example for 1D4T and 4TJX of the LNR dataset) (SI Appendix, Table S2). We can improve PatchMAN performance in such cases by loosening the filtering criteria of clashing structures, or alternatively by iterative docking starting with a shorter motif followed by subsequent elongation, in case the clashes involve mainly the peptide termini.
A new era of structural biology has opened up by Deep Learning, as strongly highlighted by Deepmind’s AF2 (20). Within this context, we recently demonstrated in another study that the peptide docking field can benefit from AF2 (21), outperforming our previously developed state-of-the-art PFPD protocol (8). The PatchMAN approach presented here performs similarly well (Fig. 2A), but not on the same set (Fig. 2D), suggesting that these approaches may be combined for further improvement. It remains to be tested how well PatchMAN will perform on structural models of the receptor, and how this work may be optimally incorporated into Deep Learning frameworks.
To summarize, we presented here a robust, quick and high-performing global peptide-docking protocol, and demonstrate that the PatchMAN approach is accurate and versatile. As such, it holds high hopes for the peptide modeling as well as peptide design. The incorporation of biological insights and concepts in the development of PatchMAN extends the implications of this work and presents a more general approach to treat peptide–protein docking and binding. It is our hope that the PatchMAN webserver (https://furmanlab.cs.huji.ac.il/patchman/) will be widely used and will contribute to the detailed study of a wide range of additional peptide-mediated interactions.
Methods
Splitting the Surface into Structural Patches.
The protein surface is defined based on solvent accessibility criteria using surface accessible area calculated using the “rolling ball” algorithm implemented in PyRosetta with ball radius = 1.35 Å (34). The surface is then split into small structural motifs by selecting the neighbors (Cα–Cα distance within 10 Å) around every second surface residue (to reduce the number of overlapping patches). Every motif is defined as one or more disjoint peptide segments, not shorter than 2 amino acids. The maximum length for a single segment is 7 residues for strands and coiled regions, and 11 residues for helices. A stretch is defined to be helical, if it has at least three consecutive helical residues based on DSSP (35).
In case of obligatory homomultimers (1NX1, 2DS8, 1OU8, 2O02, and 1CZY, 4BTA in the present study) we removed the interfaces between the monomers from the mapped surface. This prevented false positives at these inaccessible regions with strong interaction potential.
Searching for Local Structural Motif Matches Using MASTER.
Every patch is searched using the MASTER algorithm (22) against a database of nonredundant protein structures described in the original implementation of MASTER. This database includes 12,661 protein structures, generated using BLASTClust (36) at 30% sequence identity on a PDB version of 2014 (22). Briefly, MASTER aligns structural motifs containing multiple disjoint backbone segments to identify all matches within a user-specified RMSD cutoff in a dataset of protein structures. It utilizes the Kabsch algorithm for identifying the match with the lowest RMSD (30), and manages possible combinatorial explosion due to multiple segments in each motif by on-the-fly filtering of partial matches that will not answer the RMSD criteria. For the search, we used the RMSD cutoff of 1.5 Å, and took the 50 lowest, as well as the 50 highest, RMSD matches to ensure diversity.
To prevent possible bias, we removed structures solved for the receptor protein from the dataset (i.e., those annotated with the same UniProt ID) (37). We did keep other structures of homolog proteins, reflecting the real-world scenario in which some homolog templates will be available.
Generating Initial Complexes for Further Refinement.
For each of the matches we identify the residues that constitute the motif in the corresponding PDB structure. Using PyRosetta (33) we then identify the neighboring peptide stretches (Cα–Cα distance within 8 Å), and finally, we elongate peptides longer than 2 amino acids to the desired length in both directions, if possible (Fig. 1, enlargement from step 3). Using the rotation-translation matrices from the MASTER search, the peptide templates are superimposed back onto the receptor protein. We retain those peptides whose backbone does not clash with the receptor (backbone atom distance > 2 Å) and who interact with the receptor (at least 45% interacting residues with a heavy atom interaction distance within 5 Å). The peptide sequence is then threaded onto the remaining templates (using Rosetta fixed backbone design (38)).
Model Refinement.
The Rosetta FlexPepDock refinement protocol was used to refine the structures to high resolution and to discriminate near-native models from the rest [as described previously (39), one structure was generated]. For the main benchmark we included receptor backbone minimization.
Criteria for Measuring Performance.
