Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection presents an immense global health problem. Spike (S) protein of coronavirus is the primary determinant of its entry into the host as it consists of both receptor binding and fusion domain. Besides tissue tropism, and host range, coronavirus pathogenesis are primarily controlled by the interaction of S protein with the cell receptor. Moreover, the proteolytic activation of S protein by host cell proteases plays a decisive role. The host-cell proteases have shown to be involved in the proteolysis of S protein and cleaving it into two functional subunits, S1 and S2, during the maturation process. In the present study, the interaction of the S protein of SARS-CoV-2 with different host proteases like furin, cathepsin B, and plasmin has been analyzed using molecular docking and molecular dynamics (MD) simulation. Incorporation of the furin cleavage site (R-R-A-R) in the S protein of SARS-CoV-2 has been studied by mutating the individual amino acid. MD simulation results suggest the polytropic nature of the S protein. Our analysis indicated that a single amino acid substitution in the polybasic cleavage site of S protein perturb the binding of cellular proteases. This mutation study might help to generate an attenuated SARS-CoV-2. Besides, targeting host proteases by inhibitors may result in a practical approach to stop the cellular spread of SARS-CoV-2 and develop its antiviral.
Keywords: Spike protein, Proteolytic activation, Furin, Cathepsin B, Plasmin, MD simulations
1. Introduction
Coronaviruses (CoVs) possess a single-stranded positive-sense RNA genome ranging from 26 to 32 kilobases in length (Weiss and Navas-Martin, 2005). The subfamily Coronavirinae contains a significant number of avian and mammalian pathogens. The subfamily include Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Thiel, 2007). To date, six different CoV strains are known to infect humans. The CoVs infecting humans belong to genera alphacoronavirus and betacoronavirus. The alphacoronaviruses infecting humans are HCoV-229E and HCoV-NL63, while betacoronaviruses infecting humans are HCoV-HKU1, HCoV-OC43, Middle East respiratory syndrome (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV).
The recent pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) also belongs to betacoronavirus. All CoVs share correlation in their genome organization. It consists of 16 non-structural proteins (nsp1 to nsp16), encoded by open reading frame (ORF) 1a/b at the 5′ end, followed by the structural proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N), encoded by other ORFs at the 3′ end. The entry of coronavirus into the host is a compounded process of receptor binding and proteolytic cleavage of S protein into functional subunits to promote virus-host cell membrane fusion (Heald-Sargent and Gallagher, 2012). The entry can be facilitated by employing fusion directly at the cell surface receptor or through the endosomal compartment (Millet and Whittaker, 2015). The protruding S protein is responsible for the attachment of the virion to the host cell surface receptor and its fusion, enabling the release of the viral genome into the cytoplasm (Millet and Whittaker, 2015). The ectodomain of the S protein comprises two functional subunits, S1 and S2. The S1 subunit is responsible for receptor binding, while the S2 subunit maintains the fusion machinery process (Millet and Whittaker, 2015). The S protein is a class I viral fusion protein (Bosch et al., 2003) and is activated by its proteolytic cleavage by host-cell proteases (White et al., 2008). Various host-cell proteases like cathepsin B, trypsin, plasmin, elastase, and cell surface transmembrane protease/serine (TMPRSS) have been shown to cleave the S protein of SARS-CoVs to facilitate the viral attachment (Belouzard et al., 2012). It has been shown that the processing of surface glycoproteins is a prerequisite to producing comparatively more pathogenic and virulent viral particles, and inhibition of these proteases is likely to inhibit the viral infection as well (Zhou et al., 2015). Proteolytic activation unlocks the fusogenic potential of viral envelope glycoproteins, and it is a crucial step in the entry of the enveloped virus.
Recent studies have reported the incorporation of the unique furin cleavage site of Arg-Arg-Ala-Arg (R-R-A-R) between the boundary of S1 and S2 subunits in the S protein in SARS-CoV-2 (Coutard et al., 2020). Furin is a calcium-dependent protease that cleaves in an acidic environment after recognizing the specific sequence motif composed of R-X-R/K-R, where X is any amino acid residue (Izaguirre, 2019). The sequence motifs generally present at the S1/S2 junction of S protein determine many host-cell proteases' cleavage. Coronavirus has evolved for the proteolytic activation of S protein, and a large number of host proteases can have a cognate recognition domain for its cleavage.
In the present study, we showed the binding of host-cell proteases (furin, cathepsin B, and plasmin) to the S protein of SARS-CoV-2 by molecular docking. Furthermore, we analyzed the role of individual residues in binding S protein with host-cell proteases by incorporating the single point mutations in its cleavage site. The protein-protein complexes were also subjected to molecular dynamics (MD) simulations to study the conformational stability.
2. Materials and methods
2.1. Sequence analysis of S protein and its structure modeling
The SARS-CoV-2 sequences were retrieved from GenBank, accession number of each sequence used for the analysis is enlisted in (Supplementary Table S1). The sequences were aligned using the ClustalW algorithm implemented in the sequence alignment program package of MEGA-X software. A three-dimensional S protein model was generated using the SWISS-MODEL protein structure homology-modeling server (Waterhouse et al., 2018), wherein SARS-CoV-2 S glycoprotein (PDB id: 6VSB) was taken as a template model. The energy of the modeled structure was minimized by using Yasara software and finally validated using PROCHECK. All the three-dimensional structures have been visualized using PyMOL.
