Abstract
The completion of a telomere-to-telomere human reference genome (T2T-CHM13) has resolved complex regions of the genome, including repetitive and homologous regions. Here we present a high-resolution epigenetic study of previously unresolved sequences, representing entire acrocentric chromosome short arms, gene family expansions, and a diverse collection of repeat classes. This resource precisely maps CpG methylation (32.28 million CpGs), DNA accessibility, and short-read datasets (166,058 previously unresolved ChIP-seq peaks) to provide evidence of activity across previously unidentified or corrected genes and reveal clinically relevant paralog-specific regulation. Probing CpG methylation across human centromeres from six diverse individuals generated an estimate of variability in kinetochore localization. This analysis provides a framework to investigate the most elusive regions of the human genome granting insights into epigenetic regulation.
One Sentence Summary:
The T2T-CHM13 assembly enabled generation of a comprehensive epigenetic annotation of the remaining 8% of the human genome.
INTRODUCTION
The human reference genome has served as the foundation for many large-scale epigenetic initiatives (1-3) that aimed to catalog regulatory elements involved in gene activity and cellular function. However, efforts to construct a complete annotation of functional elements have been hampered by an incomplete reference genome. With recent technological advances, we are now able to study genome structure and function comprehensively across the finished, telomere-to-telomere (T2T-CHM13) human genome assembly based on the CHM13 cell line derived from a complete hydatidiform mole (4). As a result, we can now broaden the human epigenome to include 225 million basepairs (Mbp) of sequence, representing entire acrocentric chromosome short arms, gene family expansions, and a diverse collection of repeat classes.
The epigenome is influenced both by the specific genetic sequence and the sequence context, i.e. the flanking regions and placement of the loci within the complex structure and organization within the nucleus (5). The same genetic sequence can perform different functions or be regulated differently depending on the location of the sequence and its epigenetic state. This is especially relevant given possible evolutionary advantages that may be conferred by gene duplication, such as selectively silencing or activating different paralogous gene copies. These processes are hypothesized to diversify gene activity across developmental time and different tissues (6). Beyond evolutionary questions, epigenetic dysregulation of repetitive sequences can play a key role in development and human disease. A diverse set of repeat sequences, difficult to probe in the human reference genome GRCh38, have been implicated in facioscapulohumeral muscular dystrophy (FSHD) (due to deletions in D4Z4) (7), schizophrenia (linked to an expanded repeat in TAF11) (8), neuroblastoma (linked to somatic hypomethylation of SST1) (9), lung cancer (associated with CT47 expression) (10), pancreatic ductal adenocarcinomas (associated with HSat2 expression) (11) and immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF) (linked to heterochromatin abnormalities in HSat2,3) (12).
Within the improved T2T-CHM13 reference, the previously unresolved areas are highly repetitive, containing only infrequent sites of unique, mappable regions. This presents a limitation to short-read sequence mapping strategies, even with a more accurate reference and unique k-mer anchored alignments (13, 14). Emerging long-read technologies (15) offer sequence lengths capable of spanning infrequent unique markers and provide a direct measurement of the base sequence and epigenetic state on single molecules (16, 17).
RESULTS
Epigenetic profiles from a T2T genome in disease relevant loci
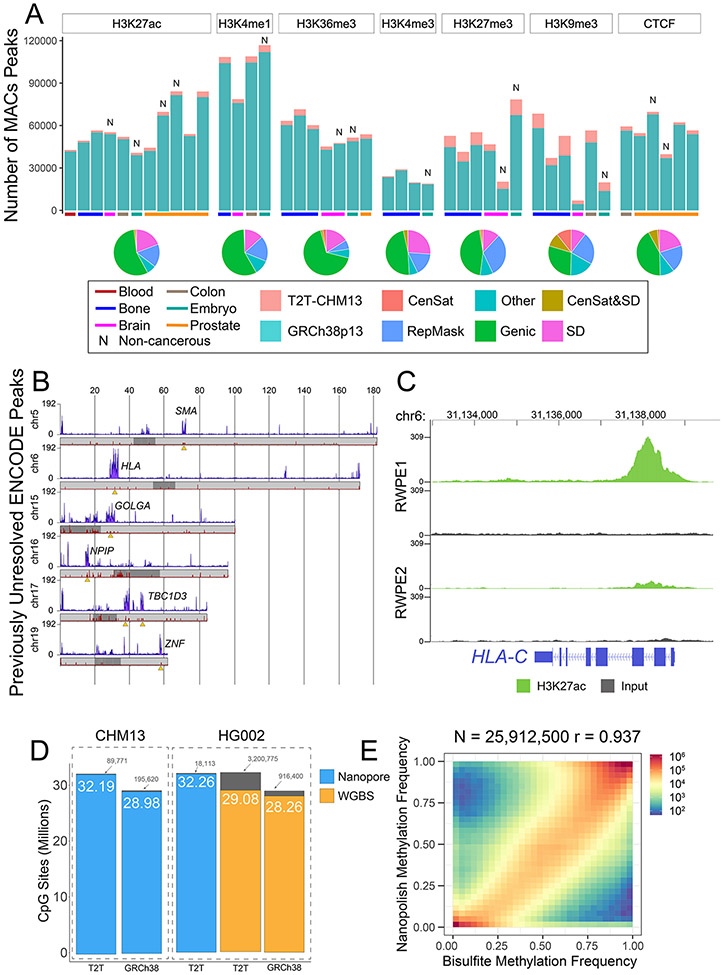
The T2T-CHM13 assembly resolves gaps and corrects misassembled or patched regions in GRCh38, leading to the introduction of nearly 225 Mbp (4). Using existing short-read epigenetic data from the ENCODE project (1) we probed previously unidentified areas of the genome. To ensure accurate mapping to these regions, we intersected ENCODE ChIP-seq alignments with unique k-mers of varying size of k (range k=50 to 100; fig. S1 and tables S1 and S2) (1, 18). On average 2.35% more reads mapped to T2T-CHM13 than GRCh38 across six different histone marks and CTCF, an important regulator of chromatin architecture (fig. S2). Reads filtered out of GRCh38 due to non-unique mapping were largely confined to the satellite DNA and segmental duplications (SDs)(fig. S3). While the total number of peaks called per sample was variable due to differences in cell type, all samples had an increase in the number of peaks called when comparing T2T-CHM13 to GRCh38 (Fig. 1A). As expected, we saw the most dramatic increase in H3K9me3 (19.4%) and H3K27me3 (15.2%) enrichment compared to GRCh38 (Table 1), consistent with the introduced peri/centromeric satellites (CenSat), SDs, and other repetitive sequences in T2T-CHM13 (Fig. 1A) that are associated with constitutive heterochromatin (19). The number of called peaks in activating marks increased as well; most notably there was a 4.9% increase in H3K36me3, a mark present across active gene bodies. Previously unresolved activating histone peaks (H3K27ac, H3K4me1, H3K36me3, and H3K4me3) and CTCF were primarily enriched in unique genic regions and in SDs (Fig. 1A).
Figure 1. Epigenetics in previously unresolved genome regions.
A) (Top) Bar plots of the number of peaks called per ENCODE sample using dynamic k-mer mapping to GRCh38 (blue) or T2T-CHM13 (salmon). (Bottom) Pie charts indicating the genomic localization of peaks found only in T2T-CHM13. B) Number of T2T-CHM13 unique ENCODE peaks across chromosomes 5, 6, 15, 16, 17, and 19 in 50kb bins (purple). Chromosome ideograms show the density of previously unannotated genes (red) with the centromere annotated as dark gray. Orange triangles denote regions of interest with a high density of previously uncalled peaks. C) ENCODE ChIP-seq read coverage at the HLA-C gene locus on chromosome 6. D) Number of CpGs with methylation profiled comparing sequencing method and reference assembly. E) Correlation of HG002 WGBS and Nanopolish methylation calls aligned to T2T-CHM13.
