Abstract
Acute pulmonary injury, or acute respiratory distress syndrome, has a high incidence in elderly individuals and high mortality in its most severe degree, becoming a challenge to public health due to pathophysiological complications and increased economic burden. Acute pulmonary injury can develop from sepsis, septic shock, and pancreatitis causing reduction of alveolar airspace due to hyperinflammatory response. Oxidative stress acts directly on the maintenance of inflammation, resulting in tissue injury, as well as inducing DNA damages. Once the DNA is damaged, enzymatic DNA repair mechanisms act on lesions in order to maintain genomic stability and, consequently, contribute to cell viability and homeostasis. Although palliative treatment based on mechanical ventilation and antibiotic using have a kind of efficacy, therapies based on modulation of DNA repair and genomic stability could be effective for improving repair and recovery of lung tissue in patients with acute pulmonary injury.
Keywords: Acute pulmonary injury, DNA repair, Hyperinflammatory response, Oxidative stress, Genomic stability
1. Introduction
At least 2 billion people in the world are exposed to some toxic component in the air, such as particles, chemicals and infectious microorganisms, making lungs vulnerable to injuries, causing respiratory incapacity and/or death [1].
Acute lung injury (ALI), described in 1967 by David R. Ashbaugh as a complex syndrome, is a syndrome that presents small variations in severity when compared to those from acute respiratory distress syndrome (ARDS) [2].Therefore, ALI causes respiratory insufficiency not fully explained by heart failure or fluid overload also bilateral opacities not fully explained by spills, pulmonary collapse or nodules. This syndrome is characterized by increased permeability, as reflected in alveolar edema, due to epithelial and endothelial cell damage and neutrophil infiltration [3].
Epidemiology of ALI presents variations considering the difficulty for obtaining a precise diagnosis, omission in the notification of cases and regions where these cases occur in different countries [4]. However, in the United States, there is an average of 79 cases per 100,000 individuals per year [4,5] with clear seasonal variations and mostly in winter. The increase in age, in range of 75–84 years, generates highest incidences reaching about 300 individuals per 100,000 per year [5], achieving 190 thousand patients hospitalized annually, which causes high costs for the health system in USA [6,7].
Inflammatory processes increase production of free radicals in lungs and defense cells, which could establish oxidative stress and, in consequence, DNA damages [8]. Free radicals generate direct damage in DNA molecule, or as second messengers, which alter gene expression at specific pathways in cellular signaling [9].
The integrity of genome is achieved after impact of different types of free radicals [10], by enzymatic DNA repair mechanism of the three excision repair pathways, base excision repair, nucleotide excision repair and mismatch repair.
2. Pathophysiology of acute lung injury
ALI and ARDS are spectra of same potentially fatal disease characterized by uncontrolled hyperinflammatory responses in lungs [11], as defined by Berlin Conference in 2012, as shown in Table 1 , their categories and mortalities. Some parameters used for definition of this disease were described by American-European Consensus Conference as a syndrome of acute respiratory failure. These parameters were characterized by bilateral pulmonary infiltrate on chest radiography, compatible with pulmonary edema, occurrence of severe hypoxemia, even as proportion of arterial oxygen to inspired oxygen fraction (PaO2/FiO2) ≤ 300 mmHg, regardless of final positive end-expiratory pressure (PEEP), pulmonary artery occlusion pressure ≤ 18 mmHg and absence of clinical or echocardiographic signs of left atrial hypertension [2,12].
Table 1.
Berlin categories and mortality for ALI/ARDS.
| Categories | Oxygenation | Mortality (%) |
|---|---|---|
| Mild | 200 mmHg < PaO2/FIO2 ≤300 mmHg with PEEP or CPAP ≥ 5 cm H2O | 27 |
| Moderate | 100 mmHg < PaO2/FIO2 ≤200 mmHg with PEEP or CPAP ≥ 5 cm H2O | 32 |
| Severe | PaO2/FIO2 ≤100 mmHg with PEEP ≥ 5 cm H2O | 45 |
Abbreviations: PEEP - positive end-expiratory pressure; CPAP - continuous positive airway pressure; PaO2 – partial pressure of arterial oxygen; FIO2 - fraction of inspired oxygen.
The ALI/ARDS risk factors were classified as direct and indirect. Direct risk factors, or direct lung injury, include pneumonia, aspiration of contents, inhalation injury, lung contusion, pulmonary vasculitis, non-pulmonary sepsis, multiple trauma, pancreatitis, non-cardiogenic shock, drug overdose and the risk factor for indirect lung injury, to acute lung injury associated with transfusion [13].
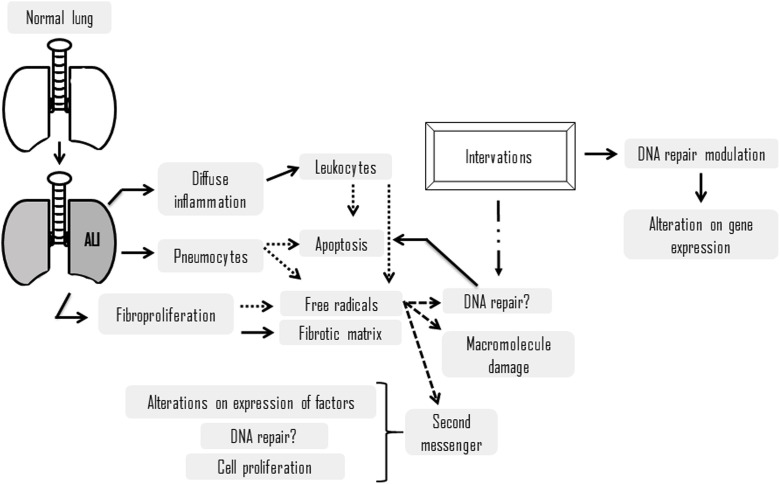
Pathogenesis at cellular and molecular levels can occur in alveolar epithelium or in microvascular endothelium [14], the first one characterizing alveolar ALI and the second one for extrapulmonary, being the most common, which could occur after sepsis, pancreatitis and/or septic shock [15]. However, the development of disease is relatively uniform with diffuse inflammation in lung tissue [14].
The inflammatory process leads to formation of alveolar and interstitial edema, which can progress to pulmonary fibrosis [14,15]. Inflammatory cells, especially neutrophils, persist with secretion of mediators, which prevent the resolution of inflammation [16]. These are the main cause of death in patients with ALI [16,17].
In sepsis conditions, inflammatory response is initiated by recognition of microbial antigens by specific receptors located on surface of macrophages, such as TLR2 and TLR4 receptors (toll-like receptor types 2 and 4, respectively) [18]. Among these microbial antigens, bacterial cell walls components such as lipopolysaccharide (LPS) of gram-negative bacteria [19]. Endothelial cells from alveolar capillaries and type I pneumocytes (alveolar epithelial cells) are impaired [16] and, with loss of normally rigid alveolar barrier, these are exposed to fluids and macromolecules, alveolar epithelium, capillary endothelium, extracellular matrix (ECM) and other cells, such as alveolar macrophages and fibroblasts [20].
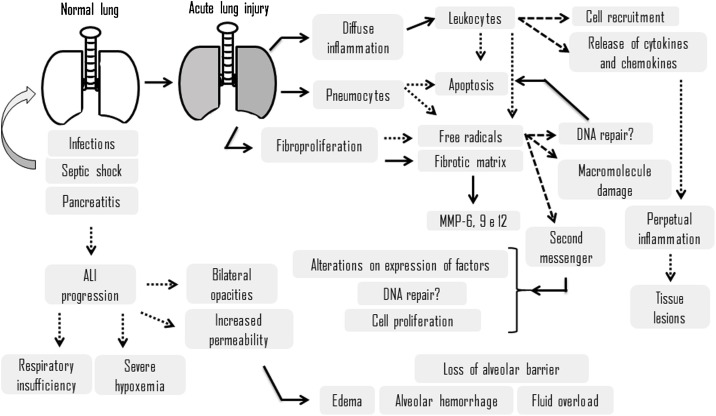
Cell-matrix interaction influences a number of cellular processes, including survival, migration, proliferation and differentiation [21]. In interstitial and alveolar space occurs accumulation of a rich protein liquid, cytokines (IL-1, IL-8 and TNF-a) as well as lipid mediators, such as leukotrienes B4, which are present in lung at onset of disease progression [22]. These proinflammatory proteins and mediators cause leukocytes, mainly neutrophils, to migrate into the interstitium and pulmonary alveoli [23], whereby exposure to microorganisms, an inflammatory process is established, neutrophils are activated, which migrate from the vessels to alveoli (Fig. 1 ). The extracellular matrix is remodeled and there is extravasation of red blood cells. Within the alveoli, intense production of proinflammatory cytokines, production of free radicals and proteases. Fibroblasts produce metalloproteinases degrading the matrix, and the pneumocytes begin to undergo apoptosis, which is associated with production and release by leukocytes. This matrix remodeling process causes reduction of alveolar airspace.
Fig. 1.
Schematic representation of factors during progression of acute lung injury and theirconsequences. Dotted lines determine a tendency toward, while dashed lines determine a consequence.
Condensed plasma proteins aggregate in air spaces with cellular debris and dysfunctional pulmonary surfactant to form hyaline membrane spirals [24]. In addition, pulmonary vascular injury occurs at the beginning of ALI, causing a vascular obstruction and fibrocellular proliferation [23].
Alveolar edema involves portions of lung leading to decreased aeration and atelectasis [22,24]. Consequently, hypoxemia develops and respiratory difficulty increases leading to dyspnea as well as exacerbation of pathophysiological changes in alveolar spaces, which are caused by microvascular occlusion, reducing pulmonary blood flow, in addition to severe hypoxemia, hypercapnia secondary to increased lung space also become prominent in ALI [25].
The progression of ALI is characterized into three sequential phases: exudative, proliferative and fibrotic. In the exudative phase, which occurs between the first and the seventh day after exposure to a risk factor, the individuals have respiratory symptoms, such as dyspnea [24]. Chest radiography images usually reveal alveolar and interstitial opacities [22]. In differential diagnosis of ALI, the most common disorders are cardiogenic pulmonary edema, diffuse pneumonia and alveolar hemorrhage [24]. In proliferative phase, comprising seventh up to twenty-first day, most individuals recover and mechanical ventilation is discontinued, despite that many individuals present dyspnea and hypoxemia yet [16]. In fact, some of these can develop progressive lung injury and early changes in pulmonary fibrosis during this phase [23]. Histologically, repair of lung tissue begins and there is a composition of alveolar exudates and change from pulmonary infiltrate of neutrophils to lymphocyte infiltrate [22]. There is a proliferation of type II pneumocytes and synthesis of new pulmonary surfactants [22]. In addition, there is presence of type III alveolar procollagen peptide, a marker of pulmonary fibrosis, associated with a prolonged clinical course and increased mortality [16]. While some individuals present lung function recovering within a period of three to four weeks after initial lung injury, others will enter the third phase, the fibrotic, requiring support by mechanical ventilation and/or supplemental oxygen [23,24]. Presence of extensive alveolar ducts and interstitial fibrosis are characteristic of this proliferative phase as well as alterations in the acinar architecture of the alveolar ducts, leading to changes similar to those seen in emphysema with large blisters [24]. There is progressive vascular occlusion and pulmonary hypertension, including risk of pneumothorax and lung space alterations with consequence in diseases and mortality increase [24].