2.2. Structure modeling of S protein mutants
Four different mutants of S protein were constructed with a single residue substitution in amino acid sequence. The mutations were incorporated with proline-to-alanine (P681A), arginine-to-alanine (R682A), arginine-to-alanine (R683A), and arginine-to-alanine (R685A). Three-dimensional structures of the mutants were generated by SWISS-MODEL protein structure homology-modeling and further validated by PROCHECK-Ramachandran plot after its energy minimization using Yasara software.
2.3. Molecular docking
The crystal structure for human plasminogen (PDB id: 1DDJ), furin (PDB id: 5MIM), and cathepsin (PDB id: 1PBH) were retrieved from the protein data bank. The interaction between the proteases and SARS-CoV-2 S protein was performed by molecular docking. Docking between receptor (proteases) and ligand (S protein) was performed using Cluspro 2.0 protein-protein docking software (Kozakov et al., 2013, 2017; Vajda et al., 2017). For furin and cathepsin B, the C-chain of S protein, and for plasmin, the A-chain of S protein was used for the docking studies. The PDB sum generator server was used to find the interacting amino acid residues spanning the proteases and S protein domain of SARS-CoV-2 (de Beer et al., 2014). Finally, the docked models were analyzed for Z-score using the DockScore tool (Malhotra et al., 2015).
2.4. Molecular dynamics simulations
The docked protein-protein complexes were subjected to MD simulations to study the conformational stability using the GROMACS v5.1.4 software program to obtain the parameters of proteins (Abraham et al., 2015; Harvey and van Gunsteren, 1993). Each system consisted of the protein-protein complex in a solvated dodecahedron box with a minimum distance of 1.2 nm from the boundary. The systems were filled with single-point charge water and subsequently neutralized by adding counter cations (Na+) or anions (Cl−) (Leszczynski and Shukla, 2012). The solvated systems were then energy minimized using the steepest descent method (Petrova and Solov’ev, 1997), followed by the equilibrium for 100 ps through NVT and NPT ensembles to optimize the orientation and system density. The final equilibrated systems were used as starting conformations to run the MD simulations for 50 ns. Finally, the output trajectories were obtained, and the estimation of Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Solvent Accessible Surface Area (SASA), and Radius of Gyration (Rg) was done using GROMACS packages. The graphs were analyzed using XMGRACE software and plotted with GraphPad Prism software.
3. Results
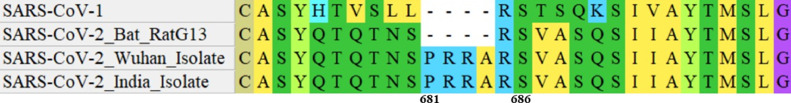
The alignment of S protein amino acid sequences of SARS-CoV-2 with other CoVs provided critical insights into a unique mutation incorporated in its S1-S2 cleavage site (Fig. 1 ). The unique incorporation of 4 amino acid residues, P681, R682, R683, and A684, were exclusive to the cleavage site of the S protein of SARS-CoV-2.
Fig. 1.
Sequence Alignment of S protein isolates. The sequence alignment of S protein of SARS-CoV-1, SARS-CoV-2 Bat RATG13 Isolate, SARS-CoV-2 India Isolate, and SARS-CoV-2 Wuhan Isolate.
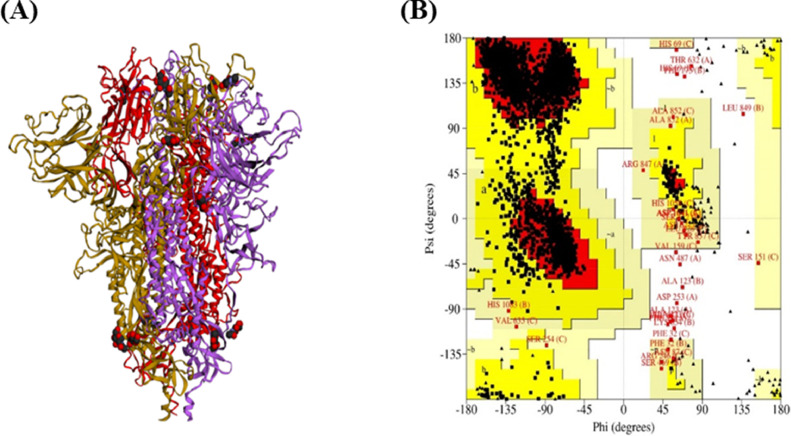
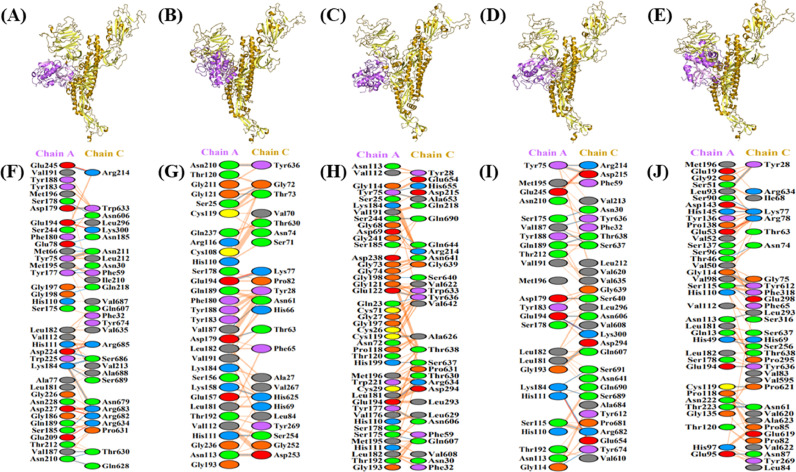
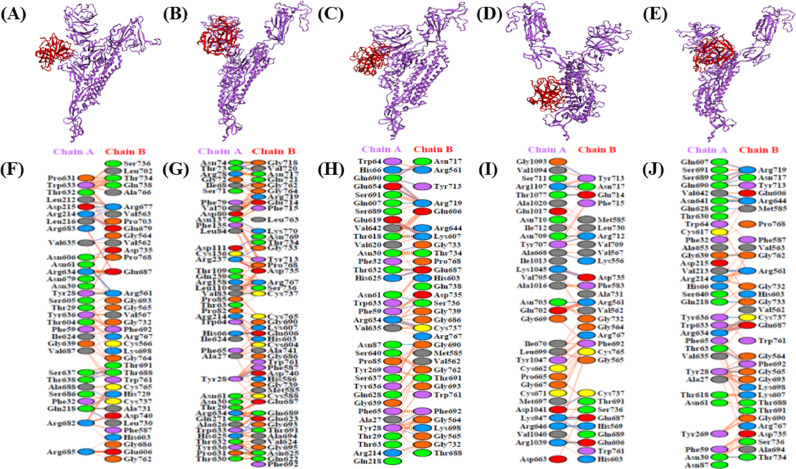