Table 1. Peaks called using ENCODE datasets.
Summary of ENCODE peak analysis showing the mark profiled, summed peak calls per mark across all datasets, the number of datasets, and the difference in peak number between references.
| Mark | Peaks called in GRCh38 |
Peaks called in CHM13 |
Difference in no. of peaks |
Increase in peaks |
No. of datasets |
|---|---|---|---|---|---|
| H3K9me3 | 194,681 | 241,497 | 46,816 | 19.40% | 6 |
| H3K27me3 | 249,945 | 294,819 | 44,874 | 15.20% | 6 |
| H3K36me3 | 373,933 | 393,224 | 19,291 | 4.90% | 7 |
| CTCF | 327,713 | 342,284 | 14,571 | 4.30% | 6 |
| H3K4me1 | 396,332 | 412,907 | 16,575 | 4.00% | 4 |
| H3K27ac | 611,645 | 632,837 | 21,192 | 3.30% | 11 |
| H3K4me3 | 88,985 | 91,724 | 2739 | 3.00% | 4 |
T2T-CHM13 increased the number of annotated genes by 5.7% (4), revealing 2,680 genes exclusive to T2T-CHM13 with no assigned ortholog in GRCh38 (18). These gene predictions require detailed study for functionality and validation. Here we generate a functional annotation of the previously unresolved genes using activating peaks (H3K4me3 or H3K27ac) from ENCODE cell lines. We annotated activating peaks from at least two ENCODE cell lines at the transcriptional start site (TSS) at 57 of these previously unresolved genes (table S3). Of these loci, most (20) were lncRNAs, including LINC01666, known for its associations with gastric cancer (20). Many (19) were pseudogenes, including FSHD region gene 1 (FRG1), which is a poorly understood candidate gene for FSHD (21). Three were protein-coding genes, including BOLBA2B, one of the most common genes associated with autism (22).
Our analysis of previously unresolved ENCODE peaks revealed enrichment of peaks for high copy number gene families (e.g GOLGA, NPIP, ZNF, and TBC1D3) (Fig. 1B). Large structural variants resolved in T2T-CHM13 explain the additional ChIP-seq mapping events (fig. S4) Epigenetic annotation at these genetic loci may lead to insights of paralog specific function in evolution (e.g. human specific neural genes) and disease (23, 24). For instance, SMN1/2 is associated with spinal muscular atrophy (SMA) and was historically one of the most difficult regions to assemble (25). At the SMN2 gene, we note peaks of the activating H3K4me3 mark at the promoter in all four ENCODE cell lines analyzed (fig. S5), indicating high transcriptional activity of the gene across tissues. SMA is a leading cause of childhood death (26) and has the potential to be treated by regulating expression through histone deacetylase inhibitors (HDACI), but understanding the disease specific epigenetic differences between paralogs has been challenging (27).
Another previously intractable region of the genome, the HLA locus, is critical for understanding a wide range of biology from immunity to neuropsychiatric disorders (28, 29). Our results reveal enrichment of ENCODE peaks across a variety of histone marks at the HLA locus (Fig. 1B and fig. S6A). Decreasing expression of HLA genes is associated with soft tissue cancers, particularly prostate cancer and can even be indicative of chemotherapy resistance (30). Comparing non-neoplastic adult human prostate epithelial cells (RWPE-1) and the c-Ki-ras transformed prostate cancer model cells from the same donor (RWPE-2) (31) we observed a decline in H3K27ac, an activating mark, at HLA gene promoters, concomitant with an increase in CTCF binding in RWPE-2 (Fig. 1C and fig. S6B). The differences in histone marks in this region indicate epigenetic dysregulation of the HLA locus in prostate cancer which may warrant further studies and inform upon potential therapies (32).
Long read sequencing to derive complete human methylomes
Methylation profiling has traditionally had special difficulties in mapping success rates to repetitive regions of the genome; such mapping inefficiencies are exaggerated by the bisulfite conversion of unmethylated cytosine to uracil, sequenced as thymine (33). Methylation profiles in T2T-CHM13 using long-read nanopore data demonstrate an increase in the genome coverage (32.8M compared to 29.17M in GRCh38, omitting chromosome Y) and surveyed more CpGs (10%, 3.18 M) when compared to short-read whole genome bisulfite sequencing (WGBS) (Fig. 1D). We called nanopore methylation data with Nanopolish (34), finding a high correlation (R=.937) both to WGBS results in regions mappable by both data types (Fig. 1E) and to the alternative nanopore methylation caller, Megalodon (R=.952) (fig. S7). Examining the difference between mapping of WGBS and nanopore methylation data, we generated short-read mappability scores in 200bp windows with a score of 0 being unmappable and 200 being highly mappable (18). We found the 165Mbp of sequence with a score of 0 (highly unmappable) is enriched in SDs and satellite DNA. Stratifying the nanopore data by read length, we found reads longer than 50 kilo-basepairs (kb) were capable of accurately determining methylation in these regions (figs. S8 and S9).
We sequenced the CHM13 cell line, representing an early developmental state and HG002, a terminally differentiated lymphoblast cell line. The sequenced cell line CHM13 and HG002 nanopore datasets surveyed 32.19M (99.7% of total CpGs) and 32.26M (99.9% of total CpGs) CpGs. As expected for differentiated cell lines, the majority of the HG002 genome is methylated (75% median methylation) with a secondary peak of unmethylated CpGs largely reflecting unmethylated CpG islands (CGIs) (figs. S10). In contrast, CHM13 is dramatically hypomethylated (36.8% median methylation) as expected from a trophoblastic cell line (35). Comparing CHM13’s methylation state to existing DNA reduced representation bisulfite sequencing (RRBS) data on early human embryos (fig. S11 andtable S4) (35), we observed that CHM13 clusters closely with cleavage and blastocyst-stage embryos as well as trophectoderm tissue.
To probe chromatin state in repetitive DNA we generated long-read nanoNOMe data on HG002, a method where we use M.CviPI methyltransferase to decorate accessible chromatin with exogenous GpC methylation (16) and call CpG and GpC methylation with Nanopolish to measure chromatin accessibility (figs. S12 and S13). With the combination of long-read epigenetic data and the complete human reference, we now describe a complete human epigenome, providing a foundation for further study.