Although there are palliative treatments, such as mechanical ventilation and antibiotic use, several host factors can influence the ALI pathogenesis, such as immunodeficiencies, genetic susceptibility, infections and environmental factors [26].
3. Animal models
Pulmonary parenchymal injury induced by an insult as sepsis results in ALI or ARDS, causing respiratory failure which can lead to death [27]. ALI pathogenesis is complex and takes days or weeks to resolve. Despite a lot of research, there is only one current therapy based on low tidal volume ventilation [28].
Animal models could be used to simulate patients with development of ALI. However, due to great difficulty for obtaining a relation of ALI characteristics in experimental animal models, even with clinical criteria established in humans that can be translated into laboratory animals [29,30], some of procedures are technically complicated and specialized equipment is required.
The experimental models based on primates are closer to complexity of ALI, but these require specialized and expensive installations [27], so the main models are still rodents, as mice and rats. Rodents are accessible, can be genetically manipulated and ALI can be induced by direct or indirect techniques [31]. In human ALI or ARDS can develop from pneumonia and sepsis, for example. These conditions are used in mice and rats models by inoculation of gram-negative bacterial endotoxin or lipopolysaccharide (LPS) [32]. These can be administered directly in lungs by intratracheal injection or inhalation [33] or intraperitoneal via [34], to mimic a systemic inflammation, such that induced by sepsis. Dejager and coworkers [35] reported that LPS administration is considered a highly controllable and simple experimental model that induces an acute response. However, LPS-mediated signaling is strictly TLR4-dependent, which could not reflect all complex human physiological responses.
Another experimental model proposed is based on lung injury induced by mechanical ventilation [36]. This experimental model correlates with lung injury induced by human ventilator of an additional stimulus or extremely high tidal volumes, but this model does not induce substantial lung injury in mice [37].
A widely experimental model used to simulate sepsis condition culminating in acute lung injury is based on cecal ligation and puncture [38]. It is considered a simple procedure, with presence of infectious focus, because it is continuous polymicrobial sepsis, which recreates the precession of human sepsis with hemodynamic and metabolic phases [35].
Wan and coworkers [39] have demonstrated that lung injury caused by cobalt nanoparticles generates oxidative stress, resulting in damage and mutations in the DNA molecule. Studies discuss several animal models of ALI with objective of assisting in choosing the most suitable experimental model for specific scientific questions, as reported by Matute-Bello and coworkers [31].
However, there is no reliable experimental model that present all human ALI features. In fact, human ALI is not based on a single event [40] and differences between human ALI and experimental models could arise from variability of interspecies innate immunity [31].
Table 2 shows the advantages and disadvantages of three of these experimental models for ALI induction according to Ballard-Croft and coworkers [41] and Sipahi & Ataly [42]. Despite these limitations, the use of experimental models is essential to study a number of human diseases, as there is no substitute for testing therapeutic interventions in individuals affected by these diseases.
Table 2.
Summary of advantages and disadvantages of three models of acute lung injury induction.
| Model | Advantage | Disadvantage |
|---|---|---|
| LPS | Good model of sepsis. | Lung injury does not mimic human ALI/ARDS (minimal intra-alveolar neutrophilic infiltrates and protein-rich alveolar edema). |
| Hyperoxia | Good model of hemorrhagic injury. | Less intra-alveolar neutrophilic infiltrates than in human ARDS. |
| Mesenteric ischemia reperfusion | Good models of sepsis. | Injury is localized primarily to the vascular and interstitial compartments of the lung. |
4. Oxidative stress in acute lung injury
Inflammation is a natural defense mechanism, in response to harmful factors, such as infections, resulting in tissue repair [43]. ALI and other acute diseases are related to increased free radical production, which could cause oxidation of lipids, proteins and nucleic acids and these are recognized as inflammatory signals [44].
Atoms and molecules containing unpaired electrons in their last electron layer are highly reactive and termed free radicals [45]. They are produced as by-products during metabolism, exposure to sunlight (ultraviolet light), ionizing radiation and toxic chemicals [45]. Among free radicals, there are reactive species of oxygen (ROS) and nitrogen (RNS) [45].
It suggests that inflammation can worsen clinical aspects due to increased oxidative stress by release of proteins oxidized from macrophages, such as peroxiredoxin 2 (PRDX2), which triggers TNF-α production in extracellular environment, indicating that these mechanisms depend on redox in oxidative cascades and can induce inflammation [44], making oxidative stress and inflammation a vicious circle.
Infectious conditions that cause lung injury due to development of sepsis, which manifests as ALI or ARDS, increase free radical levels in lung cells [16,46]. These free radicals cause oxidative stress altering the mitochondrial DNA, by generation of bulky adducts and oxidized bases, alteration of protein structure [47] and lipid peroxidation in cell membranes, as plasmatic membrane, endoplasmic reticulum and Golgi complex [48].
ROS and RNS act on NF-kB activation (nuclear kappa factor B) and AP-1 (activating protein 1) promoting cell proliferation and survival [49,50] and they are related to stabilization of HIF-1α (hypoxia induced factor 1 alpha), culminating in transcription of several genes involved in metabolism, survival and cell death, angiogenesis, invasion and metastasis [51] and in regulation of activated protein kinase by AMP serine/threonine (AMPK), which contributes to energy metabolism [52].
In conditions of increased inflammatory response, as those that occur in ALI, neutrophils and macrophages are the main cells for producing ROS [53,54]. However, there is also increased ROS production in endothelial and epithelial cells from the lung tissue [8,55].
In addition, high ROS levels in hypercapnia, acidosis and pulmonary hypertension increase oxidative stress and risk of cell death [11,16] in patients with septicemia leads to reduction of plasma volume and reduction of total antioxidant capacity [56].
Other studies also indicate that glutathione peroxidase (GPx) is an important marker for oxidative stress in ALI, correlates with PaO2/FiO2 and oxidation index initially and in disease progression correlates with PEEP (positive expiratory pressure final) used for treatment of inflammatory lung diseases [57]. Sarkele and coworkers [58] reported that patients with ALI have higher concentrations of GSSG (the oxidized form of glutathione) compared to control group, despite high levels of F2-isoprostane (a marker of lipid peroxidation), which indicates oxidative stress in patients with ALI [58].
Lee and coworkers [59] suggested that there is evidence pointing to oxidative mitochondrial damage in lung injury models and this could be considered a molecular sentinel in context of oxidative stress. These authors showed that there is an association between mitochondrial DNA damage and cell death by ROS. In another study was reported that molecular patterns associated with pro-inflammatory mitochondrial damage (TMDs), including mitochondrial deoxyribonucleic acid (mtDNA) fragments and mtDNA-associated proteins, are released into the circulation of severely diseased patients by inflammatory diseases, which could occur in acute lung injury [60]. In addition, it was showed that large numbers of mtDNA damage-associated molecular patterns (DAMPs) in blood transfusions could predict the development of ALI/ARDS [59]. Other authors have demonstrated in culture animal cells, organs and animals that mtDNA DAMPs cause endothelial injury and pulmonary edema [61,62]. Also, Ruchko and coworkers [63] demonstrated that extent of mtDNA damage determines whether endothelial cells from the pulmonary artery survive or activate an apoptotic death pathway, and this refers to mechanism by which mtDNA damage initiates an apoptotic cascade.
Under oxidative stress conditions, free radicals could act as second messengers in specific pathways of cellular signaling, immune function and signal transduction, affecting cellular homeostasis [64]. On the other hand, these free radicals can react directly with the DNA molecule, causing alterations in the nitrogenous bases, oxidizing them [65]. However, after damage, the DNA molecule can be repaired by enzymatic mechanisms [66], which is crucial for the maintenance of genome integrity [67]. Table 3 summarizes some studies that relate ALI, DNA damage and DNA repair mechanisms.
Table 3.
Summary of studies on the relationship between acute lung injury and DNA damage/DNA repair mechanisms.
| First author | Title | Reference | Observations and Conclusion |
|---|---|---|---|
| Fu H 2017 | Calcitonin gene-related peptide protects type II alveolar epithelial cells from hyperoxia-induced DNA damage and cell death | Exp Ther Med 13 (4) 1279-1284 | Exposure to 60% oxygen for 24 h predisposes to oxidative damage in cells of the alveolar epithelium, including DNA damage and apoptosis; however, the peptide related to the exogenous calcitonin gene attenuated the hyperoxic lesion and exerted a cytoprotective effect, suggesting that the positive regulation of expression may represent an alternative for the prevention of hyperoxia-induced lung injury. |
| Barker GF 2006 | DNA damage induced by hyperoxia: quantitation and correlation with lung injury | Am J Respir Cell Mol Biol 35 (3) 77-288 | Exposure to oxygen caused damage to the DNA base of lung cells as a primary effect of the attack of reactive oxygen species on DNA, which implies the ability of inhaled oxidants to alter the lung genome. Prolonged exposure led to DNA tape breaks suggesting that a side effect of activated nucleases during cell death is likely and correlated with lung disease progression, suggesting that cell injury plays a key role in oxygen pulmonary toxicity. |
| Wan R 2017 | Cobalt nanoparticles induce lung injury, DNA damage and mutations in mice | Part Fibre Toxicol 14 (1) 38 | Exposure to cobalt nanoparticles caused oxidative stress, inflammation and lung injury and cell proliferation, which resulted in DNA damage and DNA mutation with great frequency, especially in the G:C to T:A transversion, which can be explained by the greater formation of 8-OHdG induced by the nanoparticles. |
| Simmons JD 2017 | Potential contribution of mitochondrial DNA damage associated molecular patterns in transfusion products to the development of acute respiratory distress syndrome after multiple transfusions | J Trauma Acute Care Surg 82 (6) 1023-1029 | Fresh frozen plasma and platelets contain large amounts of extracellular mitochondrial DNA (mtDNA) that the amount of mAMDs associated with mtDNA administered during transfusion may be a contributor to serum mtDNA DAMP levels, and that serum levels of mtDNA DAMPs after multiple transfusions may predict the development of ARDS. Therefore, serum measurements of mtDNA DAMP may be predictive biomarkers for the evolution of ARDS. |
| Lee YL 2017 | Mitochondrial DNA Damage Initiates Acute Lung Injury and Multi-Organ System Failure Evoked in Rats by Intra-Tracheal Pseudomonas Aeruginosa | Shock 48 (1) 54-60 | mt-targeted Ogg1 suppresses bacterial-induced lung tissue mtDNA damage and ALI accompanied by attenuation of multiple organ failure and lethality is strong evidence that mitochondrial genomic integrity is a critical step driving injury propagation. |
| Sergio LPS 2019 | Low-power laser alters mRNA levels from DNA repair genes in acute lung injury induced by sepsis in Wistar rats. | Lasers Med Sci. 34 (1) 157-168 | Acute lung injury alters the mRNA levels of DNA repair genes, understanding that it would be part of the cellular response to sepsis, and after treatment with low-power infrared laser, mRNA levels of DNA repair genes are modulated in lung injury acute. |
5. Maintenance of genome after DNA damage
Since there is damage in DNA molecule, different repair pathways can be activated, one of them is specific for oxidative damages, but the other could also act on these DNA damages. Described in the 1960’s, excision repair can be divided into two: base excision repair and nucleotide excision repair [68,69]. The base excision repair pathway has a strong relation with the mismatch DNA repair pathway, which could act on oxidized bases, besides acting in recognition and repair of base numerical alterations [70].