The three-dimensional homo-trimer model of SARS-CoV-2 S protein showed 86.3% residues in most favored regions, 12.6% in additional allowed regions, 0.6% in generously allowed regions, and only 0.5% residues in disallowed regions (Fig. 2 ). After the minimization of the model selected for further analysis, the final energy was −1715047.7 kJ/mol. The wild-type S protein significantly interacted with furin protease (Fig. 3 A and F), with seven salt bridges, 13 hydrogen bonds, and 198 non-bonded contacts (Table 1 ). The cathepsin B and wild-type S protein model (Fig. 4 A and F) evinced six salt bridges, 22 hydrogen bonds, and 270 non-bonded contacts (Table 2 ). Subsequently, the plasmin model (Fig. 5 A and F) interacted with five salt bridges, 18 hydrogen bonds, and 275 non-bonded contacts (Table 3 ). Furthermore, the interactive models of proteases with each of the mutant S proteins and their interacting residues are shown in Fig. 3, Fig. 4, Fig. 5. Besides, all the other interacting residues for every model are mentioned in Supplementary Tables S2 and S3.
Fig. 2.
Structural details of target protein. The three-dimensional homo trimer model of SARS-CoV-2 S protein (A) with its analyzed Ramachandran plot (B).
Fig. 3.
Visualization of Docked Complexes and Interactive residues. The 3D-models of enzyme-protein binding complexes and residues involved. 3D-binding complexes of A-chain of enzyme furin (violet) and C-chain of wild type S protein (golden) (A), P681A substitution mutation S protein (golden) (B), R682A substitution mutation S protein(golden) (C), R683A substitution mutation S protein(golden) (D), and R685A substitution mutation S protein (golden) (E). Interacting amino acid residues of A-chain of furin enzyme and C-chain of wild type S protein (F), P681A substitution mutation S protein (G), R682A substitution mutation S protein (H), R683A substitution mutation S protein (I), and R685A substitution mutation S protein (J).
Table 1.
Data representing the interface statistics of binding complex of furin enzyme and (A) wild type S protein (B) S protein with P681A substitution mutation (C) S protein with R682A substitution mutation (D) S protein with R683A substitution mutation (E) S protein with R685A substitution mutation.
| Protein type | No. of interface residues | Interface area (A2) | No. of salt bridges | No. of H-bonds | No. of non-bonded contacts |
|---|---|---|---|---|---|
| Wild type (A) | 28:27:00 | 1348:1318 | 7 | 13 | 198 |
| P681A (B) | 33:37:00 | 1890:1992 | 3 | 17 | 251 |
| R682A (C) | 24:31:00 | 1400:1320 | 1 | 15 | 175 |
| R683A (D) | 40:36:00 | 1918:1926 | 4 | 23 | 217 |
| R685A (E) | 31:27:00 | 1525:1519 | 1 | 19 | 218 |
Fig. 4.
Visualization of Docked Complexes and Interactive residues. The 3D-models of enzyme-protein binding complexes and residues involved. 3D-binding complexes of A-chain of enzyme cathepsin B (violet) and C-chain of wild type S protein (golden) (A), P681A substitution mutation S protein (golden) (B), R682A substitution mutation S protein (golden) (C), R683A substitution mutation S protein (golden) (D), and R685A substitution mutation S protein (golden) (E). Interacting amino acid residues of A-chain of cathepsin B enzyme and C-chain of wild type S protein (F), P681A substitution mutation S protein (G), R682A substitution mutation S protein (H), R683A substitution mutation S protein (I), and R685A substitution mutation S protein (J).
Table 2.
