Abstract
Lipid bilayers play an important role in the pathological assembly of amyloidogenic proteins and peptides. This assembly yields oligomers and fibrils, which are highly toxic protein aggregates. In this study, we investigated the role of saturation in fatty acids of two phospholipids that are present in cell membranes. We found that unsaturated cardiolipin (CL) drastically shortened the lag phase of insulin aggregation. Furthermore, structurally and morphologically different aggregates were formed in the presence of unsaturated CL vs saturated CL. These aggregates exerted drastically different cell toxicity. Both saturated and unsaturated phosphatidylcholine (PC) were able to inhibit insulin aggregation equally efficiently. Similar to CL, structurally different aggregates were formed in the presence of saturated and unsaturated PC. These aggregates exerted different cell toxicities. These results show that unsaturated phospholipids catalyze the formation of more toxic amyloid aggregates comparing to those formed in the presence of saturated lipids.
Graphical Abstract
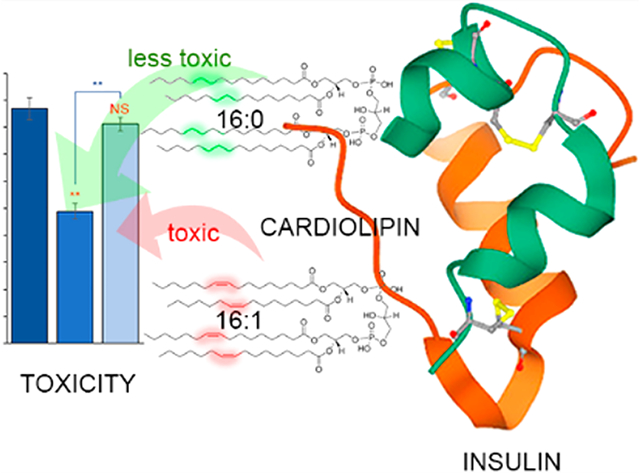
Lipid bilayers play an important role in cell and organelle compartmentalization, cell–cell interaction, endocytosis, and exocytosis.1,2 These properties are determined by the lipid composition of the bilayers that varies among different organelles and cell types.3 For instance, mitochondria possess a unique phospholipid cardiolipin (CL) that regulates electron transport complexes, carrier proteins, and phosphate kinases.4,5 At the same time, phosphatidylcholine (PC) dominates in the cell membranes of most of eukaryotic cells, including astrocytes and neurons.3,6 Membrane fluidity is further attenuated by different ratios of saturated and unsaturated fatty acids in such phospholipids. A rule of thumb is that with an increase in the concentration of unsaturated fatty acids, the membrane fluidity increases.
A growing body of evidence suggests that lipid bilayers can play an important role in the conformational changes of proteins.7,8 For instance, α-synuclein (α-Syn) is a small protein that is directly linked to Parkinson’s disease and was found to transition from unordered secondary structure to an α-helix on the surface of lipid bilayers.9,10 Galvagnion and co-workers found that lipids can lower the stability of α-Syn, promoting its aggregation into oligomers and fibrils.11–13 Our group found that lipids not only alter the rates of α-Syn aggregation but also uniquely modify the secondary structure of protein oligomers.14 The transient nature of these protein species and their morphological heterogeneity require scanning optical nanoscopy techniques, such as atomic force microscopy–infrared (AFM–IR) spectroscopy, for the elucidation of their structural organization.15–19 In AFM–IR, thermal expansions of the analyzed samples, which are induced by pulsed tunable IR light, are recorded by the scanning probe.20–22 If the laser frequency is tuned to the resonance frequency of the scanning probe, then the appearing resonance effect can be used for single-monolayer and even single-molecule sensitivity.23,24 This high sensitivity and nanometer spatial resolution were utilized to reveal the secondary structure of amyloid fibrils,15,17,18,25–27 plant epicuticular waxes,28,29 polymers,30 malaria-infected blood cells,31 bacteria,32–34 liposomes,35 and polycrystalline perovskite films.36
In addition to AFM–IR, high-speed AFM (HS-AFM),37,38 near-field scanning optical microscopy (NSOM),39 and tip-enhanced Raman spectroscopy (TERS)40–44 can be used to probe the structural organization of amyloid aggregates. For instance, HS-AFM was able to reveal structural transformations in islet amyloid precursor peptides (IAPPs) Aβ1–42 and α-Syn that led to fibril formation.37,45,46 Furthermore, HS-AFM revealed two aggregation mechanisms of Aβ1–42 that led to the formation of straight and spiral fibrils.38 Using NSOM, Kitts and Vanden Bout were able to measure the binding of fluorescent molecular probes to insulin fibrils,47 whereas Paulite and co-workers resolved the secondary structure of β2 microglobulin fibrils using this near-field scanning technique.48 Zenobi and Deckert groups pioneered TERS imaging of protein aggregates.42,43,44,49 For instance, Paulite and co-workers used TERS to determine the secondary structure of Aβ1−40 aggregates,50 whereas vandenAkker and Deckert-Gaudig probed the structural heterogeneity of IAPP and insulin fibrils, respectively.40,49
Expanding upon these findings, we want to investigate the extent to which unsaturation in the fatty acids of phospholipids alters the structure and toxicity of amyloid aggregates that were grown in their presence. We aggregated insulin, a small protein that regulates glucose metabolism, in the equimolar concentrations of saturated and unsaturated CL and PC. Insulin aggregation is associated with type 2 diabetes and injection amyloidosis.51 In a former case, insulin overproduction in the pancreas led to protein misfolding and aggregation. In the latter case, high local concentrations of insulin are created after hormone injection into the skin.52,53 This not only leads to insulin aggregation but can also catalyze the aggregation of other proteins present in cell media which may result in systemic amyloidosis.54
We first investigated whether insulin aggregation in the presence of saturated vs unsaturated lipids can have any effect on the rate of fibril formation. For this, protein was mixed in a 1:1 molar ratio with both saturated and unsaturated CL and PC. The protein–lipid solutions were mixed with ThT and kept at 37 °C under 510 rpm agitation. We found that both s- and u-CL drastically shortened the lag phase (tlag) of insulin aggregation (Figure S1). Specifically, in the presence of u-CL, tlag was 3.2 ± 0.8 h, whereas in the presence of s-CL, tlag was 5.1 ± 0.3 h. It should be noted that in the lipid-free environment, insulin exhibited a much longer tlag equal to 14.4 h. We also observed no changes in ThT intensity for insulin aggregated in the presence of both s-PC and u-PC. These findings show that PC strongly inhibits insulin aggregation.
We also found that the rate (t1/2) of insulin aggregation is different in the presence of s-CL and u-CL (Figure S1). Specifically, in the presence of u-CL, t1/2 was 6.1 ± 0.2 h, whereas in the presence of s-CL, t1/2 was 8.1 ± 0.4 h. At the same time, in the lipid-free environment, the insulin aggregation rate was t1/2 = 18.1 ± 0.4 h. These findings show that unsaturation in the fatty acids of phospholipids uniquely alters both the lag phase and the rate of insulin aggregation. Finally, ThT fluorescence measurements revealed that the intensities of the products of insulin aggregation that took place in the lipid-free environment are lower than the intensity of aggregates formed in the presence of both s-CL and u-CL. This finding suggests that insulin aggregation in the presence of lipids yields more ThT-active protein aggregates.
