We report a patient with a broad and lethal EBV infection mainly characterized by disseminated EBV-associated smooth muscle tumors. The patient was a carrier of a homozygous loss-of-function mutation in TNFSF9 that impaired expansion of EBV-specific T cells.
Abstract
Epstein–Barr virus (EBV) can infect smooth muscle cells causing smooth muscle tumors (SMTs) or leiomyoma. Here, we report a patient with a heterozygous 22q11.2 deletion/DiGeorge syndrome who developed a unique, broad, and lethal susceptibility to EBV characterized by EBV-infected T and B cells and disseminated EBV+SMT. The patient also harbored a homozygous missense mutation (p.V140G) in TNFSF9 coding for CD137L/4-1BBL, the ligand of the T cell co-stimulatory molecule CD137/4-1BB, whose deficiency predisposes to EBV infection. We show that wild-type CD137L was up-regulated on activated monocytes and dendritic cells, EBV-infected B cells, and SMT. The CD137LV140G mutant was weakly expressed on patient cells or when ectopically expressed in HEK and P815 cells. Importantly, patient EBV-infected B cells failed to trigger the expansion of EBV-specific T cells, resulting in decreased T cell effector responses. T cell expansion was recovered when CD137L expression was restored on B cells. Therefore, these results highlight the critical role of the CD137–CD137L pathway in anti-EBV immunity, in particular in the control of EBV+SMT.
Introduction
EBV is an oncogenic virus of the Gamma-herpesvirinae subfamily. During coevolution with its human host, EBV succeeded in establishing latent infection throughout lifetime through infection of B lymphocytes and by persisting in the B cell memory pool. To reach this goal, several viral proteins divert the normal B cell biology by delivering proliferative and anti-apoptotic signals. A bystander consequence of EBV persistence, thus, is the high EBV-related cancer burden in all human populations, which are mostly lymphomas of B cell origin. However, occasionally EBV also infects and persists in other cell types, including T and natural killer (NK) lymphocytes, smooth muscle cells, and epithelial cells. Like in B cells, persistence of EBV may lead to tumorigenesis of infected cells, evolving toward non-malignant and malignant T/NK lymphoproliferative diseases (LPDs), EBV+ smooth muscle tumors (SMTs, also known as leiomyoma), and nasopharyngeal and gastric carcinomas, respectively (Farrell, 2019).
The importance of the immune system to control EBV day by day is exemplified by EBV-associated diseases occurring in the context of immune deficiencies, notably primary immune deficiencies (PIDs). Patients affected by PID, AIDS, or treated with immunosuppressive drugs following organ transplantation develop mostly B cell LPDs, sometimes EBV+ SMTs, and very rarely T/NK cell LPDs. The study of PID has clarified the different layers of anti-EBV immunity, including the function of proteins and pathways selectively or particularly required for an efficient immune response to EBV (Latour and Fischer, 2019). Protein defects involved in T/NK cell cytotoxicity toward EBV-infected cells result mostly in hemophagocytic lymphohistiocytosis (HLH), an inflammatory condition caused by over-activated T cells, but rarely in the occurrence of B cell lymphoma. Defects in components required for proliferation and expansion of EBV-specific T cells cause B cell LPD in most of the cases, although HLH can occur occasionally. Recently, defects in several co-stimulatory molecules of the TNF receptor family have been associated with B-cell LPD (CD70, CD27, TNFRSF9 [CD137 or 4-1BB]), and in one case to T cell LPD (TNFRSF9), while EBV+ SMTs have been reported in patients with deficiencies in RLTPR/CARMIL2 and RASGRP1, the two intracellular signaling molecules involved in T cell activation and co-stimulation (Tangye and Latour, 2020). In these PIDs, however, the penetrance is not complete and expressivity may vary. Furthermore, the physio-pathological mechanisms underlying EBV-driven T/NK cell LPDs and EBV+ SMTs are poorly understood. Notably, they likely involve additional genetic factors to result in overt clinical diseases, as we showed recently for T/NK cell PLDs in the context of a CD137 deficiency (Rodriguez et al., 2019). In this study, the patient who developed a fatal T cell LPD was also a carrier of a homozygous loss-of-function mutation in PIK3CD, while his asymptomatic sister had only a CD137 deficiency.
Herein, we report a patient with a persistent and broad EBV infection in several cell subsets including B, T, and smooth muscle cells as the result of a 22q11.2 deletion and a homozygous mutation in TNFSF9 coding for the ligand of CD137 (CD137L). We provide evidence that CD137L is key to triggering expansion of EBV-specific T cells, highlighting the critical role of the CD137–CD137L pathway in immunity to EBV.
Results and discussion
Broad and restricted EBV susceptibility in a patient affected by disseminated EBV+ SMTs
We investigated an 18-yr-old (y.o.) female patient, born from a consanguineous family of Moroccan origin. She presented disseminated EBV+ SMTs since 9 yr of age, incidentally diagnosed during an appendicitis episode. Over a 9-yr period, SMTs successively appeared in different body areas, including the rectum, paracecal area, upper and lower limbs, external face of the left iliac bone, the spleen, lungs, and liver (Fig. 1, A and B). Tumors grew slowly and were not invasive. Immunostaining of the splenectomy revealed EBV+ SMT cells that were associated with EBV-infected B and T cells in the red pulp. Several treatment strategies were ineffective and included surgery (total splenectomy at 15 y.o. and removal of accessible tumors), chemotherapy for 6 mo at 9 y.o., and rapamycine at 15 y.o. Nonetheless, the lesions were painful and significantly restricted her mobility and quality of life. One compressive proximal bronchial lesion severely damaged lung parenchyma, which was subsequently subjected to repeated bacterial infections, eventually responsible for the fatal outcome at the age of 18 yr. In addition, she presented adenopathies in the cervical, axillar, inguinal, and mediastinal areas that remained stable since 9 y.o. and were not further investigated. EBV loads increased over time and reached high and stable levels at the age of 18 yr (Fig. 1 C). She had a persistent incomplete EBV seroconversion (negative anti–viral capsid antigen IgM and positive anti–viral capsid antigen IgG, but negative anti-EBNA IgG). No other symptoms of immune deficiency, such as viral, bacterial infections (except for those consequent of the mechanical lung lesion) or autoimmunity were noted. However, she exhibited non-hematological clinical features including delayed milestones, mild intellectual defect, growth retardation, severe amyotrophy, and palatine insufficiency. Among the five other siblings, one brother developed an EBV− B cell lymphoma at 20 y.o. successfully treated by chemotherapy. He also showed an apparent incomplete EBV seroconversion, albeit it may have been biased by the rituximab treatment he received. However, his EBV PCR was negative. The four other siblings were healthy.
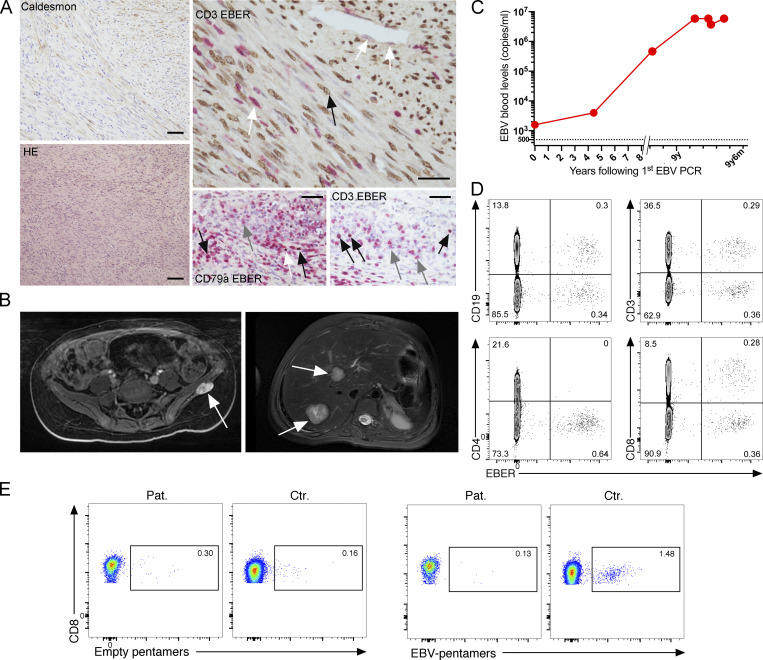
Figure 1.
Broad susceptibility to EBV in a patient affected by chronic and disseminated EBV+ smooth muscle tumors. (A) Smooth muscle tumor histology at 15 y.o. obtained by splenectomy. Left lower panel, hematoxylin–eosin (HE) staining showing fascicles of spindle-shaped smooth muscle cells with rare interspersed lymphocytes (magnification, ×100). Left upper panel, cytoplasmic staining with anti-caldesmon antibody on tumoral cells (magnification, ×200; scale, 90 μM). Right upper panel, co-staining with an EBER probe and anti-CD3 antibody reveals that tumoral cells are infected by EBV (plain black arrow) while endothelial cells and some infiltrating T cells are negative (white arrows, upper right part and lower left part, respectively; magnification, ×400; scale, 40 μM). Right lower panels, co-staining with an EBER probe and anti-CD79a or anti-CD3 antibodies in the splenic red pulp reveals both infected (black arrows) and non-infected (gray arrows) T and B cells. The white arrow indicates a CD79a-negative infected cell (magnification, ×400; scale, 30 μM). (B) Magnetic resonance images of the EBV+ smooth muscle tumors of the left iliac bone at 9 y.o. (left panel) and liver at 17 y.o. (right panel). (C) EBV blood loads in the patient at different ages (years, y; months, m) starting from the first EBV PCR at the age of 9 y.o. (D) FACS dot-plots of EBER expression in PBMCs of the patient at the age of 18 y.o. by primeflow assay coupled with anti-CD19, anti-CD3, anti-CD4, and anti-CD8. Similar findings obtained at 17 y.o. All dot-plots are gated on efluor−CD14− cells. (E) FACS dot-plots of co-staining of PBMCs of the patient at 18 y.o. and a control with anti-CD8 and HLA-A02–restricted empty pentamers (left panels) or EBV pentamers (right panels) expression in PBMCs of the patient (Pat.) and control (Ctr.). Cells gated on CD3+CD8+. No staining was observed in the CD4+ cells. Similar findings were obtained with another sample.
Study of EBV-infected cell subsets in peripheral blood of the patient by EBV-encoded small RNA (EBER) flowfish assay at 17 and 18 y.o. revealed circulating EBV-infected B cells and CD8 T cells (Fig. 1 D). EBV+ CD8 T cells were retrospectively uncovered on spleen histology as well. Immune phenotyping revealed a strong decrease of memory B cells but normal Ig level, normal T cell lymphocyte counts with decreased proportions of CD4 and CD8 naive T cells including recent naive thymic emigrants (25 cells mm−3), and decreased T cell proliferation in response to the tuberculin antigen (Table S1). Both CD4 and CD8 T effector memory cells harbored a strong expression of PD-1 (Fig. S1 A) that suggested a chronic activation of T cells. MHC-I HLA-A*02–restricted EBV-specific T cells were not detectable in peripheral blood mononuclear cells (PBMCs) of the patient who expressed HLA-A*02 molecules, while in PBMCs of HLA-A*02–positive healthy donors these cells were detected (Fig. 1 E). T cell functions and pathways involved in immune deficiencies associated with EBV susceptibility were evaluated. TCR activation signals including global tyrosine phosphorylation, NFAT2, PLC-γ1, ERK and AKT activation, and Ca++flux were tested in T cell blasts obtained from PBMCs of the patient. All were found to be comparable to those of control blasts from two healthy donors (Fig. S1, B–E). However, CD3-induced T cell proliferation was reduced when compared with control cells, while CD3-induced degranulation of CD8+ T cells was unaffected (Fig. S1, D and F). This diminution in CD3-induced proliferation may explain why the CD137-dependent and especially CD70-dependent CD3-triggered T cell proliferation were also reduced (Fig. S1, G and H). High levels of PD1 on T cells and accumulation of exhausted memory T cells may account for the decreased proliferation of patient T cells in these assays. Taken together, these defects were in favor of primary combined immunodeficiency in the patient.