5.1. Base excision repair (BER)
Oxidative damages in the DNA molecule can be caused mainly by ROS, generating damages in nitrogenous bases and simple chain breaks [71]. The increase in basal level of oxidative damages in DNA is associated with several age-related diseases, including cardiovascular diseases [72], cancer [73] and neurodegenerative diseases [74]. The main oxidized bases are 8-oxo-guanine (8-oxoG) and 5-hydroxycytosine, which unpaired with adenine and thymine, respectively, are the most commonly oxidized bases [75]. As previously described, ALI contributes for increasing of free radicals, causing oxidative stress, which in turn could damage the DNA molecule and, consequently, activate BER mechanism [76], which acts on single strand breaks, oxidized bases, and on relatively bulky adducts [77].
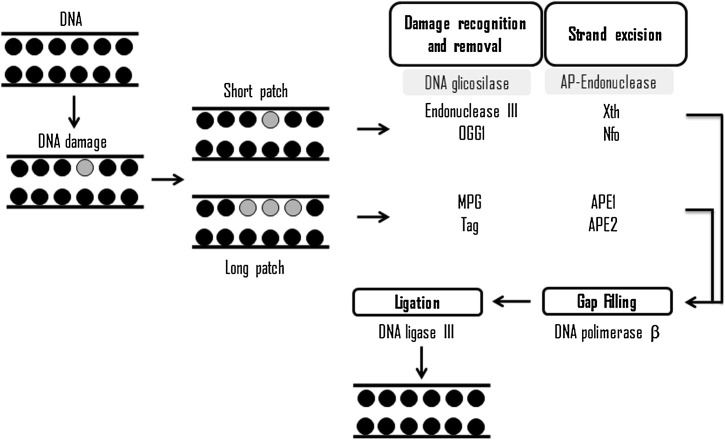
Evolutionarily conserved, BER has high functional sequence homology from bacteria to mammals presenting two pathways: short patch BER pathway to repair a single damaged base and long patch BER pathway, which is capable of removing two to eight damaged bases [76] (Fig. 2 ).
Fig. 2.
Schematic representation of base excision repair and its main proteins.
Short path (for a damaged base) and long path (for up to eight damaged bases).
Excision of a damaged base is carried out by a DNA glycosylase, which marks the beginning of this mechanism of DNA repair. One of these active glycosylases is 8-oxoguanine (OGG1), which is able to recognize the damage, promote the hydrolysis of the N-glycosyl bond and exhibits lyase activity [78]. The action of glycosylases produces an abasic site, which is recognized by an apurinic/apyrimidinic endonuclease, such as APE 1 and APE 2 (apurinic/apyrimidinic endonuclease 1 and 2, respectively), which in turn produces breaks in the 5′ and 3′ phosphodiester linkage at the abasic site [78].
PARP1 (Poly [ADP-ribose] polymerase 1) modifies the polarity of DNA strand for recruitment of other proteins required for DNA repair [79]. Rapamycin, Torin 1 and PARP1 interact with XRCC1 (X-ray repair cross-complementing group 1) in order to stimulate APE1 for cleavage of phosphate skeleton so that DNA polymerase β (POLβ) can insert a new nucleotide [80]. In addition, POLβ exhibits phosphodiesterase activity, which hydrolyzes the 5′ unmodified ends [81], permitting the subsequent connection of the new nucleotide to DNA sequence by action of the DNA ligase III, which finish the DNA repair [82].
This mechanism is considered generic and flexible, capable of recognizing and repairing a large number of damages, thus becoming fundamental for cells [80]. In addition, the choice of long or short pathways is attributed to the number of damages and the availability of ATP in the abasic site environment [83].
Sergio and coworkers [84] reported that in an experimental model for ALI, by LPS administration, there are reduced relative levels of mRNA from OGG1 gene, suggesting that this could be part of cellular response to sepsis. Lee and coauthors [59] show the association between mitochondrial DNA damages and ROS cell death is inversely related to the efficiency of a mitochondrial DNA repair, mediated by OGG1. They also reinforce that overexpression of OGG1 could have effects on nuclear DNA repair, thereby reducing sensitivity to oxidative damage and cytotoxicity accordingly. In knockout rats, there are persistent damages in the mitochondrial genome culminating in increased apoptosis.
5.2. Nucleotide excision repair (NER)
BER acts on specific oxidative damage, but in development of ALI, oxidative stress could damage the DNA molecule in lung and immune cells and consequently activate BER and NER, which effectively contributes for DNA repair [85].
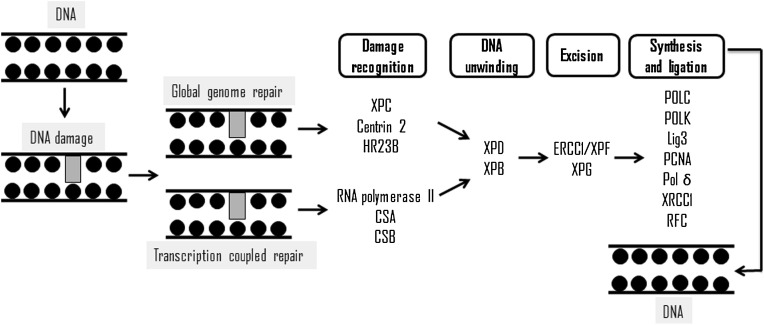
NER recognizes double strand distortion damages, such as cyclobutane pyrimidine dimers, 6-4 pyrimidine-pyrimidone chemical adducts and certain types of cross-links between the two strands of DNA [86], removing from 22 up to 30 nucleotides [87]. However, Melis and coworkers [88] reported that NER could act on oxidized bases, and the results presented by Berra and coworkers [89] showed the interaction of NER proteins with other DNA repair pathways that act on DNA damages induced by oxidative stress. This mechanism involves more than 30 enzymes in two pathways, global genome repair (GGR) and transcription-coupled repair (TCR) [69] (Fig. 3 ). Fig. 3 represents a schematic view of NER, where the major proteins are showed: PCNA (proliferating cell nuclear antigen), Lig (ligase), XP (xeroderma pigmentosum) protein, ERCC1 (excision repair cross-complementing 1), RPA (replication protein A) and lig3 (ligase III), POLK (DNA polymerase Kappa), DNA polymerase III, POL δ (DNA polymerase δ) and RFC (replication factor c). The former removes the damage in non-transcribed regions, while latter acts on transcribed genes. Another difference between GGR and TCR is proteins that initiate the repair process by identification of lesions.
Fig. 3.
Schematic representation of nucleotide excision repair and its main proteins.
XPC/HR23B complex carries out the primary recognition of damages and recruits other proteins [90], such as CSA and CSB ("Cockayne Syndrome" A and B), which act removing RNAPII (RNA polymerase II) in damaged DNA, enabling DNA repair [91].
XPE (xeroderma pigmentosum complementation group E) complex recognizes other damages not identified by XPC complex (xeroderma pigmentosum complementation group C) [89]. TFIIH (transcription factor of RNA polymerase II) is composed of ATP-dependent subunits (p62, p52, p44, p34, cdk7, cyclin H and MAT1), in addition to these subunits, 2 helicases, called XPB and XPD (xeroderma pigmentosum group B and D, respectively). TFIIH is recruited to damage site, and performs search 5′ to 3′ for a short distance before anchoring on DNA damages in cooperation with other protein components, by catalytic activity, unfolds the DNA molecule [92], allowing the assembly of other proteins.
ATP-dependent 5′-3′ helicase (XPD/ERCC2) is essential for NER to maintain the stability of entire complex formed through the interaction of C-terminal portion with p44 subunit [93]. After RPA (replication protein A), XPA and XPG (xeroderma pigmentosum group A and G, respectively) perform pre-incision and repair synthesis. The incision is performed by a heterodimer among XPF (xeroderma pigmentosum complementation group F), ERCC1 and XPG, occurring in nucleotide sequences with about 15–24 nucleotides on the 5′ side and about 2–8 nucleotides on side 3′ of the damage [94].
During 3′ cleavage, synthesis of new DNA fragment is initiated prior to 5′ incision by XPG [91], meanwhile, SIRT1 (sirtuin nuclear protein 1) plays a dual role in promoting NER by stimulating the recognition of damages by promoting the expression of XPC protein [88]. Also, SIRT1 stimulates the excision of damages, promoting the assembly of NER endonuclease in sites damaged by deacetylation of XPA protein [93]. RPA, RFC, PCNA and polymerases σ/ε perform DNA synthesis and DNA ligases (LIG1 or LIG3) binds the new fragment to DNA strand [69].
The reduction of relative levels of ERCC2 and ERCC1 gene messenger RNA in ALI model by LPS administration suggests that these proteins are part of cellular response to sepsis, demonstrating the importance of DNA repair mechanisms in ALI [84].
