Summary:
NTRK1/2/3 rearrangements have been identified as oncogenic drivers in a variety of tumors including those in the uterine cervix, and rarely, the uterine corpus. We report 2 cases of cervical sarcoma with NTRK gene rearrangements. Case 1 was a 54-yr-old woman who presented with postmenopausal bleeding and a 5.4 cm friable mass in the cervix. Microscopic examination of the tumor revealed proliferation of epithelioid and spindle cells arranged in alternating hypercellular and hypocellular areas, with subtle fibrosarcoma-like features. Coagulative tumor cell necrosis and readily recognizable mitoses (up to 40 mitotic figures per 10 high-power fields) were identified. Case 2 was a 52-yr-old woman who presented with abnormal vaginal bleeding and a 1.3 cm cervical mass. The resected cervical tumor showed proliferation of spindled cells with fascicular and storiform growth pattern, infiltrating into the smooth muscle with entrapment of normal endocervical glands. The tumor cells displayed mild cytologic atypia and low mitotic activity (1 mitotic figure per 10 high-power fields). The mixed inflammatory infiltrate was seen throughout the lesion, mimicking morphology of inflammatory myofibroblastic tumor. Immunohistochemical staining for S100 and CD34 demonstrated variable expression in case 1 and uniformly diffuse positivity in case 2. The tumor cells in both cases were focally positive for CD10, Cyclin D1, ER, and PR, and negative for AE1/AE3, desmin, SOX10, HMB-45. RNA fusion analysis identified SPECC1L-NTRK3 gene rearrangements in case 1 and TPM3-NTRK1 in case 2; DNA-based mutational analysis also revealed CDKN2A/B homozygous deletion in case 1. Despite accumulating literatures on NTRK fusion mesenchymal tumors in gynecologic pathology, these tumors are still rare and lack well-established morphologic diagnostic criteria. Diagnostic and clinical recognition of these tumors is critical given the potential patient benefit from targeted therapy.
Keywords: Cervix, NTRK fusion, Sarcoma, SPECC1L-NTRK3, TPM3-NTRK1
A pathway to precision medicine has become increasingly clear with the availability of advanced molecular techniques which are able to identify tumors with distinctive molecular alterations. The tropomyosin receptor kinase (Trk) family includes 3 transmembrane receptors (TrkA, TrkB, and TrkC) encoded by neurotrophic tyrosine receptor kinase (NTRK) genes NTRK1, NTRK2, and NTRK3 (1). Normally, these genes are predominantly expressed in neuronal tissue and have impacts on development and activity of the central and peripheral nervous system including neuronal cell differentiation, survival, and proliferation (1–3). Unlike physiological binding of neurotrophin to the extracellular portion of the Trk receptor, gene rearrangements involving C-terminus of catalytic tyrosine kinase domain of NTRK1/2/3 genes result in overexpression of Trk receptors. This leads to constitutive activation of downstream signaling cascades, particularly the PI3K/Akt/mTOR, RAS/MAPK/ERK, and PKC pathways, and subsequent carcinogenesis (4). NTRK1/2/3 rearrangements have been identified as oncogenic drivers in tumors at different anatomic sites (5–9).
Among the reported uterine sarcomas with NTRK rearrangement, TPM3-NTRK1 is the most common fusion, followed by a variety of other fusion partners including LMNA, TPR, RBPMS, EML4, and SPECC1L (10–15). As a group of tumors with distinct molecular alteration for which highly effective targeted therapy is available, recognition of these lesions by gynecologic pathologists is critical. Here, we report two cases of cervical sarcoma with NTRK-rearrangement, one with SPECC1L-NTRK3 and the other with TPM3-NTRK1 fusion. We comparatively describe their clinicopathologic features.
CASE STUDY
Clinicopathologic Findings
Case 1 was a 54-yr-old woman who presented with postmenopausal bleeding and a friable mass arising from the cervix. The biopsy specimen showed a spindle cell proliferation infiltrating around benign endocervical glands. On the basis of the morphology and immunoprofile, the diagnosis of spindle cell sarcoma with the possibility of NTRK fusion was rendered. Subsequently, she underwent radical hysterectomy, bilateral salpingo-oophorectomy and lymph node dissection. Gross examination of hysterectomy specimen revealed a 5.4×3.5×2.0 cm exophytic mass protruding from the endocervical canal with extension to the ectocervix. The mass had focal infiltrative borders but appeared to be confined to the cervix without involvement of lower uterine segment or vagina.