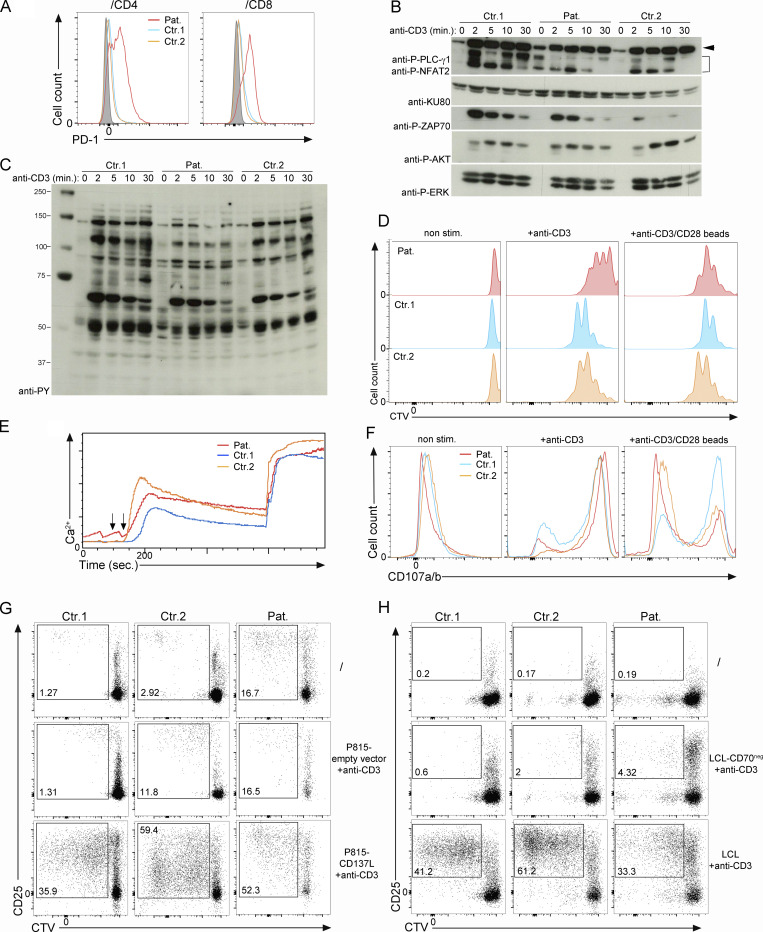
Figure S1.
Functional characterization of T cells from the patient. (A) FACS histograms of PD-1 gated on CD4 and CD8 memory T cells from PBMCs from two control donors (Ctr.1 and Ctr.2) and the CD137L-deficient patient (Pat.). Isotype control in gray. (B and C) Analysis of TCR signaling in T cell blasts from two controls donor (Ctr.1 and Ctr.2) and the CD137L-deficient patient (Pat.). Immunoblots showing the phosphorylation of proximal signaling molecules after stimulation with anti-CD3 antibodies for 0, 2, 5, 10, and 30 min. (B) Cell lysates immunoblotted with antibodies against phospho-PLC-Ɣ1 (P-PLC-Ɣ1), phospho-ERK1/2 (P-ERK), phospho-AKT (P-AKT), phospho-ZAP-70 (P-ZAP70), NFAT2 and KU80 as a loading control. Anti–P-PLC-Ɣ1 and anti-NFAT2 were tested on the same membrane. Data representative of three experiments. (C) Cell lysates immunoblotted with antibodies against tyrosine phosphorylated residues to analyze global tyrosine phosphorylation signals. Data representative of three experiments. (D–F) Analyses of T cell responses of T cell blasts from two controls (Ctr.1 and Ctr.2) and the patient (Pat.). (D) FACS histograms showing T cell blast proliferation profiles determined by dilution of CTV in response to immobilized anti-CD3 antibody at the indicated concentrations or anti-CD3/CD28–coated beads. Data representative of two experiments. (E) Flow cytometry analyses of Ca2+ flux in T cell blasts loaded with the Ca2+-sensitive fluorescent dye Indo1. Cells then stimulated with anti-CD3 (first arrow) crosslinked with rabbit anti-mouse (second arrow). Intracellular Ca2+ levels expressed in AU. Data representative of two experiments. (F) Degranulation of CD8+ T cell blasts stimulated with the indicated concentrations of anti-CD3. Cells stained with antibodies against CD107a/b (LAMP1/2), a surface-exposed marker of the secretion of lytic granules. Data representative of two experiments. (G and H) FACS dot-plots of CD137-dependent (G) or CD70-dependent (H) proliferation assays of PBMCs labeled with the CTV dye and cocultured in G with irradiated P815 cells expressing WT CD137L or an empty vector or in H with irradiated LCL cells expressing CD70 (LCL) or not (LCL-CD70neg). Irradiated LCL and P815 cells were preincubated with anti-CD3 antibody (+anti-CD3) before being added to the PBMCs. Non-stimulated PBMCs were cultured without irradiated LCL or P815 cells. Staining with anti-CD25 antibody. CD25 as activation marker and gating on CD3+ T cells. Proliferating T cells in the gates and numbers indicating proportions of cells in the gates. (G and H) Data representative of two experiments. Source data are available for this figure: SourceData FS1.
A homozygous private TNFSF9 mutation segregates with EBV susceptibility
Since EBV+ SMT has been reported in RASGRP1 and RLTPR deficiencies, RASGRP1 and RLTPR protein expressions were evaluated in lysates from T cell blasts of the patient and found to be normally expressed (data not shown). Whole exome sequencing (WES) was then performed on the patient to identify a genetic cause that could explain her peculiar susceptibility to EBV. The first finding was a de novo heterozygous 22q11.2 deletion (22q11.2del) of 3-Mb confirmed by multiplex ligation probe amplification (MLPA) that corresponds to the most common deletion causing the 22q11.2del syndrome or DiGeorge syndrome (Edelmann et al., 1999). This likely explains the intellectual defect, growth retardation, and palatine insufficiency of the patient, known to be typical features of the DiGeorge syndrome (Cancrini et al., 2014; Campbell et al., 2018). None of the other siblings presented the 22q11.2 deletion. In some cases, DiGeorge syndrome has been found to be associated with recurrent opportunistic infections and/or autoimmunity due to thymic aplasia and reduced T cell numbers, which is not the case of the patient herein. However, the patient exhibited significantly reduced numbers of naive T cells that may be the consequence of the DiGeorge syndrome. Increased but low risk of malignancy is also associated with DiGeorge syndrome (Morsheimer et al., 2017; Lambert et al., 2018). However, no SMT has been reported in large series of cancers occurring in patients with 22q11.2del, including those with symptoms of immunodeficiency (Kebudi et al., 2019; Mayor et al., 2018). Since EBV+ SMT develops during profound and broad immunodepression such as AIDS (McClain et al., 1995) or transplantation (Lee et al., 1995), we concluded that the 22.q11.2del alone could not be directly responsible for the peculiar vulnerability to EBV infection in the patient.
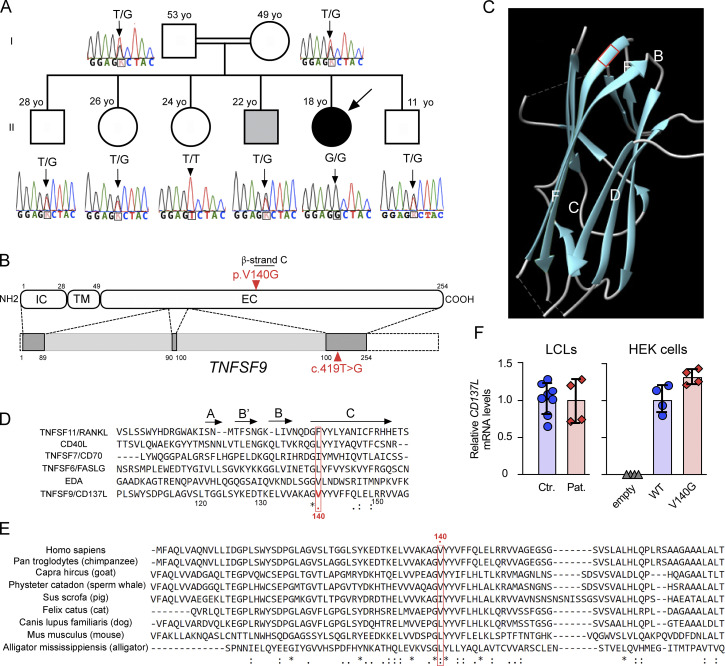
Accordingly, WES was further analyzed to identify other genetic event(s) that might explain this selective infectious susceptibility. The consanguinity of the family suggested an autosomal recessive inheritance (Fig. 2 A). Hence, we focused on rare homozygous mutations (listed in the Materials and methods). One significant variation in the gene TNFSF9 (g.9:6534730T>G, the c.419T>G), which translates into p.V140G, was identified (Fig. 2 B). This variation was not found in whole exome and genome public databases, including gnomAD and our own exome database at IMAGINE Institute that includes more than 20,600 exomes. TNFSF9 encodes CD137L protein (also known as 4-1BBL), the ligand of the T cell co-stimulatory molecule CD137/4-1BB. Importantly, homozygous loss-of-function mutations in the TNFRSF9 gene encoding CD137 (4-1BB) have been recently reported to cause EBV susceptibility (Rodriguez et al., 2019; Alosaimi et al., 2019; Somekh et al., 2019). No other family members had the homozygous c.419T>G variation. Parents and four siblings including the brother suffering from EBV− B cell lymphoma were heterozygous carriers of the variation (Fig. 2 A). One sibling was a homozygous carrier of the WT allele. Therefore, the homozygous c.419T>G mutation in TNFSF9 segregated with the EBV susceptibility in the family. Like most of the other TNF ligands, CD137L forms a trimer with a bell-shape structure (Gilbreth et al., 2018). Each monomer is composed of an intracellular, a transmembrane, and an extracellular domain. The extracellular domain consists of a TNF homology domain, critical for trimer formation and interaction with CD137. In the TNF homology domain, several β-strands form an inner sheet (A’AHCF strands) and an outer sheet (B’BGDE strands) depending on their position in the final trimer. The inner sheet is involved in trimer formation (Gilbreth et al., 2018; Li et al., 2018). The V140 amino acid is localized in the conserved C β-strand and has been previously found to be directly involved in trimer formation (Gilbreth et al., 2018; Bitra et al., 2019; Won et al., 2010; Fig. 2, C and D). The valine is not fully conserved across species, but alternative amino acids are systematically branched amino acids, such as leucine or isoleucine (Fig. 2 E). This motif is also strongly conserved in other TNFSF/TNF ligands. The importance of this residue is also highlighted when considering other TNFSF involved in autosomal recessive human diseases (RANKL, CD40L, CD70, FASLG, and EDA), in which no homozygous mutation is present in the gnomAD database for their corresponding amino acid (respectively F213, L168, I103, L188 and T300; Fig. 2 D). Altogether, these data suggested that the homozygous V140G variation in TNFSF9 was a valuable genetic candidate to explain EBV susceptibility in the patient.
Figure 2.
Identification in the patient of a private homozygous mutation in TNFSF9. (A) Family pedigree with DNA electropherograms and the TNFSF9 c.419T>G genotype of each individual. The proband is indicated by an arrow. The brother affected by an EBV− B cell lymphoma corresponds to the gray square. (B) Schematic representation of TNFSF9 intron–exon organization (intron and coding regions in light and dark gray boxes, respectively, and non-coding exon sequences in dashed boxes) and protein structure (extracellular [EC], transmembrane [TM], and intracytoplasmic [IC] domains) above. The mutation is indicated in red. (C) Ribbon representation of the 3D structure model of human CD137L/4-1BBL (Protein Data Bank accession no. 6D3N). The position of the mutated amino acid is framed by a red square, at the start of the C β-strand. The V140 is known to be directly involved in the CD137L trimerization. (D) Alignment of the human TNFSF9 sequence with that of TNFSF members, whose mutations are involved in human diseases (TNFS11/RANKL, CD40L, CD70/TNFSF7, FASLG/TNFSF6, and EDA). The position 140 of the mutated amino acid is framed by a red square. Alignments were generated using ClustalW software. "*" indicates a position that has a single, fully conserved residue; ":" indcates a position with a conserved residue and residues with strongly similar properties; "." indicates a position with a conserved residue and residues with weakly similar properties. (E) Alignment of the human TNFSF9 sequence with several other species. The position 140 of the mutated amino acid is framed by a red square. Alignments were generated using ClustalW. Observed secondary structures (β-strands) as previously described (Izawa et al., 2017) are shown above the sequences. (F) Relative mRNA CD137L expression levels from real-time PCR in control LCLs (Ctr.), in LCLs of the patient (Pat.), and in HEK cells ectopically expressing WT CD137L, CD137LV140G or not (empty vector). Levels of TNFSF9/CD137 were normalized to GAPDH. Levels relative to the mean of control or CD137LV140G quadruplet values corresponding to 1. Two control LCLs tested.
