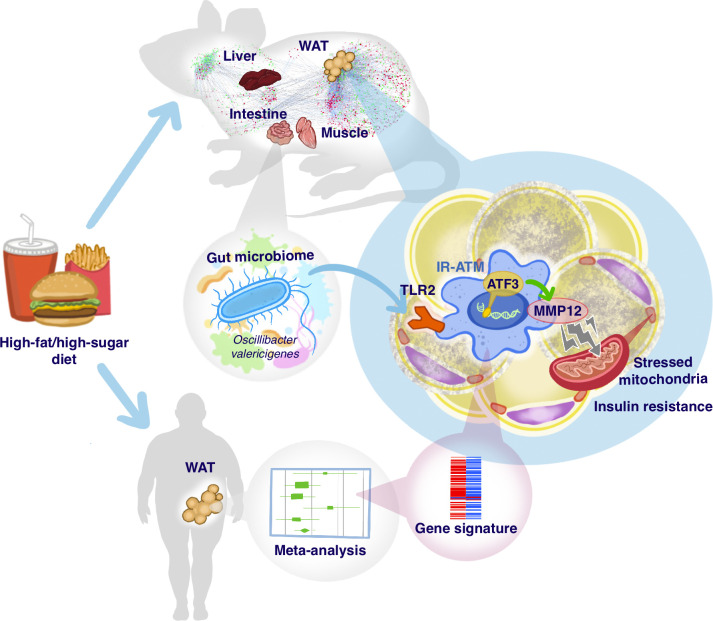
High-fat/high-sugar diet–expanded microbiota promote insulin resistance inducing Mmp12-positive adipose macrophages via TLR2/MYD88 and ATF3. Mmp12-positive adipose macrophages exhibit a transcriptomic signature associated with insulin resistance in humans.
Abstract
Microbiota contribute to the induction of type 2 diabetes by high-fat/high-sugar (HFHS) diet, but which organs/pathways are impacted by microbiota remain unknown. Using multiorgan network and transkingdom analyses, we found that microbiota-dependent impairment of OXPHOS/mitochondria in white adipose tissue (WAT) plays a primary role in regulating systemic glucose metabolism. The follow-up analysis established that Mmp12+ macrophages link microbiota-dependent inflammation and OXPHOS damage in WAT. Moreover, the molecular signature of Mmp12+ macrophages in WAT was associated with insulin resistance in obese patients. Next, we tested the functional effects of MMP12 and found that Mmp12 genetic deficiency or MMP12 inhibition improved glucose metabolism in conventional, but not in germ-free mice. MMP12 treatment induced insulin resistance in adipocytes. TLR2-ligands present in Oscillibacter valericigenes bacteria, which are expanded by HFHS, induce Mmp12 in WAT macrophages in a MYD88-ATF3–dependent manner. Thus, HFHS induces Mmp12+ macrophages and MMP12, representing a microbiota-dependent bridge between inflammation and mitochondrial damage in WAT and causing insulin resistance.
Graphical Abstract
Introduction
A combination of overconsumption of food high in simple sugars and animal fat diet and a sedentary lifestyle provoked dramatic increases in the rates of diabetes, obesity, and other metabolic diseases over recent decades. Type 2 diabetes (T2D) involves dysfunction in multiple organs, with critical events in the liver, white adipose tissue (WAT), and muscles, promoting the development of systemic insulin resistance (IR). Despite established knowledge of the disease processes occurring within each organ, their relative contribution to systemic metabolic dysregulation is less clear. Furthermore, gut microbiota are emerging as key players in multiple proposed mechanisms that affect different organs (Gurung et al., 2020; Hartstra et al., 2015; Sonnenburg and Backhed, 2016). Nevertheless, there is no consensus on which of these organs and pathways are most important for the development of IR and T2D. To recapitulate human diet–induced T2D in animal models, special diets were developed, including high-fat/high-sugar diet (HFHS), frequently referred to as western diet (WD; Small et al., 2018).
Several studies interrogating different omics data have been conducted attempting to address these questions (Hosseinkhani et al., 2021; Passaro et al., 2021). However, commonly used association analyses between diet, microbiome composition, and disease do not infer causality or attempt to establish master regulators/pathways that can be affected by inverse causality and other confounding factors. In this study, we used multiorgan network analysis and transkingdom causal inference employing statistical dependencies between differentially expressed elements (e.g., genes, microbes, and phenotypic markers) combined with causality principles (Pearl et al., 2009; Yambartsev et al., 2016), information theory (Shannon, 1953; Uda, 2020), and interventions (e.g., diet, antibiotics, etc.). This system’s approach provided us with two causal inferences: first, microbiota-dependent inflammation drives IR in WAT; second, this inflammation is caused by specific members of HFHS-enriched gut microbiota.
Specifically, we investigated pathways underlying the role of diet-modified microbiota in metabolic disease by comprehensively characterizing HFHS-induced changes in four metabolic organs in mice: intestine, liver, WAT, and muscle. We found that a novel predominant mechanism contributing to systemic IR is microbiota-dependent damage of mitochondrial oxidative phosphorylation (OXPHOS) in WAT. To establish how microbiota contributes to this process, we characterized IR-promoting microbiota-induced macrophages in WAT of mice and humans. Using network analysis coupled with loss- and gain-of-function studies, we established MMP12 as a critical effector of these cells. Finally, we identified a specific microbe (Oscillibacter valericigenes [OV]) that promotes Mmp12 expression through the production of TLR2 ligands. We demonstrate in vitro that Mmp12 is induced by stimulation of the TLR2/Myd88 signaling pathway and activation of the transcription factor (TF) ATF3.
Results
HFHS diet drives alterations predominantly in a microbiota-dependent manner
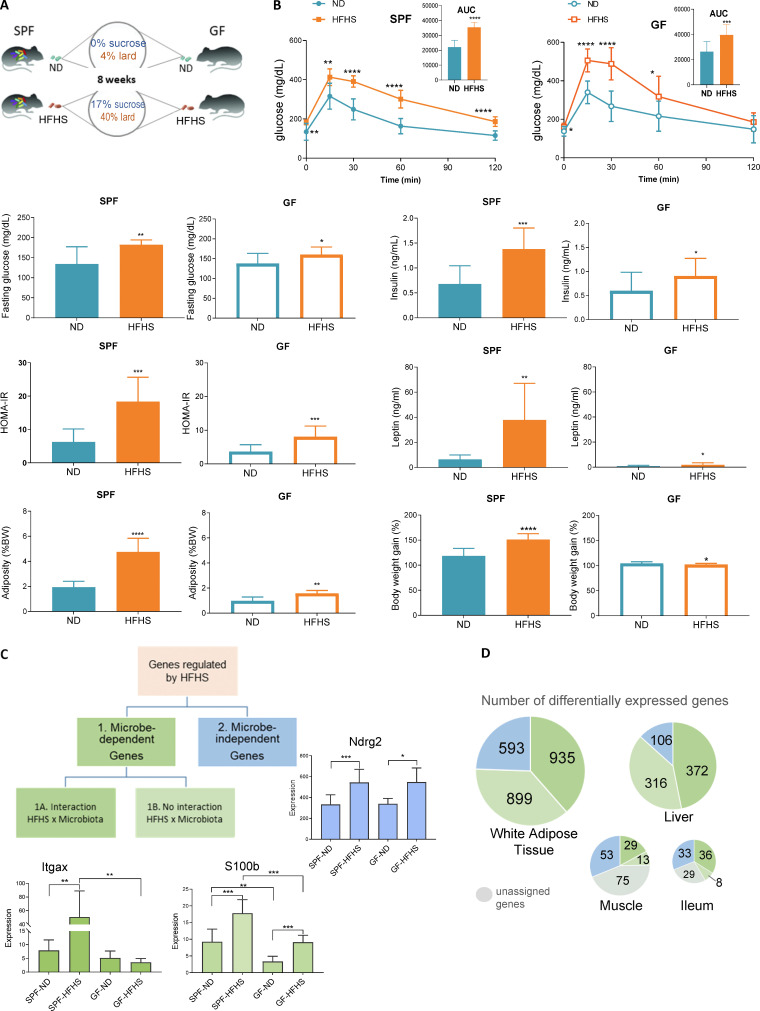
To dissect the contribution of microbiota to the effects of WD (designated herein HFHS), specific pathogen–free (SPF) and germfree (GF) C57BL/6 mice were fed a HFHS or matched normal diet (ND) for 8 wk (Fig. 1 A). As expected (Backhed et al., 2007; Rabot et al., 2010), HFHS increased glucose intolerance in GF mice and the presence of microbiota further exacerbated IR and adiposity in SPF mice (Fig. 1 B and Table S1).
Figure 1.
HFHS diet drives alterations predominantly in a microbiota-dependent manner. (A) Experimental design. SPF and GF mice were fed ND or HFHS for 8 wk (SPF mice: n = 10 per group; GF mice: n = 5–6). (B) Metabolic parameters changed by HFHS in SPF and GF mice (mean ± SD, SPF mice, n = 10; GF mice, n = 5–6, t test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (C) Schematic diagram for categorization of genes according to the regulation by diet and microbiota. (D) Pie charts showing the number of DEGs in each category in WAT, liver, muscle, and ileum.