Data signifying the interface statistics of the binding complex of cathepsin B enzyme and (A) wild type S protein (B) S protein with P681A substitution mutation (C) S protein with R682A substitution mutation (D) S protein with R683A substitution mutation (E) S protein with R685A substitution mutation.
| Protein type | No. of interface residues | Interface area (A2) | No. of salt bridges | No. of H-bonds | No. of non-bonded contacts |
|---|---|---|---|---|---|
| Wild type (A) | 37:29 | 1601:1661 | 6 | 22 | 270 |
| P681A (B) | 31:23 | 1333:1411 | 1 | 8 | 149 |
| R682A (C) | 43:31 | 1640:1664 | 1 | 12 | 224 |
| R683A (D) | 25:31 | 1519:1406 | 3 | 16 | 188 |
| R685A (E) | 36:33 | 1744:1759 | 2 | 15 | 182 |
Fig. 5.
Visualization of Docked Complexes and Interactive residues. The 3D-models of enzyme-protein binding complexes and residues involved. 3D-binding complexes of B-chain of enzyme plasmin (red) and A-chain of wild type S-protein (violet) (A) P681A substitution mutation S protein (violet) (B) R682A substitution mutation S protein (violet) (C) R683A substitution mutation S protein (violet) (D), and R685A substitution mutation S protein (violet) (E). Interacting amino acid residues of A-chain of plasmin enzyme and B-chain of wild type S protein (F), P681A substitution mutation S protein (G), R682A substitution mutation S protein (H), R683A substitution mutation S protein (I), and R685A substitution mutation S protein (J).
Table 3.
Data representing the interface statistics of the binding complex of plasmin enzyme and (A) wild type S protein (B) S protein with P681A substitution mutation (C) S protein with R682A substitution mutation (D) S protein with R683A substitution mutation (E) S protein with R685A substitution mutation.
| Protein type | No. of interface residues | Interface area (A2) | No. of salt bridges | No. of H-bonds | No. of non-bonded contacts |
|---|---|---|---|---|---|
| Wild type (A) | 31:38 | 1857:1755 | 5 | 18 | 275 |
| P681A (B) | 42:46 | 1962:2026 | 5 | 22 | 329 |
| R682A (C) | 35:32 | 1599:1702 | 3 | 19 | 213 |
| R683A (D) | 33:30 | 1497:1479 | 4 | 17 | 179 |
| R685A (E) | 33:32 | 1620:1764 | 1 | 22 | 200 |
We performed MD simulations of wild-type and mutant spike protein complexed with the host proteases for a 50 ns timescale to assess the complex stability and binding strength. Various parameters, including RMSD, RMSF, SASA, and Rg, were utilized to serve the purpose.
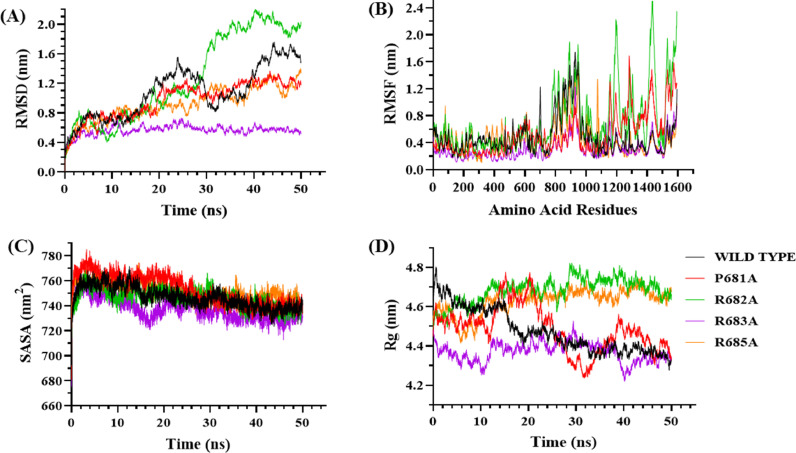
RMSD plot analysis is essential to decipher the structural stability of the protein-ligand bound complex. With furin as the acting protease, wild-type, P681A, R682A, and R685A mutant S-protein failed to achieve stability throughout the 50 ns timescale of the simulation, demonstrating continuous fluctuations. However, the R683A-furin complex attained steadiness at 17 ns and maintained it with only minor changes. The average RMSD values obtained for wild-type, P681A, R682A, R683A, and R685A with furin are 1.079, 0.969, 1.273, 0.563, and 0.916 nm, respectively (Fig. 6a). Like in the case of furin, wild-type S protein acted upon by cathepsin failed to attain stability throughout the timescale of simulation. Both P681A and R682A-cathepsin complex reached equilibrium at 43 ns and maintained it till the end of the simulation. R683A-cathepsin complex achieved stability at 42 ns and kept it throughout, while the R685A-cathepsin model failed to achieve stability. The average RMSD values for wild-type, P681A, R682A, R683A, and R685A complexed with cathepsin are 1.239, 0.983, 1.094, 0.806, and 1.06 nm, respectively (Fig. 7A). Similarly, plasmin-bound wild-type S protein started stabilizing after 26 ns and kept its equilibrium until the end with minor fluctuations. P681A-plasmin complex failed to achieve stability and continued to increase throughout the timeframe. Furthermore, the R682A-plasmin complex started stabilizing after 28 ns and maintained its strength up to 42 ns, after which it revealed considerable fluctuation until the end of the simulation. The R683A-plasmin complex started stabilizing at 29 ns and maintained its stability up to 40 ns, after which it showed minor fluctuations till the end. R685A-plasmin complex achieved stability at 36 ns and remained stable with minor volatility towards the end of the simulation. The average RMSD values obtained for wild-type, P681A, R682A, R683A, and R685A with plasmin are 1.039, 1.076, 1.258, 0.794, and 1.036 nm, respectively (Fig. 8C).