The accuracy of performance was measured as in previous studies (8). In short, the final top 1% of the models [based on the Rosetta reweighted score (40), using the Rosetta ref2015 scoring function (41)] are clustered (with 2.0 Å RMSD cutoff, as in ref. 8) and top 10 clusters representatives are analyzed. For the plots in Fig. 2, we calculated the number of structures for which the best RMSD model among these 10 representatives lies within the indicated RMSD cutoffs. All results were assessed using RMSD calculated over all interface peptide residue backbone atoms, after superposition of the receptor (i.e., rmsBB_if, as in previous studies) (e.g., ref. 8). Note that the PDB 1LVM was removed from the dataset due to an error in the previous dataset (the “unbound” structure included in the set is bound to the same peptide).
Datasets Used in This Study.
We used two datasets to assess performance of PatchMAN in this study: Initial assessment was performed on the PFPD dataset (from ref. 8) (Table 1), which includes 26 protein–peptide complexes with unique ECOD family IDs (42). For each of the complexes, the free receptor structure was used for docking simulations. To validate performance, we compiled a second, nonoverlapping set, based on the LNR set described in Tsaban et al. (21). The LNR subset includes all the complexes for which a free receptor structure is available. The criteria for choosing free structures were as follows: 1) the free receptor has the same UniProt ID as the receptor in the complex; 2) applying symmetry operations on the asymmetric unit of the free structure did not reveal any crystal contact that may bias the simulation toward the binding site; and 3) no ligands (including small molecules) were found in the proximity of the binding site. The final validation dataset includes 39 structures (SI Appendix, Table S2).
Supplementary Material
Acknowledgments
We thank Vasily Khramushin for his help in the implementation of PatchMAN. This work was supported, in whole or in part, by the Israel Science Foundation, founded by the Israel Academy of Science and Humanities Grants 717/2017 and 301/2021 (to O.S.-F.), and the United States–Israel Binational Science Foundation 2015207 (to O.S.-F.). J.K.V. is supported by a Marie Sklodowska-Curie European Training Network Grant 860517.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121153119/-/DCSupplemental.
Data Availability
All scripts, runline commands, and data analysis are provided on GitHub (https://github.com/Alisa-Kh/PatchMAN and https://github.com/Alisa-Kh/PatchMAN-figures).
References
- 1.Petsalaki E., Russell R. B., Peptide-mediated interactions in biological systems: New discoveries and applications. Curr. Opin. Biotechnol. 19, 344–350 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Babu M. M., van der Lee R., de Groot N. S., Gsponer J., Intrinsically disordered proteins: Regulation and disease. Curr. Opin. Struct. Biol. 21, 432–440 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Muttenthaler M., King G. F., Adams D. J., Alewood P. F., Trends in peptide drug discovery. Nat. Rev. Drug Discov. 20, 309–325 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Ciemny M., et al. , Protein-peptide docking: Opportunities and challenges. Drug Discov. Today 23, 1530–1537 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Raveh B., London N., Schueler-Furman O., Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins 78, 2029–2040 (2010). [DOI] [PubMed] [Google Scholar]
- 6.London N., Raveh B., Cohen E., Fathi G., Schueler-Furman O., Rosetta FlexPepDock web server--high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 39, W249-53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schueler-Furman O., London N., Modeling Peptide-Protein Interactions: Methods and Protocols (Humana Press, 2017). [DOI] [PubMed] [Google Scholar]
- 8.Alam N., et al. , High-resolution global peptide-protein docking using fragments-based PIPER-FlexPepDock. PLoS Comput. Biol. 13, e1005905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gront D., Kulp D. W., Vernon R. M., Strauss C. E. M., Baker D., Generalized fragment picking in Rosetta: Design, protocols and applications. PLoS One 6, e23294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simons K. T., Kooperberg C., Huang E., Baker D., Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 268, 209–225 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Kozakov D., Brenke R., Comeau S. R., Vajda S., PIPER: An FFT-based protein docking program with pairwise potentials. Proteins 65, 392–406 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Johansson-Åkhe I., Mirabello C., Wallner B., InterPep2: Global peptide-protein docking using interaction surface templates. Bioinformatics 36, 2458–2465 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X., Zou X., Predicting protein-peptide complex structures by accounting for peptide flexibility and the physicochemical environment. J. Chem. Inf. Model. 62, 27–39 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Zundert G. C. P., et al. , The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016). [DOI] [PubMed] [Google Scholar]