Next, using samples from SPF and GF mice, we analyzed whole-tissue transcriptomes of the intestine, as it harbors the most abundant microbial community in mammals, and of the three other major insulin-responsive organs implicated in T2D (liver, WAT, muscle). We found that WAT and liver transcriptomes were more sensitive to HFHS (2,427 and 794 differentially expressed genes [DEGs], respectively), compared to muscle and ileum (170 and 116 genes, respectively). The finding that more genes are regulated in WAT than in liver or muscle might be expected because mice that have consumed the HFHS for 8 wk represent the initial stages of T2D, with molecular changes progressing from adipose to the liver to the muscle (Kraegen et al., 1991; Perry et al., 2015). HFHS-regulated genes were classified as microbiota dependent (category 1) or microbiota independent (category 2; Fig. 1 C), while a small fraction of genes could not be assigned to either of the above categories. Microbiota-dependent genes were further classified as regulated by HFHS differently in SPF and GF mice (category 1A; e.g., Itgax) or regulated by diet in both types of mice, but whose expression was enhanced by the microbiota (category 1B; e.g., S100b; Fig. 1 C). We found that microbiota play a key role in HFHS-modulated gene expression in all four organs, with about 87% of genes in the liver, 76% in WAT, 57% in the ileum, and 44% in muscle belonging to category 1 (Fig. 1 D and Table S2).
Multiorgan network analysis identifies OXPHOS/mitochondria in WAT as a key driver of systemic metabolism
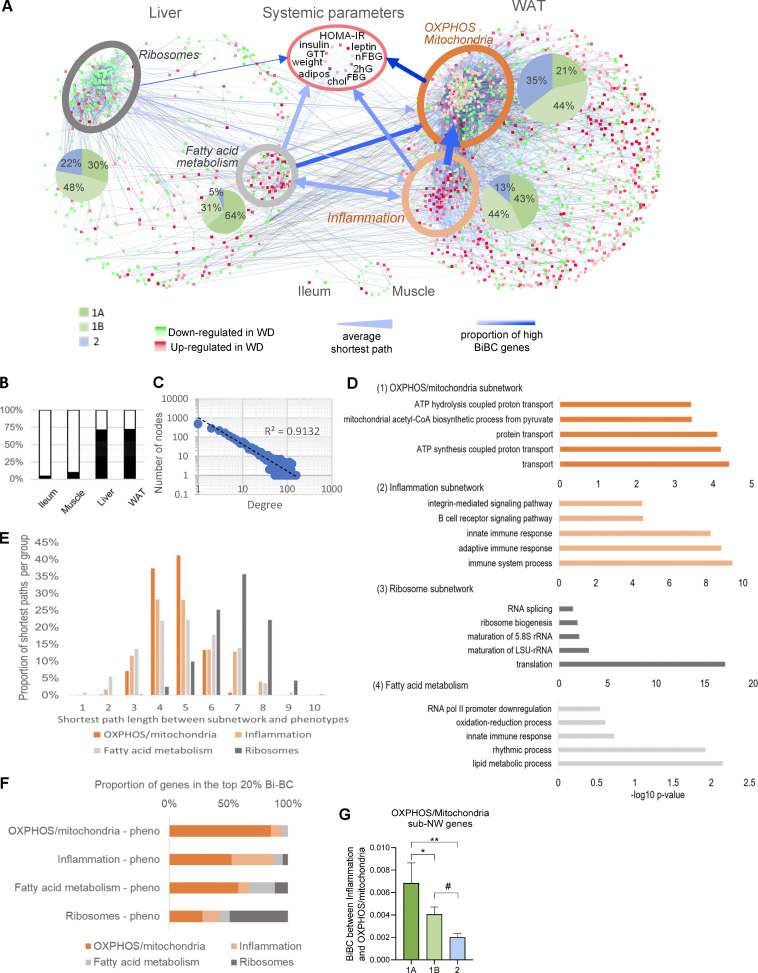
To model HFHS-induced alterations, we reconstructed a co-variation network, integrating gene expression with systemic metabolic parameters (Fig. 2 A). Approximately two thirds of DEGs in the four organs were retained in the network (Fig. 2 B and Table S3) after filtering out genes (nodes) and edges (connections between nodes of network) that did not fulfill statistical and causality criteria (Yambartsev et al., 2016). As expected, the network node degree distribution conformed with the power-law structure of regulatory biological networks (Fig. 2 C). Two subnetworks were identified in the liver and WAT (Fig. 2 A). In WAT, the larger subnetwork mainly reflected impaired mitochondrial functions, in particular OXPHOS (Fig. 2 D [1]). The smaller WAT subnetwork with mostly upregulated genes was enriched for inflammation and immune response genes (Fig. 2 D [2]). The larger liver subnetwork included mostly HFHS downregulated genes that were enriched for ribosomal components (Fig. 2 D [3]), while the other liver subnetwork, largely upregulated by HFHS, was represented by fatty acid metabolism genes (Fig. 2 D [4]). The genes controlling fatty acid metabolism in the liver and inflammation in fat were those most impacted by microbes (>85% of genes from the 1A and 1B categories; Fig. 2 A).
Figure 2.
Multiorgan network analysis identifies OXPHOS/mitochondria in WAT as a key driver of systemic metabolism. (A) Multiorgan co-variation network demonstrating connections between DEGs in liver (565 nodes), WAT (1,747 nodes), ileum (3 nodes), and muscle (16 nodes) and host systemic metabolic parameters. Subnetworks identified within WAT and liver are highlighted by ellipses. The shades of red and green colors of nodes indicate increased and decreased expression in HFHS-fed SPF mice, respectively, with color intensity indicating the level of HFHS vs. ND fold change in SPF mice. The blue and black colored edges (lines) indicate positive and negative correlations, respectively, and the color intensity of edges indicates the strength of correlation. Width of blue arrows reflect the average shortest path distance between each subnetwork; arrow color intensity represents the proportion of top-ranked genes (based on BiBC) in each of the subnetworks. The arrowheads indicate the flow of information between sub-networks showing that the inflammation subnetwork is strongly connected to the OXPHOS/mitochondria subnetwork (wide arrow), and it relies on the OXPHOS/mitochondria subnetwork to regulate the metabolic parameters (darker blue). (B) The bar plot shows percent of DEGs in each organ that were retained in the network (black) after filtering out nodes that did not fulfill statistical and causality criteria (white). (C) Power-law distribution of the number of nodes and edges in the multiorgan network. (D) Gene ontology enrichment analysis of the four subnetworks: (1) WAT OXPHOS/mitochondria, (2) WAT inflammation, (3) liver ribosomes, and (4) liver fatty acid metabolism subnetworks. The top five most significant biological processes and cellular component terms are shown along with adjusted P values. (E) Shortest path analysis. Distribution of pairwise shortest path lengths between the nodes of each subnetwork and systemic metabolic parameters (systemic phenotypes). The average shortest path is a metric evaluating closeness of one section of a network to another. The shortest paths were estimated between all four subnetworks and systemic metabolic parameters, weighting the shortest paths to account for the fact that closer nodes potentially have a bigger impact. The systemic metabolic parameters were nearly equidistant to all subnetworks except for the distant ribosomal subnetwork. The fatty acid metabolism and OXPHOS/mitochondria pathways were closest to systemic metabolic parameters overall. The y axis represents the proportion of genes, normalized per subnetwork, that are the indicated shortest path length away from systemic metabolic parameters (x axis). All comparisons between shortest path distributions of subnetworks were conducted using a chi-square test, and all were significant with a P value <0.0001. (F) BiBC as a proxy between pathway interaction. To determine the likelihood of each of these subnetworks to be the hubs of the information flow (i.e., bottlenecks) between each of the other subnetworks and systemic metabolic parameters, BiBC of each subnetwork was calculated. The proportion of genes from each subnetwork in the top 20% BiBC between that subnetwork and systemic metabolic parameters is shown. Larger proportions indicate a greater importance of the subnetwork in regulating metabolic parameters. All comparisons between subnetworks were conducted using a chi-square test, and all were significant with a P value <0.0001. This means the OXPHOS/mitochondria and ribosomal subnetworks rely little on the other identified subnetworks to reach systemic metabolic parameters. On the contrary, half of the effects of the inflammation and fatty acid metabolism subnetworks on systemic metabolic parameters are directed through the OXPHOS/mitochondria subnetwork in adipose tissue, suggesting the importance of the OXPHOS/mitochondria pathway in regulating glucose metabolism. Accordingly, the integration of the results of shortest paths and BiBC analyses showed that the OXPHOS/mitochondria pathway in adipose tissue partially mediates the effects of both fatty acid metabolism in liver and inflammation in adipose tissue on systemic metabolism. (G) Microbiota-dependent genes show higher BiBC between inflammation and OXPHOS/mitochondria than microbiota-independent genes. Comparison of BiBC values between three categories (Fig. 1 C) for genes in OXPHOS/mitochondria subnetwork. Since microbiota-driven inflammation is one of the two main pathways influencing OXPHOS/mitochondria, we compared potential regulatory capacity (using BiBC as a proxy) of interactive (cat. 1A), additive (cat. 1B), and microbiota-independent (cat. 2) components of this pathway. For this, we calculated the BiBC of each gene between the inflammation and OXPHOS/mitochondria subnetworks (Fig. 2 F). Then, we compared BiBC values of the OXPHOS/mitochondria subnetwork genes between three gene categories defining microbial contribution to their regulation (microbiota-dependent with interaction [1A], microbiota-dependent no interaction [1B], and microbiota-independent [2]). Significantly higher values of BiBC were found for category 1A genes than for 1B and 2 (mean ± SEM, P values based off ANOVA, followed by Tukey’s multiple comparisons test, *P < 0.05, **P < 0.01, #P = 0.08).