Paralog specific epigenetic regulation
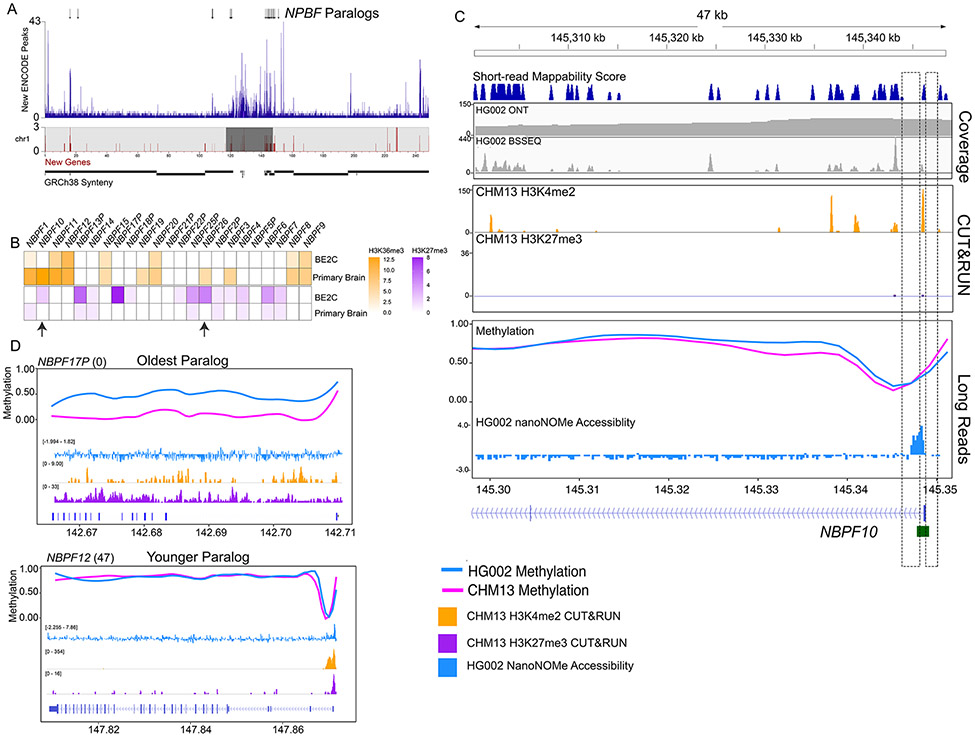
The NBPF family of genes has been implicated in the expansion of the human prefrontal cortex since our lineage diverged from apes (36). One of its copies, NBPF1 has been reported to act as a tumor suppressor in neuroblastoma where hypomethylation of CGIs has been associated with astrocytoma formation (37). Understanding the regulation of this gene family, however, has been particularly challenging because the NBPF genes correspond to large high identity duplications (>98%) that are copy number polymorphic among humans and map to gaps in the existing reference sequences (38). The fully resolved nature of T2T-CHM13 allowed us to remap ENCODE data to discover regulatory elements associated with this gene family (Fig. 2A). When comparing the balance between H3K36me3, a mark of active exons/gene bodies, and H3K27me3, a repressive mark, in samples including the BE2C cell line (neuroblastoma) and primary brain microvascular tissue (normal brain), we find that BE2C shows a higher proportion of H3K27me3 peaks (BE2C 38, Brain 8) and a lower proportion of H3K36me3 peaks (BE2C 36, Brain 89) at NPBF loci (fig. S14 andtable S5). Taking advantage of the increased resolution and more accurate NBPF copy number provided by T2T-CHM13 (39) we assayed paralog specific epigenetic changes occurring in neuroblastoma (Fig. 2B). Among the different NBPF gene copies, the largest shifts in epigenetic regulation occur at NBPF26 and NBPF10, moving from active marks in primary brain microvascular tissue to repressive marks in BE2C. These specific NBPF copies are noteworthy because they associate with human-specific duplicate genes NOTCH2NLA and NOTCH2NLR, determinants of the size and complexity of the human neocortex (40). This association identifies the functional NBPF copies, emphasizing the importance of studying paralog specific epigenetics for discovery of potential drug targets.
Figure 2. Paralog specific epigenetic regulation of the NBPF gene family.
A) Location of T2T-CHM13 previously uncalled ENCODE peaks across chromosome 1 in 50kb bins (purple). Chromosome ideograms contain the density of previously unannotated genes (red) and centromere annotations (dark gray). NBPF paralogs are indicated by black arrows (top) B) Heatmap illustrating number of peaks for H3K36me3 (orange) and H3K27me3 (purple) per NBPF paralog in ENCODE cell line BE2C (neuroblastoma) and brain tissue (Primary Brain Microvascular Tissue). Arrows indicate NBPF10 and NBPF26. C) Epigenetic data at the NBPF10 promoter and first intron (chr1:145,300,425-145,348,763). Short-read mappability score from 0-200 calculated as a 200bp region with a score of 200 being the most mappable and 0 being the least mappable. Coverage tracks (Illumina WGBS and ONT) and CUT&RUN tracks display read pileups. Long read methylation tracks show base-level methylation frequency with 0 as unmethylated and 1 as fully methylated. The long read HG002 accessibility track is a 200bp binned Z-score of nanoNOMe GpC methylation frequency. Dashed boxes highlight the promoter region which is largely unmappable with short-reads. D) (Top) Younger NBPF12 gene paralog displaying CHM13 and HG002 nanopore methylation, CHM13 H3K4me2 and H3K27me3 CUT&RUN coverage, and HG002 nanoNOMe. (Bottom) Older NBPF17P gene paralog displaying CHM13 and HG003 nanopore methylation, CHM13 H3K4me2 and H3K27me3 CUT&RUN, and HG002 nanoNOMe. Numbers in parenthesis refer to the number of PacBio Iso-seq transcripts mapped to this paralog.
Regulatory regions are excluded due to low short-read mappability scores among high identity paralogs as in the NBPF gene family (Fig. 2C andfig. S15) (18). We find that genome-wide methylation, H3K4me2, a mark of active promoters, and H3K27me3, a repressive mark, correlate with Iso-Seq coverage (transcription), and, together, can be used to systematically evaluate the functional activity of this gene family (fig. S16). We correlated this activity with the evolutionary age of the paralogs, estimated using NBPF gene paralogs from six non-human primates (NHP) from local genome assembly of the NBPF gene family from each primate (39) (fig. S17). The oldest paralog, NBPF17P, has low Iso-Seq coverage correlated with an epigenetic signature consistent with a repressive state including promoter hypermethylation and inaccessibility, enrichment of H3K27me3 and decline of H3K4me2 (Fig. 2D). In contrast, the younger paralogs, including human specific copies, have higher Iso-Seq coverage and epigenetic signatures consistent with active genes including hypomethylated and accessible promoters and enrichment of H3K4me2. Activity in the younger paralogs is more variable, with NBPF10 and NBPF20 displaying high functional activity and sharing promoters with NOTCH2NLA and NOTCH2NLB. Taken together, our results illustrate the role of epigenetics in the regulation of gene paralogs, silencing evolutionarily older paralogs while activating newer copies. This provides mechanistic insight into potentially functional genes related to human-specific cortical expansion and dysregulation in neoplasia.
Array specific epigenetic regulation of tandem repeats
Using k-mer directed ENCODE alignments to the T2T-CHM13 reference, we report epigenetic features from human centromeric regions, subtelomeres, and acrocentric short arms, which represent previously unresolved regions of the genome that are dominated by CenSat DNAs (fig. S18). Five different ENCODE lines had an enrichment of H3K9me3 in CenSat DNA, notably observed in short-read mappable regions of the acrocentric short-arms (fig. S19). Interestingly, SJCRH30 (a rhabdomyosarcoma derived line) had lower H3K9me3 enrichment in CenSat compared to the rest of the chromosome, suggesting satellite epigenetic dysregulation as a clinically relevant pathology in rhabdomyosarcoma (figs. S19 andS20A and B). This trend can be observed with more detail in an HSat3 repeat on the acrocentric arm of chromosome 15, where H3K9me3 in SJCRH30 is clearly depleted in comparison to HAP-1 (fig. S20C).
In contrast to these heterochromatic marks, we found significant enrichment of activating marks, including H3K27ac, H3K4me3, and CTCF in the telomere associated repeat (TAR) region, typically located 2kb upstream from the canonical telomeric repeat. A CTCF site in the TAR loci drives transcription of the TERRA lncRNA (41); a negative regulator of telomerase-mediated telomere elongation. We observed enrichment of CTCF in all ENCODE cell lines at the TAR loci (fig. S21A). But the subtelomeric regions are rich in SDs resulting in the TAR sequence being dispersed throughout the genome (42). When comparing telomeric TAR sequences to non-telomeric TAR sequences we do not observe statistically significant differences (Kruskal-Wallis, p-value=0.12) in sequence divergence (fig. S21B). While both telomeric and non-telomeric TAR sequences are enriched for CTCF, the non-telomeric TAR sequences are more enriched for activating chromatin marks H3K27ac and H3K4me3, suggesting differences in TERRA activity.