5.3. Mismatch repair pathway (MMR)
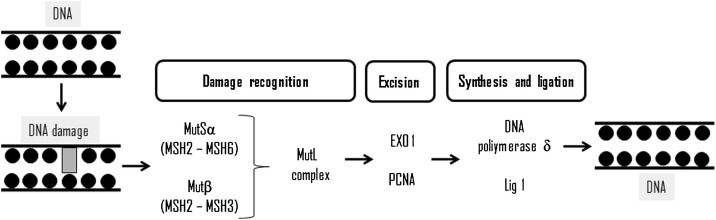
The DNA mismatch (MMR) repair pathway was initially described in Escherichia coli [95] as a system for recognizing and repairing erroneous insertions, base deletions and/or incorrect incorporations of these, which may occur during the process [96], being divided into three stages: recognition, excision and DNA synthesis of failure completion. MMR in eukaryotic cells is less understood, and more complex than in prokaryotic cells, but the steps are highly evolutionarily conserved [97]. The recognition of helix distortion by the MutSa or MutSb, next the MutL (heterodimer of MLH1 and PMS2) is recruited and form a tetrametic complex on the DNA lesions (Fig. 4 ). Next PCNA and replication factor C (RFC) is recruited and the DNA is nicked. PCNA assist in the localization of with MSH2 and MLH1, and plays a key role in DNA initiation and resynthesis in MMR [98]. Also, PCNA assists in the localization of MutSα or MutSβ in replicate DNA errors by interaction with MSH6 and MSH3 [99]. Excision occurs at the time PCNA is directed in the 3′ direction, while EXO1 (exonuclease 1) is directed in the 5′ direction, due to its role as exonuclease, where this complex performs the excision of the incompatible base [100].
Fig. 4.
Schematic representation of mismatch DNA repair pathway and its main proteins.
After that, the DNA molecule needs to be remade, so the third phase of MMR (DNA synthesis) should be accurate. Furthermore, RPA is phosphorylated after POL δ is recruited to the gapped DNA substrate. RPA facilitates MMR-associated DNA resynthesis more efficiently than unphosphorylated RPA [101]. These results are consistent with the fact that RPA-DNA complex might be required to protect nascent single strand DNA and to displace DNA-bound MutSα/MutLα [102], while a lower affinity of RPA-DNA complex might facilitate DNA resynthesis by POL δ, completing the third phase of this DNA repair pathway [101]. In addition, RPA protein has different actions in MMR: binding to heteroduplex DNA before MutSα and MutLα, stimulation of mismatch-provoked excision, protection of single strand DNA gapped regions during excision, facilitation of DNA resynthesis as well as it is phosphorylated after POL δ is recruited to the gap DNA substrate [102].
The erroneous pairing of DNA, and many other pairs of non-canonical DNA bases, tends to cause variations in the conformation of DNA strands [103], these alterations are subject to repair by the MutS protein, which negatively influences the efficiency of DNA repair [104].
It was suggested that MutSα cooperates, or slightly overlaps, functionality with BER pathway [107]. In fact, murine embryonic stem cells deficient in MSH2 present repair efficiency of 8-oxodG markedly lower than in wild-type control cells [105]. Also, it was demonstrated that MutSα reduces the frequency of transversions from G:C to A:T transversions resulting from A/8-oxodG mispairs in yeast [106]. However, Repmann and co-authors [108] reported that MMR proteins do not stimulate the repair of 8-oxoG in newly synthesized DNA.
In E. coli, defects in MutS or MutL proteins confer sensitivity to ultraviolet radiation and specifically inhibit transcription-coupled nucleotide excision repair (NER) [109]. Bertrand and co-authors [109] reported that MSH2 interacts with NER proteins, and that changes in MMR increase the sensitivity to ultraviolet in NER-deficient cells, such as MMR could play a secondary role in NER [110].
Also, MSH2 deficiency leads to hypersensitization to interstrand crosslink by ultraviolet sensitizing agents [111], and many proteins that participate in the repair of these DNA damages play important roles in NER and other DNA repair pathways [112]. In fact, the presence of DNA strand damage was determined by NER [112]. Zhao and co-authors [113] reported that MutSβ interacts directly with XPA-RPA and XPC-RAD23B, forming a higher order complex that could participate in interstrand crosslink repair.
Although evidence suggests that the MMR could be active during ALI development being associated with oxidative stress, there are no studies correlating ALI/ARDS with this DNA repair pathway. However, it is possible this DNA repair pathway plays an important role in correcting DNA damage in lung cells from ALI/ARDS patients.
6. Maintenance of mitochondrial genome after DNA damage
Mitochondria are essential eukaryotic energy-producing organelles that present their own circular genome (mtDNA), which encodes electron transport chain subunits as well as transfer and ribosomal RNAs [114,115].
Unrecognized mitochondrial DNA damaged could be harmful to mammalian cells [116], but the underlying cause is not understood yet, because the cell contains 50 up to 100 mitochondria and each mitochondrion contains 10 copies of mtDNA. It is clear that increase of free radical generation, with consequent establishment of oxidative stress, leads to activation of mtDNA repair. Lacks in repair of these DNA damages are related to aging [117] and cancer progression [118].
Since oxidative damage in mitochondria alters redox balance, causing their functional decline, cells on high demand for mitochondrially generated energy are more affected by mtDNA damage [119]. Several human pathologies have been associated with increase of free radical damage, damage in mtDNA and subsequent mitochondrial dysfunction. In fact, neurodegenerative diseases, such as Alzheimer's, Parkinson's Disease, Friedreich's Ataxia and Huntington's disease, but also diabetes mellitus, liver diseases and cardiovascular disease (atherosclerosis) could be related to mtDNA damages [120].
During inflammation, the persistent release of cytokines, chemokines, nitric oxide and ROS by resident cells attracts circulating immune cells, which in turn respond by inducing a systemic response through the activation of mtDNA-induced inflammatory pathways [121]. The release of these, can cause more mitochondrial damage, developing a vicious cycle, which reinforces the cycle leading to perpetuation of inflammation [122]. Pro-inflammatory mediators, as well as ROS produced during sepsis, directly damage the mitochondrial respiratory chain, causing damage to lipids, proteins, mtDNA and depletion of ATP, compromising mitochondrial function [122].
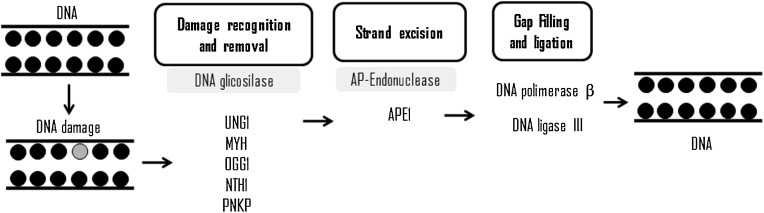
Mitochondria have components of BER pathway, which act on adducts produced by the oxidation of DNA bases by free radicals, mainly ROS [123]. As seen for the genomic DNA, the repair of mtDNA and its components are well known. The sequence is initiated by monofunctional glycosylases, which recognizes the damages, hydrolyzes the glycosidic linkage resulting in the formation of apurinic or apyrimidinic sites, among these glycosylases, UNG1 (uracil DNA glycosylase 1) and MUTYH or mutY DNA glycosylase [124]. UNG1 removes uracil formed by cytosine deamination [125], while MUTYH excises adenine error inserted during replication into the second DNA strand opposite to 8-oxodG, guanine, or cytosine [126]. After formation of apurinic/apirimidic site, AP endonucleases, such as APE1 cleaves the DNA strand, generating a single break [127], allowing mitochondrial DNA polymerase γ to remove phosphate deoxyribose due to its 5′ → 3′ deoxyribose phosphate and insert a new nucleotide in the region [128].
Another class of glycosylases is bifunctional, capable of recognizing lesions in the DNA and have additional enzymatic activity [129]. In mitochondria, glycosylase (as 8-oxoguanine DNA glycosylase, OGG1, and thymine glycol glycosylase, NTH1) were identified by the β-elimination of oxidized bases as well as enzymes able to catalyze successive β and δ deletions after removal of polynucleotide kinase 3′-phosphatase (PNKP). The final stage of mitochondrial BER is the binding of the ends catalyzed by DNA ligase 3 [123] (Fig. 5 ).
Fig. 5.
Schematic representation of mitochondrial DNA repair pathway.
Studies have shown that some enzymes involved in repair of mtDNA are actively translocated to mitochondria from cytoplasm in response to mtDNA damages, and this is typical for double-locus (nuclear and mitochondrial) enzymes, as human OGG1 and NTH1 [130]. This process occurs under oxidative stress conditions, suggesting a specific signaling initiation in the mitochondria and associated with mtDNA damages [131]. Some mtDNA repair proteins with double localization can be found in mitochondria only under conditions of oxidative stress. In fact, this was shown for APE1 [132] and Cockayne's syndrome type A and B protein (CSA and CSB) [133]. Supposedly, a signal induced by mtDNA damages moves initially from mitochondria to cytosol and, in response to this signal, the mtDNA repair proteins are translocated to mitochondria.
Since both mtDNA and nuclear DNA are often damaged at the same time, there must be regulatory pathways coordinating mitochondrial and nuclear DNA repair processes, especially since some DNA repair enzymes operate in both compartments [134]. Mitochondrial isoforms of BER glycosylases are produced by alternative splicing, which opens the possibility for regulation at the post-transcriptional level for these glycosylases [135]. Oxidative stress causes the translocation of the p53 protein to mitochondria [136]. It is known that p53 participates in nuclear DNA repair, but also this protein removes 3′-end containing 8-oxodG due to 3′-exonuclease activity in the presence of the single-stranded DNA binding protein (SSB) [137]. It is commonly believed that p53 interacts with DNA polymerase γ and regulates its activity in BER [138]. p53 was found to facilitate the removal of oxidized bases from mtDNA through interactions with OGG1 and APE1 [139]. In addition, p53 regulates gene transcription encoding OGG1 [140].
Two ERCC (ERCC6) and ERCC8 (CSA - Cockayne's syndrome type A protein) proteins related to NER were detected in mammalian mitochondria at extremely low levels, being markedly increased in cells under oxidative stress [131]. Also, CSB and CSA proteins are imported into mitochondria only during oxidative stress, where they bind to mtDNA and to some proteins involved in BER such as OGG1 and SSB [141]. However, since nuclear NER requires the involvement of other enzymes, which are not found in the mitochondria, it is possible that NER in the mitochondria differs from the that in cell nucleus or that in the mitochondria, the CSB and the CSA perform other functions, as DNA repair and transcription, similarly to their role in cell nucleus [135].
On the other hand, MMR in the mitochondria is under study yet, because only some of the essential factors involved in this DNA repair in the cell nucleus have been identified into mitochondria so far [123], but removal of non-complementary G:T and G:G pairs has been demonstrated in vitro in mammalian mitochondrial lysates. Interestingly, MMR was observed in mitochondrial extracts without MSH2, indicating that the MMR in mitochondria differs from that in cell nucleus, where MSH2 acts as a key factor [142].