To disclose the regulatory relationships between the pathways and systemic metabolic parameters, we investigated network topology reflective of information flow in the network (including shortest path [Fig. 2 E] and bipartite betweenness centrality [BiBC; Fig. 2 F and Table S4]). This analysis showed (blue arrows in Fig. 2 A) that among the four subnetworks, mitochondrial OXPHOS in WAT directly affects systemic glucose metabolism, while fatty acid pathways in the liver and inflammation in WAT primarily act indirectly through microbiota-dependent damage of mitochondrial OXPHOS in the adipose (Fig. 2, A and G).
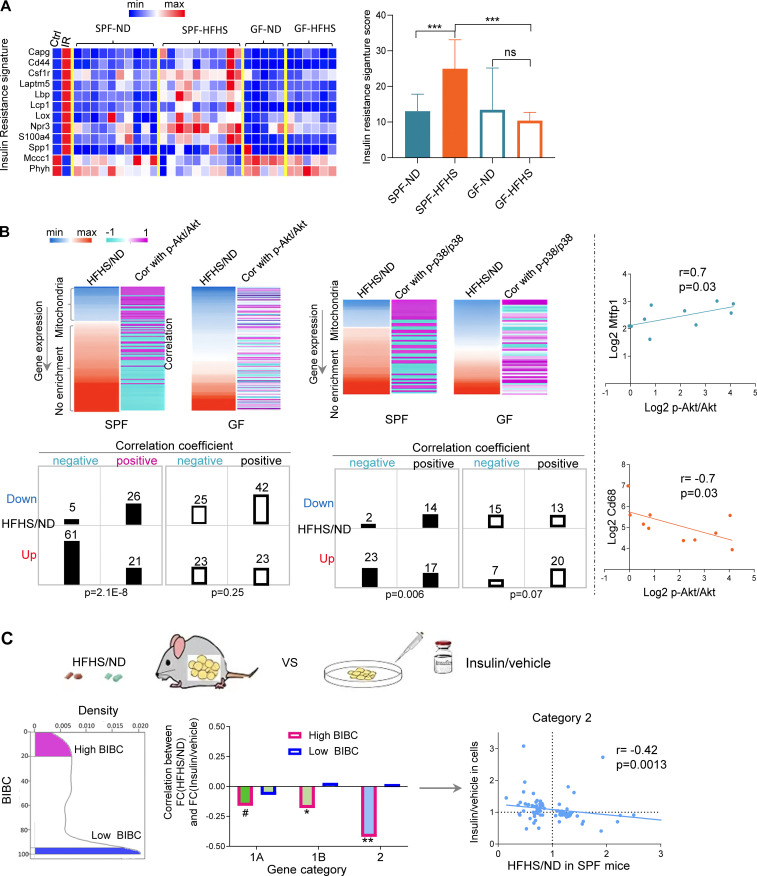
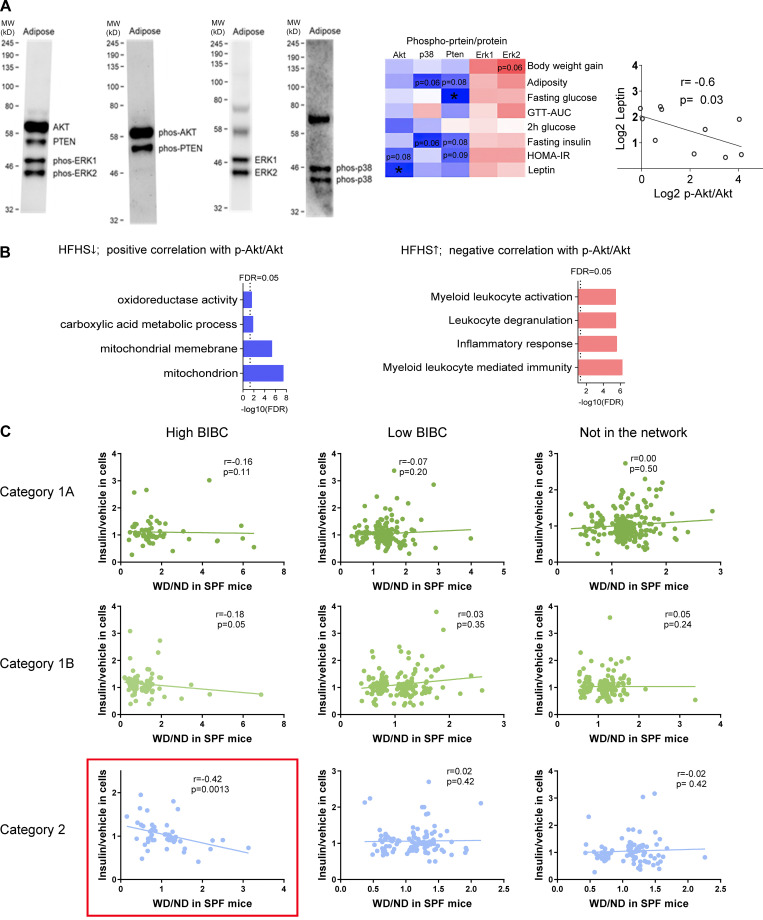
Three independent lines of evidence further supported the computational inference that IR is induced in WAT and is partially dependent on the gut microbiota. First, HFHS induced an IR gene signature (Jung et al., 2018) in WAT of SPF, but not of GF mice, indicating a significant contribution of microbiota (Fig. 3 A). Second, phosphorylated AKT and—to a lesser extent—p38 MAP kinase, both classical markers of insulin signaling (IS), negatively correlated with inflammation and positively with OXPHOS genes in WAT of SPF, but not GF mice (Fig. 3 B; Fig. S1, A and B; and Table S5). Third, genes with a high regulatory capacity (i.e., high BiBC [Dong et al., 2015] to systemic metabolic markers in the network) and upregulated by HFHS negatively correlated with the response of adipocytes to insulin in vitro (Figs. 3 C and S1 C; and Table S6). This last result indicated that a large proportion of genes inferred to mediate the negative effects of HFHS on systemic metabolism are involved in insulin sensitivity in adipocytes.
Figure 3.
Validation of network inferences revealing microbiota-dependent IR in WAT. (A) Expression levels of IR signature genes described in Jung et al. (2018) in WAT of ND or HFHS-fed SPF or GF mice (SPF mice, n = 10; GF mice, n = 5–6). (B) Heatmaps showing the fold change of gene expression (HFHS/ND) in WAT and the correlation of the gene expression levels with the abundance of phosphorylated p-Akt/Akt or p-p38/p38 in WAT in each mouse (upregulated genes by HFHS tend to negatively correlate with phosphorylated Akt and p38, while downregulated genes by HFHS tends to positively correlate with phosphorylated Akt and p38 in SPF mice; this phenomenon only holds in SPF mice). The contingency tables show the relationship between sign of correlation (indicated by + or −) and regulation by HFHS (Dn, genes downregulated by HFHS; up, genes upregulated by HFHS; +, genes positively correlated with p-Akt/Akt or p-p38/p38; −, genes negatively correlated with p-Akt/Akt or p-p38/p38, chi-square, Yates value). Scatterplots show two examples of genes demonstrating positive (Mtfp1) and negative correlation (Cd68) with p-Akt/Akt (Spearman correlation, one-tailed P value, n = 10) in WAT of SPF mice. (C) Correlation between in vivo effects of HFHS in WAT and in vitro effect of insulin in 3T3-L1 adipocyte cells on the expression of high- and low-ranking BiBC genes (left panel shows the distribution of BiBC among the DEGs in WAT; middle panel shows the correlation coefficient between fold change [HFHS/ND] in WAT and fold change [insulin/vehicle] in vitro of gene expression in each category; right panel [scatter plot] shows the correlation only in category 2 [each dot represents a gene]). Only high-ranking BiBC genes have negative correlation (Spearman correlation, one-tailed P values: *P < 0.05, **P < 0.01, #P = 0.11).
Figure S1.
Additional details of validation of network inferences revealing microbiota-dependent IR in WAT. (A) Representative examples of the multiplexed Western blots of Akt, PTEN, phosphorylated-Akt, phosphorylated-PTEN, phosphorylated-ERK1 and ERK2 in WAT. The heatmap shows Spearman correlation of IS-related proteins with metabolic parameters in HFHS-fed SPF mice (n = 7–10, one-tailed, *P < 0.05). The scatter plot shows Spearman correlation between serum leptin and p-Akt/Akt in the WAT of SPF mice (n = 10, one-tailed P = 0.03). MW, molecular weight. (B) Enrichment analysis of genes: (1) Which expression is decreased by HFHS and positively correlates with p-Akt/Akt; (2) Which expression is increased by HFHS and negatively correlates with p-Akt/Akt (FDR < 0.05). (C) Scatterplots of correlation between fold change induced by HFHS (HFHS/ND) in mice with fold change induced by insulin (insulin/vehicle) in 3T3-L1 cells for the three categories of genes regulated by HFHS (Spearman correlation, one-tailed P values). These data are related to Fig. 3 C. Source data are available for this figure: SourceData FS1.
Thus far, our experiments coupled with network analysis predict that HFHS likely acts in WAT by driving microbiota-related OXPHOS damage, leading to systemic IR.
Mmp12-positive macrophages link microbiota-dependent inflammation and IR in WAT
Since our results indicated that WAT is the most critical tissue initially involved in HFHS-driven T2D, we focused further works on this tissue. Our multiorgan network model also inferred that microbiota-dependent activation of immune/inflammation pathways leads to OXPHOS damage and consequently systemic IR.