Nanoscale imaging of Ins:CL-u revealed the presence of long fibrillar structures that were 6–8 nm in height and stretched for micrometers in length (Figure 1). Such aggregates were not observed in Ins:CL-s samples that contained relatively short (~200 nm in length) fibrils that had a height similar to that of Ins:CL-u fibrils. These findings show that u-CL promotes insulin assembly into long fibrils that cannot be formed in the presence of s-CL. It should be noted that in the lipid-free environment, insulin formed prolong fibrillar assemblies with a large distribution of lengths that were on average 12 nm in height. At the same time, we found that both s- and u-PC yielded no fibrils but rather small (4–6 nm in height) oligomers. In summary, we can conclude that s- and u-PC strongly inhibit insulin fibril formation, whereas s- and especially u-CL strongly promote it, which is in good agreement with the above-discussed ThT results.
Figure 1.
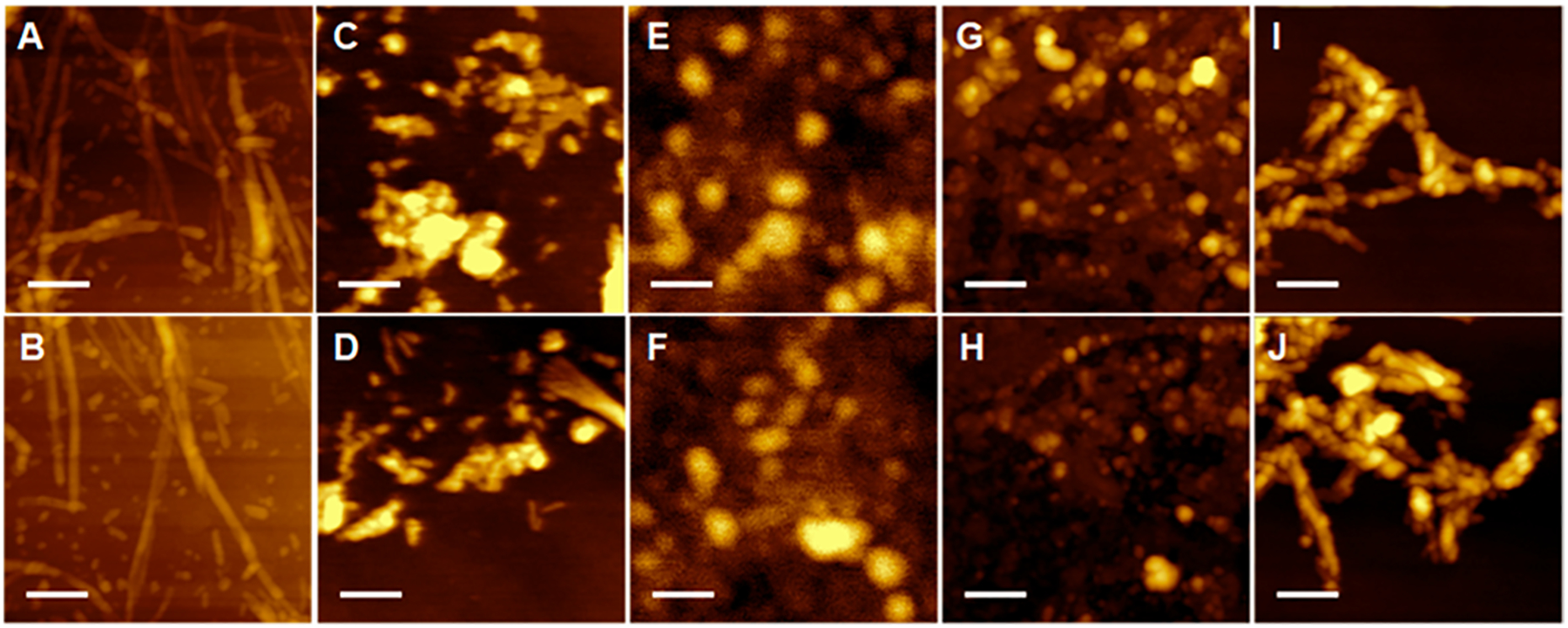
Lipids uniquely alter the morphologies of insulin aggregates. AFM images of (A and B) Ins:CL-u, (C and D) Ins:CL-s, (E and F) Ins:PC-u, (G and H) Ins:PC-s, and (I and J) insulin aggregates grown in the lipid-free environment. Scale bars are 200 nm.
We utilized CD and ATR–FTIR to examine the secondary structure of insulin aggregates grown in the presence of s-CL, u-CL, s-PC, and u-PC as well as in the lipid-free environment (Figure 2). We found that CD spectra of both Ins:PC-s and Ins:PC-u exhibited two troughs at ~210 and 223 nm. Such spectra are characteristic of proteins with a mixture of α-helical and unordered secondary structures.55,56 These findings shows that both Ins:PC-s and Ins:PC-u oligomers have predominantly α-helical and unordered secondary structures. At the same time, we observed some differences between the CD spectra collected from Ins:PC-s and Ins:PC-u. Specifically, a trough in the CD spectrum of Ins:PC-u is centered at 209 nm, whereas this trough is at 211 nm in the CD spectrum of Ins:PC-s. These spectroscopic differences point out the different secondary structure of Ins:PC-s and Ins:PC-u. The same conclusion can be made on the basis of CD spectra collected from Ins:CL-s and Ins:CL-u. Although both spectra exhibit a trough at ~223 nm, which indicates the dominance of the β-sheet in their secondary structure, this band is blue-shifted in the spectrum of Ins:CL-u (222 nm) relative to the minimum of this trough in the spectrum of Ins:CL-s (225 nm). This points out the structural differences between Ins:PC-s and Ins:PC-u aggregates. It should be noted that insulin grown in the lipid-free environment was similar to the Ins:CL-s CD spectrum with a trough at 225 nm. This suggests that the secondary structures of Ins:CL-s and insulin grown in the lipid-free environment are similar. However, FTIR spectra collected from these aggregates show substantial differences in the positions of amide I in the spectra collected from these samples. Specifically, in the spectrum collected from Ins:PC-s, amide I was found to be centered at 1631 cm−1, whereas this band was at 1628 cm−1 in the spectrum collected from insulin aggregates grown in the lipid-free environment. These findings show that the secondary structures of Ins:CL-s and insulin aggregates grown in the lipid-free environment are different.57,58 FTIR analysis of Ins:PC-s and Ins:PC-u confirmed the predominance of a mixture of α-helical and unordered protein in their secondary structure.57,59 These conclusions could be drawn on the basis of the position of amide I at ~1655 cm−1. Furthermore, in the spectrum collected from u-PC, this band was centered at 1653 cm−1, whereas in the spectrum collected from Ins:PC-s, amide I was centered at 1657 cm−1. These findings demonstrate differences in the secondary structure of Ins:PC-s and Ins:PC-u oligomers.
Figure 2.