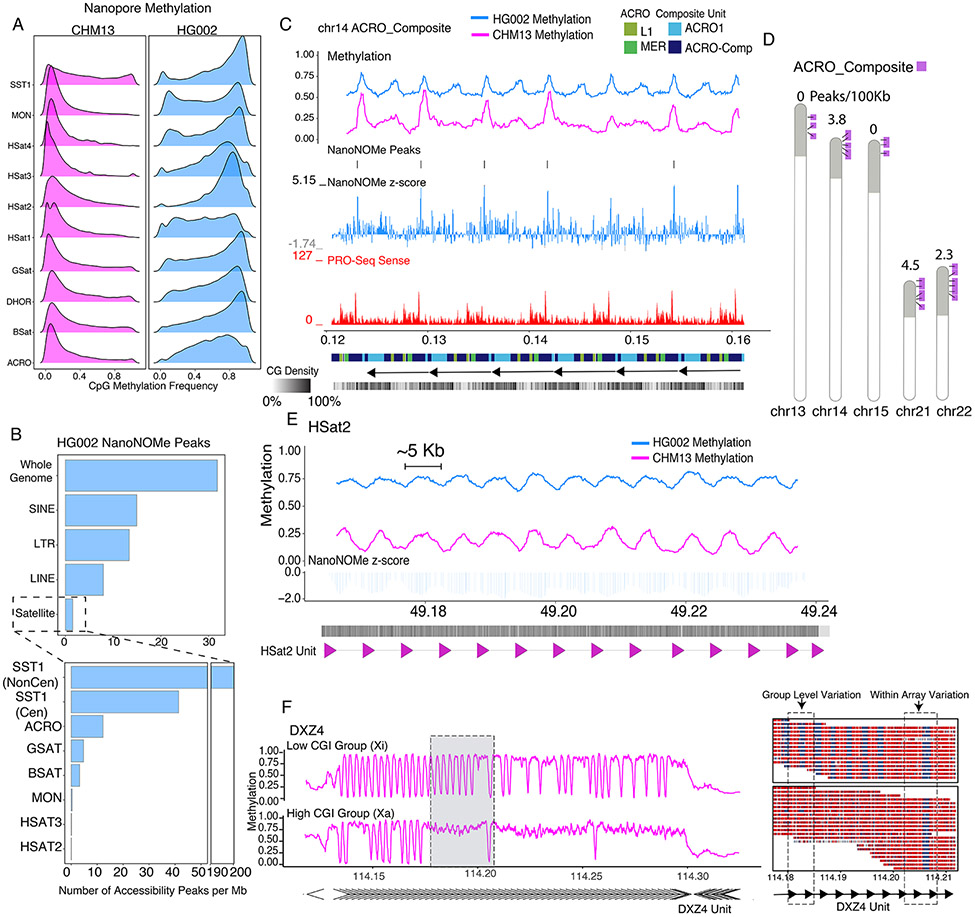
Examining nanopore CpG methylation in tandemly repeated satellite DNA elements in CHM13 and HG002 revealed hypomethylation in CHM13 compared to HG002 (Fig. 3A) (43). To assess the chromatin profile of satellite repeats we called accessibility peaks from the HG002 nanoNOMe data (18). We found that corrected for the size of the region, repeats have lower peak density than the genome as a whole. The number of nanoNOMe peaks per megabase of sequence was lower in satellite DNA (1.5), LINEs (8), SINEs (15), and LTRs (13.4) compared to the whole genome (31.8) (Fig. 3B and table S6) (44). The human satellites (HSat 2,3) and monomeric alpha satellites (MON) were largely devoid of accessibility peaks. Repetitive DNA is typically associated with densely packed heterochromatin (45); our findings are consistent with this association and transcriptional profiles from (44). However, our data allows us to investigate accessibility profiles within previously unmappable satellite repeats.
Figure 3. Context specific epigenetics in high identity tandem repeats.
A) Nanopore methylation frequency of satellite repeat classes in CHM13 and HG002. B) HG002 NanoNOMe statistically significant peak calls(18) per 1Mb of sequence in all major repeat classes compared to the whole genome (Top) and within different satellite repeats (Bottom). C) Nanopore CpG Methylation profiles, HG002 NanoNOMe accessibility peaks and Z-score (negative is inaccessible, positive is accessible), and non-kmer filtered (multimapping) PRO-Seq coverage at the ACRO_Composite repeat (chr14:121,193-162,142). Annotation tracks below are the RepeatMasker V2 annotation from (44), monomeric annotations of the ACRO_Composites and a GC density track. D) Ideogram showing the arrayed locations of the ACRO_Composite across the acrocentric chromosomes (purple) within the acrocentric short arms (gray shaded). Listed above each chromosome is the nanoNOMe ACRO_composite peak density in peaks/100kb. E) Nanopore CpG Methylation profiles and HG002 NanoNOMe accessibility Z-score of the HSat2 repeat (chr16:49,163,529-49,239,753). Annotation bars below represent CpG density and HSat2 repeat units on the bottom. F) The DXZ4 locus on CHM13 clustered into two haplotypes (low CGI methylation and high CGI methylation), based solely on promoter methylation state. (Left) Methylation frequency plot of each haplotype. (Right) Single reads from the gray highlighted region on the left with boxes highlighting CGI cluster group level epigenetic variability and intra-array level epigenetic variable between neighboring monomeric units.
Contrary to the expectation of compact chromatin and satellite DNA, we discovered enrichment of accessibility peaks in the SST1 satellite both inside the CenSat (41.4 peaks/Mbp) and in the chromosome arms (198.1 peaks/Mbp). Our peak annotations in HG002 were consistent with (44) which show higher activity in CHM13 at non-centromeric arrays on chromosomes 4 and 19 in comparison to other SST1 arrays (table S7). After the SST1, the satellite repeat with the second highest peak enrichment was the ACRO_Composite, a 7kb repeat found across 12 chromosomes, including as tandemly arrayed sequences across the five acrocentrics with high sequence identity across composite units (44). The tandemly arrayed promoter elements in the ACRO_composite give rise to a periodic bimodal methylation structure across the array (Fig. 3C). This epigenetic pattern has been proposed to be important for both the efficient transcription of non-coding RNAs and maintenance of the nearly perfect tandem arrays (46). The array has regions of increased CpG methylation which were associated with nanoNOMe peaks and transcription (CHM13 PRO-seq) (Fig. 3C). We quantified nanoNOMe peak densities across the ACRO_Composite between chromosomes and found chromosome 21 has the highest (4.5 peaks/100kb) and chromosomes 13 and 15 have the lowest (0 peaks/100kb) (Fig. 3D). The absence of nanoNOMe peaks in chromosomes 13 and 15 is correlated with low transcriptional activity (fig. S22). This high-resolution look within the acrocentric repeats suggests chromosome specific activity of the ACRO_Composite across both CHM13 and HG002, suggesting a persistent functional role for the ACRO non-coding RNA throughout early and late-stage development.
In contrast, we also observed methylation periodicity in untranscribed satellite repeats such as the HSat2, these regions were largely inaccessible as measured by nanoNOMe (Fig. 3E) (44). This periodicity in methylation corresponds to the underlying chromatin structure and echoes the genetic repeat size, suggesting the presence of functional genomic elements. Our initial epigenetic assessments of these assembled satellite sequences indicate a complicated regulatory structure stretching beyond the accepted notion that the repetitive fraction of mammalian genomes is entirely methylated and repressed by a highly condensed chromatin state (47).
Single read level analysis in satellite arrays reveals array heterogeneity
Long-reads, coupled to a complete reference assembly, confer the ability to explore methylation patterns of single molecules, each of which represents the methylation pattern of a single allele from a single cell. The X chromosome provides a unique opportunity to study these patterns because of the role of allele specific methylation in X chromosome inactivation (XCI). Female somatic tissues have a mixture of paternal or maternal X expression because the same X chromosome is not always repressed, therefore the active (Xa) and inactive (Xi) cannot be distinguished with heterozygous single nucleotide polymorphisms alone. Examining methylation state at CGIs, we clustered reads on the CHM13 X chromosome as hyper or hypomethylated (18). In order to explore whether or not the clusters represent the Xa and Xi, we first focused on genes known to be subject to XCI (XCI genes) or known to escape inactivation (escape genes) and compared our results to a clonal female lymphoblast cell line (GM12878) where the Xi is always the paternal allele (fig. S23A and B) (48). There we found the Xa to have hypomethylated promoters and hypermethylated gene bodies compared to the Xi (49). However, in CHM13 we discovered not all genes (e.g. TAF9B, PRKX) were properly regulated, with TAF9B escaping XCI and PRKX being subject to XCI, contrary to expectation. This is likely due to failure of X chromosome inactivation in androgenetic CHMs (fig. S23C and D and table S8) (50).