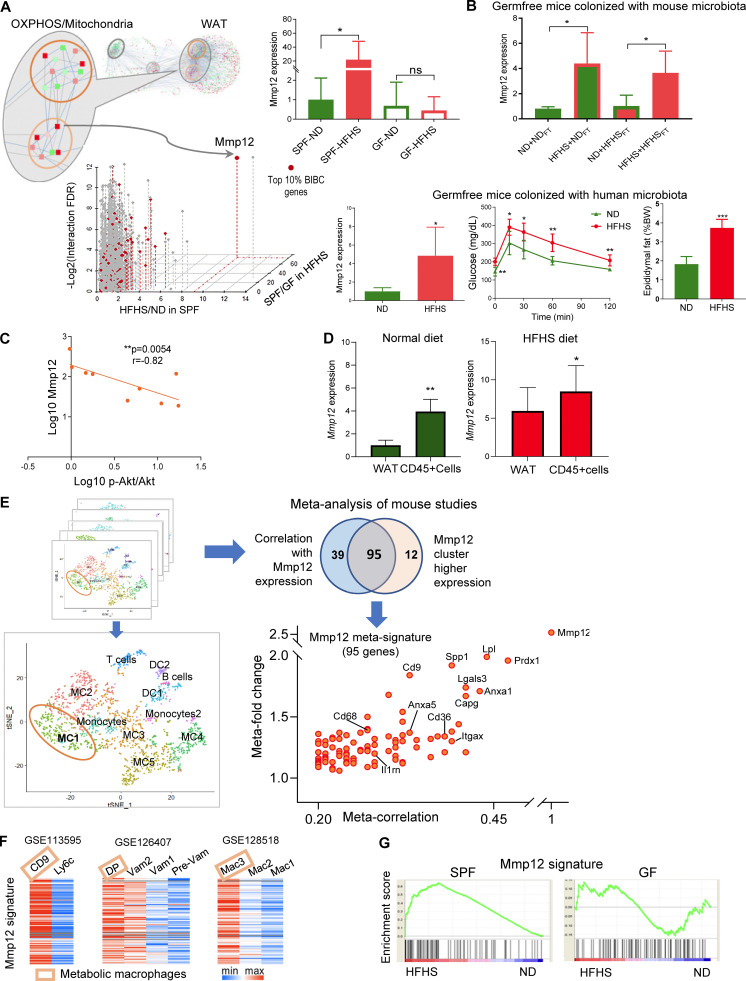
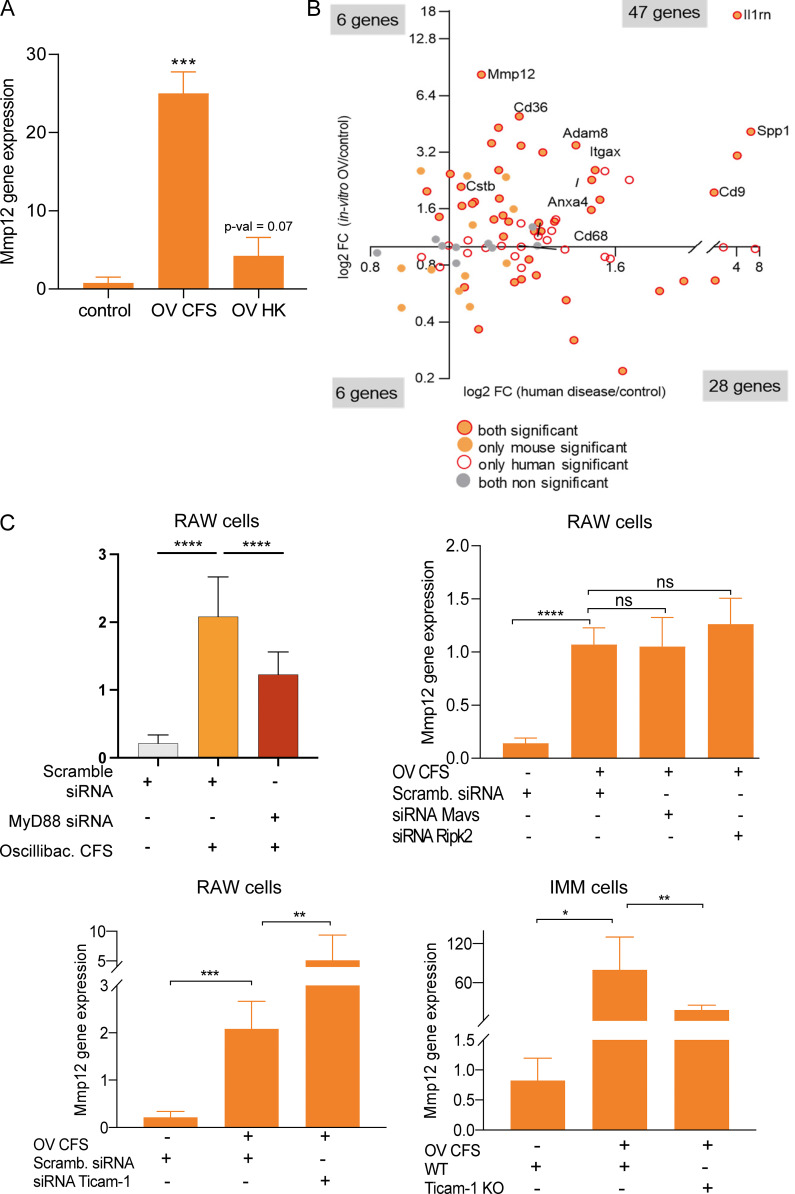
To identify microbiota-dependent genes that mediate this process, we interrogated the network for genes with high BiBC (Dong et al., 2015), reflecting their ability to mediate inflammatory damage on the OXPHOS pathway. Mmp12 was identified as the most prominent microbiota-dependent gene (category 1A) connecting inflammation and mitochondrial OXPHOS subnetworks in WAT (Fig. 4 A). Notably, HFHS induced Mmp12 expression in the presence of both mouse and human microbiota (Fig. 4 B). Mmp12 also negatively correlated with p-AKT/AKT (Fig. 4 C), further supporting its role in WAT-specific IR.
Figure 4.
Mmp12-positive macrophages link microbiota-dependent inflammation and OXPHOS damage in WAT leading to impaired glucose metabolism. (A) Identifying critical candidate genes mediating information flow from the inflammation subnetwork to the OXPHOS/mitochondria subnetwork. The scatterplot displays genes by the dimensions of: (1) BiBC between inflammatory and OXPHOS/mitochondria subnetworks (red color, top 10%); (2) fold change of gene expression induced by diet in SPF mice (HFHS/ND in SPF mice); (3) fold change of gene expression induced by microbiota in HFHS-fed mice (SPF/GF under HFHS condition); (4) log-transformed FDR of the interaction effect (diet × microbiota, two-way ANOVA). The inset bar-plot shows the intensity of Mmp12 transcripts in SPF or GF mice fed with either ND or HFHS diet (mean ± SEM, SPF mice: n = 10 per group; GF mice: n = 5, ND, n = 6, HFHS). (B) Top: Mmp12 gene expression in WAT of ND or HFHS-fed GF mice transplanted with fecal microbiota harvested from either ND or HFHS-fed SPF mice (mean ± SD, n = 4, parametric t test, one-tailed, *P < 0.05). Colors: outlines = diet and bar filling = type of microbiota used for fecal transplantation; HFHS = red, ND = green. Bottom: GF mice were transplanted with human microbiota. The figures respectively show WAT Mmp12 gene expression, glucose tolerance curve, fasting glucose levels, and epididymal fat amounts in ND or HFHS-fed mice (mean ± SD, n = 5, Mann-Whitney test, one-tailed, *P < 0.05). (C) Correlation of Mmp12 gene expression and p-Akt/Akt in adipose tissue (n = 9; Spearman correlation, one-tailed). (D) Mmp12 expression in bulk WAT and CD45+ cells isolated from WAT of SPF mice fed with ND or HFHS (mean ± SD, n = 5 for ND-fed mice, n = 7 for HFHS-fed mice, one-tailed parametric t test, *P < 0.05, **P < 0.01). (E) Meta-analysis of single cell transcriptome analysis to derive Mmp12 meta-signature in mice. Eight single-cell RNA datasets with CD45+ cells containing Mmp12+ cell subsets were used for this analysis. The meta-correlation analysis and meta-differential expression analysis from each data set (details in Materials and methods) identify a gene signature specific to subsets of Mmp12+ cells (adjusted P value <0.001); Venn diagram indicates the overlap between the two analyses, the 95 genes are shown on the scatter plot with labeling of some genes. (F) Heatmaps showing the expression of Mmp12 meta-signature genes in different types of macrophages in GSE113595, GSE126407, and GSE128518. DP, CD11c+CD64+ double-positive macrophage; VAM, vasculature associated macrophages. (G) Mmp12 meta-signature gene set is enriched by HFHS in SPF mice, but not in GF mice (SPF mice: n = 10 per group; GF mice: n = 5; ND, n = 6, HFHS).