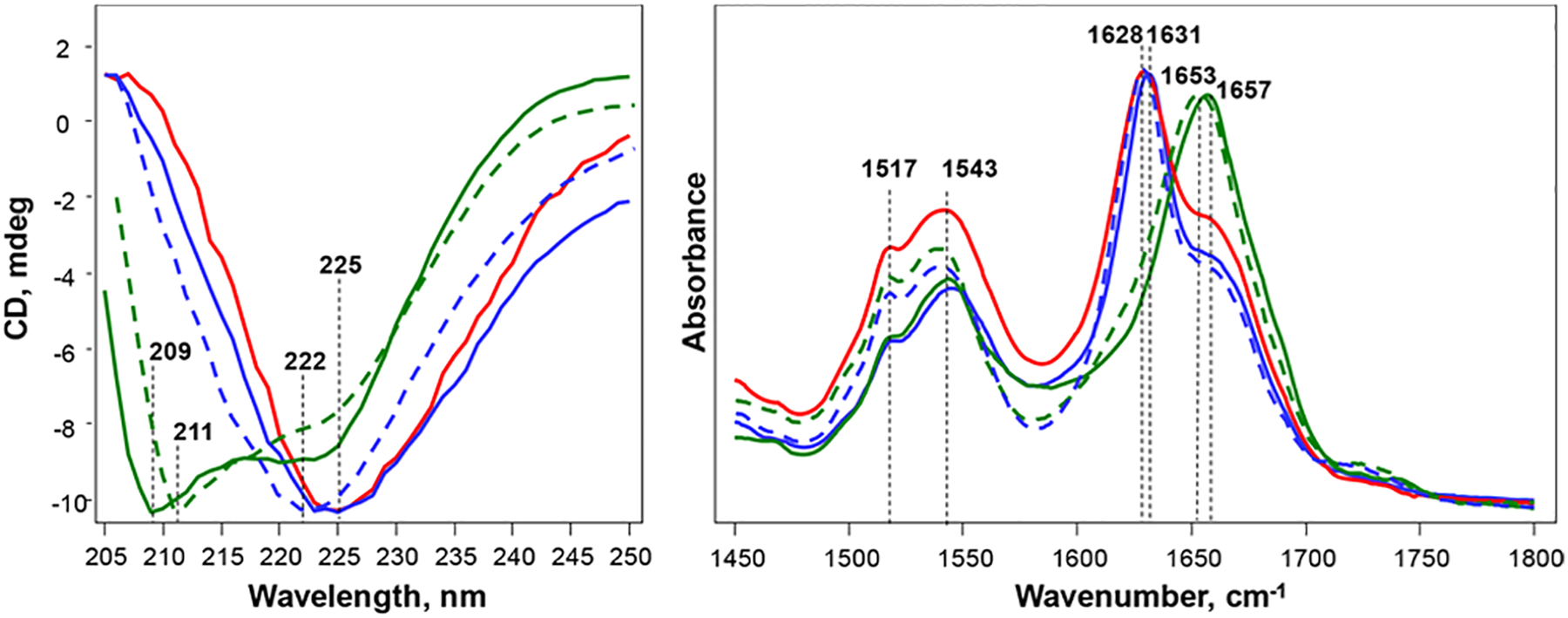
Structural analysis of insulin aggregates. (Left) CD and (right) ATR-FTIR spectra of insulin aggregates (Ins) grown in the lipid-free environment (red) as well as in the presence of Ins:CL-s (solid blue), Ins:CL-u (dashed blue), Ins:PC-s (solid green), and Ins:PC-u (dashed green).
We also utilized nanoscale infrared (AFM–IR) spectroscopy to answer two fundamentally important questions: (1) Are lipids present in the structure of Ins:PC-u, Ins:PC-s, Ins:CL-u, and Ins:CL-s? (2) To what extent do the structures of these aggregates exhibit structural homogeneity?14,17,18,60
AFM–IR analysis of individual s- and u-PC aggregates revealed the presence of two populations of oligomers in both Ins:PC-s and Ins:PC-u (Figure 3 and Figures S3–S6). AFM–IR spectra collected from population A of s-PC exhibited very intense vibrational bands at ~800 and 1000–1200 cm−1 (Figure 3). These vibrational bands correspond to C–H and PO2− vibrations, respectively.60 We have also found that these bands are present in the spectra collected from population B of Ins:PC-s; however, their intensities are substantially lower. We also found that AFM–IR spectra collected from both A and B populations of Ins:PC-s possessed vibrational bands at around 1380 and 1460 cm−1, which could be assigned to the CH and CH2 vibrations of lipids. Spectra collected from s-PC aggregates also exhibited an intense vibration at ~1734 cm−1, which can be assigned to the carbonyl vibration (C=O) of the lipid.61 These bands (CH, CH2, and C=O) were not observed in the spectra collected from insulin aggregates grown in the lipid-free environment.
Figure 3.
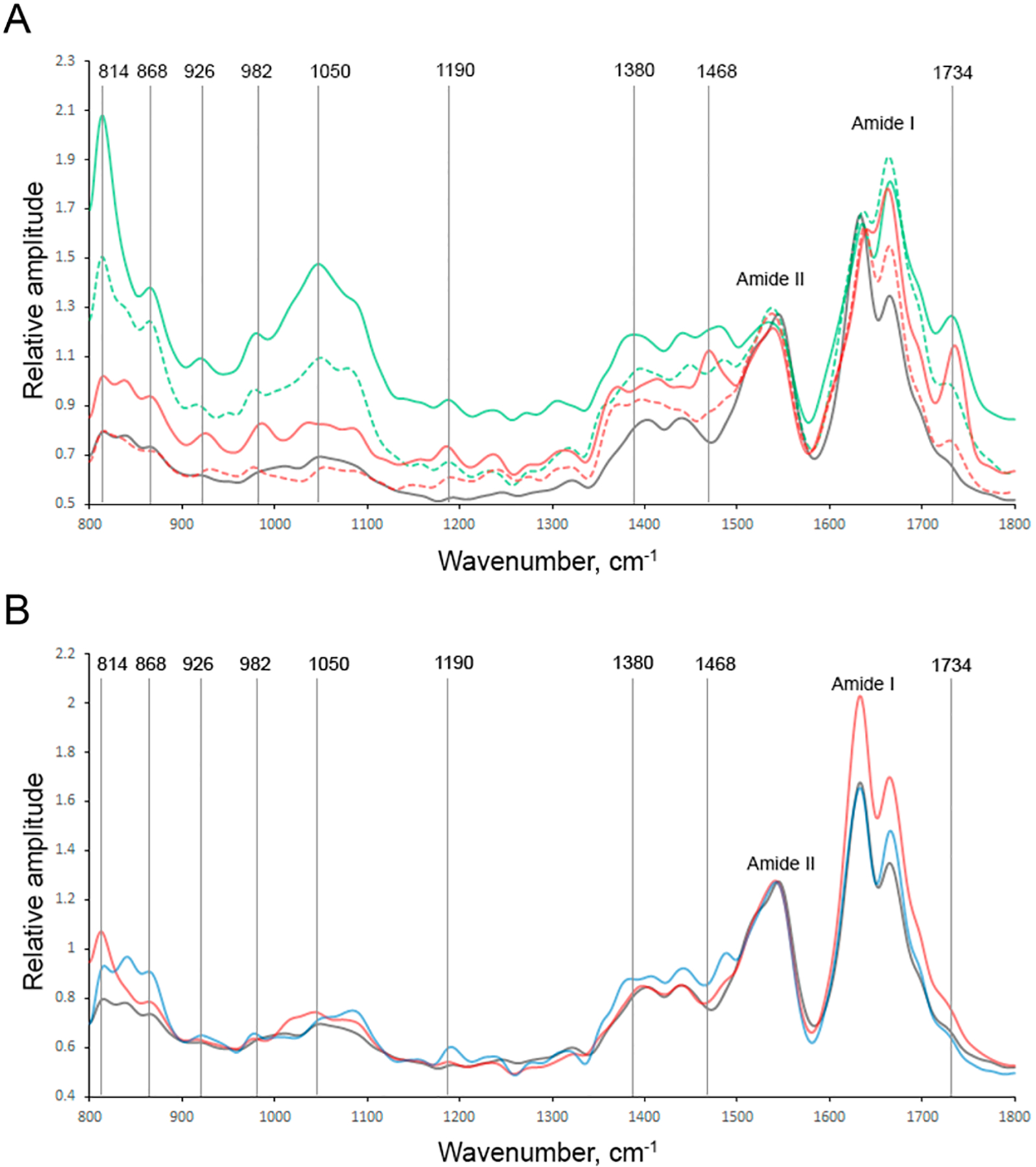
Nanoscale analysis of (A) Ins:PC-u (red) and Ins:PC-s (green) and (B) Ins:CL-u (black) and Ins:CL-s (blue) aggregates. Spectra collected from individual aggregates are shown in the SI. AFM–IR spectra of population A are shown in the corresponding solid, whereas population B is shown by the corresponding dashed lines.