The YB1 protein is another nuclear MMR factor being, different from other MMR proteins in the nucleus, partially located in mitochondria. When expression of YB1 is negatively regulated by knockdown with interfering RNA (siRNA), MMR activity in mitochondrial extracts decreases [143]. Therefore, YB1 could be involved in mtDNA repair, although other nuclear MMR participant has not been detected in mitochondria, as suggested by Zinovkina [135].
7. Therapeutic targets
ALI or ARDS continues to be a cause of morbidity and mortality in critically ill patients. Despite a better understanding of ALI pathogenesis of supportive treatment with protective pulmonary mechanical ventilation strategy remains the only treatment with survival perspective. Most clinical trials in ALI targets on mechanically ventilated patients. Therefore, new therapies are necessary, with the search for therapeutic targets, as new antibiotics for treatment of sepsis. Therapies that modulate DNA repair mechanisms could contribute to improving patients with acute lung injury or acute respiratory distress syndrome.
In an experimental model of ALI induced by intraperitoneal administration of lipopolysaccharide, pulmonary parenchyma is severely affected, with reduction of pulmonary alveoli and thickening of the interalveolar septa. However, Sergio and coworkers [145] show that after exposure to low power infrared laser (AsGaAl, 808 nm, 10 and 20 J/cm²) as an alternative and non-invasive treatment, it was observed reduction of septum thickening, increase in alveolar airspace, reduction in area and number of cells inflammatory infiltrate. In another study, exposure to low power laser at low fluences was able to alter mRNA levels from genes related to apoptosis, such as Caspase-3 and Bcl-2, preventing cell apoptosis and reducing DNA fragmentation in lung cells as well as increasing DNA fragmentation only in cells present in inflammatory infiltrate [146].
Lee and coworkers [59] demonstrated the key role of OGG1 protein on suppression of mitochondrial DNA damages in lung tissue injured by Pseudomonas aeruginosa. Also, Ye and coworkers [147] reported the role of OGG1 in pulmonary hyperoxide conditions in innate immunity. Under these conditions, increased DNA fragmentation was found by comet assay and OGG-1 deficient animals had impaired immune functions, resulting in high infiltration of polymorphonuclear cells in lung and an exacerbated inflammatory response. OGG-1, in this model, interacts with genes related to autophagy by molecular binding, which controls NF-kB nuclear translocation, leading to high levels of ROS and proinflammatory cytokines, thus suggesting as a new target for clinical interventions.
Similarly, other studies have demonstrated possibilities in the modulation of other DNA repair genes in different experimental models. It was observed alteration on mRNA ERCC1 and ERCC2 (excision repair cross-complementation group 1 and 2, respectively) levels after low power lasers exposure [148]. Also, these lasers were able to modulate ATM (ataxia-telangiectasia mutated) and p53 [149,150] and APE1 mRNA levels [150].
Exposure to oxygen (hyperoxia) could cause damages in nitrogenous bases in lung cells and this is consistent with primary effects of ROS, demonstrating that inhaled oxidants have ability to alter pulmonary genome [151]. These authors also reported DNA strand breaks in a small proportion of lung cells with prolonged oxygen exposure as well as effects of IL-6 expression on DNA damages, which modulates cell death pathways and prevents short-term tissue damage.
OGG1, ERCC1 and ERCC2 were also one of targets of studies by Sergio and coworkers [84], being suggested that DNA repair is part of cellular response to sepsis observed in an experimental model of ALI by LPS due to exposure to low power infrared laser modulates mRNA levels from BER and NER. These studies indicate that new strategies for treatment of ALI/ARDS are feasible and assessments of DNA repair mechanisms could contribute to improving clinical status in the development of ALI/ARDS (Fig. 6 ). However, studies are necessary to evaluate the eficacy of modulation of these DNA repair pathways on ALI/ARDS.
Fig. 6.
Schematic representation of intervention in DNA repair as an alternative treatment for acute lung injury.
8. Conclusion
In the development of ALI/ARDS, the inflammatory process is intense, resulting in oxidative stress mediated by free radicals in lung and immune cells, which cause damages in DNA molecule. Despite few studies have focused on oxidative damages in ALI/ARDS, DNA repair plays a crucial role for adaptation to harmful conditions, cell responses and tissue repair. In fact, BER, NER and MMR pathways are important for enabling lung or adjacent cells to survive in oxidative stress by effective repair of nuclear and mtDNA damages. Also, these pathways are closely connected, such as regulatory connections among them and genes encoding proteins involved in DNA repair could be therapeutically modulated. Thus, repair pathways involved in DNA integrity and genomic stability could be considered as targets for development of new therapies for reducing morbidity and mortality of ALI/ARDS patients.
Declaration of Competing Interest
The authors report no conflicts of interest.
Acknowledgements
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Carlos Chagas de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
References
- 1.GBD 2015 Mortality and Causes of Death Collaborators Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard G.R., Artigas A., Brigham K.L., Carlet J., Falke K., Hudson L., Lamy M., Legall J.R., Morris A., Spragg R. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am. J. Respir. Crit. Care Med. 1994;149:818–824. doi: 10.1164/ajrccm.149.3.7509706. [DOI] [PubMed] [Google Scholar]
- 3.Definition Task Force A.R.D.S., Ranieri V.M., Rubenfeld G.D., Thompson B.T., Ferguson N.D., Caldwell E., Fan E., Camporota L., Slutsky A.S. Acute respiratory distress syndrome: the berlin definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 4.McNicholasa B.A., Rooneyb G.M., Laffeyb J.G. Lessons to learn from epidemiologic studies in ARDS. Curr. Opin. Crit. Care. 2018;24:41–48. doi: 10.1097/MCC.0000000000000473. [DOI] [PubMed] [Google Scholar]
- 5.Rubenfeld G.D., Caldwell E., Peabody E., Weaver J., Martin D.P., Neff M., Stern E.J., Hudson L.D. Incidence and outcomes of acute lung injury. N. Engl. J. Med. 2005;353:1685–1693. doi: 10.1056/NEJMoa050333. [DOI] [PubMed] [Google Scholar]
- 6.Herridge M.S., Cheung A.M., Tansey C.M., Matte-Martyn A., Granados N.D., Al-Saidi F., Cooper A.B., Guest C.B., Mazer D., Mehta S., Stewart T.E., Barr A., Cook D., Slutsky A.S. One-year outcomes in survivors of the acute respiratory distress syndrome. N. Engl. J. Med. 2003;348:683–693. doi: 10.1056/NEJMoa022450. [DOI] [PubMed] [Google Scholar]
- 7.Rubenfeld G.D., Herridge M.S. Epidemiology and outcomes of acute lung injury. Chest. 2007;131:554–562. doi: 10.1378/chest.06-1976. [DOI] [PubMed] [Google Scholar]
- 8.Valavanidis A., Vlachogianni T., Fiotakis K., Loridas S. Pulmonary oxidative stress, inflammation and cancer: respirable particulate matter, fibrous dusts and ozone as major causes of lung carcinogenesis through reactive oxygen species mechanisms. Int. J. Environ. Res. Public Health. 2013;10:3886–3907. doi: 10.3390/ijerph10093886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newton A.C., Bootman M.D., Scott J.D. Second messengers, cold spring harb. Perspect. Biol. Med. 2016;8:1–14. doi: 10.1101/cshperspect.a005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barzilai A., Yamamoto K. DNA damage responses to oxidative stress. DNA Repair (Amst) 2004;3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Sun W., Wang Z.P., Gui P., Xia W., Xia Z., Zhang X.C., Deng Q.Z., Xuan W., Marie C., Wang L.L., Wu Q.P., Wang T., Lin Y. Endogenous expression pattern of resolvin D1 in a rat model of self-resolution of lipopolysaccharide-induced acute respiratory distress syndrome and inflammation. Int. Immunopharmacol. 2014;23:247–253. doi: 10.1016/j.intimp.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 12.ARDS Definition Task Force, Ranieri V.M., Rubenfeld G.D., Thompson B.T., Ferguson N.D., Caldwell E., Fan E., Camporota L., Slutsky A.S. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;20:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 13.Fanelli V., Vlachou A., Ghannadian S., Simonetti U., Slutsky A.S., Zhang H. Acute respiratory distress syndrome: new definition, current and future therapeutic options. J. Thorac. Dis. 2013;5:326–334. doi: 10.3978/j.issn.2072-1439.2013.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocco P.R., Zin W.A. Pulmonary and extrapulmonary acute respiratory distress syndrome: are they different? Curr. Opin. Crit. Care. 2003;11(2205):10–17. doi: 10.1097/00075198-200502000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Ragaller M., Richter T. Acute lung injury and acute respiratory distress syndrome. J. Emerg. Trauma Shock. 2010;3:43–51. doi: 10.4103/0974-2700.58663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ware L.B., Matthay M.A. The acute respiratory distress syndrome. N. Engl. J. Med. 2000;342:1334–1349. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- 17.Wheeler A.P., Bernard G.R. Acute lung injury and the acute respiratory distress syndrome: a clinical review. Lancet. 2007;369:1553–1564. doi: 10.1016/S0140-6736(07)60604-7. [DOI] [PubMed] [Google Scholar]
- 18.Bannered A., Gerondakis S. Coordinating TLR-activated signaling pathways in cells of the immune system. Immunol. Cell Biol. 2007;85:420–424. doi: 10.1038/sj.icb.7100098. [DOI] [PubMed] [Google Scholar]
- 19.Choi S.H., Harkewicz R., Lee J.H., Boullier A., Almazan F., Li A.C., Witztum J.L., Bae Y.S., Miller Y.I. Lipoprotein accumulation in macrophages via toll-like receptor-4-dependent fluid phase uptake. Circ. Res. 2009;104:1355–1363. doi: 10.1161/CIRCRESAHA.108.192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunsmore S.E., Rannels D.E. Extracelular matrix biology in the lung. Am. J. Physiol. 1996;270:L3–L27. doi: 10.1152/ajplung.1996.270.1.L3. [DOI] [PubMed] [Google Scholar]