In WAT, Mmp12 is predominantly expressed in CD45+ hematopoietic cells (Fig. 4 D). Using meta-analysis of eight publicly available single-cell RNA sequencing (scRNA-seq) datasets (Table S7), we established a 95-gene signature of Mmp12+ CD45+ cells (Figs. 4 E and S2 A; and Table S8) and confirmed its expression in macrophages (Fig. S2 B; Lee et al., 2014; Martinez-Santibanez et al., 2015). This new Mmp12 meta-signature comprehensively defined metabolic macrophages described by three independent reports (Hill et al., 2018; Jaitin et al., 2019; Silva et al., 2019; Fig. 4 F). Although these studies used different sets of markers, they are all part of our newly discovered meta-signature together with other genes that were previously implicated in IR in WAT (Cd9 [Hill et al., 2018], Cd36 [Kennedy et al., 2011], Lgals3 [Li et al., 2016; Pejnovic et al., 2013], and Itgax [Patsouris et al., 2008]). This signature identifies a unique cell population—IR-associated adipose tissue macrophages (IR-ATMs; Fig. 4 E)—that was strongly increased by HFHS in SPF, but not in GF mice (Figs. 4 G and S2 C). These data demonstrate that HFHS increases not only Mmp12 expression but also IR-ATMs in WAT in a microbiota-dependent manner (Figs. 4 G and S2 C). Altogether, these results suggest that the Mmp12 meta-signature is characteristic of WAT-infiltrating, metabolically active macrophages with a negative impact on glucose metabolism.
Figure S2.
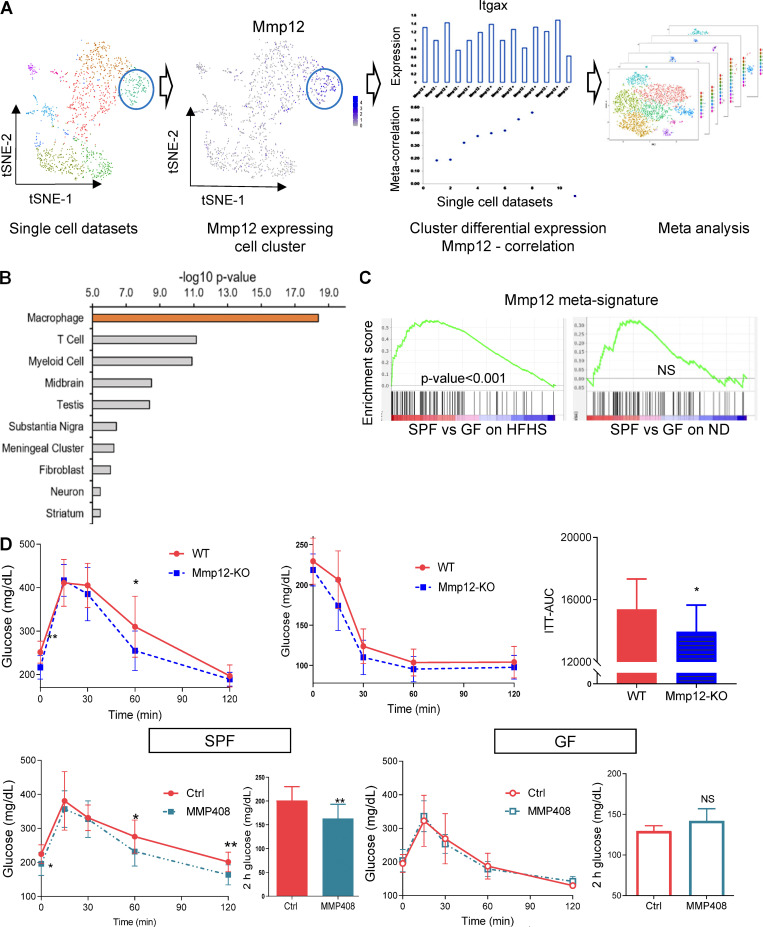
Mmp12/IR-ATM signature and Mmp12 blockade in mice. (A) Overview of the meta-analysis of single cell transcriptome analysis to derive Mmp12 meta-signature. The eight chosen datasets with CD45+ cells expressed Mmp12 in a subset of the cells. The meta-correlation analysis and meta-differential expression analysis from each dataset (detailed in Materials and methods) led to identification of a gene signature specific to these subsets of Mmp12+ cells. (B) The cell type identification using the Mmp12 meta-signature genes with scQuery showing highly significant macrophages as the primary cell type with this signature. (C) Mmp12 meta-signature enrichment in (left) WAT of SPF mice compared to GF mice under HFHS condition (P value <0.001) and (right) WAT of SPF mice compared to GF mice under ND condition (not significant). (D) Top: GTT (left) and insulin tolerance test (right) in HFHS-fed wild type and Mmp12 knockout SPF mice (mean ± SEM; n = 10; one-tailed parametric t test, *P < 0.05, **P < 0.01). Bottom: GTT and 2 h (120 min) blood glucose (measured after 2 h of intraperitoneal glucose injection) in HFHS-fed SPF or GF mice injected with Mmp12 inhibitor, MMP408 (mean ± SEM, one-tailed parametric t test, n = 12 for SPF mice, n = 5 for GF mice (two-tailed), *P < 0.05, **P < 0.01).
MMP12 inhibits the OXPHOS/mitochondria pathway in WAT, worsening systemic glucose metabolism
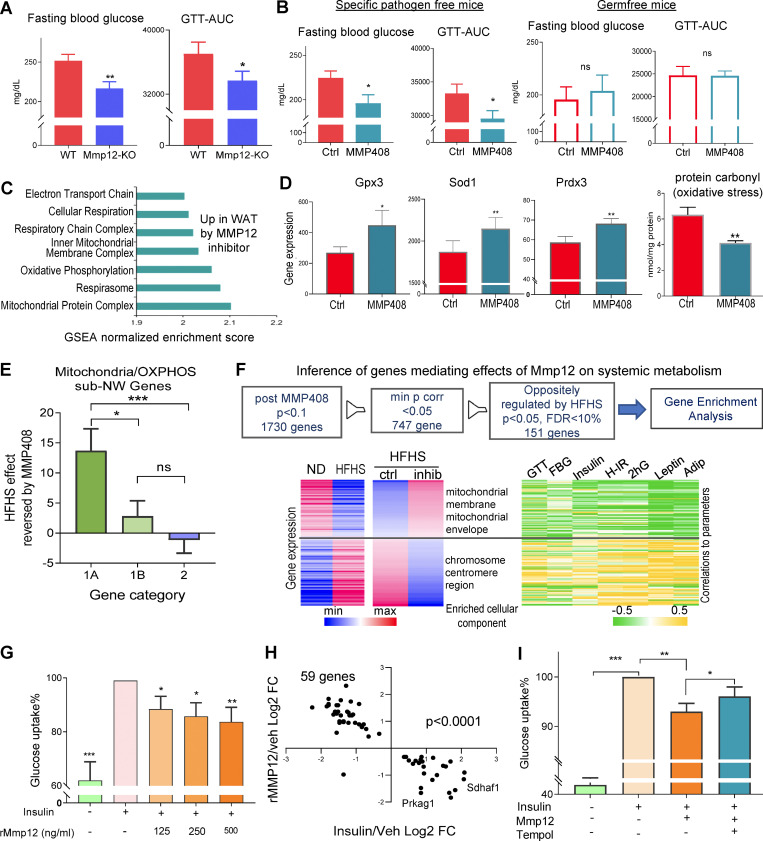
While several genes in the IR-ATM signature were previously associated with ATM metabolic potential, our analysis predicted that MMP12 is an effector molecule contributing to impairment of mitochondrial OXPHOS in adipocytes and systemic glucose metabolism. Accordingly, we observed that genetic deletion of Mmp12 or inhibition of MMP12 protein activity by the specific inhibitor MMP408 (Li et al., 2012; Li et al., 2009; Long et al., 2015) in HFHS-fed SPF mice improved fasting glucose levels, glucose tolerance, and IR (Fig. 5, A and B, left panel; and Fig. S2 D). The most pronounced transcriptional effect of MMP408 was the restoration of mitochondria-related pathways (Fig. 5 C), including increased expression of genes responsible for various mitochondrial functions (Ndufaf5, Acacb, Maob, Lpin1, Nat8l, and Mrpl38; Table S9), and of antioxidative genes (Fig. 5 D). This was accompanied by a decrease in carbonyl protein, a marker of oxidative stress (Fig. 5 D, right panel). MMP12 inhibition had a significantly higher impact on microbiota-dependent than on microbiota-independent genes in the OXPHOS/mitochondria subnetwork (Fig. 5 E), and improved glucose metabolism in HFHS-fed SPF mice but not in GF mice (Figs. 5 B and S2 D). Finally, mitochondrial/OXPHOS-related gene expression that was decreased by HFHS and increased by MMP408 negatively correlated with markers of metabolic disease (Fig. 5 F and Table S10). These results confirm the negative impact of MMP12 on glucose metabolism and OXPHOS/mitochondria as well as its microbiota dependence.
Figure 5.