An analysis of the amide I band of the spectra collected from A and B populations of Ins:PC-s revealed substantial differences in their secondary structure. Specifically, population A possessed 38% parallel and 20% antiparallel β-sheets and 42% unordered protein secondary structure, whereas these structures occupied 35, 11, and 54% of population B, respectively. Thus, oligomers of population B contained a substantially greater amount of the unordered protein secondary structure and a significantly smaller amount of the antiparallel β-sheet than oligomers of population A. It should be noted that additional nanoscale imaging analysis is required to fully elucidate the relative abundance of population A and B aggregates as well as the possible additional heterogeneity of protein aggregates in these samples. This work is currently underway in our laboratory.
We also observed two populations of oligomers in Ins:PC-u (Figure 3). AFM–IR spectra collected from population A exhibited vibrational bands at ~800, 1000–1200 and ~1734 cm−1, which had significantly lower intensities than the corresponding bands in the spectra of Ins:PC-s aggregates. This suggests that these oligomers have a significantly smaller amount of lipid present in their structure. It should be noted that population B had a very small amount of lipid, if any, present in the structure. Similar conclusions could be drawn on the basis of the intensities of CH and CH2 vibrations of lipids (1380 and 1460 cm−1) in the spectra of populations A and B of u-PC. Populations A of Ins:PC-s and Ins:PC-u exhibited very similar protein secondary structures. However, population A and population B oligomers of Ins:PC-u appeared to be significantly different. Specifically, population B Ins:PC-u possessed a significantly greater amount of the parallel β-sheet (60%) and a significantly smaller amount of the antiparallel β-sheet (9%) than oligomers of population A (40% parallel β-sheet and 39% antiparallel β-sheet). These results demonstrate that AFM–IR can be used to reveal the polymorphism of amyloid oligomers, which cannot be accessed using conventional CD and FTIR.
AFM–IR analysis of Ins:CL-u and Ins:CL-s aggregates did not reveal any internal structural heterogeneity that was observed for both Ins:PC-u and Ins:PC-s oligomers. We also found that AFM–IR spectra collected from both Ins:CL-u and Ins:CL-s exhibit a set of vibrational bands at 800–900 and 1000–1200 cm−1, which indicates the presence of lipids in their structures. However, we found that the relative intensities of these bands in the spectra collected from Ins:CL-u and Ins:CL-s were different. This points out the different local environments of CL in these aggregates. Thus, although CL is present in both Ins:CL-u and Ins:CL-s, unsaturation of the fatty acids of this lipid drastically alters the binding between the lipid and the protein. The same conclusions can be drawn on the basis of the analysis of the intensity of the ~1734 cm−1 vibration. We have shown that the intensity of this band drastically changes upon protein–lipid interaction.
A detailed analysis of amide I in the spectra collected from Ins:CL-u and Ins:CL-s revealed significant differences in their secondary structures (Figure S7). Specifically, Ins:CL-u contained a substantially greater amount of unordered protein secondary structure (35%) and a significantly lower amount of the antiparallel β-sheet (13%) than Ins:CL-s aggregates (25% unordered and 23% antiparallel β-sheet). It should be noted that all aggregates discussed above that were grown in the presence of u-PC, s-PC, u-CL, and s-CL exhibit distinctly different secondary structure profiles compared to those of insulin aggregates grown in the lipid-free environment.
The question to ask is whether observed structural differences have any biological significance. To answer this question, we investigate the extent to which Ins:CL-u, Ins:CL-s, Ins:PC-u, and Ins:PC-s exert cell toxicity and cause ROS stress in the mice midbrain N27 cell line (Figure 4).
Figure 4.