Microscopically, the hysterectomy specimen showed an exophytic cervical tumor composed of spindle cell proliferation with focal infiltrating pattern (Fig. 1A) and entrapped benign endocervical glands (Fig. 1B). Under low power, the tumor displayed alternating hypercellular and hypocellular areas of epithelioid and spindle cell proliferation (Fig. 1C). Focal hemangiopericytoma-like (Fig. 1D) and fibrosarcoma-like morphology (Fig. 1E) was present. The hypercellular areas were predominantly comprised of epithelioid cells with moderate nuclear atypia, coarse chromatin, small nucleoli, and moderate amount of eosinophilic cytoplasm (Fig. 2A). The hypocellular areas mainly consisted of spindle cells arranged in fascicles and cords with a myxoid background (Fig. 2B). Readily recognizable mitoses (Fig. 2C, up to 40 mitotic figures per 10 high-power fields) and coagulative tumor cell necrosis (Fig. 2D) were identified.
FIG. 1.
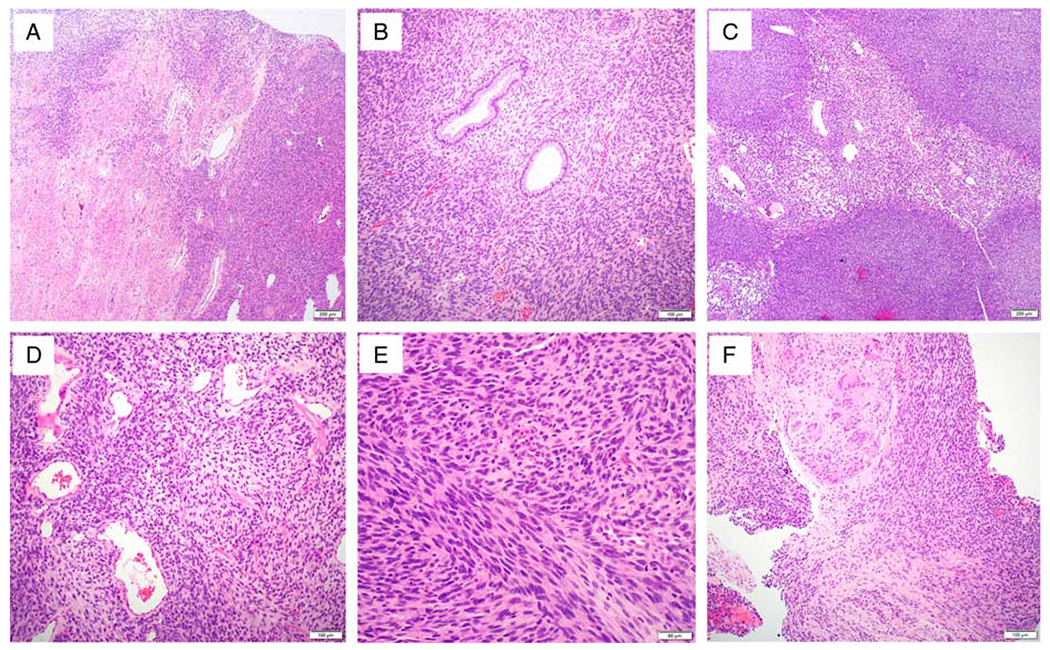
Histopathologic findings of case 1. The cervical tumor (SPECC1L-NTRK3 gene fusion) shows an exophytic spindle cell tumor with focal infiltrating pattern (A) and entrapped benign endocervical glands (B). The tumor cells are arranged in alternating hypercellular and hypocellular areas (C). The tumor focally exhibits subtle features of hemangiopericytoma-like vasculature (D) and fibrosarcoma-like morphology (E). Local recurrence involving vagina, adjacent to the prior procedure site (F).
FIG. 2.