- 21.Berrier A.L., Yamada K.M. Cell-matrix adhesion. J. Cell. Physiol. 2008;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 22.Matthay M.A., Zimmerman G.A. Acute lung injury and the acute respiratory distress syndrome: four decades of inquiry into pathogenesis and rational management. Am. J. Respir. Cell Mol. Biol. 2005;33:319–327. doi: 10.1165/rcmb.F305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusushima K., King L.S., Aggarwal N.R., De Gorordo A., D’Alessio F.R., Kubo K. Acute lung injury review. Intern. Med. 2009;48:621–630. doi: 10.2169/internalmedicine.48.1741. [DOI] [PubMed] [Google Scholar]
- 24.Matthay M.A., Zemans R.L. The acute respiratory distress syndrome: pathogenesis and treatment. Annu. Rev. Pathol. 2011;6:147–163. doi: 10.1146/annurev-pathol-011110-130158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercat A., Richard J.C., Vielle B., Jaber S., Osman D., Diehl J.L., Lefrant J.Y., Prat G., Richecoeur J., Nieszkowska A., Gervais C., Baudot J., Bouadma L., Brochard L. Expiratory Pressure (Express) Study Group, Positive end-expiratory pressure setting in adults with acute lung injury and acute respiratory distress syndrome: a randomized controlled trial. JAMA. 2008;299:646–655. doi: 10.1001/jama.299.6.646. [DOI] [PubMed] [Google Scholar]
- 26.Sethi S., Murphy T.F. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med. 2008;359:2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- 27.Bastarache J.A., Blackwell T.S. Development of animal models for the acute respiratory distress syndrome. Dis. Model. Mech. 2009;2:218–223. doi: 10.1242/dmm.001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beloncle F., Lorente J.A., Esteban A., Brochard L. Update in acute lung injury and mechanical ventilation. Am. J. Respir. Crit. Care Med. 2014;198:1187–1193. doi: 10.1164/rccm.201402-0262UP. [DOI] [PubMed] [Google Scholar]
- 29.Aeffner F., Bratasz A., Flano E., Powell K.A., Davis I.C. Postinfection A77-1726 treatment improves cardiopulmonary function in H1N1 influenza-infected mice. Am. J. Respir. Cell Mol. Biol. 2012;47:543–551. doi: 10.1165/rcmb.2012-0112OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traylor Z.P., Aeffner F., Davis I.C. Influenza A H1N1 induces declines in alveolar gas exchange in mice consistent with rapid post-infection progression from acute lung injury to ARDS. Influenza Other Respir. Viruses. 2013;7:472–479. doi: 10.1111/j.1750-2659.2012.00414.x. https://doi:0.1111/j.1750-2659.2012.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matute-Bello G., Frevert C.W., Martin T.R. Animal models of acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;295:L379–L399. doi: 10.1152/ajplung.00010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raetz C.R., Ulevitch R.J., Wright S.D., Sibley C.H., Ding A., Nathan C.F. Gram-negative endotoxin: an extraordinary lipid with profound effects on eukaryotic signal transduction. FASEB J. 1991;5:2652–2660. doi: 10.1096/fasebj.5.12.1916089. [DOI] [PubMed] [Google Scholar]
- 33.Liu F., Li W., Pauluhn J., Trübel H., Wang C. Lipopolysaccharide-induced acute lung injury in rats: comparative assessment of intratracheal instillation and aerosol inhalation. Toxicology. 2013;8:158–166. doi: 10.1016/j.tox.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 34.Rabelo M.A.E., Lucinda L.M.F., Reboredo M.M., da Fonseca L.M.C., Reis F.F., Fazza T.F., Brega D.R., de Paoli F., de Souza da Fonseca A., Pinheiro B.V. Acute lung injury in response to intratracheal instillation of lipopolysaccharide in an animal model of emphysema induced by elastase. Inflammation. 2018;41:174–182. doi: 10.1007/s10753-017-0675-5. [DOI] [PubMed] [Google Scholar]
- 35.Dejager L., Pinheiro I., Dejonckheere E., Libert C. Cecal ligation and puncture: the gold standard model for polymicrobial sepsis? Trends Microbiol. 2011;19:198–208. doi: 10.1016/j.tim.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Wolthuis E.K., Vlaar A.P., Choi G., Roelofs J.J., Juffermans N.P., Schultz M.J. Mechanical ventilation using non-injurious ventilation settings causes lung injury in the absence of pre-existing lung injury in healthy mice. Crit. Care. 2009;13:R1. doi: 10.1186/cc7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson M.R., Choudhury S., Goddard M.E., O’Dea K.P., Nicholson A.G., Takata M. High tidal volume upregulates intrapulmonary cytokines in an in vivo mouse model of ventilator-induced lung injury. J. Appl. Physiol. 2003;95:1385–1393. doi: 10.1152/japplphysiol.00213.2003. [DOI] [PubMed] [Google Scholar]
- 38.Rittirsch D., Huber-Lang M.S., Flierl M.A., Ward P.A. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 2009;4:31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan R., Mo Y., Zhang Z., Jiang M., Tang S., Zhang Q. Cobalt nanoparticles induce lung injury, DNA damage and mutations in mice. Part. Fibre Toxicol. 2017;14:13–38. doi: 10.1186/s12989-017-0219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ware L.B. Modeling human lung disease in animals. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;294:L149–L150. doi: 10.1152/ajplung.00472.2007. [DOI] [PubMed] [Google Scholar]
- 41.Ballard-Croft C., Wang D., Sumpter L.R., Zhou X., Zwischenberger J.B. LArge-animal models of acute respiratory distress syndrome. Ann. Thorac. Surg. 2012;93:1331–1339. doi: 10.1016/j.athoracsur.2011.06.107. [DOI] [PubMed] [Google Scholar]
- 42.Sipahi E.Y., Atalay F. Experimental models of acute lung injury. J. Transl. Inter. Med. 2015;2:154–159. doi: 10.5152/ejp.2014.49091. [DOI] [Google Scholar]
- 43.Abdulkhaleq L.A., Assi M.A., Abdullah R., Zamri-Saad M., Taufiq-Yap Y.H., Hezmee M.N.M. The crucial roles of inflammatory mediators in inflammation: a review. Vet. World. 2018;11:627–635. doi: 10.14202/vetworld.2018.627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salzano S., Checconi P., Hanschmann E.M., Lillig C.H., Bowler L.D., Chan P., Vaudry D., Mengozzi M., Coppo L., Sacre S., Atkuri K.R., Sahaf B., Herzenberg L.A., Herzenberg L.A., Mullen L., Ghezzi P. Linkage of inflammation and oxidative stress via release of glutathionylated peroxiredoxin-2, which acts as a danger signal. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12157–12162. doi: 10.1073/pnas.1401712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baran C.P., Zeigler M.M., Tridandapani S., Marsh C. The role of ROS and RNS in regulating life and death of blood monocytes. Curr. Pharm. Des. 2004;10:855–866. doi: 10.2174/1381612043452866. [DOI] [PubMed] [Google Scholar]
- 46.Chow C.W., Herrera Abreu M.T., Suzuki T., Downey G.P. Oxidative stress and acute lung injury. Am. J. Respir. Cell Mol. Biol. 2003;29:427–431. doi: 10.1165/rcmb.F278. [DOI] [PubMed] [Google Scholar]
- 47.Guo C., Sun L., Chen X., Zhang D. Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen. Res. 2013;21:2003–2014. doi: 10.3969/j.issn.1673-5374.2013.21.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao S.S., Kaufman R.J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox Signal. 2014;21:396–413. doi: 10.1089/ars.2014.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabe Y., Ando K., Hirao S., Yoshida M., Handa H. Redox regulation of NF-κB activation: distinct redox regulation between the cytoplasm and the nucleus. Antioxid. Redox Signal. 2005;7:395–403. doi: 10.1089/ars.2005.7.395. [DOI] [PubMed] [Google Scholar]
- 50.Pantano C., Reynaert N.L., van der Vliet A., Janssen-Heininger Y.M. Redox-sensitive kinases of the nuclear factor-kappa B signaling pathway. Antioxid. Redox Signal. 2006;8:1791–1806. doi: 10.1089/ars.2006.8.1791. [DOI] [PubMed] [Google Scholar]
- 51.Schaur R.J. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol. Aspects Med. 2003;24:149–159. doi: 10.1016/S0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 52.Irrcher I., Ljubicic V., Hood D.A. Interactions between ROS and AMP kinase activity in the regulation of PGC-1α transcription in skeletal muscle cells. Am. J. Physiol. Cell Physiol. 2009;296:C116–C123. doi: 10.1152/ajpcell.00267.2007. [DOI] [PubMed] [Google Scholar]
- 53.Brigham K.L. Role of free radicals in lung injury. Chest. 1986;89:859–863. doi: 10.1378/chest.89.6.859. [DOI] [PubMed] [Google Scholar]
- 54.Rinaldo J.E. Mediation of ARDS by leukocytes: clinical evidence and implications for therapy. Chest. 1986;89:590–593. doi: 10.1378/chest.89.4.590. [DOI] [PubMed] [Google Scholar]
- 55.Victor V.M., Rocha M., Solá E., Bañulus C., Garcia-Malpartida K., Hernández-Mijares A. Oxidative stress, endothelial dysfuncion and atherosclerosis. Curr. Pharm. Des. 2009;15:2988–3002. doi: 10.2174/138161209789058093. [DOI] [PubMed] [Google Scholar]
- 56.Alonso de Vega J.M., Díaz J., Serrano E., Carbonell L.F. Oxidative stress in critically ill patients with systemic inflammatory response syndrome. Crit. Care Med. 2002;30:1782–1786. doi: 10.1097/00003246-200208000-00018. [DOI] [PubMed] [Google Scholar]
- 57.Sarkele M., Sabelnikovs O., Vanags I., Ozolina A., Skesters A., Silova A. The role of oxidative stress markers in acute respiratory distress syndrome. Acta Chir. Lat. 2013;13:22–26. doi: 10.2478/chilat-2014-0005. [DOI] [Google Scholar]
- 58.Sarkele M., Ozolina A., Sabelnikovs O., Skesters A., Silova A., Vanags I. The activity of oxidative stress markers in acute respiratory distress syndrome. Proc. Lat. Acad. Sci. 2014;68:1–11. doi: 10.2478/prolas-2014-0032. [DOI] [Google Scholar]