Decreased CD137L protein expression in cells of the patient
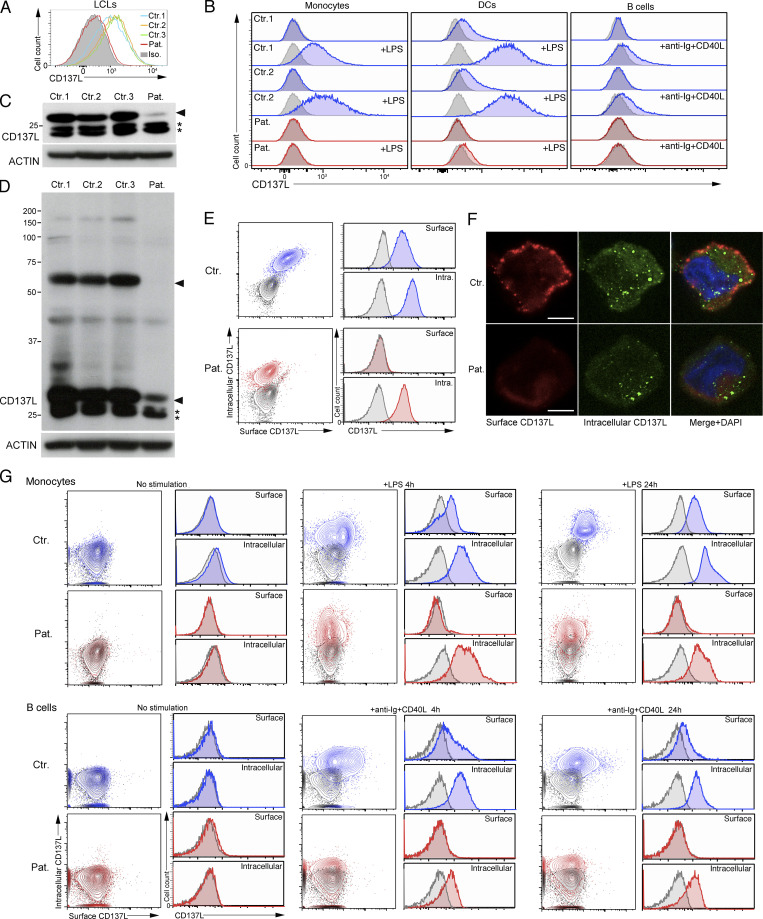
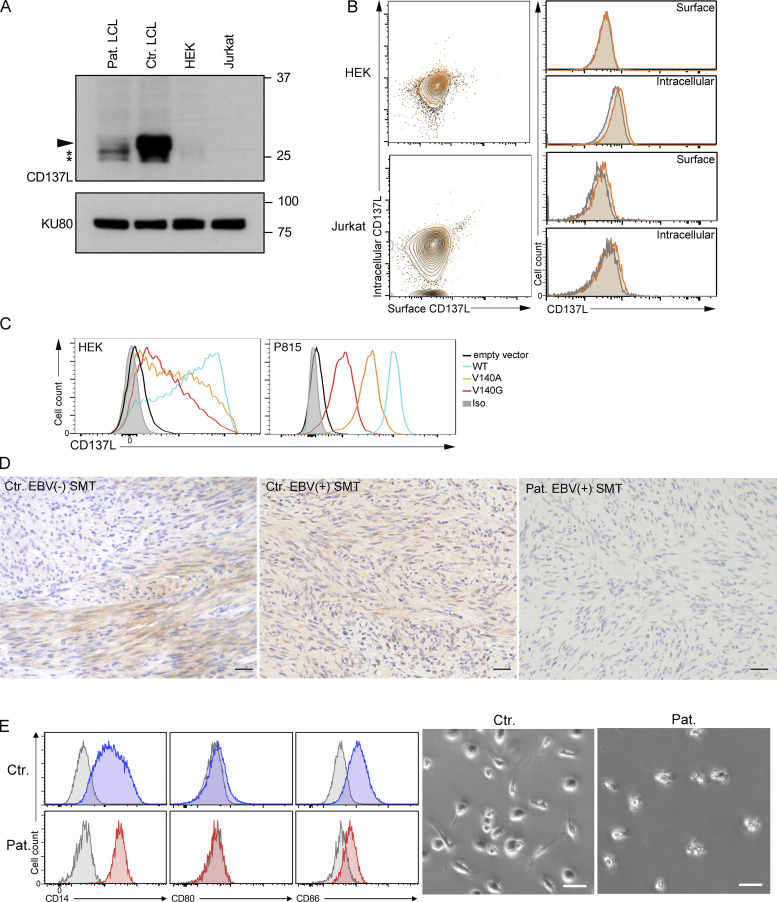
First, we found that c.419T>G (p.V140G) variation had no effect on mRNA expression in EBV-transformed B cells (also termed lymphoblastoid cell lines or LCLs) of the patient or in human embryonic kidney (HEK) cells transduced with an expression vector containing the cDNA coding the CD137LV140G mutant (Fig. 2 F). We next evaluated the effect of the V140G variation on CD137L expression. In the few studies in which CD137L expression was analyzed by flow cytometry or Western blot (WB), CD137L was not expressed on most of hematopoietic cells, except in a small fraction (1–2%) of resting monocytes (Bukczynski et al., 2004) and dendritic cells (DCs; Laderach et al., 2003; Lee et al., 2003). Confirming these previous observations, we repeatedly did not observe any extracellular staining by flow cytometry in resting PBMCs or when activated by PMA + Iono or Pokeweed, a mitogen specific to B cells (data not shown). However, we detected CD137L expression on LCLs (Fig. 3 A), B cells activated by anti-Ig + CD40L, monocytes, and GM-CSF + IL-4 monocyte-derived DCs activated by LPS from PBMCs of healthy controls (Fig. 3 B), whereas non-stimulated B cells, monocytes, and DCs did not express CD137L. In contrast, there was no detectable CD137L expression on LCLs, activated monocytes, DCs, and B cells of the patient. To confirm these data, CD137L expression was further analyzed by WB. Under denaturing and non-denaturing conditions, specific bands with apparent molecular sizes corresponding to monomers and dimers of CD137L, respectively, were detected in lysates from control LCLs (Fig. 3, C and D). These bands were very faint or absent in the lysate from LCLs of the patient. However, two lower bands were still present in the lysate of patient LCLs. These lower bands likely corresponded to immature CD137L proteins potentially lacking post-translational modifications or/and degraded CD137L products not able to form dimers, since they were not detected in lysates of HEK or Jurkat cells that did not express CD137L (Fig. S2, A and B). Similar immature or degraded CD137L products were detectable in PBMCs of the patient stimulated with PMA + ionomycin or Pokeweed (data not shown). Thus, these data were in favor of residual intracellular expression of CD137L in LCLs of the patient. To test this hypothesis, we assessed intracellular and extracellular expressions of CD137L by flow cytometry or by confocal fluorescence microscopy. As expected, no intracellular and surface CD137L expression was found in HEK and Jurkat cells (Fig. S2 B). In control LCLs, both intracellular and surface CD137L expression was detectable (Fig. 3, E and F). In contrast, in LCLs of the patient, no surface CD137L expression was detected, while there was detectable but reduced intracellular CD137L expression (compared to that found in control LCLs). Similar reduced intracellular expression of CD137L with no surface CD137L expression was also observed in LPS-activated monocytes and anti-Ig + CD40L–activated B cells of the patient (Fig. 3 G). Thus, surface (or membrane) CD137L expression is strongly reduced or almost absent in cells of the patient. These data also suggest that CD137L trimerization or dimerization is required for cell-surface expression. Confirming the effect of the V140G mutation on CD137L expression, when ectopically introduced in HEK cells or in P815 murine mastocytoma cells, the CD137LV140G mutant resulted in a weak CD137L expression compared to the ectopic expression of the WT CD137L (Fig. S2 C). We also tested the V140A mutant that was previously predicted to impair trimer formation in silico (Gilbreth et al., 2018; Bitra et al., 2019). Like the CD137LV140G, the CD137LV140A showed decreased expression in these two cell lines. However, the diminution of CD137LV140G expression was more severe than that of the CD137LV140A. Taken together, therefore, these results indicate that the V140G mutation severely impairs CD137L expression.
Figure 3.
CD137L is expressed in the context of EBV infection and strongly decreased in cells of the patient. (A and B) FACS histograms of CD137L expression. Isotype control in gray. (A) EBV-infected cell lines (LCLs) from the patient (Pat.) and three controls (Ctr.1, Ctr.2, and Ctr.3). (B) Monocytes, differentiated DCs, and B cells activated or not in the presence of LPS for 24 h or anti-Ig + CD40L for 4 h from PBMCs of the patient (Pat., in red) and two healthy controls (Ctr.1 and Ctr.2, in blue). Isotype control in gray. (C and D) WBs of lysates from LCLs lines of panel A, under denaturing (C) and non-denaturating (D) conditions, showing the loss of CD137L expression and dimer formation in the patient. CD137L is indicated by a plain arrowhead and immature/degraded CD137L products by asterisks (*). Anti-ACTIN antibody as the loading control (lower blots). Size markers (in kD) on the left. (E and F) Surface and intracellular CD137L expression in control LCLs (Ctr.) and LCLs of the patient (Pat.). (E) Dot-plots and FACS histograms with Ctr. in blue, Pat. in red, and isotype controls in gray. (F) Fluorescence microscopy of surface (in red) and intracellular (in green) CD137L staining of fixed cells. Merged and DAPI staining in right panels. Magnification ×800, scale, 4 μM. (G) Same as B and E. Surface and intracellular CD137L expression of activated monocytes (upper panels) or B cells (lower panels) for 4 h, 24 h, or not (No stimulation) from a healthy control (Ctr.) and the patient (Pat.). (A) One representative experiment of four. (B) One representative experiment of two. (C and D) One representative experiment of three. (E) One representative experiment of two. (F) Images representative of one cell among >20 analyzed cells. Source data are available for this figure: SourceData F3.
Figure S2.
The V140G mutation (CD137LV140G) results in decreased CD137L expression in patient cells and impaired reversed signaling in patient monocytes. (A) WBs of lysates from LCLs of one control (Ctr. LCL), LCLs of the patient (Pat. LCL), HEK, and Jurkat cell lines. CD137L is indicated by a plain arrowhead and immature/degraded CD137L products by asterisks (*). Anti-KU80 antibody as the loading control (lower blot). Size markers (in kD) on the right. (B) Surface and intracellular CD137L expression in HEK and Jurkat cells. Dot-plots and FACS histograms with CD137L staining in yellow/orange and isotype controls in gray. (C) FACS histograms of CD137L expression in HEK 293-T (left) and P815 cells (right) ectopically expressing WT CD137L, the mutant CD137L V140A and V140G proteins, or transfected with an empty vector. (D) Analysis of CD137L expression on EBV+ and EBV− SMT tissues of two individuals and the patient (Pat.). Magnification ×400, scale, 40 μM. Similar findings with two EBV+ and two EBV− SMT tissues from different individuals. (E) Analysis of CD137L reverse signaling in CD14+ monocytes of one healthy control (Ctr.) and the patient (Pat.) that were treated with CD137-Fc protein for 7 d, and surface markers were analyzed by flow cytometry (left panels). Enhanced CD86 and CD80 and decreased CD14 expression in treated control monocytes are characteristics of monocytes differentiating into DCs. Cells from a control showed spindle shaped morphology associated with DC differentiation (right panels). Scale, 50 μM. (A) One representative experiment of four. Source data are available for this figure: SourceData FS2.