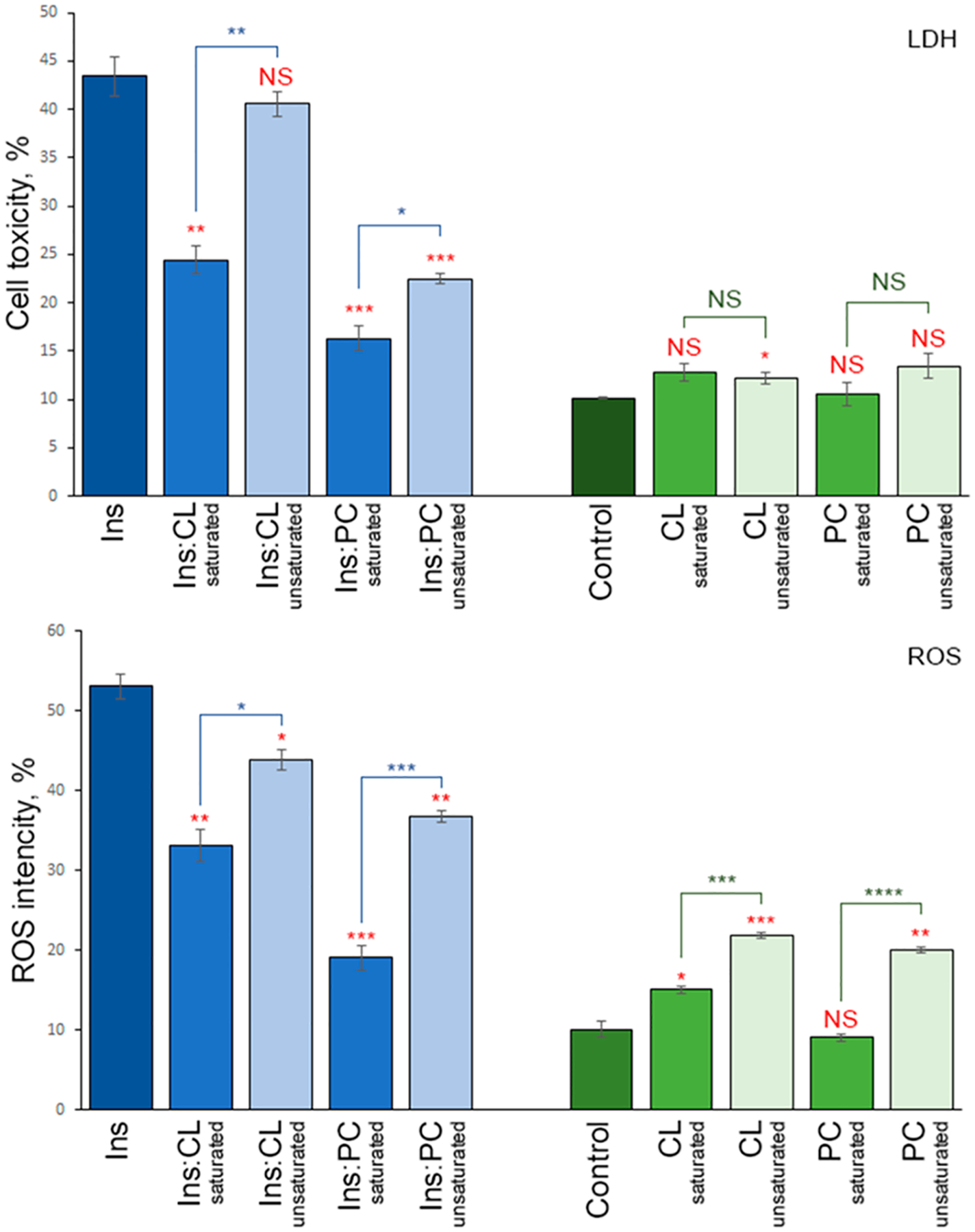
Insulin aggregates grown in the presence of saturated lipids possess cell toxicity different from that of the aggregates grown in the presence of unsaturated lipids. Histograms of (top) LDH and (bottom) ROS assays of Ins, Ins:CL-s, Ins:CL-u, Ins:PC-s, and Ins:PC-u (blue bars) and saturated and unsaturated lipids (green bars) themselves. For LDH and ROS production, error bars represent the means of three replicates. Red asterisks (*) show the significance of the level of difference between Ins and Ins aggregates grown in the presence of lipids as well as between lipid samples and the control. Blue asterisks show the significance of the level of difference between protein samples with saturated lipids and unsaturated lipids. NS is a nonsignificant difference, and *p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001.
The LDH test indicated that Ins:CL-u exerted significantly higher cell toxicity compared to Ins:CL-s. The same conclusion can be drawn for Ins:PC-u and Ins:PC-s oligomers. We also found that the toxicity of Ins:CL-s, Ins:PC-u, and Ins:PC-s is significantly lower than the toxicity exerted by insulin aggregates grown in the lipid-free environment. It should be noted that lipids themselves did not exert any significant cell toxicity. These findings show that lipids can uniquely alter the toxicity of insulin aggregates. Furthermore, insulin aggregates that were formed in the presence of unsaturated lipids exert significantly higher cell toxicity than the aggregates formed in the presence of saturated lipids. It should be noted that lipids themselves, except u-CL, exerted an insignificant LHD response on N27 cells relative to the control (Figure 4). Finally, we point out that the LDH assay detects cell necrosis. Therefore, mode advanced toxicity assays should be utilized in the future to reveal the exact mechanism of the amyloid toxicity of cells.
Similar conclusions could be drawn about the ROS levels exerted by insulin aggregates formed in the presence of unsaturated vs saturated lipids. Specifically, Ins:CL-u exerted significantly higher ROS production compared to that of Ins:CL-s, whereas ROS levels were found to be higher for Ins:PC-u than for Ins:PC-s. It should be noted that u-PC u-CL and s-CL themselves exerted significantly higher levels of ROS in N27 cells compared to their saturated analogs. At the same time, s-PC itself exerts significantly higher ROS levels in N27 cells compared to the ROS levels observed in the control.
The lipid profile of cell and organelle membranes varies across different organs and tissues. For instance, the concentration of major lipids, such as PC, cholesterol, phosphatidylserine, and ceramide, is different in the membranes of astrocytes, microglia, neurons, and oligodendrocytes. Similar variations in the lipid concentrations are also observed for different brain regions: frontal cortex, midbrain, and stratum. The lipid profile of these brain areas changes upon neurodegeneration. Furthermore, fragments of lipid membranes are detected in Lewy bodies, which are extracellular deposits that are formed in Parkinson’s disease. These and other pieces of experimental evidence suggest that lipids can be involved in the destructive biochemical processes that lead to abrupt protein aggregation and amyloid fibril formation, a hallmark of a large group of neurodegenerative diseases.
Our results show that lipids uniquely alter the rates of insulin aggregation. These properties depend on the chemical structure of the lipid. Specifically, negatively charged CL (with a net charge of −2) drastically accelerates the rate of insulin aggregation while simultaneously shortening the lag phase. Furthermore, u-CL has a stronger effect on the observed changes in the rate and lag phase than its saturated analog (s-CL). However, zwitterionic PC (both s- and u-PC) strongly suppresses insulin aggregation.
Our findings show that lipids not only alter the rates of protein aggregation but also uniquely modify the morphology and secondary structures of the formed aggregates. Specifically, in the presence of both u- and s-PC, insulin yields only spherical oligomers, whereas in the presence of u-CL, insulin assembles into long fibrils. None of these structures are evident upon insulin aggregation in the lipid-free environment. The utilization of CD and FTIR revealed major structural differences between PC and CL aggregates. If the former are primarily composed of α-helix and unordered protein secondary structures, then the latter are dominated by the parallel β-sheet. It should be noted that structural differences are also observed between s- and u-PC as well as between s-and u-CL. These findings show that the lipid structure and unsaturation in fatty acids of phospholipids determine the structure and morphology of amyloid aggregates.
We infer than this effect arises from interactions that are developed between charged amino acid residues of proteins and polar headgroups of lipids. It was previously shown that lysine and glutamic acid residues on the N-terminus (amino acids 1–60) of α-Syn exhibit strong electrostatic interactions with headgroups of phospholipids. It was also hypothesized that such interactions trigger α-Syn aggregation.62 Furthermore, NMR and fluorescence methods revealed that lipid–protein interactions are also facilitated by hydrophobic interactions between nonpolar amino acid residues of the protein and the fatty acid tails of lipids.63,64
Our findings also reveal significant differences in ROS production and cell toxicity induced by insulin aggregates formed in the presence of saturated and unsaturated phospholipids. Specifically, toxicities exerted by unsaturated PC and CL on the N27 cell line are significantly higher than the toxicities of their saturated analogs. One can expect that the toxicity of insulin aggregates may correlate with the amount of β-sheet structure. Specifically, insulin fibrils that were grown in the lipid-free environment possess ~82% of both parallel and antiparallel β-sheets (Figure S7), whereas Ins:PC-u and Ins:PC-s aggregates possess ~65 and ~49% of the β-sheet, respectively. These β-sheet distributions correlate with toxicities exerted by these aggregates (Figure 4). However, this conclusion cannot be drawn for Ins:CL-s and Ins:CL-u aggregates. Our results show that Ins:CL-u and Ins:CL-s have very similar amounts of both parallel and antiparallel β-sheets (66% for Ins:CL-u and 74% for Ins:CL-s), which are only slightly lower than the amount of β-sheets in insulin fibrils (82% for Ins) that were grown in the lipid-free environment (Figure S7). However, the toxicity of Ins is approximately twice the toxicity of Ins:CL-s and nearly the same as the toxicity of Ins:CL-u (Figure 4). Thus, we do not observe the relationship between the toxicity of Ins:CL aggregates and the amount of β-sheet in their structure.