- 59.Lee Y.L., Obiako B., Gorodnya O.M., Ruchko M.V., Kuck J.L., Pastukh V.M., Wilson G.L., Simmons J.D., Gillespie M.N. Mitochondrial DNA damage initiates acute lung injury and multi-organ system failure evoked in rats by Intra-Tracheal Pseudomonas aeruginosa. Shock. 2017;48:54–60. doi: 10.1097/SHK.0000000000000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simmons J.D., Lee Y.L., Mulekar S., Kuck J.L., Brevard S.B., Gonzalez R.P., Gillespie M.N., Richards W.O. Elevated levels of plasma mitochondrial DNA DAMPs are linked to clinical outcome in severely injured human subjects. Ann. Surg. 2013;258:591–598. doi: 10.1097/SLA.0b013e3182a4ea46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., Brohi K., Itagaki K., Hauser C.J. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun S., Sursal T., Adibnia Y., Zhao C., Zheng Y., Li H., Otterbein L.E., Hauser C.J. Mitochondrial DAMPs increase endothelial permeability through neutrophil dependent and independent pathways. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruchko M., Gorodnya O., LeDoux S.P., Alexeyev M.F., Al-Mehdi A.B., Gillespie M.N. Mitochondrial DNA damage triggers mitochondrial dysfunction and apoptosis in oxidant-challenged lung endothelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2005;288:L530–L535. doi: 10.1152/ajplung.00255.2004. [DOI] [PubMed] [Google Scholar]
- 64.Lander H.M. An essential role for free radicals and derived species in signal transduction. FASEB J. 1997;11:118–124. doi: 10.1096/fasebj.11.2.9039953. [DOI] [PubMed] [Google Scholar]
- 65.Halliwell B., Gutteridge J.M.C. fifth ed. Oxford University Press; USA: 2007. Free Radicals in Biology and Medicine. [Google Scholar]
- 66.Cadet J., Bourdat A.G., D’Ham C., Duarte V., Gasparutto D., Romieu A., Ravanat J.L. Oxidative base damage to DNA: specificity of base excision repair enzymes. Mutat. Res. 2000;462:121–128. doi: 10.1016/S1383-5742(00)00022-3. [DOI] [PubMed] [Google Scholar]
- 67.Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberg T. second ed. ASM Press; Washington, DC: 2005. DNA Repair and Mutagenesis. [Google Scholar]
- 68.Setlow R.B., Carrier W.L., Bollum F.J. Nuclease-resistent sequences in ultravioleta-irradiated deoxyribonucleic acid. Biochim. Biophys. Acta. 1964;91:446–461. doi: 10.1016/0926-6550(64)90075-1. [DOI] [PubMed] [Google Scholar]
- 69.Costa R.M.A., Chiganças V., Galhardo R.S., Carvalho H., Menck C.F. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85:1083–1099. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 70.Bridge G., Rashid S., Martin S.A. DNA mismatch repair and oxidative DNA damage: implications for cancer biology and treatment. Cancers (Basel) 2014;6:1597–1614. doi: 10.3390/cancers6031597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trzeciak A.R., Mohanty J.G., Jacob K.D., Barnes J., Ejiogu N., Lohani A., Zonderman A.B., Rifkind J.M., Evans M.K. Oxidative damage to DNA and single strand break repair capacity: relationship to other measures of oxidative stress in a population cohort. Mutat. Res. 2012;736:93–103. doi: 10.1016/j.mrfmmm.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collins A.R., Gedik C.M., Olmedilla B., Southon S., Bellizzi M. Oxidative DNA damage measured in human lymphocytes: large differences between sexes and between countries, and correlations with heart disease mortality rates. FASEB J. 1998;12:1397–1400. doi: 10.1096/fasebj.12.13.1397. [DOI] [PubMed] [Google Scholar]
- 73.Malins D.C., Johnson P.M., Wheeler T.M., Barker E.A., Polissar N.L., Vinson M.A. Age-related radical-induced DNA damage is linked to prostate cancer. Cancer Res. 2001;61:6025–6028. [PubMed] [Google Scholar]
- 74.Mórocz M., Kálmán J., Juhász A., Sinkó I., McGlynn A.P., Downes C.S., Janka Z., Raskó I. Elevated levels of oxidative DNA damage in lymphocytes from patients with Alzheimer’s disease. Neurobiol. Aging. 2002;23:47–53. doi: 10.1016/S0197-4580(01)00257-3. [DOI] [PubMed] [Google Scholar]
- 75.van Loon B., Markkanen E., Huübscher U. Oxygen as a friend and enemy: how to combat the mutational potential of 8-oxo-guanine. DNA Repair. 2010;9:604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 76.Dianov G.L., Hübscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41:3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klungland A., Bjelland S. Oxidative damage to purines in DNA: role of mammalian Ogg1. DNA Repair (Amst) 2007;6:481–488. doi: 10.1016/j.dnarep.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 78.Krokan H.E., Bjoras M. Base excision repair, cold spring harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 80.El-Khamisy S.F., Masutani M., Suzuki H., Caldecott K.W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucl. Acids. Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Podlutsky A.J., Dianova I.I., Podust V.N., Bohr V.A., Dianov G.L. Human DNA polymerase beta initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. EMBO J. 2001;20:1477–1482. doi: 10.1093/emboj/20.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caldecott K.W. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/S1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 83.Petermann E., Zieler M., Oei S.L. ATP-dependent selection between single nucleotide and long patch base excision repair. DNA Repair (Amst) 2003;2:1101–1114. doi: 10.1016/S1568-7864(03)00117-4. [DOI] [PubMed] [Google Scholar]
- 84.da Silva Sergio L.P., Thomé A.M.C., da Silva Neto Trajano L.A., Vicentini S.C., Teixeira A.F., Mencalha A.L., de Paoli F., de Souza da Fonseca A. Low-power laser alters mRNA levels from DNA repair genes in acute lung injury induced by sepsis in Wistar rats. Lasers Med. Sci. 2019;43:157–168. doi: 10.1007/s10103-018-2656-9. [DOI] [PubMed] [Google Scholar]
- 85.Clancy S. DNA damage & repair: mechanisms for maintaining DNA integrity. Nat. Educ. 2008;1:103. https://www.nature.com/scitable/topicpage/dna-damage-repair-mechanisms-for-maintaining-dna-344 [Google Scholar]
- 86.Zhu Q., Wani A.A. Nucleotide excision repair: finely tuned molecular orchestra of early pre-incision events. Photochem. Photobiol. 2016;93:166–177. doi: 10.1111/php.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scharer O.D. Nucleotide excision repair in eukaryotes, cold spring harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Melis J.P., van Steeg H., Luijten M. Oxidative DNA damage and nucleotide excision repair. Antioxid. Redox Signal. 2013;18:2409–2419. doi: 10.1089/ars.2012.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berra C.M., Oliveira C.S., Garcia C.C.M., Rocha C.R.R., Lerner L.K., Lima L.C.A., Baptista M.S., Menck C.F.M. Nucleotide excision repair activity on DNA damage induced by photoacivated methylene blue. Free Radic. Biol. Med. 2013;61:343–356. doi: 10.1016/j.freeradbiomed.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 90.Krasikova Y.S., Rechkunova N.I., Maltseva E.A., Petruseva I.O., Silnikov V.N., Zatsepin T.S., Oretskaya T.S., Schärer O.D., Lavrik O.I. Interaction of nucleotide excision repair factors XPC-HR23B, XPA, and RPA with damaged DNA. Biochemistry (Mosc.) 2008;73:886–896. doi: 10.1134/S0006297908080063. [DOI] [PubMed] [Google Scholar]
- 91.Petruseva I.O., Evdikimov N.A., Lavrik O.I. Molecular mechanism of global genome nucleotide excision repair. Acta Naturae. 2014;6:23–34. https://www.ncbi.nlm.nih.gov/pubmed/24772324 [PMC free article] [PubMed] [Google Scholar]
- 92.Sugasawa K., Akagi J., Nishi R., Iwai S., Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: directional binding of the XPC complex and DNA strand scanning. Mol. Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 93.Ahmad A., Enzlin J.H., Bhagwat N.R., Wijgers N., Raams A., Appledoorn E., Theil A.F., Hoejimakers J.H., Vermeulen W., Jaspers N.G.J., Schärer O.D., Niedernhofer L.J. Mislocalization of XPF-ERCC1 nuclease contributes to reduced DNA repair in XP-F patients. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fan W., Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol. Cell. 2010;39:247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 95.Lahue R.S., Au K.G., Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 96.Hsieh P., Yamane K. DNA mismatch repair: molecular mechanism, cancer, and aging. Mech. Ageing Dev. 2009;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kunkel T.A., Erie D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 98.Gu L., Hong Y., McCulloch S., Watanabe H., Li G.M. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 1998;26:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shell S.S., Putnam C.D., Kolodner R.D. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol. Cell. 2007;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dzantiev L., Constantin N., Genschel J., Iyer R.R., Burgers P.M., Modrich P. A defined human system that supports bidirectional mismatch-provoked excision. Mol. Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 101.Lindsey-Boltz L.A., Reardon J.T., Wold M.S., Sancar A. In vitro analysis of the role of replication protein A (RPA) and RPA phosphorylation in ATR-mediated checkpoint signaling. J. Biol. Chem. 2012;19:36123–36131. doi: 10.1074/jbc.M112.407825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guo S., Zhang Y., Yuan F., Gao Y., Gu L., Wong I., Li G.M. Regulation of replication protein A functions in mismatch repair by phosphorylation. J. Biol. Chem. 2006;281:21607–21616. doi: 10.1074/jbc.M603504200. [DOI] [PubMed] [Google Scholar]
- 103.Yuan F., Gu L., Guo S., Wang C., Li G.M. Evidence for involvement of HMGB1 protein in human DNA mismatch repair. J. Biol. Chem. 2004;279:20935–20940. doi: 10.1074/jbc.M401931200. [DOI] [PubMed] [Google Scholar]
- 104.Mazurek A., Johnson C.N., Germann M.W., Fishel R. Sequence context effect for hMSH2-hMSH6 mismatch-dependent activation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4177–4182. doi: 10.1073/pnas.0808572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.DeWeese T.L., Shipman J.M., Larrier N.A., Buckley N.M., Kidd L.R., Groopman J.D., Cutler R.G., te Riele H., Nelson W.G. Mouse embryonic stem cells carrying one or two defective Msh2 alleles respond abnormally to oxidative stress inflicted by low-level radiation. Proc. Natl. Acad. Sci. U. S. A. 1998;95:11915–11920. doi: 10.1073/pnas.95.20.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ni T.T., Marsischky G.T., Kolodner R.D. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. Cerevisiae. Mol. Cell. 1999;4:439–444. doi: 10.1016/S1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 107.Gu Y., Parker A., Wilson T.M., Bai H., Chang D.Y., Lu A.L. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 108.Repmann S., Olivera-Harris M., Jiricny J. Influence of oxidized purine processing on strand 27 directionality of mismatch repair. J. Biol. Chem. 2015;290:9986–9999. doi: 10.1074/jbc.M114.629907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bertrand P., Tishkoff D.X., Filosi N., Dasgupta R., Kolodner R.D. Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc. Natl. Acad. U. S. A. 1998;95:14278–14283. doi: 10.1073/pnas.95.24.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rochette P.J., Bastien N., McKay B.C., Therrien J.P., Drobetsky E.A., Drouin R. Human cells bearing homozygous mutations in the DNA mismatch repair genes hMLH1 or hMSH2 are fully proficient in transcription-coupled nucleotide excision repair. Oncogene. 2002;21:5743–5752. doi: 10.1038/sj.onc.1205641. [DOI] [PubMed] [Google Scholar]