Moving this analysis into repetitive regions, we analyzed DXZ4, a satellite that acts as a major epigenetic regulator of XCI (51). This 165kb macrosatellite repeat contains 3kb monomeric units, each with a bidirectional CGI promoter and a CTCF site that is hypomethylated on the Xi and hypermethylated on the Xa in healthy cells (52, 53). Single-read clustering revealed two distinct clusters of reads, one with higher methylation across the repeat, and the other with lower methylation across the repeat (Fig. 3F). This analysis revealed a surprising level of heterogeneity in methylation of monomers within the array. We hypothesize this variation is a result of the aberrant XCI state of CHM13, as intra-array variation was not observed in the Xa at DXZ4 in HG002 (fig. S24). Observing epigenetic differences between monomers of satellite repeats could grant insights into human disease, granting detailed mechanistic understanding of satellite dysregulation. From this analysis, we demonstrate that we can cluster reads using methylation alone to identify heterogeneous populations and intra-array epigenetic variation even in the absence of heterozygous genetic variants.
Methylation Maps of Human Centromeres Reveal Complex Epigenetic Patterns
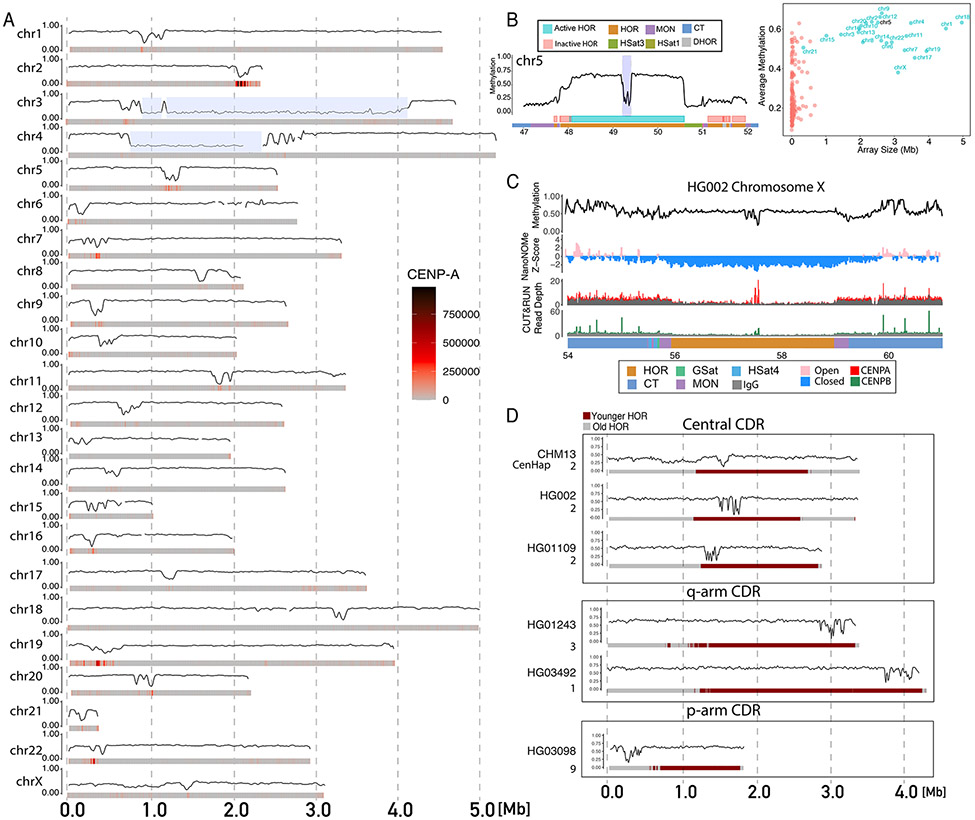
Human centromeres are composed of alpha satellite DNA, with an AT-rich ~171bp repeat unit (or ‘monomer’). The largest arrays of alpha satellites in the human genome are further organized in chromosome specific, higher-order repeats (HORs), or larger, multi-monomeric repeat units (54). Centromeres can contain multiple distinct alpha satellite HOR arrays which can be classified into active and inactive HORs (55, 56). The HORs within active arrays have specialized epigenetic regulation that are important in establishing and maintaining centromere identity (56, 57). Centromere protein A (CENP-A), is an H3-variant enriched in centromeric nucleosomes and marks sites of kinetochore assembly (58). In HOR arrays notable hypomethylation co-localizing with CENP-A enrichment at chromosomes X and 8 have been described (13, 14). We extended this finding to all CHM13 centromeres––terming this hypomethylation the centromeric dip region (CDR) (Fig. 4A and table S9). We found that CDRs were present only in active HORs (fig. S25) and that active HORs were larger in size and had higher mean methylation frequency than inactive HORs, as exemplified by the chromosome 5 centromere (Fig. 4B). These results underscore the importance of methylation in proper centromere regulation and kinetochore assembly.
Figure 4: Epigenetic maps within human centromeres.
A) Smoothed methylation frequency in 10kb bins of the active HOR array for all CHM13 chromosomes. CENP-A enrichment from CUT&RUN data shown as a heatmap under each plot. Chromosomes 3 and 4 have a HSat1 repeat (blue highlight) that breaks up the live HOR array. B) (Left) CHM13 methylation in the centromeric region of chromosome 5. Smoothed methylation frequency is plotted in 10 kb bins. HOR arrays are annotated as blue (“active”) and pink (“inactive”). (Right) Scatter plot of average methylation within each HOR array versus size in Mbp. C) Methylation, nanoNOMe accessibility, CENP-A and CENP-B CUT&RUN data across the chromosome X centromeric array on HG002. Smoothed methylation and accessibility are plotted in 15kb bins, CUT&RUN is plotted as raw read counts with input shaded gray. Bottom bar annotates satellite regions indicating the location of the HOR, MON, GSat, HSat4 and CT regions. D) Methylation in the active HOR array across diverse individuals. Coriell cell line sample ID and cenhap group annotated to left. HORs are annotated as red (younger) and gray (older) computed on the basis of sequence divergence.
To investigate if CDRs were confined only to early developmental samples, we examined HG002 nanopore sequencing data to probe centromere methylation in an adult differentiated cell line. However, the high level of HOR array variability, and the resulting inability to confidently phase and map reads from diploid chromosomes prevented us from using the T2T-CHM13 HOR reference for HG002 reads, as evidenced by the anomalous coverage we observe for HG002 alignments in the HOR arrays (fig. S26) (59). Instead, we took advantage of the haploid nature of the HG002 X chromosome and used a HG002 specific X centromere reference (4, 56). Here, in this data, we clearly observe a CDR (Fig. 4C). Furthermore, using nanoNOMe, the CDR was coordinated in this sample with a highly inaccessible region. When we examined the size of the inaccessible regions in the HOR versus the surrounding pericentromeric and centromeric transition (CT) regions, we found the HORs were enriched in dinucleosomes compared to these other regions (fig. S27). Finally, looking at CUT&RUN CENP-A and Centromere protein B (CENP-B) data, we observe a significant peak of CENP-A and CENP-B binding at the CDR. This is coordinated with a marked hypomethylation of the CENP-B motif within CDRs as opposed to outside the CDRs (fig. S28); methylation is known to reduce CENP-B binding (60). Taken together, this highlights the potential functional importance of the CDR for kinetochore formation.