The difference in the cell toxicity and ROS production between insulin aggregates that possess saturated and unsaturated CL and PC can be explained by the higher chemical reactivity of unsaturated fatty acids present in the phospholipids. A growing body of evidence shows that under ambient conditions, unsaturated lipids are prone to oxidation, which yields the formation of peroxides and hydroxylated analogs of lipids. This evidence explains the high ROS levels caused by unsaturated lipids themselves (Figure 4). Our findings show that the presence of such unsaturated lipids in insulin aggregates increases their toxicity. At the same time, the presence of their saturated analogs helps to reduce the toxicity of the aggregates relative to the toxicity of insulin aggregates that were grown in the lipid-free environment.
In summary, our experimental findings show that u-CL drastically shortens the lag phase of insulin aggregation compared to the s-CL present at the same lipid-to-protein ratio. Furthermore, we observed the inverse relationship between the quantity of β-sheets and the kinetics of fibril formation. Specifically, insulin fibrils that were grown in the lipid-free environment possess ~82% of both parallel and antiparallel β-sheets (Figure S7), whereas their tlag is 14.4 h. Ins:CL-u and Ins:CL-s possess 66 and 74% β-sheets (Figure S7) with tlag values of 6.1 and 8.1 h, respectively. However, this inverse relationship does not hold true for Ins:CL aggregates. These findings suggest that the lipid environment rather than β-sheet-driven templating is the major driving force of insulin aggregation in the lipid environment.
We also found that structurally and morphologically different aggregates were formed in the presence of s- vs u-CL. Furthermore, insulin aggregates grown in the presence of u-CL exerted higher cell toxicity comparing to the aggregates that were formed in the presence of the saturated phospholipid. At the same time, both Ins:PC-u and Ins:PC-s were able to inhibit insulin aggregation with equivalent efficiency. Similar to CL, structurally different aggregates were formed in the presence of s- and u-PC. These aggregates exerted different cell toxicities. These results show that unsaturated phospholipids catalyze the formation of more toxic amyloid aggregates compared to those formed in the presence of saturated lipids. These findings help to explain the underlying molecular determinant of toxicity and the structural variability of amyloid aggregates that are formed under pathological conditions.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the National Institute of Health for the provided financial support (R35GM142869).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications Web site. The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpclett.2c00559.
DLS results of LUVs analyses; raw AFM–IR spectra collected from individual Ins:PC-u, Ins:PC-s, Ins:CL-u, and Ins:CL-s aggregates; and the results of amide I fitting in the averaged AFM–IR spectra reported in Figure 3 (PDF)
Transparent Peer Review report available (PDF)
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jpclett.2c00559
Contributor Information
Mikhail Matveyenka, Department of Biochemistry and Biophysics, Texas A&M University, College Station, Texas 77843, United States.
Stanislav Rizevsky, Department of Biochemistry and Biophysics, Texas A&M University, College Station, Texas 77843, United States; Department of Biotechnology, Binh Duong University, Thu Dau Mot 820000, Vietnam.
Dmitry Kurouski, Department of Biochemistry and Biophysics and Department of Biomedical Engineering, Texas A&M University, College Station, Texas 77843, United States.
REFERENCES
- (1).van Meer G; Voelker DR; Feigenson GW Membrane Lipids: Where They Are and How They Behave. Nat. Rev. Mol. Cell Biol 2008, 9, 112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Fahy E; Subramaniam S; Murphy RC; Nishijima M; Raetz CR; Shimizu T; Spener F; van Meer G; Wakelam MJ; Dennis EA Update of the Lipid Maps Comprehensive Classification System for Lipids. J. Lipid Res 2009, 50 (Suppl), S9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Fitzner D; Bader JM; Penkert H; Bergner CG; Su M; Weil MT; Surma MA; Mann M; Klose C; Simons M Cell-Type- and Brain-Region-Resolved Mouse Brain Lipidome. Cell Rep 2020, 32, 108132. [DOI] [PubMed] [Google Scholar]
- (4).Pope S; Land JM; Heales SJ Oxidative Stress and Mitochondrial Dysfunction in Neurodegeneration; Cardiolipin a Critical Target? Biochim. Biophys. Acta 2008, 1777, 794–9. [DOI] [PubMed] [Google Scholar]
- (5).Falabella M; Vernon HJ; Hanna MG; Claypool SM; Pitceathly RDS Cardiolipin, Mitochondria, and Neurological Disease. Trends Endocrinol. Metab 2021, 32, 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Michaelson DM; Barkai G; Barenholz Y Asymmetry of Lipid Organization in Cholinergic Synaptic Vesicle Membranes. Biochem. J 1983, 211, 155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sonntag Y; Musgaard M; Olesen C; Schiott B; Møller JV; Nissen P; Thogersen L Mutual Adaptation of a Membrane Protein and Its Lipid Bilayer During Conformational Changes. Nat. Commun 2011, 2, 304. [DOI] [PubMed] [Google Scholar]
- (8).Kayed R; Sokolov Y; Edmonds B; McIntire TM; Milton SC; Hall JE; Glabe CG Permeabilization of Lipid Bilayers Is a Common Conformation-Dependent Activity of Soluble Amyloid Oligomers in Protein Misfolding Diseases. J. Biol. Chem 2004, 279, 46363–6. [DOI] [PubMed] [Google Scholar]
- (9).Jiang Z; de Messieres M; Lee JC Membrane Remodeling by Alpha-Synuclein and Effects on Amyloid Formation. J. Am. Chem. Soc 2013, 135, 15970–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jo E; McLaurin J; Yip CM; St George-Hyslop P; Fraser PE Alpha-Synuclein Membrane Interactions and Lipid Specificity. J. Biol. Chem 2000, 275, 34328–34334. [DOI] [PubMed] [Google Scholar]
- (11).Alza NP; Iglesias Gonzalez PA; Conde MA; Uranga RM; Salvador GA Lipids at the Crossroad of Alpha-Synuclein Function and Dysfunction: Biological and Pathological Implications. Front. Cell Neurosci 2019, 13, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Galvagnion C The Role of Lipids Interacting with α-Synuclein in the Pathogenesis of Parkinson’s Disease. J. Parkins. Dis 2017, 7, 433–450. [DOI] [PubMed] [Google Scholar]