Fig. 6.
Molecular Dynamics Simulation of Furin and S protein. Root Mean Square Deviation (RMSD) analysis graph (A), Root Mean Square Fluctuations (RMSF) analysis graph (B), Radius of Gyration (Rg) analysis graph (C), and Solvent Accessible Surface Area (SASA) analysis graph (D).
Fig. 7.
Molecular Dynamics Simulation of Cathepsin B and S protein. Root Mean Square Deviation (RMSD) analysis graph (A), Root Mean Square Fluctuations (RMSF) analysis graph (B), Radius of Gyration (Rg) analysis graph (C), and Solvent Accessible Surface Area (SASA) analysis graph (D).
Fig. 8.
Molecular Dynamics Simulation of Plasmin and S protein. Root Mean Square Deviation (RMSD) analysis graph (A), Root Mean Square Fluctuations (RMSF) analysis graph (B), Radius of Gyration (Rg) analysis graph (C), and Solvent Accessible Surface Area (SASA) analysis graph (D).
RMSF of the complex were analyzed to assess the flexibility and rigidity of bound complexes. We analyzed the C-alpha atom of the residues of the bound complexes to infer the fluctuations of each atom across the backbone. Furin-bound wild-type and mutant S-protein complexes showed considerably greater atomic fluctuations than plasmin-bound complexes. The average RMSF values obtained for wild-type, P681A, R682A, R683A, and R685A with furin are 0.482, 0.515, 0.711, 0.309, and 0.429 nm respectively (Fig. 6B). For cathepsin interacting with wild-type and mutant S-protein, atomic fluctuations were prominent in the residue region from 650 to 1375. A higher degree of fluctuations was observed for the wild-type S protein. The average RMSF values obtained for wild-type, P681A, R682A, R683A, and R685A with cathepsin are 0.644, 0.421, 0.456, 0.394, and 0.547 nm, respectively (Fig. 7B). Atomic fluctuations were observed throughout the simulation for the plasmin-bound complexes, with a peak in the RMSF value at around 700 amino acid position of S protein for all the plasmin-bound complexes. The average RMSF values obtained for wild-type, P681A, R682A, R683A, and R685A with cathepsin are 0.485, 0.583, 0.555, 0.386, and 0.526 nm, respectively (Fig. 8B).
SASA values were further analyzed to estimate the solvent behavior of wild-type and mutant S-protein complexed with proteases. Furin-interacting wild-type, P681A, R682A, R683A, and R685A complexes revealed average SASA values of 746.65, 752.75, 746.54, 737.15, and 751.31 nm2, respectively (Fig. 6C). Cathepsin bound wild-type, P681A, R682A, R683A, and R685A complexes showed average SASA values of 690.52, 681.29, 684.74, 669.59, and 683.38 nm2, respectively (Fig. 7C). Furthermore, plasmin-bound wild-type, P681A, R682A, R683A, and R685A showed average SASA values of 672.97, 680.86, 671.77, 682.42, and 677.09 nm2, respectively (Fig. 8C).
Rg values were also analyzed to interpret the compactness and rigidity of the S-protein-protein complexes. Wild-type S protein bound to furin started stabilizing at 20 ns of the simulation and maintained it till the end except for minor fluctuations at around 32–40 ns timeframe. P681A-furin complex failed to achieve stability throughout the process. R682A-furin complex achieved stability at 30 ns which is maintained till the end of the simulation. R683A-furin complex attained strength at 14 ns and kept it till 38 ns, after which it again attained stability at 46 ns up till the end. Similarly, the R685A-furin complex achieved stability at 19 ns and maintained it until the end with minor fluctuations. The average Rg values obtained for furin complexed wild-type, P681A, R682A, R683A, and R685A are 4.48, 4.49, 4.68, 4.37, and 4.63 nm, respectively (Fig. 6D). The wild-type and P681A bound to cathepsin started stabilizing at 45 ns in the simulation. R682A-cathepsin complex started stabilizing at 20 ns and maintained it till the end. Both R683A and R685A-cathepsin models did not stabilize throughout the simulation timescale. The average Rg values obtained for cathepsin complexed wild-type, P681A, R682A, R683A, and R685A are 4.53, 4.34, 4.21, 4.5, and 4.49 nm, respectively (Fig. 7D). Plasmin bound to wild-type S protein showed minor fluctuations at the start and started attaining equilibrium at around 12 ns till the end. P681A with plasmin reached stability at 20 ns and maintained it up to 28 ns, after which it showed a slight increase in its value and achieved stability at 37 ns and kept it, with a slight decrease at 46 ns timescale. R682A-plasmin complex achieved stability at 19 ns and kept it throughout with minor fluctuations. Similarly, the R683A-plasmin complex achieved stability at 14 ns and maintained it throughout, though slightly increasing towards the end. R685A-plasmin complex showed significant volatility and did not seem to attain considerable strength till the end of the simulation. The average Rg values obtained for wild-type, P681A, R682A, R683A, and R685A with plasmin are 4.28, 4.65, 4.48, 4.48, and 4.81 nm, respectively (Fig. 6, Fig. 7, Fig. 8D ).