- 15.de Vries S. J., Rey J., Schindler C. E. M., Zacharias M., Tuffery P., The pepATTRACT web server for blind, large-scale peptide-protein docking. Nucleic Acids Res. 45 (W1), W361–W364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurcinski M., Badaczewska-Dawid A., Kolinski M., Kolinski A., Kmiecik S., Flexible docking of peptides to proteins using CABS-dock. Protein Sci. 29, 211–222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kundrotas P. J., Zhu Z., Janin J., Vakser I. A., Templates are available to model nearly all complexes of structurally characterized proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 9438–9441 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H., Heo L., Lee M. S., Seok C., GalaxyPepDock: A protein-peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Res. 43 (W1), W431–W435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanhee P., et al. , Protein-peptide interactions adopt the same structural motifs as monomeric protein folds. Structure 17, 1128–1136 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsaban T., et al. , Harnessing protein folding neural networks for peptide-protein docking. Nat. Commun. 13, 176 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J., Grigoryan G., Rapid search for tertiary fragments reveals protein sequence-structure relationships. Protein Sci. 24, 508–524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar M., et al. , ELM-the eukaryotic linear motif resource in 2020. Nucleic Acids Res. 48 (D1), D296–D306 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar M., et al. , Structural basis for UFM1 transfer from UBA5 to UFC1. Nat. Commun. 12, 5708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson M. A., Reczek D., Bretscher A., Karplus P. A., Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell 101, 259–270 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Bradshaw W. J., et al. , The structure of the FERM domain and helical linker of human moesin bound to a CD44 peptide. Protein Data Bank. 10.2210/pdb6TXS/pdb. Accessed 22 February 2022. [DOI]
- 27.Khramushin A., et al. , Modeling beta-sheet peptide-protein interactions: Rosetta FlexPepDock in CAPRI rounds 38-45. Proteins 88, 1037–1049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali M., Khramushin A., Yadav V. K., Furman-Schueler O., Ivarsson Y., Defining binding motifs and dynamics of the multi-pocket FERM domain from ezrin, radixin, moesin and merlin. bioRxiv [Preprint] (2020). https://www.biorxiv.org/content/10.1101/2020.11.23.394106v2 (Accessed 22 February 2022).
- 29.Berman H. M., et al. , The protein data bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.S. K. Hong, K.-H. Kim, E. J. Song, E. E. Kim, Structural basis for the interaction between the IUS-SPRY domain of RanBPM and DDX-4 in germ cell development. J. Mol. Biol. 428, 4330–4344 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Verschueren E., Vanhee P., Rousseau F., Schymkowitz J., Serrano L., Protein-peptide complex prediction through fragment interaction patterns. Structure 21, 789–797 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W., A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32, 922–923 (1976). [Google Scholar]
- 33.Peterson L. X., Roy A., Christoffer C., Terashi G., Kihara D., Modeling disordered protein interactions from biophysical principles. PLoS Comput. Biol. 13, e1005485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhury S., Lyskov S., Gray J. J., PyRosetta: A script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabsch W., Sander C., Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
- 36.National Center for Biotechnology Information (NCBI), Documentation of the BLASTCLUST-algorithm. ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html. Accessed 1 July 2014.
- 37.UniProt Consortium, UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49 (D1), D480–D489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leaver-Fay A., Snoeyink J., Kuhlman B., “On-the-fly rotamer pair energy evaluation in protein design” in Bioinformatics Research and Applications, Măndoiu I., Sunderraman R., Zelikovsky A., Eds. (Springer, Berlin, 2008), pp. 343–354. [Google Scholar]
- 39.Alam N., Schueler-Furman O., Modeling peptide-protein structure and binding using Monte Carlo sampling approaches: Rosetta FlexPepDock and FlexPepBind. Methods Mol. Biol. 1561, 139–169 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Raveh B., London N., Zimmerman L., Schueler-Furman O., Rosetta FlexPepDock ab-initio: Simultaneous folding, docking and refinement of peptides onto their receptors. PLoS One 6, e18934 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alford R. F., et al. , The Rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 13, 3031–3048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng H., et al. , ECOD: An evolutionary classification of protein domains. PLoS Comput. Biol. 10, e1003926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stolt P. C., et al. , Origins of peptide selectivity and phosphoinositide binding revealed by structures of disabled-1 PTB domain complexes. Structure 11, 569–579 (2003). [DOI] [PubMed] [Google Scholar]
- 44.O’Neal C. J., Jobling M. G., Holmes R. K., Hol W. G. J., Structural basis for the activation of cholera toxin by human ARF6-GTP. Science 309, 1093–1096 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Muthurajan U. M., et al. , Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 23, 260–271 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts, runline commands, and data analysis are provided on GitHub (https://github.com/Alisa-Kh/PatchMAN and https://github.com/Alisa-Kh/PatchMAN-figures).