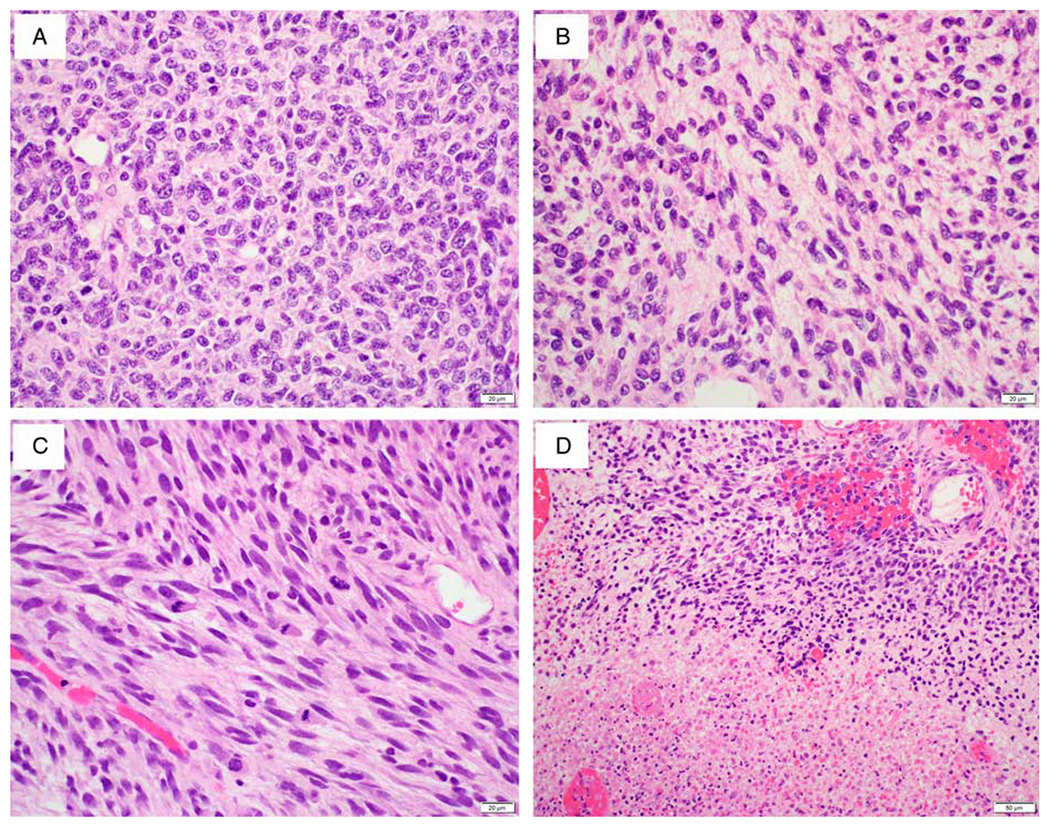
Histopathologic findings of case 1. The hypercellular areas in the tumor are predominantly comprised of epithelioid cells (A). The alternating hypocellular areas show more spindled cells arranged in fascicles and cords with a myxoid background (B). Readily recognizable mitoses (C) and coagulative tumor cell necrosis (D) are present.
Case 2 was a 52-yr-old woman who presented with abnormal vaginal bleeding and a cervical mass, phenotypically similar to that of case 1. Biopsy was performed and molecular testing of this specimen revealed TPM3-NTRK1 gene fusion. The patient underwent radiation followed by a radical hysterectomy, bilateral salpingo-oophorectomy and lymph node dissection. The hysterectomy specimen showed a relatively well-demarcated 1.3 × 1.3 × 0.9 cm tan-pink mass lesion, involving the cervix. Grossly, the mass appeared to be confined to the cervix without extension to lower uterine segment or vagina. Upon sectioning, the mass showed a smooth, glistening, and tan-pink cut surface.
Microscopic examination of the resected cervical tumor revealed a mass infiltrating into the smooth muscle (Fig. 3A) with entrapment of normal endocervical glands (Fig. 3B). The tumor was characterized as a spindle cell proliferation arranged in disordered fascicles (Fig. 3C) with focal storiform growth pattern (Fig. 3D). The tumor cells displayed mild cytologic atypia and low mitotic activity (1 mitotic figure per 10 high-power fields). The mixed inflammatory infiltrate including lymphocytes, eosinophils and plasma cells, was seen throughout the lesion (Figs. 3E, F). The thick-wall blood vessels focally showed a rim of collagen.