Taking this a step further, using Human Pangenome Reference Consortium (HPRC) data, we leveraged the assembled X chromosomes of four additional diverse male samples representing individuals included in the 1000 Genomes Project (Fig. 4D) (56, 61). All arrays showed a distinct CDR in the X chromosome, with positional variability in the CDR location across individuals. Furthermore, CDR position was shared between individuals with more closely related centromere-spanning haplotypes (cenhap) assignments.
Cenhaps are long haplotypes which include centromere arrays due to reduced recombination in CenSat regions (56, 62). Three of the samples, CHM13 (European), HG002 (European) and HG01109 (Puerto Rican) are within cenhap group 2, and all contain a centrally positioned CDR within an evolutionarily “younger” region of the HOR, as defined in (56). Two of the samples, HG01243 (Puerto Rican) and HG03492 (Pakistani) are within cenhap groups 3 and 1, which are shown to be phylogenetically related, (ie. sharing a clade with cenhaps 1-4) (56), and have a CDR positioned more towards the q-arm side of the centromere within the evolutionarily younger region of the HOR array. Finally, one of the samples, HG03098 (African), from the more distantly related cenhap group 9, has a CDR positioned towards the p-arm of the centromere, and notably in an older (more diverged) region of the HOR array (supporting the previous observation of an epiallele in the region using available short-read datasets) (56). Thus, we demonstrate the use of CDRs to identify epigenetic variability within human centromeres, variations which may influence the centromere function during cell division. These variations show the critical importance of epigenetic profiling in the centromere, finding variation between individuals in a discrete, epigenetically defined region of the centromere.
DISCUSSION
This work provides a comprehensive view of epigenetic organization of a complete human genome, uncovering complex epigenetic patterns in the previously unresolved 8% of the human genome. Functional annotation of these intractable regions has not been overlooked due to their lack of importance, but rather due to technological limitations. Our study opens these regions to explore their epigenome, leaving no region of the genome unreachable. Here, with the combination of a complete genome assembly and the technological advances in epigenetic profiling presented herein, we make drastic strides in functional genome assessment, expanding ENCODE (1) to include 3-19% more peak calls and increasing the number of CpG methylation calls by 10%. Long-read epigenetic methods—here focusing on nanopore methylation and chromatin accessibility—can resolve single molecule epigenetic patterns within these regions, providing a foundational assessment of these areas. Long-read methylomes of distinctive developmental time points surveyed more than 99% of CpGs, establishing the CHM13 and HG002 methylomes as the most complete human methylomes to date (3). With these datasets, we profiled the additional 225Mbp of sequence and 2,680 gene annotations.
Of the previously unresolved genes, we found 57 with evidence of active promoters, including H3K4me3 or H3K27ac marks, in more than one cell type. We found 82 genes with a single cell type supporting active promoters, providing evidence that these previously unresolved gene annotations are functionally active across tissues; with more data from different tissue types we may identify even more functional genes. More generally we found that evolutionarily older gene paralogs were epigenetically repressed—similar to the epigenetic silencing of transposons—conferring genome stability and thus influencing genome evolution (63, 64).
Examining satellite DNA, we integrated short and long-read datasets to interrogate complete satellite arrays, revealing that these regions vary in epigenetic and transcriptional activity despite high sequence identity, highlighting the importance of the local chromosome environment as a modulator of epigenetics. Repetitive DNA on the acrocentric short arms is known to play a role in nucleolar formation, however the previous absence of these regions from the human reference has hampered research (65). Our findings suggest that rather than acting in unison, the repeat families on these individual acrocentric chromosomes all have their own epigenetic identity, likely contributing to unique functional roles in genome integrity and organization.
One of the features of our single-molecule epigenetic data is our ability to investigate single-molecule patterns of epigenetics. We use methylation alone to cluster reads in repetitive areas devoid of heterozygous polymorphisms; this includes the DXZ4 array where the methylation signature is critical to X chromosome inactivation (66, 67). With the increase in resolution, our results show methylation variability between the clustered populations and intra-array epigenetic variation within adjacent monomers in the same array. As satellite arrays are known to be hypervariable in the human population and linked to several human diseases, these results highlight the importance of long-read single molecule epigenetic studies for understanding disease pathology.
Finally, the T2T-CHM13 genome assembly has opened exploration of the human centromere, enabling us to probe the epigenetic elements that define centromeric chromatin. We extended our original discovery of the CDR in chromosome 8 and chromosome X to all chromosomes, and found that CDRs denote the position of centromere associated proteins (CENP-A and CENP-B, in the HG002 genome) in differentiated cells (HG002, a lymphoblast). This provides evidence of CDRs outside of early developmental CHMs and emphasizes their importance in kinetochore positioning and epigenetic regulation of chromosome segregation. Expanding our CDR analysis to male X chromosomes representing diverse haplotypes, we uncovered variability in the localization of the CDR within the X HOR array. Such variability in active centromeric arrays has been explored through the presence of epi-alleles (68); however, we have been able to demonstrate the use of CDRs to precisely predict kinetochore site localization within an active array and report across individuals representing diverse ancestry. When combined with findings in other organisms, e.g. maize (69) and medaka (70), this suggests the CDR is a conserved, functionally important feature of complex centromeres across vertebrate and plant lineages. Proper kinetochore formation is an essential process for eukaryotic cell division, a process that occurs in humans 330 billion times per day to sustain life. Our results lead to two major conclusions about the CDR: 1) CDR location on a given array is fixed in early development and maintained upon differentiation and 2) there is a single stable CDR in each centromere. Our initial profile provides a multitude of avenues for future research, including how CDR position influences meiotic and mitotic stability, disease, and aneuploidy.
Our results act as a foundational study, expanding studies of the genome through the use of the complete reference. There remain significant challenges to further exploring the epigenome in a larger and more diverse sample set to achieve optimal sequence alignment, especially amongst structurally variable repetitive regions, e.g. HORs. Efforts by the HPRC (71) to generate fully phased diploid genome assemblies will enable population-scale exploration of these areas. Limitations of short-read sequencing in unique regions can be supplemented by developing long-read epigenetic methods currently under rapid development (16, 17). We are on the precipice of exploration into duplicated and repetitive portions of the genome; further development of long-read epigenetic profiling across different populations and disease states will reveal more about regulation within the genome’s most elusive regions.
METHODS SUMMARY
Methylation processing:
Nanopore reads were obtained from (13, 14, 72). Ultra-long nanopore reads were aligned to the CHM13 reference (4) with Winnowmap-v2.0 (73) with a k-mer size of 15. BAM files were filtered for primary alignments with SAMtools (v1.9), analysis of centromeric regions was done on reads >50kb. To measure CpG methylation in nanopore data we used Nanopolish (v0.13.2) with an LLR cutoff of −1.5/1.5 (34). HG002 bisulfite FASTQs were collected from an ONT open data repository https://labs.epi2me.io/gm24385-5mc. Paired-end FASTQs were aligned with Bismark (v0.22.2) (74). For Nanopolish to Megalodon comparisons, Megalodon was run with the r9.4.1_450bps 5mC model with thresholding set as default.
NanoNOMe:
HG002 cells grown in culture and treated according to methods outlined in (16). Purified gDNA was prepared for nanopore sequencing following the protocol in the genomic sequencing by ligation kit LSK-SQK109 (ONT). To measure CpG and GpC methylation in nanopore data we used Nanopolish (v0.13.2) on the nanonome branch https://github.com/jts/nanopolish/tree/nanonome (34). We set an LLR threshold of −1/1 for GpC methylation calls and −1.5/1.5 for CpG methylation calls.