- (13).Galvagnion C; Brown JW; Ouberai MM; Flagmeier P; Vendruscolo M; Buell AK; Sparr E; Dobson CM Chemical Properties of Lipids Strongly Affect the Kinetics of the Membrane-Induced Aggregation of Alpha-Synuclein. Proc. Natl. Acad. Sci. U. S. A 2016, 113, 7065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Dou T; Zhou L; Kurouski D Unravelling the Structural Organization of Individual Alpha-Synuclein Oligomers Grown in the Presence of Phospholipids. J. Phys. Chem. Lett 2021, 12, 4407–4414. [DOI] [PubMed] [Google Scholar]
- (15).Rizevsky S; Kurouski D Nanoscale Structural Organization of Insulin Fibril Polymorphs Revealed by Atomic Force Microscopy-Infrared Spectroscopy (Afm-Ir). Chembiochem 2020, 21, 481–485. [DOI] [PubMed] [Google Scholar]
- (16).Ruggeri FS; Charmet J; Kartanas T; Peter Q; Chia S; Habchi J; Dobson CM; Vendruscolo M; Knowles TPJ Microfluidic Deposition for Resolving Single-Molecule Protein Architecture and Heterogeneity. Nat. Commun 2018, 9, 3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ruggeri FS; Flagmeier P; Kumita JR; Meisl G; Chirgadze DY; Bongiovanni MN; Knowles TPJ; Dobson CM The Influence of Pathogenic Mutations in Alpha-Synuclein on Biophysical and Structural Characteristics of Amyloid Fibrils. ACS Nano 2020, 14, 5213–5222. [DOI] [PubMed] [Google Scholar]
- (18).Ruggeri FS; Longo G; Faggiano S; Lipiec E; Pastore A; Dietler G Infrared Nanospectroscopy Characterization of Oligomeric and Fibrillar Aggregates During Amyloid Formation. Nat. Commun 2015, 6, 7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhou L; Kurouski D Structural Characterization of Individual Alpha-Synuclein Oligomers Formed at Different Stages of Protein Aggregation by Atomic Force Microscopy-Infrared Spectroscopy. Anal. Chem 2020, 92, 6806–6810. [DOI] [PubMed] [Google Scholar]
- (20).Dazzi A; Glotin F; Carminati R Theory of Infrared Nanospectroscopy by Photothermal Induced Resonance. J. Appl. Phys 2010, 107, 124519. [Google Scholar]
- (21).Dazzi A; Prater CB Afm-Ir: Technology and Applications in Nanoscale Infrared Spectroscopy and Chemical Imaging. Chem. Rev 2017, 117, 5146–5173. [DOI] [PubMed] [Google Scholar]
- (22).Kurouski D; Dazzi A; Zenobi R; Centrone A Infrared and Raman Chemical Imaging and Spectroscopy at the Nanoscale. Chem. Soc. Rev 2020, 49, 3315–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ruggeri FS; Mannini B; Schmid R; Vendruscolo M; Knowles TPJ Single Molecule Secondary Structure Determination of Proteins through Infrared Absorption Nanospectroscopy. Nat. Commun 2020, 11, 2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lu F; Jin MZ; Belkin MA Tip-Enhanced Infrared Nanospectroscopy Via Molecular Expansion Force Detection. Nat. Photonics 2014, 8, 307–312. [Google Scholar]
- (25).Ruggeri FS; Benedetti F; Knowles TPJ; Lashuel HA; Sekatskii S; Dietler G Identification and Nanomechanical Characterization of the Fundamental Single-Strand Protofilaments of Amyloid Alpha-Synuclein Fibrils. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 7230–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ruggeri FS; Vieweg S; Cendrowska U; Longo G; Chiki A; Lashuel HA; Dietler G Nanoscale Studies Link Amyloid Maturity with Polyglutamine Diseases Onset. Sci. Rep 2016, 6, 31155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ramer G; Ruggeri FS; Levin A; Knowles TPJ; Centrone A Determination of Polypeptide Conformation with Nanoscale Resolution in Water. ACS Nano 2018, 12, 6612–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Farber C; Li J; Hager E; Chemelewski R; Mullet J; Rogachev AY; Kurouski D Complementarity of Raman and Infrared Spectroscopy for Structural Characterization of Plant Epicuticular Waxes. ACS Omega 2019, 4, 3700–3707. [Google Scholar]
- (29).Farber C; Wang R; Chemelewski R; Mullet J; Kurouski D Nanoscale Structural Organization of Plant Epicuticular Wax Probed by Atomic Force Microscope Infrared Spectroscopy. Anal. Chem 2019, 91, 2472–2479. [DOI] [PubMed] [Google Scholar]
- (30).Dazzi A Photothermal Induced Resonance. Application to Infrared Spectromicroscopy. In Thermal Nanosystems and Nano-materials; Volz S, Ed.; Springer: Berlin, 2009; Vol. 118, pp 469–503. [Google Scholar]
- (31).Perez-Guaita D; Kochan K; Batty M; Doerig C; Garcia-Bustos J; Espinoza S; McNaughton D; Heraud P; Wood BR Multispectral Atomic Force Microscopy-Infrared Nano-Imaging of Malaria Infected Red Blood Cells. Anal. Chem 2018, 90, 3140–3148. [DOI] [PubMed] [Google Scholar]
- (32).Dazzi A; Prazeres R; Glotin F; Ortega JM; Al-Sawaftah M; de Frutos M Chemical Mapping of the Distribution of Viruses into Infected Bacteria with a Photothermal Method. Ultramicroscopy 2008, 108, 635–641. [DOI] [PubMed] [Google Scholar]
- (33).Mayet C; Deniset-Besseau A; Prazeres R; Ortega JM; Dazzi A Analysis of Bacterial Polyhydroxybutyrate Production by Multimodal Nanoimaging. Biotechnol. Adv 2013, 31, 369–374. [DOI] [PubMed] [Google Scholar]
- (34).Kochan K; Perez-Guaita D; Pissang J; Jiang JH; Peleg AY; McNaughton D; Heraud P; Wood BR Vivo Atomic Force Microscopy-Infrared Spectroscopy of Bacteria. J. Royal Soc. Interface 2018, 15, 20180115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wieland K; Ramer G; Weiss VU; Allmaier G; Lendl B; Centrone A Nanoscale Chemical Imaging of Individual Chemo-therapeutic Cytarabine-Loaded Liposomal Nanocarriers. Nano Res 2019, 12, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Strelcov E; Dong Q; Li T; Chae J; Shao Y; Deng Y; Gruverman A; Huang J; Centrone A Ch3nh3pbi3 Perovskites: Ferroelasticity Revealed. Sci. Adv 2017, 3, No. e1602165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Banerjee S; Sun Z; Hayden EY; Teplow DB; Lyubchenko YL Nanoscale Dynamics of Amyloid Beta-42 Oligomers as Revealed by High-Speed Atomic Force Microscopy. ACS Nano 2017, 11, 12202–12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Watanabe-Nakayama T; Ono K; Itami M; Takahashi R; Teplow DB; Yamada M High-Speed Atomic Force Microscopy Reveals Structural Dynamics of Amyloid Beta1–42 Aggregates. Proc. Natl. Acad. Sci. U. S. A 2016, 113, 5835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Bhattacharya M; Mukhopadhyay S Nanophotonics of Protein Amyloids. Nanophotonics 2014, 3, 51–59. [Google Scholar]