CD137L expression is upregulated in the context of EBV infection
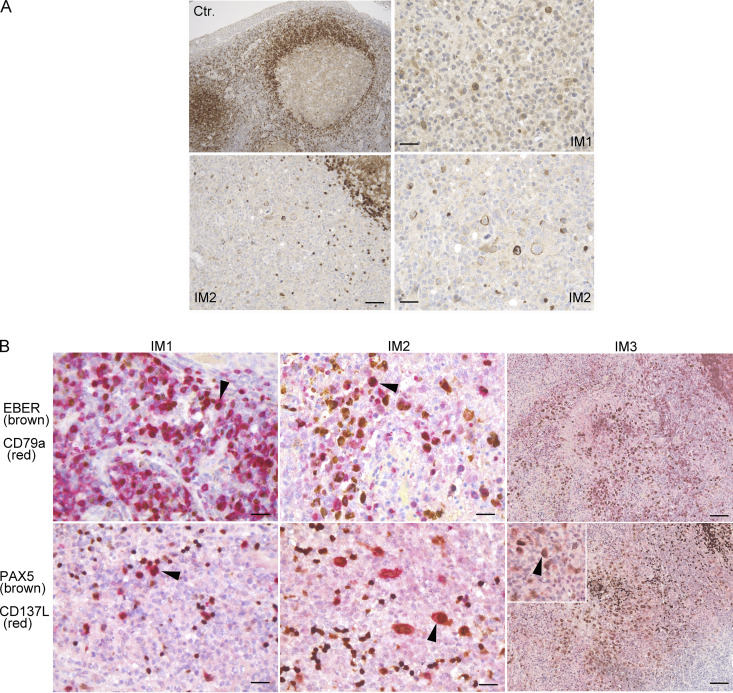
The expression of CD137L in LCLs and on activated B cells could suggest that CD137L is upregulated on B cells during infection by EBV. Along these lines, we previously found that expression of CD70, the ligand of CD27, another key co-stimulatory TNF receptor involved in the expansion of EBV-specific T cells is specifically induced in B cells upon EBV infection (Izawa et al., 2017). Furthermore, expression of the viral EBV LMP1 protein in mice has been shown to induce CD137L and CD70 expression in B cells, resulting in potent cytotoxic CD4 and CD8 T cell responses, by enhancing antigen presentation and T cell co-stimulation (Choi et al., 2018). Hence, we investigated a potential relationship between EBV-infected tissues/cells and CD137L expression. Histology of tonsils of one control individual with follicular lymphoid hyperplasia and three patients with infectious mononucleosis (IM) were examined (Fig. 4 A). In the control case with follicular lymphoid hyperplasia, there were anti-CD137L antibody stained cells in the mantle-zone area with an intracellular pattern, which were likely B cells (Fig. 4 A). In the three patients with IM (IM1, IM2, and IM3), large cells in inter-follicular areas were marked by anti-CD137L antibody with an extracellular pattern. These cells corresponded to EBV+ B cells as they were positively co-stained with EBER, anti-PAX5, and anti-CD79a antibodies in three patients with IM (Fig. 4 B). Therefore, these data indicate that extracellular/membrane CD137L expression is induced in B cells in the context of EBV infection. We also tested CD137L expression on EBV+ and EBV− SMT tissues that appeared mostly intracellular, while no or very weak CD137L expression was detected in the EBV+ SMT of the patient (Fig. S2 D).
Figure 4.
CD137L is expressed on B cells of tonsils from individuals with EBV IM. (A) Analysis of CD137L expression on tonsils of one individual with follicular lymphoid hyperplasia (Ctr.) and two individuals with IM (IM1 and IM2). Magnification ×100 (upper left; scale, 120 μM), ×200 (lower left; scale, 60 μM), and ×400 (upper and lower right; scale, 30 μM). CD137L is expressed in mantle area cells from follicular lymphoid hyperplasia and at the membrane of large cells scattered in interfollicular areas during IM. (B) Co-staining of tonsils of three individuals with IM (IM1, IM2, and IM3) with an EBER probe (nuclear brown staining) and anti-CD79a (red staining; upper panels) or anti-PAX5 (nuclear brown staining) and CD137L (red staining; lower panels) antibodies that show EBV-infected B cells expressing CD137L. Magnification ×400 (scale, 50 μM) for left and middle panels, ×200 (scale, 90 μM), and ×400 in the insert for right panels. Arrowheads indicate representative co-stained cells. (A and B) Each image is representative of at least five different images/sections.
CD137LV140G strongly impairs T cell proliferation and EBV-specific T cell expansion
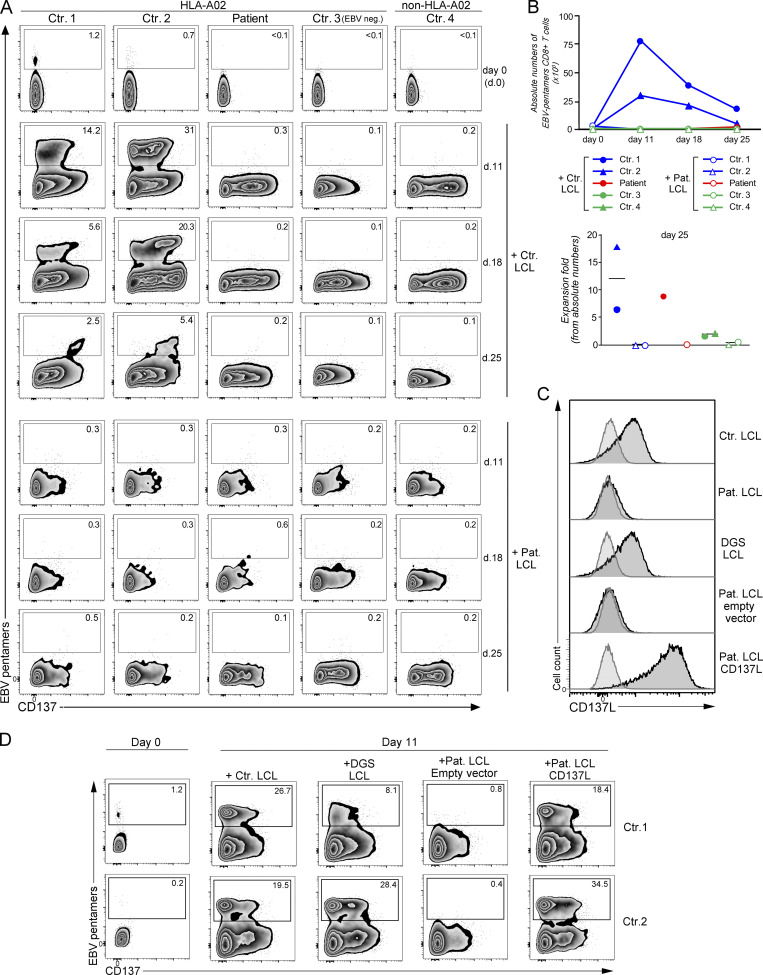
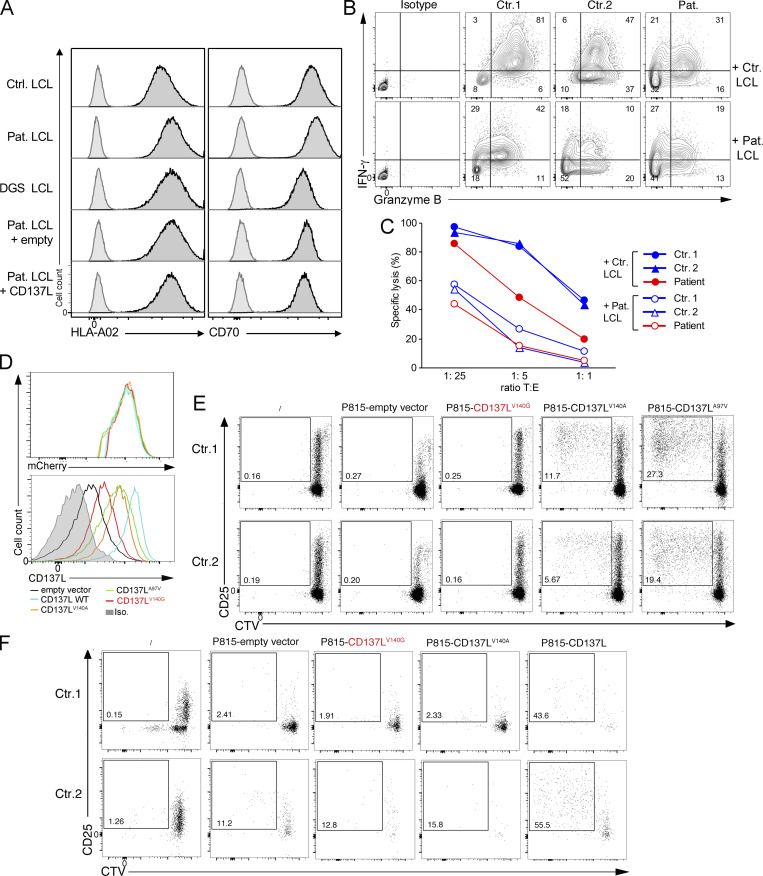
We and others previously showed that CD137 expressed on activated T cells plays a key role to activate T cell proliferation and in particular to expand EBV-specific T cells (Rodriguez et al., 2019; Somekh et al., 2019). Thus, LCLs of the patient (LCLs CD137LV140G) that expressed HLA-A*02 molecules were tested for their capacity to trigger allogenic HLA-A matched expansion of HLA-A*02–restricted EBV-specific T cells when co-cultured with PBMCs of healthy controls expressing HLA-A*02 molecules (Fig. 5, A and B). EBV-specific T cells derived from PBMCs of two HLA-A*02 controls (Ctr.1 and Ctr.2) vigorously expanded between day 0 and day 11 in the presence of control LCLs, while expansion decreased on days 18 and 25. CD137 expression in these EBV-specific T cells was particularly marked on day 18. In a striking contrast, EBV-specific T cells from the two HLA-A*02 controls (Ctr.1 and Ctr.2) failed to expand (until day 25) when incubated with patient LCLs, while both control and patient LCLs expressed similar levels of HLA-A*02 molecules and CD70 (Fig. 5 C and Fig. S3 A). As expected, no such expansion was observed from PBMCs of an unmatched non–HLA-A*02 control (Ctr.4; Fig. 5 A) or from PBMCs of an EBV-naive HLA-A*02 control (EBV neg. Ctr. 3; Fig. 5 A). When incubated with control LCLs or their autologous LCLs (patient LCLs), EBV-specific T cells did not expand from PBMCs of the patient on days 11 and 18 (both in proportions and absolute numbers; Fig. 5, A and B), reflecting the absence of detectable EBV-specific T cells on day 0 in the PBMCs of the patient (also shown in Fig. 1 E). However, when calculated in absolute numbers, few EBV-specific T cells accumulated on day 25 in the co-culture with the control LCLs, but not in the co-culture with patient LCLs showing a relative fold expansion similar to controls (Fig. 5 B, lower panel). This is likely explained by the presence of few EBV-specific cells in the PBMCs of the patient on day 0 under the threshold of detection. Thus, this confirmed that T cells of the patient had no intrinsic defect and had a preserved capacity to expand. On day 18, IFN-γ production and cell cytotoxicity by expanded T cells were analyzed by re-stimulation with LCLs. Only co-cultures with control LCL in which EBV-specific T cells had expanded contained T cells with significant cytotoxic activity and capacity to produce both IFN-γ and granzyme B toward LCLs (Fig. S3, B and C). These results indicate that LCLs of the patient have an impaired capacity to stimulate expansion of EBV-specific T cells likely caused by the loss of CD137L expression associated with the CD137LV140G mutant. These observations are also consistent with the absence of detectable EBV-specific T cells among PBMCs of the patient who are not able to normally expand in vivo (Fig. 1 E and Fig. 5 A).
Figure 5.
The V140G mutation in CD137L impairs T cell proliferation and expansion of EBV-specific T cells. (A) FACS dot-plots gated CD3+CD8+ cells of co-cultures of PBMCs of HLA-A02* healthy controls (Ctr.1, Ctr.2, and Ctr.3), non–HLA-A02* (HLA-A24/A31; Ctr.4), and the CD137L-deficient patient who is HLA-A02* (Pat.) with irradiated CD137L-deficient/patient LCL (Pat. LCL) or control HLA-A02* LCL (Ctr. LCL) at day 0, 11, 18, and 25 showing EBV-specific T cells co-stained with EBV-HLA-A02* pentamers and anti-CD137 antibody. CD137 as an activation marker. Ctr.3 has not been infected by EBV and had a negative EBV serology (EBV neg.). EBV-specific T cells in the gates and numbers indicating proportions of cells in gates. (B) Upper panel, graph of absolute counts of EBV+-specific T cells calculated from A. Lower panel, fold of expansion at day 25 calculated from absolute counts in the upper panel. (C) FACS histograms of CD137L expression on control HLA-A02* LCL (Ctr. LCL), CD137L-deficient/patient LCL (Pat. LCL), LCL from a patient with a DiGeorge syndrome (DGS LCL), LCL of the patient transfected with an empty vector (Pat. LCL empty vector), or a vector containing cDNA coding CD137L (Pat. LCL CD137L). Anti-CD137L and isotype control staining corresponding to histograms with black line with deep gray and gray line with pale gray, respectively. (D) Same as in A. PBMCs from two HLA-A02* healthy controls (Ctr.1 and Ctr. 2) co-cultured with the different LCL lines shown in panel C. EBV-specific T cells have been analyzed at day 11. (C) One representative experiment of three. (A and D) One representative experiment of two.