FIG. 3.
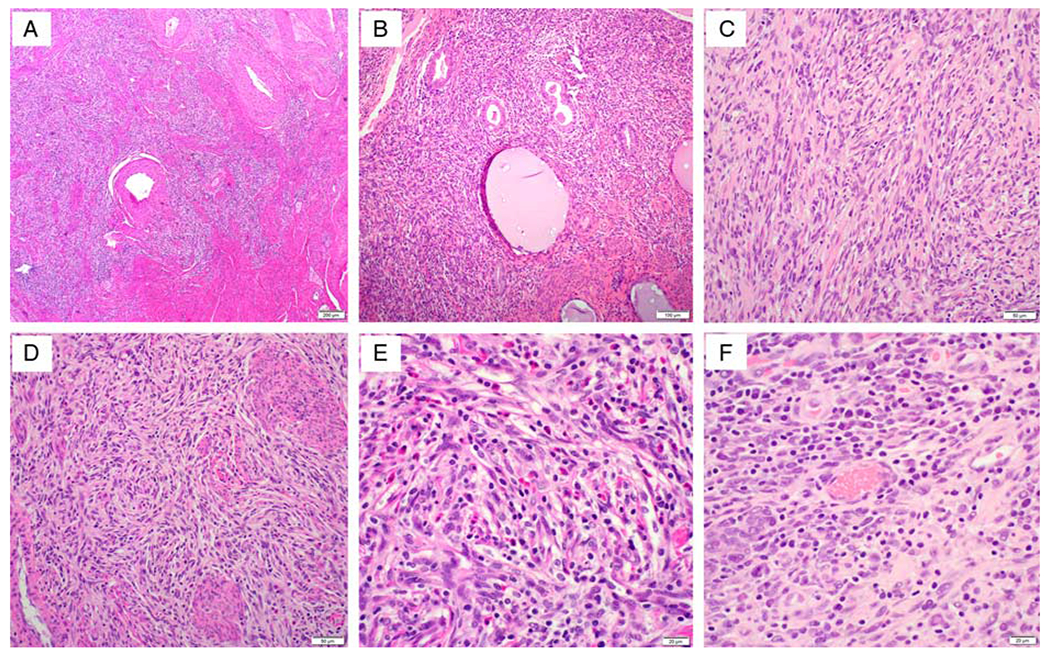
Histopathologic findings of case 2. Section from resected cervical tumor (TPM3-NTRK1 gene fusion) shows a mass lesion infiltrating into the smooth muscle (A) with entrapment of normal endocervical glands (B). The tumor is characterized as a spindle cell proliferation arranged in disordered fascicles (C) with focal storiform growth pattern (D). The cytology of tumor cells is relatively bland and the mixed inflammatory infiltrate including lymphocytes, eosinophils and plasma cells, is seen throughout the lesion (E, F).
Immunohistochemical Study
Immunohistochemical staining for case 1 was performed on both biopsy and hysterectomy specimens and demonstrated variable expression of S100 and CD34, ranging from focal to diffuse (Figs. 4A–E). The tumor cells exhibited focal expression of Cyclin D1, CD10 (15% of tumor cells with moderate intensity), ER (5% of tumor cells with strong intensity), and PR (20% of tumor cells with weak intensity); stains for AE1/AE3, desmin, SOX10, and HMB-45 were negative. P16 staining was completely negative with internal positive control (Fig. 4F).
FIG. 4.