Methylation clustering:
Methylation clustering was performed across the CHM13 X chromosome on all CpG islands (CGI) that overlap an annotated promoter of a protein-coding gene. Within the CGI, reads with an average methylation > .2 were considered methylated and reads with an average methylation < .2 were considered unmethylated. Reads were only considered if they spanned the entirety of the CG islands and were longer than 5kb. Clustered reads were then intersected with known escape and XCI genes from (51). The same clustering procedure was performed at the DXZ4 locus.
CUT&RUN:
CUT&RUN was performed as detailed in (75) with some variations. For library preparation, NEBNext Ultra II End repair/A-tailing and Ligation kits were used as indicated by the manufacturer, with 1.5 pg of Spike-in Yeast DNA added (obtained from the Henikoff lab). Marker-assisted mapping of CUT&RUN data (CHM13 CENP-A, CHM13 H3K4me2, CHM13 H3K27me3, HG002 CENP-A, HG002 CENP-B) to a sample specific reference (CHM13 to T2T-CHM13 or HG002 to CHM13 autosomes (chromosomes 1-22), HG002 T2T chromosome X and GRCh38 chromosome Y) was performed according to the methods outlined in (56).
ENCODE Dynamic k-mer assisted mapping:
We selected several ChIP-seq datasets generated as part of the ENCODE project (1) choosing datasets with at least 100bp paired-end sequencing data and at least one matching input control. These criteria yielded 96 total sequencing libraries (table S9). Reads were mapped with Bowtie2 (v2.4.1) (76), alignments were filtered using SAMtools (v1.10) (77) and PCR duplicates were identified and removed with Picard tools (v2.22.1, http://broadinstitute.github.io/picard). Alignments were then filtered for unique k-mers. Specifically, for each alignment, reference sequences aligned with template ends were compared to a database of k-mers unique in the whole genome. For each end of the paired-end sequencing reads, the k-mer length was determined by finding the largest multiple of 5 less than or equal to the aligned reference sequence length. Peak calls were made using MACS2 (v2.2.7.1) (78) with default parameters and estimated genome sizes 3.03x109 and 2.79x109 for chm13v1 and GRCh38p13, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Rachael Workman, Samantha Sholes and Annelise Royles for reading and editing the manuscript, Isac Lee for engaging in discussion about epigenetics and chromatin state and Circulomics for their help in ultra high molecular weight DNA extraction. CHM13hTERT cells were obtained for research use via a material transfer agreement with the University of Pittsburgh.
Funding:
This study was supported by grants from the NIH R01HG009190 (W.T.), F32 GM134558 (G.A.L.), R24 DK106766-01A1 (M.C.S.), U24HG010263 (M.C.S.), 1R01HG011274-01 and 1U01HG010971 (K.H.M.) This work was supported, in part, by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (S.K., A.R., and A.M.P.). This work was supported, in part, by the Damon Runyon Postdoctoral Fellowship and PEW Latin American Fellowship (G.V.C.) and a HHMI Hanna Gray Fellowship (N.A.).
Footnotes
Competing interests: WT has two patents (8,748,091 and 8,394,584) licensed to Oxford Nanopore Technologies. KHM and WT have received travel funds to speak at symposia organized by Oxford Nanopore. KHM is a SAB member of Centaura, Inc.
Data and Materials Availability:
- Nanopolish methylation calls are available on zenodo (79)
- HG002 nanoNOMe data can be accessed on Sequence Read Archive with BioProject Accession number PRJNA725525
- CUT&RUN data on CHM13 and HG002 can be accessed on Sequence Read Archive with BioProject Accession PRJNA559484 and PRJNA752795
- All other datasets used in this study are properly cited with accessions referenced in the methods and materials
- Code for all CHM13 and HG002 CpG methylation and GpC methylation available: https://github.com/timplab/T2T-Epigenetics and zenodo (79)
- ENCODE analysis pipeline available: https://github.com/msauria/T2T_Encode_Analysis and zenodo (79)
- Mappability analysis (MUK) available: https://github.com/msauria/T2T_MUK_Analysis and zenodo (79)
- SatFire figures: https://mrvollger.github.io/SafFire/ and zenodo (80)
REFERENCES
- 1.ENCODE Project Consortium, An integrated encyclopedia of DNA elements in the human genome. Nature. 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekker J et al. , The 4D nucleome project. Nature. 549, 219–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roadmap Epigenomics Consortium et al. , Integrative analysis of 111 reference human epigenomes. Nature. 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurk S et al. , The complete sequence of a human genome. bioRxiv (2021), p. 2021.05.26.445798. [Google Scholar]
- 5.Jost D, Vaillant C, Epigenomics in 3D: importance of long-range spreading and specific interactions in epigenomic maintenance. Nucleic Acids Res. 46, 2252–2264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fedoroff NV, Presidential address. Transposable elements, epigenetics, and genome evolution. Science. 338, 758–767 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Gabellini D, Green MR, Tupler R, Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 110, 339–348 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Bruce HA et al. , Long tandem repeats as a form of genomic copy number variation: structure and length polymorphism of a chromosome 5p repeat in control and schizophrenia populations. Psychiatr. Genet 19, 64–71 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thoraval D et al. , Demethylation of repetitive DNA sequences in neuroblastoma. Genes Chromosomes Cancer. 17, 234–244 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Chen Y-T et al. , Identification of a new cancer/testis gene family, CT47, among expressed multicopy genes on the human X chromosome. Genes Chromosomes Cancer. 45, 392–400 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Ting DT et al. , Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 331, 593–596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassan KM, Norwood T, Gimelli G, Gartler SM, Hansen RS, Satellite 2 methylation patterns in normal and ICF syndrome cells and association of hypomethylation with advanced replication. Hum. Genet 109, 452–462 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Logsdon GA et al. , The structure, function and evolution of a complete human chromosome 8. Nature, 101–107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miga KH et al. , Telomere-to-telomere assembly of a complete human X chromosome. Nature. 585, 79–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Logsdon GA, Vollger MR, Eichler EE, Long-read human genome sequencing and its applications. Nat. Rev. Genet 21, 597–614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee I et al. , Simultaneous profiling of chromatin accessibility and methylation on human cell lines with nanopore sequencing. Nat. Methods 17, 1191–1199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stergachis AB, Debo BM, Haugen E, Churchman LS, Stamatoyannopoulos JA, Single-molecule regulatory architectures captured by chromatin fiber sequencing. Science. 368, 1449–1454 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Materials and methods are available as supplementary materials.