4. Discussion
It is known that the sequence motifs present between the boundary of S1 and S2 subunits of S protein in SARS-CoV determine the active binding and cleavage site for host-cell proteases (White et al., 2008). The multiple sequence alignment results suggested the incorporation of four additional polybasic amino acid residues at the S1/S2 site, as reported previously (Coutard et al., 2020). These amino acids signify the involvement of furin as host protease in the proteolytic processing of S protein. The polybasic cleavage sites for host cell proteases directly impact the viral pathogenicity and host range (Nao et al., 2017). Several other viruses, including the Zika virus (Nambala and Su, 2018), dengue virus (Yu et al., 2008), avian influenza virus (Alexander and Brown, 2009), and Newcastle disease virus (Kumar and Kumar, 2014; Mohamed et al., 2011) have also been shown to modulate their pathogenicity by acquiring basic amino acids containing cleavage sites. The docking results of different host-cell proteases revealed their binding to the S protein through the formation of salt bridges and hydrogen bond linkages. The analysis of the residues involved in the interaction shows their presence in the binding complex. To emphasize the role of these additional amino acid residues in the binding of host-cell proteases to the S protein, we sequentially mutated the basic amino acids to alanine. Further, we analyzed each mutant's binding efficiency with respective host protease by molecular docking and confirmed by MD simulations.
Firstly, we determined all the residues involved in binding host-cell proteases, including furin, cathepsin B, and plasmin, with wild-type S protein. Our docking study showed crucial details regarding the interaction and binding of the proteases at the proteolytic cleavage site of the SARS-CoV-2 S protein. The enzyme-protein interaction is sufficiently compromised when a single residue is mutated at the cleavage site of the protein. The dwindled and reduced number of salt bridges and hydrogen bond interactions in all mutant models from the wild-type model signify a weaker enzyme-protein binding at the mutated cleavage site.
The proteomic identification of protease cleavage sites profiling of cathepsin B has revealed a strong cleavage site specificity for amino acid residue glycine and a partial preference for phenylalanine (Biniossek et al., 2011). The action of pH-independent cysteine protease cathepsin B is vital for the entry, hence establishing infection of SARS-CoV (Gierer et al., 2013). Plasmin is a crucial enzyme in fibrinolysis, and its natural substrate is fibrinogen and fibrin. The cleavage of the influenza virus by plasmin is well characterized (Berri et al., 2013; Goto et al., 2001; LeBouder et al., 2010). The role of plasmin has been studied in the A/WSN/1993 H1N1 influenza virus, where the hemagglutinin (HA) cleavage site of the virus governs the spread of infection in plasmin dependent manner (Sun et al., 2010). The pivotal role of plasmin in the pathogenicity of influenza virus was explained by the distribution of mini-plasmin and plasmin fragments in epithelial cells of bronchioles (Murakami et al., 2001). The SARS-CoV-2 is characterized by hyperfibrinolysis, as evident by high D-dimers and breakdown product of fibrinolysis; however, it has been reported that plasmin can cleave the S protein of SARS-CoV in vitro (Ji et al., 2020; Kam et al., 2009).
The docking studies convey the functional inference of these incorporated residues on the binding of proteases to the cleavage site of S protein. On adequately mutating one amino acid residue at the cleavage site of S protein, the binding of the protein is sufficiently hindered, and the residues of this site are not involved in the interaction with the proteases. The proteolytic cleavage, hence the activation of S protein, is amply controlled. All viral fusion protein undergoes a structural transition and finally attain a compact, low-energy structure. These conformational changes brought viral and cellular host membranes nearby, which induces fusion, followed by the pore formation that allows viral genetic material to enter the cell (White et al., 2008). This is exemplified by the HA protein of highly pathogenic avian influenza virus, where conversion of the monobasic site, cleaved by a trypsin-like protease to a polybasic site, allows cleavage by ubiquitously expressed furin-like proteases to facilitate the spread of the virus and to make it more virulent (Klenk and Garten, 1994; Lazarowitz and Choppin, 1975). Our results of substitution mutation R682A of S protein showed a minor interaction with furin protease compared to its wild-type homolog. Also, substitution mutation P681A of S exhibits little interaction with cathepsin; similarly, the substitution mutation of R685A of S displays minimum interaction with plasmin protease. Mutation of P682A in the wild type S protein results in the best mutant model that blocks the enzyme active site for cellular proteases, owing to a minimum number of salt bridges and hydrogen-bond formation. The finding suggested that the PRRARS amino acid motif in wild type S protein is responsible for its proper binding with furin, cathepsin, and plasmin, and mutation of these residues impairs the interaction. Endoproteolytic cleavage, usually at arginine, is a common post-translational modification for activating several proteins, such as peptide hormones and growth factors (Klenk and Garten, 1994). Docking studies revealed that changes at these sites might weaken the interaction between the S protein and the host-cell proteases.