- (40).Deckert-Gaudig T; Kämmer E; Deckert V Tracking of Nanoscale Structural Variations on a Single Amyloid Fibril Tip-Enhanced Raman Scattering. J. Biophotonics 2012, 5, 215–219. [DOI] [PubMed] [Google Scholar]
- (41).Krasnoslobodtsev AV; Deckert-Gaudig T; Zhang Y; Deckert V; Lyubchenko YL Polymorphism of Amyloid Fibrils Formed by a Peptide from the Yeast Prion Protein Sup35: Afm and Tip-Enhanced Raman Scattering Studies. Ultramicroscopy 2016, 165, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Kurouski D; Deckert-Gaudig T; Deckert V; Lednev IK Structure and Composition of Insulin Fibril Surfaces Probed by Ters. J. Am. Chem. Soc 2012, 134, 13323–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Kurouski D; Deckert-Gaudig T; Deckert V; Lednev IK Surface Characterization of Insulin Protofilaments and Fibril Polymorphs Using Tip-Enhanced Raman Spectroscopy (Ters). Biophys. J 2014, 106, 263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Lipiec E; Perez-Guaita D; Kaderli J; Wood BR; Zenobi R Direct Nanospectroscopic Verification of the Amyloid Aggregation Pathway. Angew. Chem., Int. Ed 2018, 130, 8655–8660. [DOI] [PubMed] [Google Scholar]
- (45).Zhang Y; Hashemi M; Lv Z; Williams B; Popov KI; Dokholyan NV; Lyubchenko YL High-Speed Atomic Force Microscopy Reveals Structural Dynamics of Alpha-Synuclein Monomers and Dimers. J. Chem. Phys 2018, 148, 123322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Kakinen A; et al. Single-Molecular Heteroamyloidosis of Human Islet Amyloid Polypeptide. Nano Lett 2019, 19, 6535–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Kitts CC; Vanden Bout DA Near-Field Scanning Optical Microscopy Measurements of Fluorescent Molecular Probes Binding to Insulin Amyloid Fibrils. J. Phys. Chem. C 2009, 113, 12090–12095. [DOI] [PubMed] [Google Scholar]
- (48).Paulite M; Fakhraai Z; Li IT; Gunari N; Tanur AE; Walker GC Imaging Secondary Structure of Individual Amyloid Fibrils of a Beta2-Microglobulin Fragment Using near-Field Infrared Spectroscopy. J. Am. Chem. Soc 2011, 133, 7376–83. [DOI] [PubMed] [Google Scholar]
- (49).vandenAkker CC; Deckert-Gaudig T; Schleeger M; Velikov KP; Deckert V; Bonn M; Koenderink GH Nanoscale Heterogeneity of the Molecular Structure of Individual Hiapp Amyloid Fibrils Revealed with Tip-Enhanced Raman Spectroscopy. Small 2015, 11, 4131–9. [DOI] [PubMed] [Google Scholar]
- (50).Paulite M; Blum C; Schmid T; Opilik L; Eyer K; Walker GC; Zenobi R Full Spectroscopic Tip-Enhanced Raman Imaging of Single Nanotapes Formed from Beta-Amyloid(1–40) Peptide Fragments. ACS Nano 2013, 7, 911–20. [DOI] [PubMed] [Google Scholar]
- (51).D’Souza A; Theis JD; Vrana JA; Buadi F; Dispenzieri A; Dogan A Localized Insulin-Derived Amyloidosis: A Potential Pitfall in the Diagnosis of Systemic Amyloidosis by Fat Aspirate. Am. J. Hematol 2012, 87, E131–2. [DOI] [PubMed] [Google Scholar]
- (52).Gupta Y; Singla G; Singla R Insulin-Derived Amyloidosis. Indian J. Endocrinol. Metab 2015, 19, 174–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Shikama Y; Kitazawa J; Yagihashi N; Uehara O; Murata Y; Yajima N; Wada R; Yagihashi S Localized Amyloidosis at the Site of Repeated Insulin Injection in a Diabetic Patient. Int. Med 2010, 49, 397–401. [DOI] [PubMed] [Google Scholar]
- (54).Iwaya K; et al. Toxicity of Insulin-Derived Amyloidosis: A Case Report. BMC Endocr. Dis 1991, 19, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Kurouski D; Luo H; Sereda V; Robb FT; Lednev IK Rapid Degradation Kinetics of Amyloid Fibrils under Mild Conditions by an Archaeal Chaperonin. Biochem. Biophys. Res. Commun 2012, 422, 97–102. [DOI] [PubMed] [Google Scholar]
- (56).De Simone A; Naldi M; Tedesco D; Milelli A; Bartolini M; Davani L; Widera D; Dallas ML; Andrisano V Investigating in Vitro Amyloid Peptide 1–42 Aggregation: Impact of Higher Molecular Weight Stable Adducts. ACS Omega 2019, 4, 12308–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Kurouski D; Lombardi RA; Dukor RK; Lednev IK; Nafie LA Direct Observation and Ph Control of Reversed Supramolecular Chirality in Insulin Fibrils by Vibrational Circular Dichroism. Chem. Commun 2010, 46, 7154–6. [DOI] [PubMed] [Google Scholar]
- (58).Sarroukh R; Goormaghtigh E; Ruysschaert JM; Raussens V Atr-Ftir: A “Rejuvenated” Tool to Investigate Amyloid Proteins. Biochim. Biophys. Acta 2013, 1828, 2328–38. [DOI] [PubMed] [Google Scholar]
- (59).Barth A Infrared Spectroscopy of Proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [DOI] [PubMed] [Google Scholar]
- (60).Dou T; Li Z; Zhang J; Evilevitch A; Kurouski D Nanoscale Structural Characterization of Individual Viral Particles Using Atomic Force Microscopy Infrared Spectroscopy (Afm-Ir) and Tip-Enhanced Raman Spectroscopy (Ters). Anal. Chem 2020, 92, 11297–11304. [DOI] [PubMed] [Google Scholar]
- (61).Rizevsky S; Matveyenka M; Kurouski D Nanoscale Structural Analysis of a Lipid-Driven Aggregation of Insulin. J. Phys.s Chem. Lett 2022, 13, 2467–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Viennet T; et al. Structural Insights from Lipid-Bilayer Nanodiscs Link Alpha-Synuclein Membrane-Binding Modes to Amyloid Fibril Formation. Commun. Biol 2018, 1, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Giasson BI; Murray IV; Trojanowski JQ; Lee VM A Hydrophobic Stretch of 12 Amino Acid Residues in the Middle of Alpha-Synuclein Is Essential for Filament Assembly. J. Biol. Chem 2001, 276, 2380–6. [DOI] [PubMed] [Google Scholar]
- (64).Ueda K; Fukushima H; Masliah E; Xia Y; Iwai A; Yoshimoto M; Otero DA; Kondo J; Ihara Y; Saitoh T Molecular Cloning of Cdna Encoding an Unrecognized Component of Amyloid in Alzheimer Disease. Proc. Natl. Acad. Sci. U. S. A 1993, 90, 11282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