Figure S3.
Impaired T cell responses and NK cell expansion toward LCL of the patient or P815 expressing CD137LV140G. (A) FACS histograms of HLA-A02 and CD70 expression on control HLA-A02* LCL (Ctr. LCL), CD137L-deficient/patient LCL (Pat. LCL), LCL from a patient with a DiGeorge syndrome (DGS LCL), LCL of the patient transfected with an empty vector (Pat. LCL + empty) or a vector containing cDNA coding CD137L (Pat. LCL + CD137L). Anti-CD137L and isotype control staining corresponding to histograms with black line with deep gray and gray line with pale gray, respectively. (B) FACS dot plots of IFN-γ and Granzyme B expression of EBV-specific T cell expansions at day 25 (from co-cultures in Fig. 5) that have been re-stimulated with irradiated Ctr. LCL or Pat. LCL before intracellular co-staining with anti–IFN-γ and Granzyme B antibodies. Double IFN-γ+ GranzymeB+ cells are shown in gates and numbers indicating proportions of cells in gates. (C) Graphs of cell lysis assays of EBV-specific T cell expansions at day 28 (from co-cultures in Fig. 5) as effector cells and control LCLs (Ctr. LCL) or LCLs of the patient (Pat. LCL) as target cells that were mixed at the indicated target cells/effector cells ratio in the longitudinal axis. (D) FACS histograms of mCherry (upper panel) or CD137L (lower panel) in P815 cells ectopically expressing WT CD137L, CD137L-V140G, V140A, A97V mutants, or infected with an empty vector. (E) FACS dot-plots of proliferation assays of PBMCs labeled with the CTV dye and cocultured or not (no P815) for 6 d with irradiated P815 cell lines expressing CD137L V140G, V140A, A97V (as positive control), or an empty vector in the presence of anti-CD3 antibody. Staining with anti-CD25 antibody as activation marker and gating on CD3+ cells. Proliferating T cells in the gate with numbers corresponding to the proportions of cells. (F) FACS dot-plots of proliferation assays of PBMCs labeled with the CTV dye and cocultured or not (no P815) for 6 d with irradiated P815 cell lines expressing CD137L, CD137L V140G and V140A, or an empty vector. Staining with anti-CD25 antibody. CD25 as activation marker. Cells gated on CD3−/CD56+ cells. Proliferating NK cells in the gate with numbers corresponding to the proportions of cells. (A) One representative experiment of two. (D) One representative experiment of four. (E) One representative experiment of three. (F) Data representative of three experiments.
To formally prove that the CD137LV140G mutant causing loss of CD137L expression in LCLs impaired the proliferation of EBV-specific T cells, expression of CD137L was restored in LCLs of the patient by introducing WT CD137L using a lentiviral expression vector, and then cells were tested for their ability to expand EBV-specific T cells. Corrected LCLs of the patient expressing WT CD137L recovered the ability to trigger expansion of EBV-specific T cells in contrast to non-corrected CD137L-negative LCLs transduced with an empty vector (Fig. 5, C and D). Excluding a role of the DiGeorge syndrome or 22q11.2 deletion in the inability of LCLs of the patient to induce expansion of EBV-specific T cells, HLA-A*02–expressing LCLs derived from a patient with DiGeorge syndrome allowed the expansion of EBV-specific T cells as efficiently as control LCLs (Fig. 5, C and D, and Fig. S3 A).
Furthermore, supporting the loss-of-function effect of the CD137LV140G mutant, P815 cells that ectopically expressed CD137LV140G failed to induce T cell proliferation when co-cultured with PBMCs in the presence of anti-CD3 antibody, in contrast to P815 cells expressing CD137LA97V that triggered substantial T cell proliferation (Fig. S3, D and E). CD137LA97V is a rare heterozygous variant identified in our exome data with unknown functional significance that was used as a surrogate of WT CD137L in these experiments. P815 expressing CD137LV140A, a mutant previously reported to cause impaired trimer formation, showed a reduced capacity to induce T cell proliferation (Gilbreth et al., 2018; Bitra et al., 2019). CD137LV140A and CD137LA97V exhibited comparable but decreased expression when compared to WT CD137L, while CD137LV140G expression was markedly diminished as shown before (Fig. S3 D, lower panel). Of note, all cell lines expressed similar levels of mCherry expression indicating that equal amounts of each pLVX-mCherrry vector encoding the different mutants were delivered to the cells (Fig. S3 D, upper panel). Furthermore, we also repeatedly observed proliferation of few CD3− CD56+ NK cells when PBMCs were co-cultured with P815-expressing WT CD137L in absence of anti-CD3 antibody, which was neither observed in co-cultures with P815-expressing CD137LV140G nor CD137V140A (Fig. S3 F). Therefore, these findings demonstrate that the V140G mutation in CD137L impairs CD137-dependent proliferation of T and NK cells, explaining in all probability the high susceptibility to EBV of the patient.
We report herein a novel primary immune deficiency caused by a homozygous loss-of-function mutation in TNFSF9 coding for the ligand of CD137 that is associated with a broad susceptibility to EBV infection. This contrasts with patients with CD137-deficiency in whom the EBV infection is restricted to B cells (Rodriguez et al., 2019), excepted in one who exhibited EBV-infected T cells causing chronic active EBV infection (Alosaimi et al., 2019; Somekh et al., 2019). The patient reported here had EBV-infected B, T, and smooth muscle cells and did not suffer from other infections. Concomitant EBV infection of B, T, and smooth muscle cells has been previously reported in one patient with a hypomorphic IL2RG mutation; however, infectious susceptibility in this case was not restricted to EBV and associated with prevalent bacterial infections (Tanita et al., 2019). In our patient, the 22.q11.2del syndrome may have caused some immunodeficiency contributing to the EBV susceptibility in the setting of CD137L deficiency. Indeed, incomplete penetrance has been reported in several TNF-R co-stimulation defects associated with EBV LPD, such as CD137, CD70, and CD27 deficiencies (Ghosh et al., 2020). Notably, one-third of CD137-deficient individuals are asymptomatic (Rodriguez et al., 2019; Alosaimi et al., 2019; Somekh et al., 2019). In one CD137-deficient symptomatic individual, a PI3KCD homozygous loss-of-function mutation was identified as a supplementary event involved in the development of an overt systemic T cell chronic active EBV infection, indicating that additional genetic factors are required to develop EBV susceptibility in the context of a CD137 deficiency (Rodriguez et al., 2019). Likewise, EBV predisposition in the context of CD137L deficiency herein could have been uncovered by a second genetic event, in the present case, the 22q11.2del. Interestingly, EBV+ LPD has been reported in two patients with 22.q11.2del (Cancrini et al., 2014; Sullivan, 2019), who presented EBV+ diffuse large B cell lymphoma and systemic EBV+ T cell lymphoma of childhood (Stevens et al., 2017; Itoh et al., 2011). However, these phenotypes have not been observed in large series of patients with 22.q11.2del (Cancrini et al., 2014; Sullivan, 2019). It is tempting to speculate that additional germline genetic factors underlie EBV+ LPD in these two patients.
It has been well established that T cells lacking co-stimulation provided by CD137 failed to expand and to constitute a sufficient antigen-specific memory pool to efficiently control viral infection (Bukczynski et al., 2004; Fuse et al., 2007; Laderach et al., 2002). We showed that CD137L-deficient LCLs did not support expansion of EBV-specific T cells consistent with the absence of detectable circulating EBV-specific T cells in the patient. Taken together, these observations likely explain the defective immune control of EBV-infected B cells, but also EBV-infected T cells and EBV+ SMT. Indeed, CD137L is expressed on hematological malignancies including T cell lymphomas that expressed high levels of CD137L. Furthermore, intra-tumor lymphoid cells that lack CD137L would not be able to bring co-stimulation to T cells, as exemplified in human lymphomas (Houot et al., 2009). Finally, smooth muscle cells in vessels have been reported to express CD137L during inflammatory conditions (Seko et al., 2004), as well as smooth muscle and endothelial cells of intra-tumor vessels (Broll et al., 2001; Wang et al., 2008). Our preliminary data also suggest that CD137L is expressed in SMT cells (Fig. S2). Similar to T and B cells, CD137L expressed on smooth muscle cells could directly activate T cell and NK cell responses. In addition, engagement of CD137 and CD137L induces integrin expression on endothelial cells (Palazón et al., 2011; Olofsson Peder et al., 2008; Drenkard et al., 2007) that may recruit T and NK cells. However, it is possible that the primary EBV infection of B cells or latter EBV reactivations in B cells are key to expand and constitute a pool of EBV-specific T cells necessary for a second time to control EBV-infected smooth muscle cells. Our results also open up the possibility of a role of other antigen-presenting cells (other than infected B cells) such as monocytes and DCs that express CD137L when activated (Fig. 3 B). Moreover, CD137L reverse signaling in monocytes has been shown to induce DC differentiation (Kwajah and Schwarz, 2010). Our data suggest that CD137L reverse signaling in monocytes of the patient is impaired (Fig. S2 D), supporting that this defect could also contribute to immunodeficiency. However, humanized mice models have suggested that DCs are not required to control EBV infection (Gujer et al., 2019). Interestingly, RLTPR and RASGRP1 deficiencies also predisposed to EBV infection and occurrence of EBV+ SMT in some patients. RLTPR and RASGRP1, which are two intracellular signaling molecules, might be involved in the CD137–CD137L pathway. Further studies are warranted to test this hypothesis.
Like the CD70–CD27 pathway, CD137–CD137L may also be particularly important for tumoral immune surveillance of abnormal B lymphocytes independently of EBV infection as CD137L-deficient mice develop spontaneous B cell lymphoma (Middendorp et al., 2009). One brother of the patient was a heterozygous carrier of the TNFSF9 mutation, and he developed an EBV− B cell lymphoma. The TNFSF9 heterozygosity might have participated in the lymphoma occurrence in association with additional undetermined genetic factors.
The preservation of CD137-induced T cell proliferation in the patient suggests that agonist CD137 antibodies that previously demonstrated potent effects on anti-tumor and viral responses (Chester et al., 2018) could be an attractive targeted therapy to treat EBV-driven proliferative disorders including EBV+ SMT, and potentially SMT in contexts irrespective of EBV, as our data suggest that EBV− SMT also expressed CD137L (Fig. S2). In conclusion, by means of the identification of a CD137L-deficient patient, we further underlined the importance of the CD137–CD137L pathway in the control of EBV infection, and more particularly when EBV infects smooth muscle cells.
Materials and methods
Ethics
Written informed consent forms were obtained from all humans in this study in accordance with Declaration of Helsinki and local legislation and ethical guidelines from the Comité de Protection des Personnes de l’Ile de France II, Hôpital Necker-Enfants Malades, Paris and the Institut national de la santé et de la recherche médicale institutional review board. Blood from healthy donors was obtained at the Etablissement du Sang Français under approved protocols (convention 15/EFS/012).