- 19.Janssen A, Colmenares SU, Karpen GH, Heterochromatin: Guardian of the Genome. Annu. Rev. Cell Dev. Biol 34, 265–288 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Yuan Z-H, Hou X-H, Shi M-H, Jiang R, LINC01116 promotes the proliferation and inhibits the apoptosis of gastric cancer cells. Eur. Rev. Med. Pharmacol. Sci 24, 1807–1814 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Gabellini D et al. , Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature. 439, 973–977 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Giannuzzi G et al. , The Human-Specific BOLA2 Duplication Modifies Iron Homeostasis and Anemia Predisposition in Chromosome 16p11.2 Autism Individuals. The American Journal of Human Genetics. 105 (2019), pp. 947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Z et al. , Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet 39, 1361–1368 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Maggiolini FAM et al. , Genomic inversions and GOLGA core duplicons underlie disease instability at the 15q25 locus. PLoS Genet. 15, e1008075 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmutz J et al. , The DNA sequence and comparative analysis of human chromosome 5. Nature. 431, 268–274 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Prior TW, Perspectives and diagnostic considerations in spinal muscular atrophy. Genet. Med 12, 145–152 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Hauke J et al. , Survival motor neuron gene 2 silencing by DNA methylation correlates with spinal muscular atrophy disease severity and can be bypassed by histone deacetylase inhibition. Human Molecular Genetics. 18 (2009), pp. 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruz-Tapias P, Castiblanco J, Anaya J-M, in Autoimmunity: From Bench to Bedside (El Rosario University Press, 2013), pp. 271–284. [PubMed] [Google Scholar]
- 29.Sekar A et al. , Schizophrenia risk from complex variation of complement component 4. Nature. 530, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsukahara T et al. , Prognostic significance of HLA class I expression in osteosarcoma defined by anti-pan HLA class I monoclonal antibody, EMR8-5. Cancer Science. 97 (2006), pp. 1374–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS, Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 18, 1215–1223 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Souri Z et al. , HDAC Inhibition Increases HLA Class I Expression in Uveal Melanoma. Cancers. 12, 3690 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karimzadeh M, Ernst C, Kundaje A, Hoffman MM, Umap and Bismap: quantifying genome and methylome mappability. Nucleic Acids Res. 46, e120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson JT et al. , Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Guo H et al. , The DNA methylation landscape of human early embryos. Nature. 511, 606–610 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Suzuki IK et al. , Human-Specific NOTCH2NL Genes Expand Cortical Neurogenesis through Delta/Notch Regulation. Cell. 173, 1370–1384.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu X et al. , CpG island hypermethylation in human astrocytomas. Cancer Res. 70, 2718–2727 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sudmant PH et al. , Diversity of human copy number variation and multicopy genes. Science. 330, 641–646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vollger MR, Guitart X, Dishuck PC, Mercuri L, Segmental duplications and their variation in a complete human genome. bioRxiv (2021), p. 2021.05.26.445678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiddes IT et al. , Human-Specific NOTCH2NL Genes Affect Notch Signaling and Cortical Neurogenesis. Cell. 173, 1356–1369.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng Z et al. , A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 31, 4165–4178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ambrosini A, Paul S, Hu S, Riethman H, Human subtelomeric duplicon structure and organization. Genome Biol. 8, R151 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li C et al. , DNA methylation reprogramming of functional elements during mammalian embryonic development. Cell Discov. 4, 41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoyt SJ et al. , From telomere to telomere: the transcriptional and epigenetic state of human repeat elements. bioRxiv (2021), p. 2021.07.12.451456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yunis JJ, Yasmineh WG, Heterochromatin, satellite DNA, and cell function. Structural DNA of eucaryotes may support and protect genes and aid in speciation. Science. 174, 1200–1209 (1971). [DOI] [PubMed] [Google Scholar]
- 46.Jiang C, Liao D, Striking bimodal methylation of the repeat unit of the tandem array encoding human U2 snRNA (the RNU2 locus). Genomics. 62, 508–518 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Nishibuchi G, Déjardin J, The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosome Res. 25, 77–87 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Cotton AM et al. , Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet 24, 1528–1539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hellman A, Chess A, Gene body-specific methylation on the active X chromosome. Science. 315, 1141–1143 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Chen X et al. , Loss of X Chromosome Inactivation in Androgenetic Complete Hydatidiform Moles With 46, XX Karyotype. Int. J. Gynecol. Pathol 40, 333–341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bansal P, Kondaveeti Y, Pinter SF, Forged by DXZ4, FIRRE, and ICCE: How Tandem Repeats Shape the Active and Inactive X Chromosome. Front Cell Dev Biol. 7, 328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chadwick BP, DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Research. 18 (2008), pp. 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giacalone J, Friedes J, Francke U, A novel GC–rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat. Genet 1, 137–143 (1992). [DOI] [PubMed] [Google Scholar]
- 54.Willard HF, Waye JS, Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet. 3, 192–198 (1987). [Google Scholar]
- 55.Shepelev VA et al. , Annotation of suprachromosomal families reveals uncommon types of alpha satellite organization in pericentromeric regions of hg38 human genome assembly. Genom Data. 5, 139–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Altemose N et al. , Complete genomic and epigenetic maps of human centromeres. bioRxiv (2021), p. 2021.07.12.452052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allshire RC, Karpen GH, Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet 9, 923–937 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Hooser AA et al. , Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci 114, 3529–3542 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Miga KH, Centromeric Satellite DNAs: Hidden Sequence Variation in the Human Population. Genes. 10, 352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka Y, Kurumizaka H, Yokoyama S, CpG methylation of the CENP-B box reduces human CENP-B binding. FEBS Journal. 272 (2004), pp. 282–289. [DOI] [PubMed] [Google Scholar]
- 61.1000 Genomes Project Consortium et al. , A global reference for human genetic variation. Nature. 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langley SA, Miga KH, Karpen GH, Langley CH, Haplotypes spanning centromeric regions reveal persistence of large blocks of archaic DNA. Elife. 8, e42989 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lippman Z et al. , Role of transposable elements in heterochromatin and epigenetic control. Nature. 430, 471–476 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Badyaev AV, Epigenetic resolution of the “curse of complexity” in adaptive evolution of complex traits. J. Physiol 592, 2251–2260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Sluis M et al. , Human NORs, comprising rDNA arrays and functionally conserved distal elements, are located within dynamic chromosomal regions. Genes Dev. 33, 1688–1701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Darrow EM et al. , Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl. Acad. Sci. U. S. A 113, E4504–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lemmers RJLF et al. , Cis D4Z4 repeat duplications associated with facioscapulohumeral muscular dystrophy type 2. Hum. Mol. Genet 27, 3488–3497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA, Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 26, 1301–1311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koo D-H, Han F, Birchler JA, Jiang J, Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res. 21, 908–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ichikawa K et al. , Centromere evolution and CpG methylation during vertebrate speciation. Nat. Commun 8, 1833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miga KH, Wang T, The Need for a Human Pangenome Reference Sequence. Annu. Rev. Genomics Hum. Genet 22, 81–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shafin K et al. , Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol 38, 1044–1053 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jain C, Rhie A, Hansen N, Koren S, Phillippy AM, A long read mapping method for highly repetitive reference sequences. bioRxiv (2020), p. 2020.11.01.363887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krueger F, Andrews SR, Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thakur J, Henikoff S, Unexpected conformational variations of the human centromeric chromatin complex. Genes Dev. 32, 20–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Danecek P et al. , Twelve years of SAMtools and BCFtools. Gigascience. 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y et al. , Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gershman A et al. , Code repositories used for T2T Epigenetics (2022; 10.5281/zenodo.6025533). [DOI] [Google Scholar]
- 80.Vollger MR, Lo A, mrvollger/SafFire: Version used for T2T (2022; https://zenodo.org/record/5911863). [Google Scholar]
- 81.Zeileis A, Grothendieck G, zoo: S3 Infrastructure for Regular and Irregular Time Series. Journal of Statistical Software, Articles. 14, 1–27 (2005). [Google Scholar]
- 82.Sobecki M et al. , MadID, a Versatile Approach to Map Protein-DNA Interactions, Highlights Telomere-Nuclear Envelope Contact Sites in Human Cells. Cell Rep. 25, 2891–2903.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hansen KD, Langmead B, Irizarry RA, BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 13, R83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25 (2009), pp. 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller JR et al. , Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 24, 2818–2824 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davis CA et al. , The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 46, D794–D801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kokot M, Dlugosz M, Deorowicz S, KMC 3: counting and manipulating k-mer statistics. Bioinformatics. 33, 2759–2761 (2017). [DOI] [PubMed] [Google Scholar]
- 88.Ramírez F et al. , deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rice P, Longden I, Bleasby A, EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- 91.Kirsche M, Das A, Schatz MC, Sapling: accelerating suffix array queries with learned data models. Bioinformatics. 37, 744–749 (2021). [DOI] [PubMed] [Google Scholar]
- 92.Katoh K, Standley DM, MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bouckaert R et al. , BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol 15, e1006650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marques-Bonet T, Ryder OA, Eichler EE, Sequencing primate genomes: what have we learned? Annu. Rev. Genomics Hum. Genet 10, 355–386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Manuel M et al. , Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science. 354, 477–481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL, Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kovaka S et al. , Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.