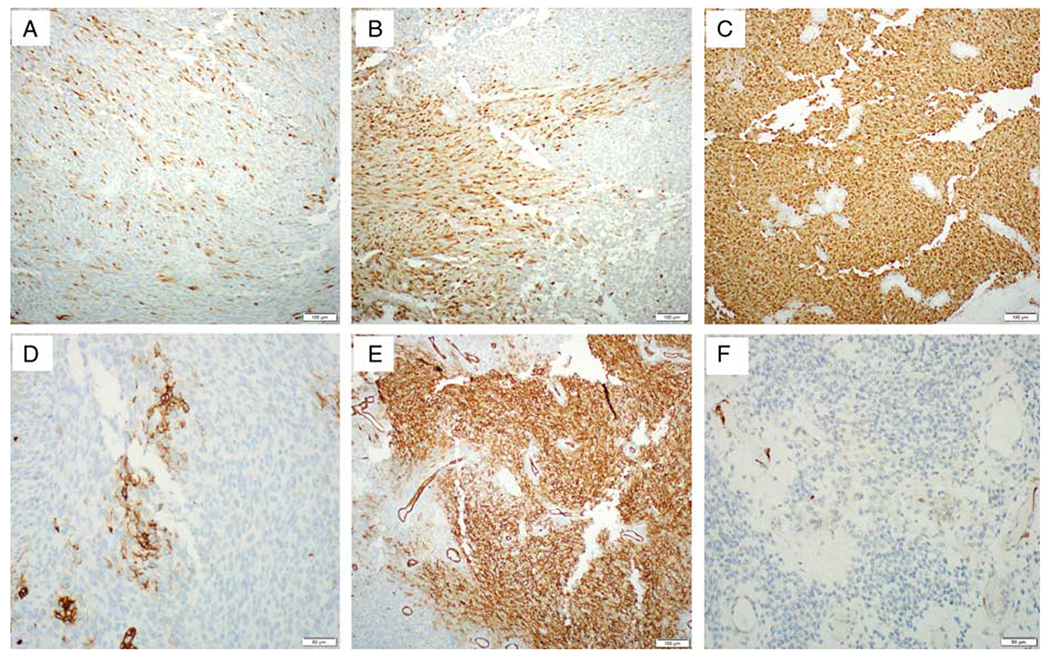
Immunohistochemical findings of case 1. S100 and CD34 immunostains show a variable staining pattern, ranging from focal to diffuse (S100, A–C; CD34, D, E). Loss of CDKN2A/B in the tumor cells in case 1 is confirmed by a negative immunostaining for p16 with positive internal control (F).
Likewise, immunohistochemical staining for case 2 was also performed on both biopsy and hysterectomy specimens and demonstrated uniformly diffuse expression of S100 and CD34 throughout the tumor (Figs. 5A, B) and focal expression of Cyclin D1, CD10 (20% of tumor cells with weak to moderate intensity), ER (15% of tumor cells with moderate intensity), and PR (10% of tumor cells with weak intensity). Stains for AE1/AE3, desmin, SOX10, and HMB-45 were negative. For this case, since some morphologic features indicated the possibility of inflammatory myofibroblastic tumor (IMT), immunostain for ALK was performed and demonstrated a negative pattern. Unlike a completely negative staining in case 1, stain for p16 in this case showed focal expression.
FIG. 5.
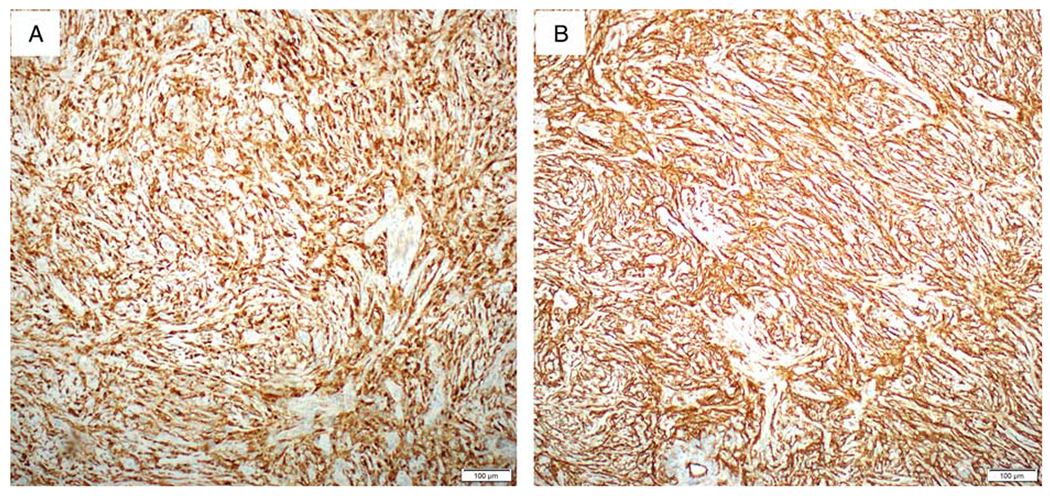
Immunohistochemical findings of case 2. The tumor cells are uniformly diffusely positive for S100 (A) and CD34 (B).