WES
Genomic DNA was extracted from whole blood according to the standard methods, and WES and data analysis were performed as previously described (Izawa et al., 2017). An autosomal recessive inheritance pattern was applied to filter WES data with a focus on homozygous variations. 18 genetic variants with potential deleterious Combined Annotation Dependent Depletion (CADD) scores and low allele frequency (<9.31e−4) with no homozygotes in public WES databases including our own databases from our institute were identified. These include with their GRCh37 genomic positions, protein positions, CADD scores, and allele frequency (AF) in GnomAD: ACAD9 (g.3:128627086G>A, p.R466H, CADD 24, 6.37e-5), CAPN12 (g.19:39227954C>G, p.G402R, CADD24, no AF), COL14A1 (g.8-121301974G>A, p.R1402Q, CADD 23, 1.38e-4), FBXO32 (g.8:124546959C>T, p.S71N, CADD 19, no AF), FLVCR2 (g.14:76045679G>A, p.A122T, CADD 23, 2.97e-4), GPRC6A (g.6:117113495A>C, p.I864S, CADD 21, no AF), HIVEP2 (g.6:143095538G>A, p.T113I, CADD 8, 1.2e-5, 1), KCNT1 (g.9:138646997G>A, p.M174I, 21, 3.67e-4), PAPLN (g.14:73732154G>A, p.E1041K, CADD 25, 7.52e-4), PRODH2 (g.19:36297981delG, L290SfsTer31, no CADD, 3.05e-4), RALGDS (g.9:135985782G>A, P130L, CADD23, 2.84e-5, 8), RINL (g.19:39361220G>C, p.D338E, CADD 1, no AF), TNFSF9 (g.19:6534731, p.V140G, CADD16, no AF), VARS1 (g.6:31747270G>A, p.A1111V, CADD 15, 1.6e-4), VASH2 (g.1:213147403G>A, p.R285Q, CADD 26, 9.31e-4), ZBTB43 (g.9:129595263T>C, p.S159P, CADD 20, 2.13e-5), ZDHHC12 (g.9:131483695A>T, F190Y, CADD 19, 7.46e-5) ZFP36L2 (g.2:43451931C>G, p.G338R, CADD 19, 3.26e-5). Based on tissue expression, functional and disease association data, and information from the literature and public databases, the genetic variation in TNFSF9 was considered to be the best variation candidate. MLPA was performed with SALSA MLPA Probemix P250 DiGeorge according to manufacturer’s instructions (MRC Holland). Genomic DNA regions flanking the TNFSF9 mutation were amplified by using the forward primer 5′-CTAAGCTAAGTGCATGCTTTCC-3′ and reverse primer 5′-GGCCCTGGAAACCGAAGG-3′ with Q5 HighFidelity DNA Polymerase (NEB) according to the manufacturer’s recommendations, gel-purified with a High Pure PCR Product Purification Kit (Roche), sequenced with a BigDye terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), and analyzed on a 3500xL Genetic Analyzer (Applied Biosystems). All collected sequences were analyzed by using a DNADynamo (BlueTractorSoftware). Exome sequencing data is not publicly available due to ethical restrictions, including the possibility of compromising privacy.
Gene expression analysis
Total RNA was extracted using a Pure Link RNA Mini kit (Thermo Fisher Scientific) and cDNA was synthesized using a first-strand synthesis system (SuperScript II; Thermo Fisher Scientific). Real-time quantitative PCR was performed using a real-time PCR system (ViiA 7; Thermo Fisher Scientific). The following Taqman probes and primers were used: Hs00169409 for TNFSF9 and Hs99999905 for GAPDH (Applied Biosystems). Relative expression levels of TNFSF9 were normalized to GAPDH and analyzed by the ΔΔCt method.
Cell culture and stimulation
PBMCs were isolated by Ficoll-Paque (Lymphoprep; Proteogenix) density gradient centrifugation, washed, and resuspended at a density of 106 cells per ml in complete Panserin 401 medium (Pan Biotech) containing 5% human male AB serum (BioWest), 100 U/ml of penicillin, and 100 mg/ml of streptomycin (Gibco; Thermo Fischer Scientific). T cell blasts were expanded by incubating PBMCs for 72 h with 2.5 mg/ml of PHA (Sigma-Aldrich). Dead cells were then removed by Ficoll–Paque density-gradient (Lymphoprep; Proteogenix) centrifugation, and T cell blasts were cultured in complete Panserin 401 medium culture supplemented with 100 IU/ml of recombinant human IL-2 (R&D Systems). HEK293T cells were cultured in complete Dulbecco modified Eagle medium, high glucose, GlutaMAX supplement, pyruvate medium containing 10% heat-inactivated fetal calf serum (Gibco), 100 U/ml of penicillin, and 100 mg/ml of streptomycin (Gibco). LCL and P815 cell lines were cultured in complete RPMI 1640 GlutaMax medium (Invitrogen) containing 10% heat-inactivated fetal calf serum (Gibco), 100 U/ml penicillin, and 100 µg/ml streptomycin (Gibco).
Fresh PBMCs were stimulated for 72 h with 100 ng/ml of PMA (Sigma-Aldrich) and 1 μg/ml ionomycin (Sigma-Aldrich) or 1 μg/ml of Pokeweed (Sigma-Aldrich). In Fig. 3, B and G, PBMCs were stimulated for 24 h with 1 μg/ml LPS (Sigma-Aldrich) or 4 h with 10 μg/ml of anti-Ig (Invitrogen) and 5 μg/ml of CD40L (R&D Systems). Monocyte-derived DCs were generated in a culture of monocyte with 1,000 U/ml of IL-4 (Proteintech) and 800 U/ml GM-CSF (Proteintech) for 7 d. Monocyte-derived DCs were stimulated for 24 h with 1 μg/ml LPS. DCs maturation/activation was confirmed by enhanced expression of CD80 and CD86 expression analyzed by flow cytometry.
Flow cytometry RNA assay
Primeflow EBER assay was performed as previously described (Fournier et al., 2020). Briefly, PBMCs were washed in staining buffer (PBS-CSF 2%) before seeding in 96 round-bottom well plate. Membrane staining was performed prior to Primeflow RNA assay. 1 µl/106 cells of membrane-targeting antibodies and 1/2,000 fixable viability dye eFluor 450 were used with PBS-CSF 2% in a total volume of 150 µl per well. The following validated antibodies were used: anti-CD3 (SK7), anti-CD4 (SK3 or OKT4), anti-CD8 (HIT8a), anti-CD14 (M5E2), anti-CD16 (3G8), anti-CD19 (HIB19), anti-CD56 (B159 or 5.1H11), and Fixable Viability Dye eFluor 450. These antibodies were conjugated to FITC, PE, phycoerythrin-cyanin5 (PE-Cy5), PE-Cy7, Brilliant Violet 421 (BV421), BV711, or BV785. RPL13A and Bacillus cereus specific probes (Alexa Fluor 488 [Type 4, AF488] and Alexa Fluor 647 [Type 1, AF647] respectively) were used respectively as a positive and negative control for EBER probe staining in the same well stained only with eFluor 450. EBER target probe Alexa Fluor 647 (Type 1, AF647) was used to stain EBV-infected cells previously stained with extracellular antibodies. In situ hybridization for EBER, RPL13A, and Bacillus was performed using probes and reagents supplied with Primeflow RNA Assay kit as described by the manufacturer (eBioscience).
Immunochemistry
Biopsies were fixed in 10% neutral buffered formalin, embedded in paraffin, and stained with hematoxylin–eosin. Immunohistochemical staining and in situ hybridization were performed on an automated stainer (Bond Max; Leica Biosystems). The presence of EBV was demonstrated by in situ hybridization with a probe for small RNA-encoding regions 1 and 2 (EBER) probe (Dako). Antibodies used were: anti-CD3 (polyclonal rabbit, CD3ε, A0452; Dako Agilent), anti-CD79a (monoclonal mouse IgG1k, JCB117; Dako), anti-caldesmon (monoclonal mouse IgG, clone h-CD [M3557]; Dako), anti-PAX5 (clone DAK-PAX5; 1/30; Dako), and anti-4-1BBL/CD137L (polyclonal rabbit IgG, ab64912; Abcam).
Constructs
A cDNA of TNFSF9 (NM_003811.4) was obtained by RT-PCR from RNA of HUT-78 cells with the forward primer 5′-CACCATGGAATACGCCTCTGACGC-3′ and the reverse primer 5′-TTATTCCGACCTCGGTGAAGGG-3′ and inserted into the pCR 2.1-TOPOTA vector (Invitrogen) according to manufacturer’s instructions. The c.419T>G (p.V140G) and c.419T>C (p.V140A) mutants were obtained with a Q5 Site-Directed Mutagenesis Kit (NEB) with the forward primer 5′-AAGGCTGGAGGCTACTATGTCTTC-3′ and reverse primer 5′-GGCCACCACCAGCTCCTT-3′, and forward primer 5′-AAGGCTGGAGCCTACTATGTCTTCTTTCAAC-3′ and reverse primer 5′-GGCCACCACCAGCTCCTT-3′, respectively. Coding sequences were confirmed by Sanger sequencing and subcloned into a bicistronic lentiviral vector encoding the mCherry protein as a reporter gene (pLVX-EF1alpha-IRES-mCherry Vector; ClonTech).
Transfections
HEK293T cells were transfected with empty pLVX-mCherry vector or pLVX-mCherry vector encoding WT, p.V140G, p.V140A with lipofectamin 2000 (Invitrogen) according to the supplier’s instructions. Cell lysates were performed 48 h after transfection.
Lentiviral gene transfer
Transduction of P815 cell lines with empty pLVX-mCherry or pLVX-mCherry vector encoding WT, p.V140G, p.A97V, and p.V140A CD137L were performed by lentivirus-mediated gene transfer. LCLs from the patient were transduced with pLVX-mCherry or pLVX-mCherry vector encoding WT CD137L. All cell lines were found after transduction to express similar levels of mCherry expression indicating that equal amounts of each pLVX-mCherrry vectors encoding WT, p.V140G, p.A97V, or p.V140A CD137L was delivered to the cells (Fig. S3 D, upper panel). HEK293T cells were cotransfected with the lentiviral plasmid together with the plasmids pVSVG and pGag-pol at a molar ratio of 1:1:1 with Lipofectamine 2000 (Invitrogen) according to the supplier’s instructions. Supernatants containing virus particles were harvested 36 h after transfection and freshly used for infection of P815.
Immunoblotting
Cells were washed in PBS and lysed in lysis buffer containing 1% NP-40 (NP-40 alternative; Calbiochem), 50 mM Tris, pH 8, 150 mM NaCl, 20 mM EDTA, 1 mM Na3VO4, 1 mM NaF, complete protease inhibitor cocktail (Roche), and phosphatase inhibitor cocktails 2 and 3 (Sigma-Aldrich). Cell lysates were clarified by centrifugation at 15,000 g for 5 min. Supernatants were transferred to new tubes, and protein concentrations were determined by Coomassie protein assay reagent (Thermo Fischer Scientific). Under denaturating conditions, proteins were denatured by boiling for 10 min with sample buffer (125 mM Tris, pH 6.8, 3% SDS, 10% glycerol, 5% 2β-mercaptoethanol, and 0.01% bromophenol blue). Under non-denaturating conditions, proteins were mixed with sample buffer (125 mM Tris, pH 6.8, 3% SDS, 10% glycerol, no 2β-mercaptoethanol, and 0.01% bromophenol blue) and kept on ice. Proteins were then separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). Membranes were blocked with milk or BSA buffer before incubation with antibodies. The following antibodies were used for immunoblotting: anti-phosphorylated ERK1/2 (Phospho-p44/42 MAPK, 20G11, catalog no. 4376S), anti-phosphorylated PLCγ-1 (D6M9S, catalog no. 140008S), anti-phosphorylated tyrosine (P-Tyr-100, catalog no. 9411S), anti-phosphorylated ZAP-70 (catalog no. 2704S), anti-NFAT2 (D15F1, catalog no. 8032), anti-KU80 (catalog no. 2753S) all from Cell Signaling Technology; anti–β-actin (A2066; Sigma-Aldrich); and anti-CD137L (EPR1172Y, catalog no. ab68185) from Abcam. Membranes were then washed and incubated with secondary anti-mouse (GE Healthcare) or anti-rabbit (Cell Signaling Technology) horseradish peroxidase–linked antibodies. Pierce ECL Western blotting substrate was used for revelation. For reprobing, membranes were incubated with Restore Western Blot Stripping Buffer (Thermo Fischer Scientific) before blocking.