MMP12 inhibits the OXPHOS/mitochondrial pathways in WAT and worsens systemic glucose metabolism in mice. (A) Fasting blood glucose and area under the curve (AUC) from GTT in Mmp12 knockout or littermate wild-type SPF mice fed with HFHS (mean ± SEM, n = 10, per group, paired one-tailed t test, *P < 0.05, **P < 0.01). (B) Fasting blood glucose and GTT-AUC in HFHS-fed wild-type SPF and GF mice injected with MMP12 inhibitor, MMP408 (mean ± SEM, n = 12 per group, one-tailed parametric t test, *P < 0.05). (C) Over-represented gene ontology terms in WAT of SPF mice treated with MMP12 inhibitor (MMP408) vs. vehicle (FDR < 0.05). (D) Expression of antioxidant genes Gpx3, Sod1, and Prdx3 in WAT in mice treated with MMP12 inhibitor (MMP408) vs. vehicle (mean ± SEM, one-tailed parametric t test; n = 12; *P < 0.05, **P < 0.01). Right: Oxidative stress (protein carbonyl) in WAT in mice treated with MMP12 inhibitor (MMP408) vs. vehicle (mean ± SEM, one-tailed parametric t test; n = 8; *P < 0.05, **P < 0.01). (E) Average effects of MMP408 reversing impact of HFHS on genes in the mitochondrial/OXPHOS subnetwork (chi-square test, Yates, P = 0.012). Categories: 1A, microbiota dependent, interaction with HFHS; 1B, microbiota dependent, no interaction with HFHS; 2, microbiota independent. (F) Top: Inference of genes mediating effects of Mmp12 on systemic metabolism: after MMP408 treatment, expression of 1,730 genes was changed (P < 10%). After applying the filter of minimal P values for correlation with various metabolic parameters, there are 747 genes left (P < 5%). Finally, among this, 151 genes were oppositely regulated by HFHS (FDR < 10% P < 5%). Bottom: Heatmaps representing 151 adipose gene expression changes induced by HFHS and reversed by MMP12 blockade and associated with systemic metabolic parameters (n = 12 per group). (G) Insulin-stimulated glucose uptake is impaired in recombinant MMP12 treated 3T3-L1 cells (mean ± SEM, n = 5, the control group was set at 100%, one-sample parametric t test, hypothetical value 100, one-tailed, *P < 0.05, **P < 0.01). (H) Regulation of genes by recombinant MMP12 vs. regulation by insulin in differentiated 3T3-L1 cells (DEGs, two-tailed t test P value <0.05; chi-square test, Yates P < 0.0001). x axis: fold change insulin/vehicle (n = 3); y axis: rMMP12/vehicle (n = 5). (I) Glucose uptake inhibited by recombinant MMP12 is restored by Tempol in differentiated 3T3-L1 cells (one-tailed t test P value < 0.023; n = 9 independent experiments; mean ± SD).
Mitochondrial OXPHOS damage in adipocytes leads to an increase in the reactive oxygen species and is linked to systemic IR (Kusminski et al., 2012). Thus, we sought to identify whether adipocytes represent the target cells of MMP12. Treatment of adipocytes in vitro with activated recombinant MMP12 reduced insulin-stimulated glucose uptake by about 50% (Fig. 5 G) and reversed the insulin-induced transcriptional profile in adipocytes (Fig. 5 H and Table S11). On one hand, our in vivo data showed that MMP12 blockade leads to a reduction of oxidative markers (Fig. 5 D). On the other hand, mitochondrial OXPHOS dysregulation increases oxidative stress (Kusminski et al., 2012). Thus, we used an antioxidant (Tempol; Wilcox, 2010) to reduce oxidative stress in adipocytes treated with recombinant MMP12 (rMMP12) and observed partial restoration of insulin-stimulated glucose uptake in the presence of the antioxidant (Fig. 5 I).
Previous attempts to investigate the role of MMP12 in glucose metabolism using Mmp12 KO mice showed contradictory results, with some studies reporting phenotypes consistent with our results (Amor et al., 2016; Bauters et al., 2013; Lee et al., 2014; Martinez-Santibanez et al., 2015). In contrast to earlier research that used only KO mice, our study employed two “loss of function” (Mmp12 KO and chemical inhibitor) perturbations and one “gain of function” (rMMP12) perturbation, and showed full consistency across all three types of perturbations. The discrepancies between previous studies are likely to be explained by microbiota-dependent Mmp12 expression in WAT (Fig. 4, A and B) and the confounding effect of large differences between microbiota composition in mice of the same genetic background housed in different animal facilities (Rausch et al., 2016).
MMP12-dependent molecular signature in WAT is associated with IR in humans
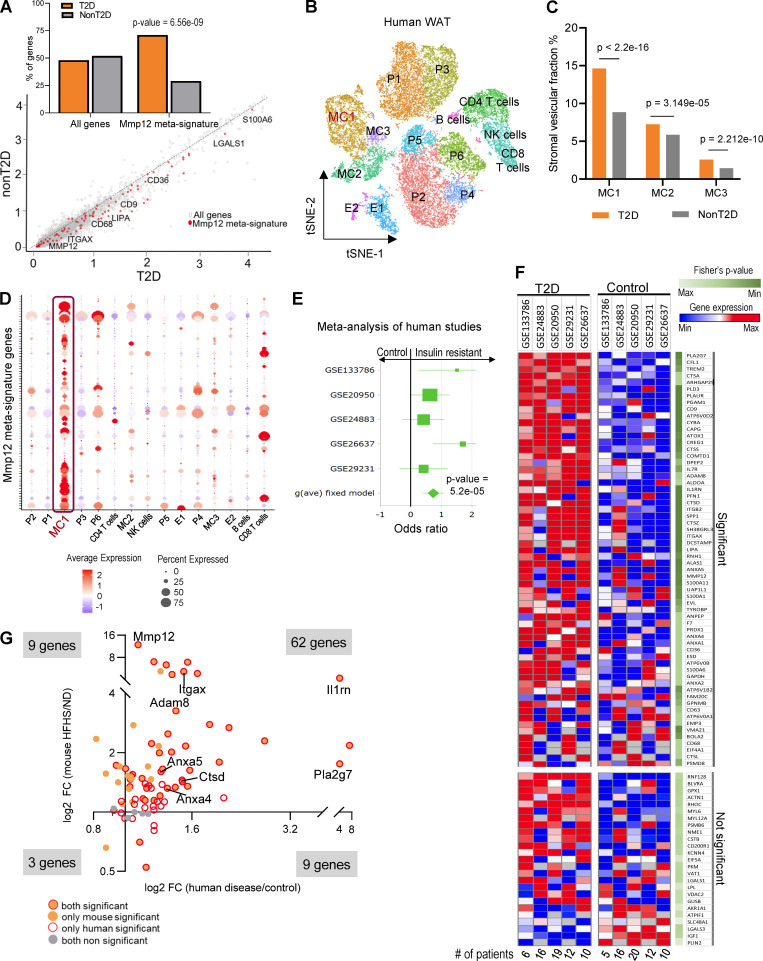
To verify whether these findings in experimental animals translate into human pathology, we tested the Mmp12/IR-ATM meta-signature in patients with T2D. We first performed scRNA-seq analysis of the stromal vascular fraction (SVF) of WAT from patients with obesity and with or without T2D (Vijay et al., 2020). The IR-ATM meta-signature was overexpressed in WAT from obese individuals with T2D compared with those without T2D (Fig. 6 A), and predominantly in the MC1, a macrophage subtype with the highest increase in patients with T2D (Fig. 6, B–D; and Fig. S3 A). Meta-analysis of total WAT transcriptomes from five available patient cohorts (Hardy et al., 2011; Jain et al., 2013; Klimcakova et al., 2011; Soronen et al., 2012; Yang et al., 2019; three with similar body mass index in patients and controls) showed a consistent increase of Mmp12/IR-ATM signature (65/83 genes) in individuals with IR (Fig. 6, E and F; Fig. S3 B; and Table S12) and also highly overlapping with the results in mice (Fig. 6 G).
Figure 6.
MMP12-IR-ATM molecular signature in WAT is associated with IR in humans. (A) Scatter plot of log-transformed average gene expression in human visceral adipose tissue SVF (GSE129363), with a fit line and Mmp12 signature genes marked in red. Inset graph shows the percentage of genes in each group with Mmp12 meta-signature significantly higher in T2D (n = 5) vs. non-T2D (n = 9) obese patients (two-sided, Wilcoxon rank sum test with continuity correction, P < 0.0001). (B) t-SNE plot of the human scRNA-seq from adipose tissue SVF (GSE129363). Cell type names are annotated in the plots based on identified cell type specific marker genes. MC, myeloid cells; P, progenitor cells; E, endothelial cells. (C) Percentage distribution of human SVF of myeloid cell (MC) clusters in the scRNA-seq samples. Significant enrichment of MC1 cells are seen in T2D patient samples as compared to other clusters (Pearson’s chi-squared test, ****P < 0.0001). (D) Dot plot of average expression and percentage of cells with Mmp12 meta-signature enrichment in MC1 cluster cells among all clusters of the adipose tissue SVF in humans. (E) Forest plot of Hedge’s g effect size in meta-analysis of five human studies of whole tissue adipose transcriptome for the Mmp12 meta-signature genes. Studies were obtained from GEO and contained gene expression data of individuals with/without T2D. The P value shown is from the fixed effect model. See materials and methods for exclusion criteria. (F) Heatmap showing the expression of Mmp12 IR-ATM genes from meta-analysis of the human adipose tissues of insulin resistant (IR) and non-IR patients (Fisher’s P values <0.05 considered significant), each column represents mean gene expression in corresponding group of patients. (G) Fold change (FC) of Mmp12 meta-signature in human adipose tissue by diseases (T2D/control) and by diet in the WAT of mouse (HFHS/ND, n = 10 per group) (Fisher’s P value < 0.05 for human and parametric, one-tailed P value <0.05 for mouse, binomial test P = 2.2E−16).