Molecular Analysis
To identify somatic mutations as well as diagnostic and actionable fusions, the tumor in case 1 (hysterectomy specimen) was sequenced using the Foundationone Heme platform (Foundation Medicine, Cambridge, MA) and the tumor in case 2 (biopsy specimen) was sequenced using Caris DNA and RNA sequencing platform (Caris Life Sciences, Phoenix, AZ). The former analysis utilizes DNA sequencing to interrogate the entire coding sequence of 406 genes, selected introns of 31 genes involved in rearrangements, and utilizes RNA sequencing to interrogate 265 genes that are known to be somatically altered in human hematologic malignancies and/or sarcomas including a broad range of gene fusions. The median exon coverage/sequencing depth for case 1 is ~870X. The Caris molecular profiling platform utilizes whole-exome DNA sequencing and whole transcriptome RNA sequencing to test point mutations, copy number alterations and gene fusions of ~22,000 genes. The average depth of coverage for DNA sequencing is ~1000X for 720+ clinical and research genes. A SPECC1L-NTRK3 gene fusion in case 1 and a TPM3-NTRK1 gene fusion in case 2 were detected, respectively. In case 1, the primary tumor also harbored CDKN2A/B homozygous deletion with a microsatellite-stable status with no tumor mutational burden (0 mutation/Mb). Loss of CDKN2A/B in the tumor cells was confirmed by a negative immunostaining for p16 with positive internal control (Fig. 4F). Several variants of unknown significance were also detected in this tumor: FGFR4, FLYWCH1, HNF1A, KMT2A (MLL), MED12, NF1, TNFRSF17, TSC2, and WHSC1 (MMSET). In case 2, a possible pathogenic variant c.594+2_594+5delTGAG in the exon 7 of ERCC2 gene was detected. The tumor displayed a microsatellite-stable status and low tumor mutational burden (1 mutation/Mb). Similarly, several variants of unknown significance were detected in this tumor: NFE2L2, NPM1, PIK3CB, PMS2, PRKACA, PTPN11, and RB1. Deletion of CDKN2A/B was not detected in this case.
Follow-up
After 8 mo of hysterectomy, the patient of case 1 presented with local recurrence involving vagina and a portion of bladder, adjacent to the prior procedure site (Fig. 1F). Although no gross residual tumor was present after re-excision, microscopic sections showed margin involvement that was unresectable due to limitation of anatomical location. Since the tumor harbored an NTRK fusion, she has been treated with NTRK inhibitor larotrectinib by tumor board recommendation. After hysterectomy, the patient of case 2 remained no evidence of disease during 6-mo follow-up.
DISCUSSION
We report 2 cases of spindle cell sarcoma of the uterine cervix with NTRK fusions. As newly described tumors in the female genital tract, NTRK-rearranged uterine sarcomas represent a subset of uterine sarcomas with distinct clinicopathologic features. In the female genital tract, these tumors predominantly affect premenopausal women in their 20s to late 40s, with a median age of 32 yr (15), and occasionally postmenopausal patient (12). The age of our patients (54 and 52 yr) is notably older than typically seen.
On the basis of the morphology, there is a concern the tumor in case 1 may behave like a high-grade sarcoma. At the molecular level, this tumor harbored a SPECC1L-NTRK3 gene rearrangement which, until now, has been described in only 3 uterine mesenchymal tumors (11,12,16). The tumor in case 2 harbored a gene fusion involving TPM3-NTRK1, the most common translocation reported in the NTRK tumors of the uterine cervix. Similar to other NTRK tumors, TPM3-NTRK1 fusion tumors can exhibit variable morphology. In this case, the tumor cells displayed features concerning for a differential diagnosis of IMT. In fact, IMTs with ETV6-NTRK3 gene rearrangement without ALK fusion have been reported in lung and uterus (17,18). However, it is difficult to ascertain whether these tumors truly represent IMTs or should be placed in the category of NTRK fusion tumors.