Flow cytometry
Cell membrane staining and the flow cytometry-based phenotypic analyses were performed according to standard flow cytometry methods. The following validated antibodies were used: anti-CD3 (UCHT1), anti-CD4 (OKT4), anti-CD8 (RPA-T8), anti-CD14 (M5E2), anti-CD19 (HIB19), anti-CD25 (BC96), anti-CD27 (O323), anti-CD38 (HIT-2), anti-CD56 (HCD56), anti-CD57 (NK.1), anti-CD80 (2D10), anti-CD86 (IT2.2), anti-CD107a/b (H4A3/HAB4), anti-CD137 (4B4-1), anti-CD137L (5F4), anti-PD-1 (MIH4) anti-CD31 (WM59), anti-CD161 (HP-3G10), anti-TCR Vα7.2 (3C10), and anti-IgM (MHM88; all purchased from BioLegend); anti-CD16 (3G8), anti-CD45RA (HI100), anti-CD45RO (UCHL1), anti-CD197/CCR7 (3D12), anti-TCRαβ (OT31), and anti-IgD (IA6-2; BD Biosciences); anti-CD21 (BL13), anti-TCRγδ (IMMY510), anti-TCR Vα24 (C15), anti-TCR Vβ11 (X21; Beckman Coulter), and anti-CD209 (REA617; Miltenyi Biotech). These antibodies were conjugated to FITC, PE, PE-Cy5, PE-Cy5.5, PE-Cy7, peridininchlorophyll (PerCP), PerCP-Cy5.5, APC, APC-Cy7, APC–violet 7 (APC-Vio7), BV421, BV510, BV605, BV650, BV711, or BV785. CD70-APC and CD137L were from BioLegend. HLA-A2-PE was from BD Pharmingen. For intracellular staining, surface CD137L was stained with anti–CD137L-PE mAb (BioLegend). Then, cells were fixed and permeabilized with a BD cytofix/cytoperm kit (BD Biosciences) and stained with anti–CD137L-APC mAb (BioLegend). Expression of CD137L was analyzed in cells gated on CD3+ (T cells), CD19+ (B cells), or CD14+ (monocytes). All data were collected on an LSRFortessa X-20 cytometer (BD Biosciences).
Proliferation assays
T cell blasts were washed and cultured without IL-2 for 72 h to synchronize the cells. T cell blasts or PBMCs were labeled with Tag-it Violet Proliferation and Cell Tracking Dye (BioLegend) according to the manufacturer’s instructions. Cells were then cultured for 4 d in complete Panserin 401 medium alone or in the presence of 0.1, 1, or 10 μg/ml of immobilized anti-CD3 antibody (clone OKT3 [eBioscience]) or Dynabeads Human T-Activator CD3/CD28 (Invitrogen). Cells were surface-stained for CD3, CD4, CD8, and CD25 detection and analyzed by flow cytometry (LSRFortessa X-20 [BD Biosciences]). The proliferation index was calculated by using FlowJo software (TreeStar). Proliferation assays with cocultures of T cells with the P815 cells or LCL cells have been previously described elsewhere (Izawa et al., 2017; Wen et al., 2002). Briefly, irradiated P815 expressing or not expressing CD137L or irradiated EBV B cell lines (LCLs) expressing CD70 were preincubated with soluble 0.25 μg/ml anti-CD3 antibody, washed, and cocultured with PBMCs labeled with Cell Tracking Dye.
Calcium flux analysis
Ca2+ influx was assessed by real-time flow cytometry, as previously described (Martin et al., 2014). Briefly, cells were loaded with 5 mM Indo-1 AM (Molecular Probes) in the presence of 2.5 mM probenecid (PowerLoad; Molecular Probes), washed, and surface-stained for CD4 and CD8 detection. Cells were analyzed in real-time with a FACS ARIA II flow cytometer (BD Biosciences). During acquisition, 1 mg/ml of anti-CD3 antibody was added to the cells, followed by 10 μg/ml of F(ab′)2 rabbit–anti-mouse IgG crosslinker (Jackson Immunoresearch), and finally incubated with a calcium ionophore (1 mM ionomycin [Sigma-Aldrich]). Changes in the intracellular calcium concentration are quantified by a shift in the indo-1 emission peak from 485 nm (indo-blue) for unbound dye to 405 nm (indo-violet) when the indo-1 molecule is bound to calcium. Data were analyzed by using the kinetic tool of FlowJo software. Intracellular Ca2+ levels correspond to the normalized ratio of 405- to 485-nm indo-1 emission peaks.
Degranulation assay
T cell blasts were stimulated for 4 h with 0.03, 0.3, 3, and 30 μg/ml of immobilized anti-CD3 in the presence of PE-conjugated anti–LAMP-1/2 (H4A3 and H4B4 [BD Biosciences]) as previously described (Martin et al., 2014). Cells were then washed and stained for surface expression of CD3 and CD8 and analyzed by flow cytometry.
Expansion of EBV-specific T cells
The expansion of EBV-specific T cells and their detection were described elsewhere (Izawa et al., 2017). In brief, PBMCs were co-cultured with 40-Gy irradiated autologous or HLA-A matched LCLs at a 40:1 ratio (PBMCs: LCLs). Cells were re-stimulated with irradiated LCLs at a ratio of 4:1 on days 9, 16, and 23. IL-2 (50 U/ml) was added from day 16 of culture. For the detection of EBV-specific CD8+ T cells, cells were incubated with unlabeled Pro5 MHC Pentamer mix followed by Pro5 fluorotag APC, anti–CD3-BV785 (Proimmune), anti–CD4-BV421, anti–CD8-BV510, anti–CD19-BV711 (from BioLengend), and anti–CD137-PE (from BD Pharmingen). Pentamer mix contains HLA A*02:01/YLLEMLWRL (residues 125–133 from LMP1), HLA A*02:01/FLYALALLL (residues 356–364 from LMP2), HLA A*02:01/CLGGLLTMV (residues 259–267 from LMP2), and HLA A*02:01/GLCTLVAML (residues 259–267 from BMLF-1). Stained cells were analyzed by flow cytometry. Three LCLs from DiGeorges patients were retrieved from our hospital and other places. Only one was HLA-A02*.
Intracellular cytokine staining
EBV-specific T cell lines on day 23 were stimulated with LCLs overnight in the presence of brefeldin A. Cells were fixed, permeabilized, and stained with anti–CD3-BV785, anti–IFN-γ-PE/Cy7, anti–granzyme B-APC (from BioLengend), and anti–CD8-BV510 (from SONY). Stained cells were analyzed by flow cytometry.
Cytotoxicity assay
For the cytotoxicity assay, EBV-specific T cell lines on day 28 were used. CD8+ T cells were isolated by means of negative selection using MACS system (Miltenyi Biotech) and labeled with CellTrace Violet (CTV). The effector cells were cultured with LCLs at different effector-to-target ratios (1:1, 5:1, and 25:1) for 4 h. Cells were stained for propidium iodide (PI) and analyzed by flow cytometry. The target cells corresponding to CTV-negative cells were selected and FSC-low and PI-low apoptotic bodies and debris were excluded. PI-positive were considered as target dying apoptotic cells and PI-negative living target cells. Specific lysis (percentage) was calculated according to the formula: 100 × (sample lysis − basal lysis)/(100 − basal lysis). Sample lysis and basal lysis mean the percentages of apoptotic cells in the presence and absence of effector cells, respectively.
Confocal microscopic analysis
Surface CD137L was stained with anti-CD137L mAb (EPR1172Y; Abcam) followed by Alexa Fluor 488–conjugated secondary antibody (Invitrogen). Then, cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and incubated with anti-CD137L antibody (EPR1172Y; Abcam) followed by Alexa Fluor 546–conjugated secondary antibody (Invitrogen). Cell nuclei were stained with DAPI/49-6-Diamidino-2-phenylindole. Images were acquired with a Leica SP8 STED confocal microscope (Leica Microsystems).
CD137L reverse signaling
Monocytes were isolated using CD14 microbeads and a magnetic cell separator (Miltenyi Biotech). Cells were cultured in a plate coated with CD137-Fc protein (10 μg/ml; R&D Systems) for 7 d as previously described (Kwajah and Schwarz, 2010).
Online supplementary material
Fig. S1 shows the functional characterization of T cells from PBMCs and blasts of the patient. Fig. S2 shows the expression of CD137L in LCL and SMT cells of the patient and CD137L reverse signaling in monocytes of the patient. Fig. S3 shows T and NK cell responses toward LCL of the patient or P815 cells expressing different mutants of CD137L. Table S1 shows the immunological parameters of the patient.
Supplementary Material
shows the immunological parameters of the patient.
contains original blots for Fig. 3.
contains original blots for Fig. S1.
contains original blots for Fig. S2.
Acknowledgments
We thank the patient and her family for their trust and contribution, and the healthy donors for blood gifts. We acknowledge Graham Davies (Great Ormond Street Hospital for Children NHS Foundation Trust, UK) for his help finding HLA-A02 LCLs from a DiGeorge patient. We thank Alain Fischer for discussion and support.
S. Latour is a senior scientist at the Centre National de la Recherche Scientifique (France). B. Fournier was supported by the Fondation pour la Recherche Médicale (FDM20170638301). This work was supported by grants from the Ligue Contre le Cancer-Equipe Labélisée (France; to S. Latour), Institut National de la Santé et de la Recherche Médicale (France), the Agence Nationale de Recherche (France; ANR-18-CE15-0025-01 to S. Latour and ANR-10-IAHU-01 to Institut Imagine), the Institut National du Cancer (PLBIO-2020, PEDIAC consortium INCa_15670) the Société Française de Lutte contre les Cancers et Leucémies de l’Enfant et de l’Adolescent, AREMIG (France; to S. Latour), and the Fédération Enfants et Santé (France; to S. Latour). Exome sequencing was funded by the Rare Diseases Foundation (France; to S. Latour).
Author contributions: B. Fournier and A. Hoshino designed and performed experiments and analyzed the data. C. Bachelet, J. Bruneau, T. Molina, M. Fusaro, C. Lenoir, and C. Soudais performed experiments and analyzed the data. B. Fournier, R. Lévy, R. Klifa, M. Castelle, S. Blanche, D. Moshous, B. Neven, and C. Picard provided and analyzed clinical and immunological evaluation data. A.-S. Defachelles identified the patient and provided clinical data. S. Latour and B. Fournier wrote the manuscript. S. Latour designed and supervised the research.