- 111.Wu Q., Christensen L.A., Legerski R.J., Vasquez K.M. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Noll D.M., Mason T.M., Miller P.S. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao J., Jain A., Iyer R.R., Modrich P.L., Vasquez K.M. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res. 2009;37:4420–4429. doi: 10.1093/nar/gkp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Falkenberg M., Larsson N.G., Gustafsson C.M. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 116.Tann A.W., Boldogh I., Meiss G., Qian W., Housten B.V., Mitra S., Szczesny B. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5′-EXO/endonuclease) in their repair. J. Biol. Chem. 2011;286:31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pinto M., Moraes C.T. Mechanisms linking mtDNA damage and aging. Free Radic. Biol. Med. 2015;85:250–258. doi: 10.1016/j.freeradbiomed.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ishikawa K., Takenaga K., Akimoto M., Koshikawa N., Yamaguchi A., Imanishi H., Nakada K., Honma Y., Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 119.Singh G., Pachouri U.C., Khaidem D.C., Kundu A., Chopra C., Singh P. Mitochondrial DNA damage and diseases. F1000Res. 2015;4:176–182. doi: 10.12688/f1000research.6665.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Housten B.V., Hunter S.E., Meyer J.N. Mitochondrial DNA damage induced autophagy, cell death, and disease. Front Biosci (Landmark) 2016;21:42–54. doi: 10.2741/4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nakayama H., Otsu K. Mitochondrial DNA as an inflammatory mediator in cardiovascular diseases. Biochem. J. 2018;475:839–852. doi: 10.1042/BCJ20170714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Escames G., López L.C., García J.A., García-Corzo L., Ortiz F., Acuña-Castroviejo D. Mitochondrial DNA and inflammatory diseases. Hum. Genet. 2011;131:161–173. doi: 10.1007/s00439-011-1057-y. [DOI] [PubMed] [Google Scholar]
- 123.Kazak L., Reyes A., Holt I.J. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat. Rev. Mol. Cell Biol. 2012;13:659–671. doi: 10.1038/nrm3439. [DOI] [PubMed] [Google Scholar]
- 124.Wallace S.S. Base excision repair: a critical player in many games. DNA Repair (Amst.) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nakabeppu Y. Regulation of intracellular localiza tion of human MTH1, OGG1, and MYH proteins for repair of oxidative DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:75–94. doi: 10.1016/S0079-6603(01)68091-7. [DOI] [PubMed] [Google Scholar]
- 126.Parker A., Gu Y., Lu A.L. Purification and characterization of a mammalian homolog of Escherichia coli MutY mismatch repair protein from calf liver mito chondria. Nucleic Acids Res. 2000;28:3206–3215. doi: 10.1093/nar/28.17.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mitra S., Izumi T., Boldogh I., Bhakat K.K., Chattopadhyay R., Szczesny B. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair (Amst.) 2007;6:461–469. doi: 10.1016/j.dnarep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 128.Kroeger K.M., Hashimoto M., Kow Y.W., Greenberg M.M. Cross-linking of 2-deoxyribonolactone and its β-elimination product by base excision repair enzymes. Biochemistry. 2003;42:2449–2455. doi: 10.1021/bi027168c. [DOI] [PubMed] [Google Scholar]
- 129.Dou H., Theriot C.A., Das A., Hegde M.L., Matsumoto Y., Boldogh I., Hazra T.K., Bhakat K.K., Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen: the potential for replication associated repair of oxidized bases in mammalian genomes. J. Biol. Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 130.Shinmura K., Yokota J. The OGG1 gene encodes a repair enzyme for oxidatively damaged DNA and is involved in human carcinogenesis. Antioxid. Redox Signal. 2001;3:597–609. doi: 10.1089/15230860152542952. [DOI] [PubMed] [Google Scholar]
- 131.Swartzlander D.B., Griffiths L.M., Lee J., Degtyareva N.P., Doetsch P.W., Corbett A.H. Regulation of base excision repair: Ntg1 nuclear and mitochondrial dynamic localization in response to genotoxic stress. Nucleic Acids Res. 2010;38:3963–3974. doi: 10.1093/nar/gkq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Frossi B., Tell G., Spessotto P., Colombatti A., Vitale G., Pucillo C. H2O2 induces translocation of APE/Ref1 to mitochondria in the Raji Bcell line. J. Cell. Physiol. 2002;193:180–186. doi: 10.1002/jcp.10159. [DOI] [PubMed] [Google Scholar]
- 133.Stevnsner T., Muftuoglu M., Aamann M.D., Bohr V.A. The role of Cockayne syndrome group B (CSB) protein in base excision repair and aging. Mech. Ageing Dev. 2008;129:441–448. doi: 10.1016/j.mad.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Boesch P., WeberLotfi F., Ibrahim N., Tarasenko V., Cosset A., Paulus F., Lightowlers R.N., Dietrich A. A, DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim. Biophys. Acta Mol. Cell Res. 2011;1813:186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 135.Zinovkina L.A. Mechanisms of mitochondrial DNA repair in mammals. Biochemistry (Moscow) 2018;83:233–249. doi: 10.1134/s0006297918030045. [DOI] [PubMed] [Google Scholar]
- 136.Yoshida Y., Izumi H., Torigoe T., Ishiguchi H., Itoh H., Kang D., Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. http://cancerres.aacrjournals.org/content/63/13/3729 [PubMed] [Google Scholar]
- 137.Wong T.S., Rajagopalan S., Townsley F.M., Freund S.M., Petrovich M., Loakes D., Fersht A.R. Physical and functional interactions between human mito chondrial singlestranded DNAbinding protein and tumour suppressor p53. Nucleic Acids Res. 2009;37:568–581. doi: 10.1093/nar/gkn974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bakhanashvili M., Grinberg S., Bonda E., Simon A.J., MoshitchMoshkovitz S., Rahav G. p53 in mitochondria enhances the accuracy of DNA synthesis. Cell Death Differ. 2008;15:1865–1874. doi: 10.1038/cdd.2008.122. [DOI] [PubMed] [Google Scholar]
- 139.Achanta G., Huang P. Role of p53 in sensing oxidative DNA damage in response to reactive oxygen species generating agents. Cancer Res. 2004;64:6233–6239. doi: 10.1158/0008-5472.CAN-04-0494. [DOI] [PubMed] [Google Scholar]
- 140.Chatterjee A., Mambo E., Osada M., Upadhyay S., Sidransky D. The effect of p53RNAi and p53 knockout on human 8oxoguanine DNA glycosylase (hOgg1) activity. FASEB J. 2006;20:112–114. doi: 10.1096/fj.04-3423fje. [DOI] [PubMed] [Google Scholar]
- 141.Kamenisch Y., Fousteri M., Knoch J., von Thaler A.K., Fehrenbacher B., Kato H., Becker T., Dolle M.E.T., Kuiper R., Majora M., Schaller M., van der Horst G.T.J., van Steeg H., Rocken M., Rapaport D., Krutmann J., Mullenders L.H., Berneburg M. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J. Exp. Med. 2010;207:379–390. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mason P.A., Matheson E.C., Hall A.G., Lightowlers R.N. Mismatch repair activity in mammalian mitochondria. Nucleic Acids Res. 2003;31:1052–1058. doi: 10.1093/nar/gkg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.de Souza-Pinto N.C., Mason P.A., Hashiguchi K., Weissman L., Tian J., Guay D., Lebel M., Stevnsner T.V., Rasmussen L.J., Bohr V.A. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst) 2009;8:704–719. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sergio L.P.S., Trajano L.A.S.N., Thomé A.M.C., Mencalha A.L., Paoli F., Fonseca A.S. Low power infrared laser modifies the morphology of lung affected with acute injury induced by sepsis. Laser Phys. 2018;28 doi: 10.1088/1555-6611/aab44d. [DOI] [Google Scholar]
- 146.Sergio L.P.S., Thomé A.M.C., Trajano L.A.S.N., Mencalha A.L., da Fonseca A.S., de Paoli F. Photobiomodulation prevents DNA fragmentation of alveolar epithelial cells and alters the mRNA levels of caspase 3 and Bcl-2 genes in acute lung injury. Photochem. Photobiol. Sci. 2018;17:975–983. doi: 10.1039/C8PP00109J. [DOI] [PubMed] [Google Scholar]
- 147.Ye Y., Lin P., Zhang W., Tan S., Zhou X., Li R., Pu Q., Koff J.L., Dhasarathy A., Ma F., Deng X., Jiang J., Wu M. DNA repair interacts with autophagy to regulate inflammatory responses to pulmonary hyperoxia. J. Immunol. 2017;198:2844–2853. doi: 10.4049/jimmunol.1601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sergio L.P.S., Campos V.M., Vicentini S.C., Mencalha A.L., Paoli F., Fonseca A.S. Low-intensity red and infrared lasers affect mRNA expression of DNA nucleotide excision repair in skin and muscle tissue. Lasers Med. Sci. 2016;31:429–435. doi: 10.1007/s10103-016-1870-6. [DOI] [PubMed] [Google Scholar]
- 149.Trajano L.A.S.N., Trajano E.T.L., Sergio L.P.S., Teixeira A.F., Mencalha A.L., Stumbo A.C., Fonseca A.S. Photobiomodulation effects on mRNA levels from genomic and chromosome stabilization genes in injured muscle. Lasers Med. Sci. 2018;33:1513–1519. doi: 10.1007/s10103-018-2510-0. [DOI] [PubMed] [Google Scholar]
- 150.Trajano L.A.S.N., Sergio L.P.S., Silva C.L., Carvalho L., Mencalha A.L., Stumbo A.C., Fonseca A.S. Low-level laser irradiation alters mRNA expression from genes involved in DNA repair and genomic stabilization in myoblast. Laser Phys. Lett. 2016;13 doi: 10.1088/1612-2011/13/7/075601. (6p) [DOI] [Google Scholar]
- 151.Barker G.F., Manzo N.D., Cotich K.L., Shone R.K., Waxman A.B. DNA damage induced by hyperoxia: quantitation and correlation with lung injury. Am. J. Respir. Cell Mol. Biol. 2006;35:277–288. doi: 10.4049/jimmunol.1601001. [DOI] [PMC free article] [PubMed] [Google Scholar]