The results of MD simulations are essential in better assessing the protein-ligand complex stability and energy dynamics. Estimating RMSD is crucial to infer structural stability. With reference to the wild-type, R683A-protein depicts the lowest RMSD values compared to all other mutants for all three proteases, thereby serving as the best mutant model for stable interaction with the proteases. R682A mutant shows higher RMSD values than the wild type on interacting with furin and plasmin. R682A-cathepsin model did not reveal a higher RMSD value than the wild-type but is the highest compared to other mutant models. Hence, it can be inferred from the average RMSD values, that the R682A-protein be the best model for inefficient binding and proteolytic cleavage by the host proteases.
The average RMSF for the backbone of the R683A-protein accommodating all the three proteases shows the lowest value compared to other mutant and wild-type S-protein. Hence, average RMSF values, like RMSD, also reveal the mutant model R683A to be the best for the stable binding of proteases. With furin as the binding protease, P681A and R682A models show higher RMSF values than the wild-type S protein, with R682A having an even higher RMSF value than P681A. For cathepsin, average RMSF values for all mutant proteins are lower than the wild type, signifying less flexibility during the MD simulations. Among all the mutant S-proteins, R685A showed the maximum RMSF value. In the case of plasmin, all except the R683A protein showed higher RMSF values than the wild-type S protein. Like in the case of furin, P681A and R682A proteins show sufficiently higher average RMSF values, with P681A having the higher value among the two models. It can be deduced that, for furin and plasmin, P681A and R682A served as the best mutant models to disturb the binding of these two proteases, while the R685A mutant model is the best in staggering the binding of cathepsin.
Higher SASA values specify the enlargement of protein volume, and low fluctuations are expected throughout the simulation. In the case of furin and cathepsin, R683A mutant model shows the lowest average SASA value compared to other mutant and wild-type S proteins. Thus, it can be congruously inferred that binding furin and cathepsin to the R683A mutant model could sufficiently reduce protein expansion. In the case of plasmin, only the R682A protein has a lower average SASA value than the wild-type S protein. In the case of furin, P681A and R685A proteins show higher average SASA values than the wild-type S protein, with P681A having the highest value. Furin exposes P681A protein the most to the solvent on binding. In addition, with cathepsin, all mutant S-proteins except R683A show comparable average SASA values to the wild-type S-protein. All mutant S-proteins offer lower average SASA values than the wild-type S protein signifying a more compact and rigid complex. R682A protein has the highest average SASA value among the four mutants. Furthermore, in the case of plasmin, the R683A protein shows a considerably higher average SASA value than the wild-type S-protein. At all-time points during the simulation, the R683A protein shows higher SASA values than the wild-type S protein.
Furthermore, the Rg analysis is crucial to assess the global compactness of the system. R683A protein in the case of furin and R682A in the case of cathepsin shows the lowest average Rg value among all the mutant models compared to the wild-type S protein, thus serving as the respective model to achieve stable conformation and global compactness of the system for the two proteases. In the case of plasmin, however, all the mutant models have higher average Rg values than the wild-type S protein. For furin, R682A protein reveals a comparatively higher average Rg value from the wild-type. While for cathepsin, all mutant models show lower average Rg values than the wild-type S protein, with R685A showing the highest average Rg value among the four mutant models and almost comparable to the wild-type S protein. In addition, the R685A-cathepsin bound model also reveals a higher degree of fluctuations throughout the simulation. Unlike in the case of cathepsin, with plasmin, all the four mutant models show higher average Rg values than the wild-type S-protein. However, the highest average Rg value is recorded for the R685A protein, and wherein considerable minor fluctuations were also seen for this mutant model throughout the simulation.
Our analysis taking various parameters, RMSD, RMSF, SASA, and Rg, helped us decisively infer that the R683A mutant S-protein is the best mutant model for the stable and efficient binding of the three proteases: furin, cathepsin, and plasmin. Furthermore, we could also deduce that the R682A model serves as the best mutant model for the inefficient and insecure binding of furin and plasmin. Both R682A and R685A models showed high potential of unstable protease binding for cathepsin B.
These single amino acid substitutions in the cleavage site helped us understand the role of individual residue in the binding complex. The participation of these crucial amino acids at the boundary of S1 and S2 subunits in the proteolytic processing step will provide a unique opportunity to develop a lower pathogenic strain of SARS-CoV-2, which can further be used for vaccine development studies. Considering the fact that this in silico analysis proves that the single amino acid substitutions can help make an attenuated form of virus, further in vitro work is needed to validate the findings.
CRediT authorship contribution statement
Kamal Shokeen: Conceptualization, Data curation, Formal analysis, Methodology, Software, Visualization, Writing – original draft. Shambhavi Pandey: Conceptualization, Data curation, Formal analysis, Methodology. Manisha Shah: Conceptualization, Data curation, Formal analysis, Methodology. Sachin Kumar: Conceptualization, Methodology, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
The virus research in our laboratory is currently supported by the Department of Biotechnology, India (BT/PR24308/NER/95/644/2017 and BT/PR41246/NER/95/1685/2020) and the Department of Health Research, Government of India (NER/71/2020-ECD-I).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198845.
Appendix. Supplementary materials
References
- Abraham, M.J., Murtola, T., Schulz, R., Páll, S., Smith, J.C., Hess, B., Lindahl, E.J.S., 2015. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. 1, 19-25.