References
- Alosaimi, M.F., Hoenig M., Jaber F., Platt C.D., Jones J., Wallace J., Debatin K.-M., Schulz A., Jacobsen E., Möller P., et al. 2019. Immunodeficiency and EBV-induced lymphoproliferation caused by 4-1BB deficiency. J. Allergy Clin. Immunol. 144:574–583.e5. 10.1016/j.jaci.2019.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitra, A., Doukov T., Destito G., Croft M., and Zajonc D.M.. 2019. Crystal structure of the m4-1BB/4-1BBL complex reveals an unusual dimeric ligand that undergoes structural changes upon 4-1BB receptor binding. J. Biol. Chem. 294:1831–1845. 10.1074/jbc.RA118.006297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broll, K., Richter G., Pauly S., Hofstaedter F., and Schwarz H.. 2001. CD137 expression in tumor vessel walls.High correlation with malignant tumors. Am. J. Clin. Pathol. 115:543–549. 10.1309/e343-kmyx-w3y2-10ky [DOI] [PubMed] [Google Scholar]
- Bukczynski, J., Wen T., Ellefsen K., Gauldie J., and Watts T.H.. 2004. Costimulatory ligand 4-1BBL (CD137L) as an efficient adjuvant for human antiviral cytotoxic T cell responses. Proc. Natl. Acad. Sci. USA. 101:1291–1296. 10.1073/pnas.0306567101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, I.M., Sheppard S.E., Crowley T.B., McGinn D.E., Bailey A., McGinn M.J., Unolt M., Homans J.F., Chen E.Y., Salmons H.I., et al. 2018. What is new with 22q? An update from the 22q and You Center at the Children’s Hospital of Philadelphia. Am. J. Med. Genet. A. 176:2058–2069. 10.1002/ajmg.a.40637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancrini, C., Puliafito P., Digilio M.C., Soresina A., Martino S., Rondelli R., Consolini R., Ruga E.M., Cardinale F., Finocchi A., et al. 2014. Clinical features and follow-up in patients with 22q11.2 deletion syndrome. J. Pediatr. 164:1475–1480.e2. 10.1016/j.jpeds.2014.01.056 [DOI] [PubMed] [Google Scholar]
- Chester, C., Sanmamed M.F., Wang J., and Melero I.. 2018. Immunotherapy targeting 4-1BB: Mechanistic rationale, clinical results, and future strategies. Blood. 131:49–57. 10.1182/blood-2017-06-741041 [DOI] [PubMed] [Google Scholar]
- Choi, I.K., Wang Z., Ke Q., Hong M., Qian Y., Zhao X., Liu Y., Kim H.J., Ritz J., Cantor H., et al. 2018. Signaling by the Epstein-Barr virus LMP1 protein induces potent cytotoxic CD4+ and CD8+ T cell responses. Proc. Natl. Acad. Sci. USA. 115:E686–E695. 10.1073/pnas.1713607115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard, D., Becke F.M., Langstein J., Spruss T., Kunz‐Schughart L.A., Tan T.E., Lim Y.C., and Schwarz H.. 2007. CD137 is expressed on blood vessel walls at sites of inflammation and enhances monocyte migratory activity. FASEB J. 21:456–463. 10.1096/fj.05-4739com [DOI] [PubMed] [Google Scholar]
- Edelmann, L., Pandita R.K., and Morrow B.E.. 1999. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am. J. Hum. Genet. 64:1076–1086. 10.1086/302343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell, P.J. 2019. Epstein–Barr virus and cancer. Annu. Rev. Pathol. 14:29–53. 10.1146/annurev-pathmechdis-012418-013023 [DOI] [PubMed] [Google Scholar]
- Fournier, B., Boutboul D., Bruneau J., Miot C., Boulanger C., Malphettes M., Pellier I., Dunogué B., Terrier B., Suarez F., et al. 2020. Rapid identification and characterization of infected cells in blood during chronic active Epstein-Barr virus infection. J. Exp. Med. 217:e20192262. 10.1084/jem.20192262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse, S., Bellfy S., Yagita H., and Usherwood E.J.. 2007. CD8+ T cell dysfunction and increase in murine gammaherpesvirus latent viral burden in the absence of 4-1BB ligand. J. Immunol. 178:5227–5236. 10.4049/jimmunol.178.8.5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S., Köstel Bal S., Edwards E.S.J., Pillay B., Jimenez-Heredia R., Erol Cipe F., Rao G., Salzer E., Zoghi S., Abolhassani H., et al. 2020. Extended clinical and immunological phenotype and transplant outcome in CD27 and CD70 deficiency. Blood. 136:2638–2655. 10.1182/blood.2020006738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujer, C., Murer A., Müller A., Vanoaica D., Sutter K., Jacque E., Fournier N., Kalchschmidt J., Zbinden A., Capaul R., et al. 2019. Plasmacytoid dendritic cells respond to Epstein-Barr virus infection with a distinct type I interferon subtype profile. Blood Adv. 3:1129–1144. 10.1182/bloodadvances.2018025536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbreth, R.N., Oganesyan V.Y., Amdouni H., Novarra S., Grinberg L., Barnes A., and Baca M.. 2018. Crystal structure of the human 4-1BB/4-1BBL complex. J. Biol. Chem. 293:9880–9891. 10.1074/jbc.RA118.002803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houot, R., Goldstein M.J., Kohrt H.E., Myklebust J.H., Alizadeh A.A., Lin J.T., Irish J.M., Torchia J.A., Kolstad A., Chen L., and Levy R.. 2009. Therapeutic effect of CD137 immunomodulation in lymphoma and its enhancement by Treg depletion. Blood. 114:3431–3438. 10.1182/blood-2009-05-223958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, S., Ohno T., Kakizaki S., and Ichinohasama R.. 2011. Epstein-Barr virus–positive T-cell lymphoma cells having chromosome 22q11.2 deletion: An autopsy report of DiGeorge syndrome. Hum. Pathol. 42:2037–2041. 10.1016/j.humpath.2010.03.014 [DOI] [PubMed] [Google Scholar]
- Izawa, K., Martin E., Soudais C., Bruneau J., Boutboul D., Rodriguez R., Lenoir C., Hislop A.D., Besson C., Touzot F., et al. 2017. Inherited CD70 deficiency in humans reveals a critical role for the CD70–CD27 pathway in immunity to Epstein-Barr virus infection. J. Exp. Med. 214:73–89. 10.1084/jem.20160784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebudi, R., Kiykim A., and Sahin M.K.. 2019. Primary immunodeficiency and cancer in children: A review of the literature. Curr. Pediatr. Rev. 15:245–250. 10.2174/1573396315666190917154058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwajah, M.M.S., and Schwarz H.. 2010. CD137 ligand signaling induces human monocyte to dendritic cell differentiation. Eur. J. Immunol. 40:1938–1949. 10.1002/eji.200940105 [DOI] [PubMed] [Google Scholar]
- Laderach, D., Movassagh M., Johnson A., Mittler R.S., and Galy A.. 2002. 4-1BB co-stimulation enhances human CD8+ T cell priming by augmenting the proliferation and survival of effector CD8+ T cells. Int. Immunol. 14:1155–1167. 10.1093/intimm/dxf080 [DOI] [PubMed] [Google Scholar]
- Laderach, D., Wesa A., and Galy A.. 2003. 4-1BB-ligand is regulated on human dendritic cells and induces the production of IL-12. Cell Immunol. 226:37–44. 10.1016/j.cellimm.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Lambert, M.P., Arulselvan A., Schott A., Markham S.J., Crowley T.B., Zackai E.H., and McDonald‐McGinn D.M.. 2018. The 22q11.2 deletion syndrome: Cancer predisposition, platelet abnormalities and cytopenias. Am. J. Med. Genet. A. 176:2121–2127. 10.1002/ajmg.a.38474 [DOI] [PubMed] [Google Scholar]
- Latour, S., and Fischer A.. 2019. Signaling pathways involved in the T-cell-mediated immunity against Epstein-Barr virus: Lessons from genetic diseases. Immunol. Rev. 291:174–189. 10.1111/imr.12791 [DOI] [PubMed] [Google Scholar]
- Lee, E.S., Locker J., Nalesnik M., Reyes J., Jaffe R., Alashari M., Nour B., Tzakis A., and Dickman P.S.. 1995. The association of Epstein–Barr virus with smooth-muscle tumors occurring after organ transplantation. N. Engl. J. Med. 332:19–25. 10.1056/NEJM199501053320104 [DOI] [PubMed] [Google Scholar]
- Lee, P.-K., Chang C.-J., and Lin C.-M.. 2003. Lipopolysaccharide preferentially induces 4-1BB ligand expression on human monocyte-derived dendritic cells. Immunol. Lett. 90:215–221. 10.1016/j.imlet.2003.08.002 [DOI] [PubMed] [Google Scholar]
- Li, Y., Tan S., Zhang C., Chai Y., He M., Zhang C.W.-H., Wang Q., Tong Z., Liu K., Lei Y., et al. 2018. Limited cross-linking of 4-1BB by 4-1BB ligand and the agonist monoclonal antibody utomilumab. Cell Rep. 25:909–920.e4. 10.1016/j.celrep.2018.09.073 [DOI] [PubMed] [Google Scholar]
- Martin, E., Palmic N., Sanquer S., Lenoir C., Hauck F., Mongellaz C., Fabrega S., Nitschké P., Esposti M.D., Schwartzentruber J., et al. 2014. CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature. 510:288–292. 10.1038/nature13386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, P.C., Eng K.H., Singel K.L., Abrams S.I., Odunsi K., Moysich K.B., Fuleihan R., Garabedian E., Lugar P., Ochs H.D., et al. 2018. Cancer in primary immunodeficiency diseases: Cancer incidence in the United States Immune Deficiency Network Registry. J. Allergy Clin. Immunol. 141:1028–1035. 10.1016/j.jaci.2017.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain, K.L., Leach C.T., Jenson H.B., Joshi V.V., Pollock B.H., Parmley R.T., DiCarlo F.J., Chadwick E.G., and Murphy S.B.. 1995. Association of Epstein–Barr virus with leiomyosarcomas in young people with AIDS. N. Engl. J. Med. 332:12–18. 10.1056/NEJM199501053320103 [DOI] [PubMed] [Google Scholar]
- Middendorp, S., Xiao Y., Song J.-Y., Peperzak V., Krijger P.H.L., Jacobs H., and Borst J.. 2009. Mice deficient for CD137 ligand are predisposed to develop germinal center–derived B-cell lymphoma. Blood. 114:2280–2289. 10.1182/blood-2009-03-208215 [DOI] [PubMed] [Google Scholar]
- Morsheimer, M., Whitehorn T.F.B., Heimall J., and Sullivan K.E.. 2017. The immune deficiency of chromosome 22q11.2 deletion syndrome. Am. J. Med. Genet. A. 173:2366–2372. 10.1002/ajmg.a.38319 [DOI] [PubMed] [Google Scholar]
- Olofsson, P.S., Söderström L.A., Wågsäter D., Sheikine Y., Ocaya P., Lang F., Rabu C., Chen L., Rudling M., Aukrust P., et al. 2008. CD137 is expressed in human atherosclerosis and promotes development of plaque inflammation in hypercholesterolemic mice. Circulation. 117:1292–1301. 10.1161/CIRCULATIONAHA.107.699173 [DOI] [PubMed] [Google Scholar]
- Palazón, A., Teijeira A., Martínez-Forero I., Hervás-Stubbs S., Roncal C., Peñuelas I., Dubrot J., Morales-Kastresana A., Pérez-Gracia J.L., Ochoa M.C., et al. 2011. Agonist anti-CD137 mAb act on tumor endothelial cells to enhance recruitment of activated T lymphocytes. Cancer Res. 71:801–811. 10.1158/0008-5472.CAN-10-1733 [DOI] [PubMed] [Google Scholar]
- Rodriguez, R., Fournier B., Cordeiro D.J., Winter S., Izawa K., Martin E., Boutboul D., Lenoir C., Fraitag S., Kracker S., et al. 2019. Concomitant PIK3CD and TNFRSF9 deficiencies cause chronic active Epstein-Barr virus infection of T cells. J. Exp. Med. 216:2800–2818. 10.1084/jem.20190678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seko, Y., Sugishita K., Sato O., Takagi A., Tada Y., Matsuo H., Yagita H., Okumura K., and Nagai R.. 2004. Expression of costimulatory molecules (4-1BBL and fas) and major histocompatibility class I chain-related A (MICA) in aortic tissue with Takayasu’s arteritis. J. Vasc. Res. 41:84–90. 10.1159/000076437 [DOI] [PubMed] [Google Scholar]
- Somekh, I., Thian M., Medgyesi D., Gülez N., Magg T., Duque A.G., Stauber T., Lev A., Genel F., Unal E., et al. 2019. CD137 deficiency causes immune dysregulation with predisposition to lymphomagenesis. Blood. 134:1510–1516. 10.1182/blood.2019000644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, T., van der Werff Ten Bosch J., De Rademaeker M., Van Den Bogaert A., and van den Akker M.. 2017. Risk of malignancy in 22q11.2 deletion syndrome. Clin. Case Rep. 5:486–490. 10.1002/ccr3.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, K.E. 2019. Chromosome 22q11.2 deletion syndrome and DiGeorge syndrome. Immunol. Rev. 287:186–201. 10.1111/imr.12701 [DOI] [PubMed] [Google Scholar]
- Tangye, S.G., and Latour S.. 2020. Primary immunodeficiencies reveal the molecular requirements for effective host defense against EBV infection. Blood. 135:644–655. 10.1182/blood.2019000928 [DOI] [PubMed] [Google Scholar]
- Tanita, K., Hoshino A., Imadome K.-I., Kamiya T., Inoue K., Okano T., Yeh T.-W., Yanagimachi M., Shiraishi A., Ishimura M., et al. 2019. Epstein-Barr virus-associated γδ T-cell lymphoproliferative disorder associated with hypomorphic IL2RG mutation. Front. Pediatr. 7:15. 10.3389/fped.2019.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Zhang P., Zhang Q., Wang X., Li J., Ma C., Sun W., and Zhang L.. 2008. Analysis of CD137 and CD137L expression in human primary tumor tissues. Croat. Med. J. 49:192–200. 10.3325/cmj.2008.2.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, T., Bukczynski J., and Watts T.H.. 2002. 4-1BB ligand-mediated costimulation of human T cells induces CD4 and CD8 T cell expansion, cytokine production, and the development of cytolytic effector function. J. Immunol. 168:4897–4906. 10.4049/jimmunol.168.10.4897 [DOI] [PubMed] [Google Scholar]
- Won, E.-Y., Cha K., Byun J.-S., Kim D.-U., Shin S., Ahn B., Kim Y.H., Rice A.J., Walz T., Kwon B.S., and Cho H.-S.. 2010. The structure of the trimer of human 4-1BB ligand is unique among members of the tumor necrosis factor superfamily. J. Biol. Chem. 285:9202–9210. 10.1074/jbc.M109.084442 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the immunological parameters of the patient.
contains original blots for Fig. 3.
contains original blots for Fig. S1.
contains original blots for Fig. S2.