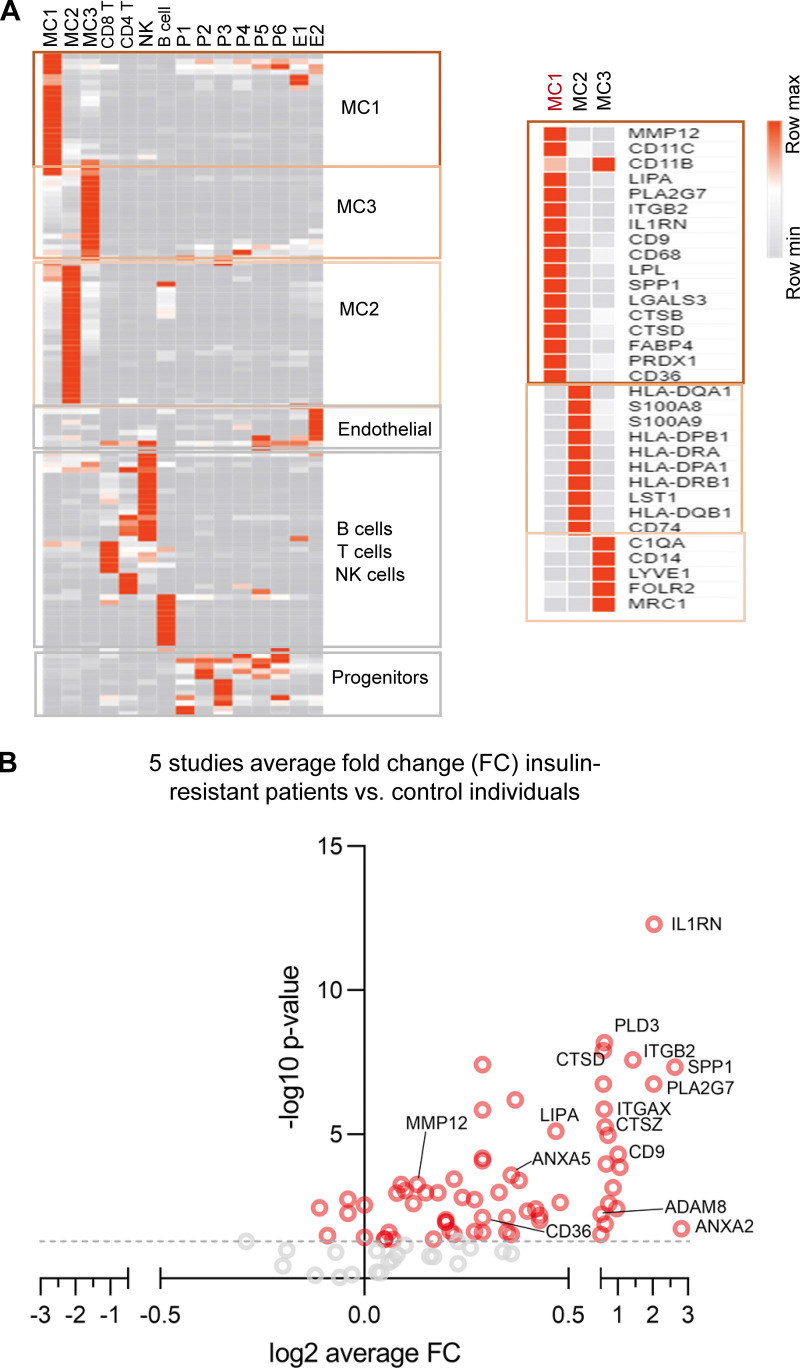
Figure S3.
MMP12/IR-ATM signature in patients with T2D. (A) Left heatmap shows the cluster markers in the human adipose tissue SVF. The color range is depicted as row max in orange to least in gray. The right heatmap shows the single cell gene expression of cluster specific markers MC1, MC2, and MC3 respectively. (B) Volcano plot of the Mmp12 meta-signature genes fold change (FC, insulin resistant patients/control) from meta-analysis of the human studies of whole tissue transcritomes.
OV induces Mmp12/IR-ATM meta-signature in macrophages
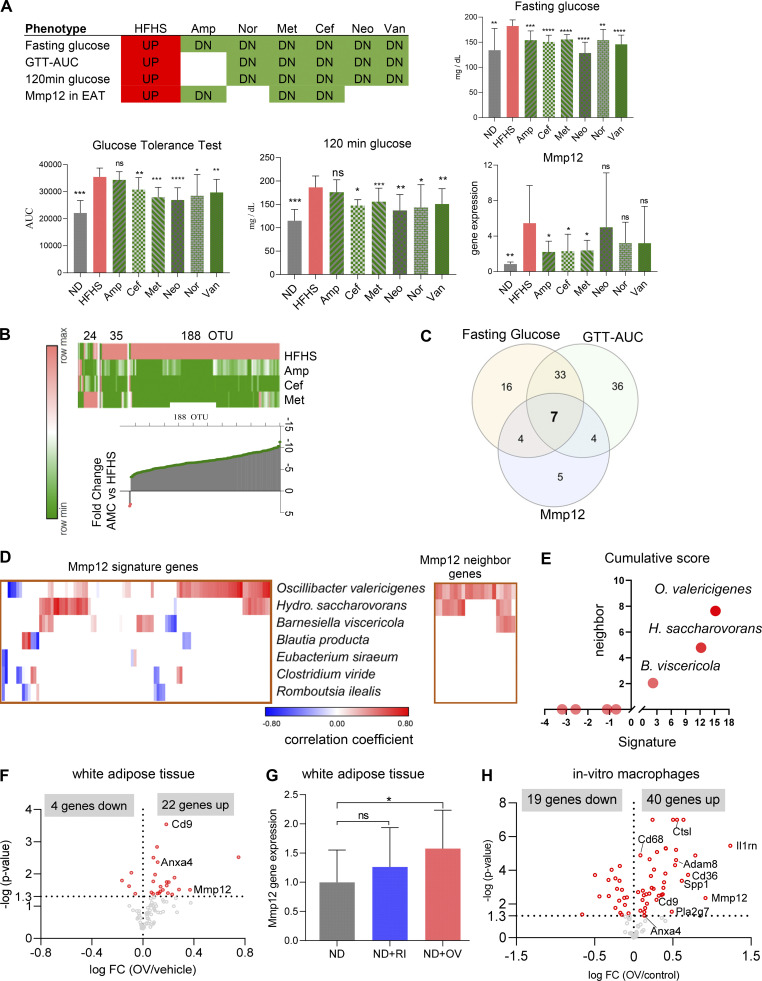
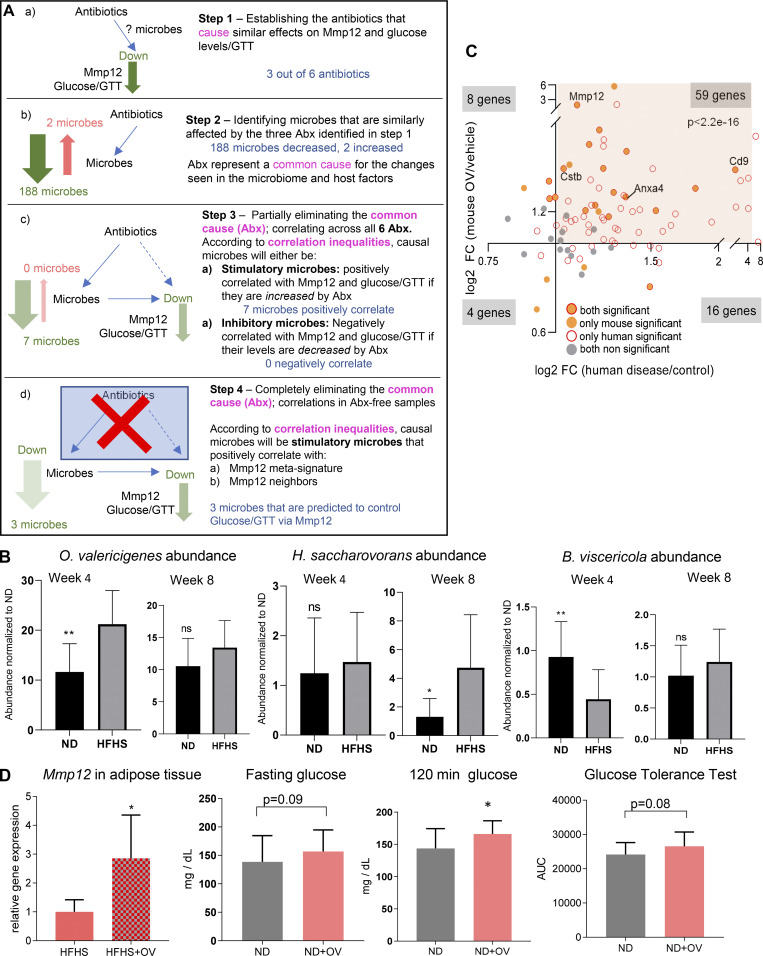
To identify which bacterial taxa are responsible for the induction of Mmp12 in WAT, we performed transkingdom causal inference including a combination of standard techniques (such as instrumental variables and mediation analysis) incorporating additional aspects of principles of causality such as common cause (Pearl et al., 2009; Reichenbach et al., 1991), Simpson Paradox (Blyth, 1972), and correlation inequalities (Yambartsev et al., 2016). Specifically, we first perturbed microbiota of HFHS-fed SPF mice with six antibiotics for 4 wk. We next assessed the effects on the microbiome, Mmp12 expression in WAT and glucose metabolism markers (Fig. 7 A). Using these data, we inferred (see details of inference in Figs. S4 A and 7, B–D) that among 263 different taxa, three bacteria (Figs. 7 E and S4 B) were potential inducers of Mmp12 in HFHS-fed mice, with OV being the candidate with the largest impact (Fig. 7 E). To test this prediction, we first supplemented ND- and HFHS-fed mice for 4 wk with OV. We found that OV supplementation to mice increased the expression of several genes of the IR-ATM signature (Fig. 7 F), including Mmp12 in WAT (Fig. 7 G), with ∼70% similarity to human T2D (Fig. S4 C). OV supplementation also showed a trend of worsened glucose tolerance in mice with 2-h serum glucose levels being significantly increased (Fig. S4 D). Interestingly, another HFHS-associated bacterium (Romboutsia ilealis), reportedly a negative regulator of glucose metabolism (Rodrigues et al., 2021), was eliminated at the last step of the analysis (Fig. 7, D and E). Accordingly, supplementation of R. ilealis did not induce Mmp12 expression in mouse WAT (Fig. 7 G), indicating some level of specificity for OV.