- Alexander D.J., Brown I.H. History of highly pathogenic avian influenza. Rev. Sci. Tech. 2009;28(1):19–38. doi: 10.20506/rst.28.1.1856. [DOI] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berri F., Rimmelzwaan G.F., Hanss M., Albina E., Foucault-Grunenwald M.L., Le V.B., Vogelzang-van Trierum S.E., Gil P., Camerer E., Martinez D., Lina B., Lijnen R., Carmeliet P., Riteau B. Plasminogen controls inflammation and pathogenesis of influenza virus infections via fibrinolysis. PLoS Pathog. 2013;9(3) doi: 10.1371/journal.ppat.1003229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biniossek M.L., Nagler D.K., Becker-Pauly C., Schilling O. Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S. J. Proteome Res. 2011;10(12):5363–5373. doi: 10.1021/pr200621z. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T.A., Berka K., Thornton J.M., Laskowski R.A. PDBsum additions. Nucleic Acids Res. 2014;42:D292–D296. doi: 10.1093/nar/gkt940. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Kramer-Kuhl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., von Hahn T., Simmons G., Hofmann H., Pohlmann S. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87(10):5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H., Wells K., Takada A., Kawaoka Y. Plasminogen-binding activity of neuraminidase determines the pathogenicity of influenza A virus. J. Virol. 2001;75(19):9297–9301. doi: 10.1128/JVI.75.19.9297-9301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey T.S., van Gunsteren W.F., Angeletti R.H. Techniques in Protein Chemistry IV. Academic Press; 1993. The application of chemical shift calculation to protein structure determination by NMR; pp. 615–622. [Google Scholar]
- Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4(4):557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre G. The proteolytic regulation of virus cell entry by furin and other proprotein convertases. Viruses. 2019;11(9):837. doi: 10.3390/v11090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H.L., Zhao R., Matalon S., Matthay M.A. Elevated plasmin(ogen) as a common risk factor for COVID-19 susceptibility. Physiol. Rev. 2020;100(3):1065–1075. doi: 10.1152/physrev.00013.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.W., Okumura Y., Kido H., Ng L.F., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One. 2009;4(11):e7870. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk H.D., Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2(2):39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- Kozakov D., Beglov D., Bohnuud T., Mottarella S.E., Xia B., Hall D.R., Vajda S. How good is automated protein docking? Proteins. 2013;81(12):2159–2166. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12(2):255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C.S., Kumar S. Species based synonymous codon usage in fusion protein gene of Newcastle disease virus. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0114754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz S.G., Choppin P.W. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975;68(2):440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- LeBouder F., Lina B., Rimmelzwaan G.F., Riteau B. Plasminogen promotes influenza A virus replication through an annexin 2-dependent pathway in the absence of neuraminidase. J. Gen. Virol. 2010;91(Pt 11):2753–2761. doi: 10.1099/vir.0.023804-0. [DOI] [PubMed] [Google Scholar]
- Leszczynski J., Shukla M.K. Springer; 2012. Practical Aspects of Computational Chemistry. [Google Scholar]
- Malhotra S., Mathew O.K., Sowdhamini R. DOCKSCORE: a webserver for ranking protein-protein docked poses. BMC Bioinform. 2015;16:127. doi: 10.1186/s12859-015-0572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed M.H., Kumar S., Paldurai A., Samal S.K. Sequence analysis of fusion protein gene of Newcastle disease virus isolated from outbreaks in Egypt during 2006. Virol. J. 2011;8:237. doi: 10.1186/1743-422X-8-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Towatari T., Ohuchi M., Shiota M., Akao M., Okumura Y., Parry M.A., Kido H. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur. J. Biochem. 2001;268(10):2847–2855. doi: 10.1046/j.1432-1327.2001.02166.x. [DOI] [PubMed] [Google Scholar]
- Nambala P., Su W.C. Role of Zika virus prM protein in viral pathogenicity and use in vaccine development. Front. Microbiol. 2018;9:1797. doi: 10.3389/fmicb.2018.01797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nao N., Yamagishi J., Miyamoto H., Igarashi M., Manzoor R., Ohnuma A., Tsuda Y., Furuyama W., Shigeno A., Kajihara M., Kishida N., Yoshida R., Takada A. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. mBio. 2017;8(1) doi: 10.1128/mBio.02298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova S.S., Solov’ev A.D. The origin of the method of steepest descent. Hist. Math. 1997;24(4):361–375. [Google Scholar]
- Sun X., Tse L.V., Ferguson A.D., Whittaker G.R. Modifications to the hemagglutinin cleavage site control the virulence of a neurotropic H1N1 influenza virus. J. Virol. 2010;84(17):8683–8690. doi: 10.1128/JVI.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V. Caister Academic Press; Norfolk: 2007. Coronaviruses: Molecular and Cellular Biology; p. UK2007. [Google Scholar]
- Vajda S., Yueh C., Beglov D., Bohnuud T., Mottarella S.E., Xia B., Hall D.R., Kozakov D. New additions to the ClusPro server motivated by CAPRI. Proteins. 2017;85(3):435–444. doi: 10.1002/prot.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69(4):635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Zhang W., Holdaway H.A., Li L., Kostyuchenko V.A., Chipman P.R., Kuhn R.J., Rossmann M.G., Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319(5871):1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Nunneley J.W., Barnard D., Pohlmann S., McKerrow J.H., Renslo A.R., Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.