As oncogenic drivers, it can be expected that various NTRK fusions may confer different transforming capacity that give rise to tumors with various biological behavior and phenotype. However, our result and published data demonstrated that, even with the same driver gene fusion, the tumor may behave in clinically distinct manners. In theory, rare additional genetic alterations, which might be acquired during tumor progression and related to tumor behavior, can occur in the NTRK fusion tumors. In fact, CDKN2A/B deletion is considered one of most common genetic changes in these tumors. In one study, it has been found that 4 of 8 soft tissue sarcoma patients with NTRK fusion also harbored CDKN2A/B deletion (19). Another study showed that all 7 uterine NTRK-tumors harbored CDKN2A loss (4 homozygous and 3 heterozygous) (13). Consistent with results, CDKN2A/B homozygous deletion has been detected in primary tumor of case 1 for which the tumor harbored SPECC1L-NTRK3 fusion. CDKN2A is a tumor suppressor gene that encodes the proteins p16INK4A and p14ARF that regulate cell cycle progression. Located on chromosome 9p21, CDKN2A shows frequent homogenous deletion in a wide range of human cancers. The significance of CDKN2A deletion in NTRK fusion tumors remains unclear and warrants further investigation.
Immunohistochemically, pediatric mesenchymal tumors with NTRK1 or NTRK2 gene rearrangement commonly exhibited cytoplasmic staining for pan-Trk, whereas tumors with NTRK3 gene rearrangement showed nuclear +/− cytoplasmic staining (20). Although pan-Trk is a relatively sensitive and specific marker for NTRK gene rearrangements in soft tissue mesenchymal tumors, 6% of leiomyosarcomas and up to 91% of high-grade endometrial stromal sarcomas displayed cytoplasmic and/or nuclear pan-Trk staining of variable extent and intensity, indicating its nonspecificity in gynecologic sarcomas (10,21). It has also been reported that NTRK fusion tumors commonly co-express S100 and CD34, but lack SOX10 expression (15). In fact, the tumor cells in our both cases were focally to diffusely positive for S100 and CD34, but negative for SOX10. As such, it has been proposed that S100 and/or CD34 immunohistochemistry can be used as screening tools for NTRK fusion tumors. Interestingly, some studies have demonstrated that S100 and CD34 co-expression can be seen in fusion tumors involving non-NTRK genes such as RAF1 and BRAF (22). Therefore, an NTRK fusion-associated neoplasm can be unequivocally diagnosed by molecular or cytogenetic test, rather than solely based on the morphology and immunoprofile.
NTRK fusion tumors can arise in varying sites including lower female genital tract, and recognition of NTRK fusion tumors are of clinical significance given the availability of FDA-approved highly effective Trk inhibitors such as larotrectinib and entrectinib. It has been demonstrated that high response rates (> 75%) have been achieved for patients with NTRK fusion tumors treated with these Trk inhibitors, regardless of tumor histology and location (23). A recent study reported a larotrectinib-treated patient with cervical sarcoma harboring SPECC1L-NTRK3 gene rearrangement (11). Likewise, our first patient has been treated with lacotrectinib since 2mo ago and the clinical response is under observation.
In summary, we report 2 cases of NTRK fusion sarcomas of the uterine cervix with SPECC1L-NTRK3 and TPM3-NTRK1 gene rearrangement, respectively. It is important to recognize that these 2 cases, like other NTRK fusion mesenchymal tumors, display different morphologic features, posing a diagnostic challenge for practicing pathologists. Despite accumulating literatures on NTRK fusion mesenchymal tumors in gynecologic pathology, these tumors are still rare and lack well-established morphologic diagnostic criteria. Continuous awareness and recognition of these uterine tumors is important given the potential patient benefit from targeted therapy.
Acknowledgments
This study is supported by a Clinician Scientist Award at The Johns Hopkins University School of Medicine (D.X.) and partially supported by the Pilot Project Award by the Cervical Cancer SPORE program at Johns Hopkins (D.X.).
Footnotes
The authors declare no conflict of interest.
REFERENCES
- 1.Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018;15:731–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikoletopoulou V, Lickert H, Frade JM, et al. Neurotrophin receptors TrkA and TrkC cause neuronal death whereas TrkB does not. Nature 2010;467:59–63. [DOI] [PubMed] [Google Scholar]
- 3.Rubin JB, Segal RA. Growth, survival and migration: the Trk to cancer. Cancer Treat Res 2003;115:1–18. [DOI] [PubMed] [Google Scholar]
- 4.Amatu A, Sartore-Bianchi A, Bencardino K, et al. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann Oncol 2019;30(suppl 8):viii5–viii15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon JP, Linkov I, Rosado A, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol 2020;33:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siozopoulou V, Smits E, De Winne K, et al. NTRK fusions in sarcomas: diagnostic challenges and clinical aspects. Diagnostics (Basel) 2021;11:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong DD, Vargas AC, Bonar F, et al. NTRK-rearranged mesenchymal tumours: diagnostic challenges, morphological patterns and proposed testing algorithm. Pathology 2020;52:401–9. [DOI] [PubMed] [Google Scholar]
- 8.Antonescu CR. Emerging soft tissue tumors with kinase fusions: an overview of the recent literature with an emphasis on diagnostic criteria. Genes Chromosomes Cancer 2020;59:437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croce S, Hostein I, McCluggage WG. NTRK and other recently described kinase fusion positive uterine sarcomas: a review of a group of rare neoplasms. Genes Chromosomes Cancer 2021;60:147–59. [DOI] [PubMed] [Google Scholar]
- 10.Chiang S, Cotzia P, Hyman DM, et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol 2018;42:791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabban JT, Devine WP, Sangoi AR, et al. NTRK fusion cervical sarcoma: a report of three cases, emphasising morphological and immunohistochemical distinction from other uterine sarcomas, including adenosarcoma. Histopathology 2020;77:100–11. [DOI] [PubMed] [Google Scholar]
- 12.Hodgson A, Pun C, Djordjevic B, et al. NTRK-rearranged cervical sarcoma: expanding the clinicopathologic spectrum. Int J Gynecol Pathol 2021;40:73–77. [DOI] [PubMed] [Google Scholar]
- 13.Croce S, Hostein I, Longacre TA, et al. Uterine and vaginal sarcomas resembling fibrosarcoma: a clinicopathological and molecular analysis of 13 cases showing common NTRK-rearrangements and the description of a COL1A1-PDGFB fusion novel to uterine neoplasms. Mod Pathol 2019;32:1008–22. [DOI] [PubMed] [Google Scholar]
- 14.Boyle W, Williams A, Sundar S, et al. TMP3-NTRK1 rearranged uterine sarcoma: a case report. Case Rep Womens Health 2020;28:e00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang S. S100 and Pan-Trk staining to report NTRK fusion-positive uterine sarcoma: Proceedings of the ISGyP Companion Society Session at the 2020 USCAP Annual Meeting. Int J Gynecol Pathol 2021;40:24–27. [DOI] [PubMed] [Google Scholar]
- 16.Gatalica Z, Xiu J, Swensen J, et al. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol 2019;32:147–53. [DOI] [PubMed] [Google Scholar]
- 17.Alassiri AH, Ali RH, Shen Y, et al. ETV6-NTRK3 is expressed in a subset of ALK-negative inflammatory myofibroblastic tumors. Am J Surg Pathol 2016;40:1051–61. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi A, Kurosawa M, Uemura M, et al. Anaplastic lymphoma kinase-negative uterine inflammatory myofibroblastic tumor containing the ETV6-NTRK3 fusion gene: a case report. J Int Med Res 2018;46:3498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doebele RC, Davis LE, Vaishnavi A, et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov 2015;5:1049–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudzinski ER, Lockwood CM, Stohr BA, et al. Pan-Trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. Am J Surg Pathol 2018;42:927–35. [DOI] [PubMed] [Google Scholar]
- 21.Momeni-Boroujeni A, Mohammad N, Wolber R, et al. Targeted RNA expression profiling identifies high-grade endometrial stromal sarcoma as a clinically relevant molecular subtype of uterine sarcoma. Mod Pathol 2021;34:1008–16. [DOI] [PubMed] [Google Scholar]
- 22.Suurmeijer AJH, Dickson BC, Swanson D, et al. A novel group of spindle cell tumors defined by S100 and CD34 co-expression shows recurrent fusions involving RAF1, BRAF, and NTRK1/2 genes. Genes Chromosomes Cancer 2018;57:611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med 2018;378:731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