Figure 7.
OV is a primary inducer of Mmp12 IR-ATM signature in mouse adipose and in macrophages in vitro. (A) Fasting glucose, AUC-GTT, 120 min blood glucose and Mmp12 expression in antibiotic-treated mice on HFHS (n = 10, two independent experiments with n = 5 each; two-tailed, parametric, unpaired t test, one-tailed, unpaired t test for Mmp12 gene expression, mean ± SD). Neo, neomycin; Van, vancomycin; Nor, norfloxacin. (B) Top: Heatmap showing microbiota perturbation following the three antibiotics treatment. Bottom: Median fold change of the abundance of 188 OTUs between mice treated with antibiotics (Amp, Cef, Met) and control mice, all fed HFHS. (n = 10, two independent experiments with n = 5 each; two-tailed, parametric, unpaired t test.) (C) Number of OTUs correlated positively with fasting glucose, GTT-AUC, and/or WAT Mmp12 gene expression measured by qPCR (mean ± SD, n = 10 mice per group). (D) Correlation of the seven common microbes with the expression of Mmp12 signature genes and Mmp12 neighbor genes in the gene regulation network in WAT. (E) Cumulative sum of correlation coefficients of seven OTUs with Mmp12/IR-ATM signature (x axis) and Mmp12 gene neighbors (y axis). (F) Volcano plot showing fold change (FC, log2 transformed) and P values of Mmp12 signature genes in OV-supplemented mouse WAT (n = 10 per group, one-tailed, parametric, P < 0.05). (G) Mmp12 gene expression in WAT measured by qPCR (mean ± SD, n = 10 for OV, n = 5 for R ilealis (RI), one-tailed, unpaired, parametric t test, normalized to control diet, *P < 0.05). (H) Volcano plot showing fold change (FC, log2 transformed) and P values of Mmp12 signature genes in OV CFS–treated macrophages (n = 4 independent experiments, parametric t test, two-tailed P value <0.05).
Figure S4.
Inference and validation of microbes inducing Mmp12/IR-ATMs. (A) Transkingdom causal inference analysis workflow for identifying candidate microbes that induce Mmp12 expression/IR-ATM signature and worsen glucose metabolism. Abx, antibiotics. (B) Abundance of OV, H. saccharovorans, and B. viscericola in ND- or HFHS-fed mice at week 4 and week 8 (mean ± SD, parametric, unpaired, one-sided t test, *P < 0.05, **P < 0.01). (C) Fold change (FC) of Mmp12 signature genes in diabetic human subjects and in OV supplemented mouse WAT (Fisher’s P value < 0.05 for human data, and one-tailed, parametric P value <0.05 for mouse data). (D) WT SPF mice were treated with OV or PBS. The figures show WAT Mmp12 expression (HFHS condition, n = 5), fasting glucose level, 120 min glucose, and glucose tolerance-AUC (total n = 10, two independent experiments with n = 5 each), respectively (mean ± SD, one-tailed t test, *P < 0.05).
To test whether OV can stimulate Mmp12 in macrophages directly or whether it requires cell–cell contact or circulating factors, we treated a macrophage cell line (RAW 264.7 cells) with heat-killed (HK) OV or cell-free supernatant (CFS). We found that the CFS from OV induced higher expression of Mmp12 (Fig. S5 A) and a large part of the IR-ATM meta-signature (Fig. 7 H) with significant overlap to observations in humans with IR (Fig. S5 B). These results offer another indication that macrophage-related inflammation in WAT of insulin-resistant patients is driven by microbiota.
Figure S5.
Testing innate adaptor molecules used by OV to stimulate Mmp12 expression and IR-ATM signature. (A) Mmp12 gene expression in RAW cells treated with HK OV (mean ± SD, n = 3, two-tailed, unpaired, parametric t test, ***P < 0.001). (B) Mmp12 meta-signature gene expression change in human diabetic subjects compared to healthy controls and in macrophage cell line treated with OV compared to vehicle (Fisher’s P value < 0.05 for human data and two-tailed parametric P < 0.05 for macrophage cells). (C) Mmp12 gene expression in RAW cells treated with OV and (top right) siRNAs Myd88 (n = 10). Top right: siRNAs Mavs, Ripk2 (mean ± SD, n = 3, two-tailed, unpaired, parametric t test, ***P < 0.001). Top left: Mmp12 gene expression in RAW cells treated with siRNAs Ticam-1. Bottom right: Ticam-1−/− IMM cells (right; mean ± SD, n = 5 independent experiments, one-tailed, unpaired, parametric t test, ****P < 0.0001).
TLR2-induced Mmp12 expression in macrophages is Atf3 dependent
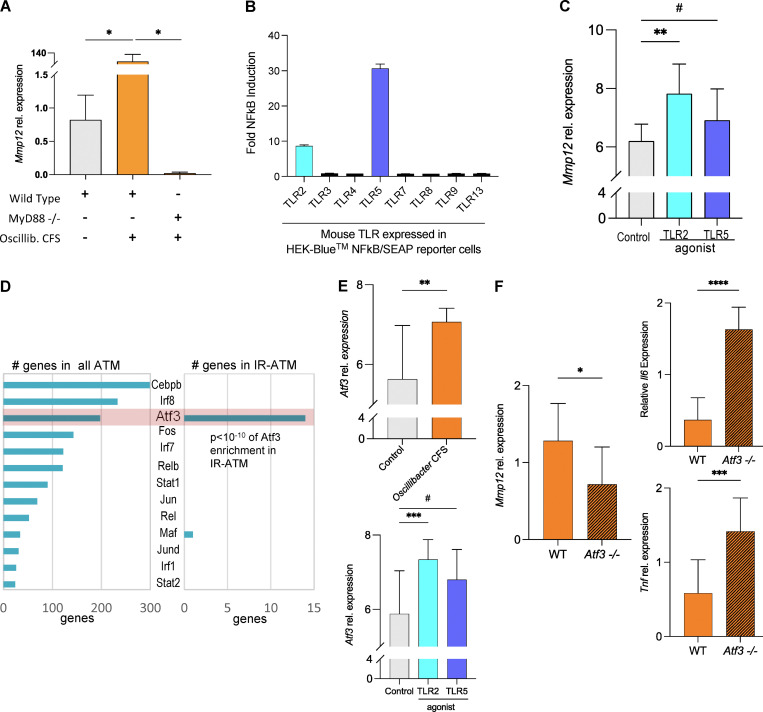
To identify the mechanisms of Mmp12 induction by OV, we first tested (via knockdowns and/or knockouts) four major adaptor molecules (Myd88, Ticam-1, Mavs, Ripk2) that mediate signaling downstream of receptors of innate immunity and inflammasome activation. These experiments revealed that Mmp12 regulation by OV is Myd88 dependent (Figs. 8 A and S5 C). Next, we screened OV supernatant for TLR-ligands and found that this bacterium produces Tlr2 and Tlr5 agonists (Fig. 8 B). The follow-up studies demonstrated that Tlr2 agonist strongly induced Mmp12 in the macrophage cell line, while Tlr5 ligand shows only a trend in the same direction (Fig. 8 C).
Figure 8.
OV-derived Tlr2 ligands induce Mmp12 expression in macrophages in Myd88-and Atf3-dependent manner. (A) Relative Mmp12 gene expression measured by qPCR in WT IMM or Myd88−/− IMM treated with OV CFS (median normalized data shown as mean ± SD, n = 4 independent experiments, one-tailed Mann-Whitney test). (B) Mouse TLR agonist screen conducted by InvivoGen using single mouse TLR expressing HEK-Blue NFkB/SEAP reporter cells treated with OV CFS. Data shown as fold induction (CFS treated/control), mean ± SD of n = 3. (C) Mmp12 expression measured by qPCR in WT IMM cells treated with a TLR2 agonist, TLR5 agonist, or control. Data are median normalized and log2-transformed, shown as mean ± SD, n = 8 independent experiments, one-tailed Mann–Whitney tests were used, #P = 0.1284. (D) Atf3 as one of the top TFs regulating IR-ATMs signature. Left: The ATMs scRNA-seq GRN analysis using SCENIC predicted 38 TFs for the adipose macrophages. Right: Among the predicted TFs in IR-ATMS, Atf3 is shown to regulate 14 genes from the signature including Mmp12. (E) Atf3 expression in OV CFS (top)– or TLR2 or TLR5 agonist (bottom)–treated IMM cells. Data are median normalized and log2 transformed, shown as mean ± SD, n = 8 independent experiments, two-tailed Mann–Whitney test used, #P = 0.2113. (F) Mmp12, Il-6, and Tnf gene expression in WT (orange) and Atf3−/− (orange with lines) IMM cells treated with OV CFS. Data are median normalized relative expression levels in OV CFS–treated cells divided by PYG media (control)–treated cells, mean ± SD, n = 9 independent experiments, two-tailed (Mmp12) or one-tailed (Il6, Tnf) unpaired parametric t test used. For all experiments above: #P < 0.25, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
While the classical view of Myd88-dependent signaling is focused on NF-κB activation, we investigated if any other factor might be involved in Mmp12 transcription in IR-ATMs. Analyzing scRNA-seq data (Aibar et al., 2017), we inferred ATF3 to be one of the top TFs regulating the IR-ATM signature (Fig. 8 D), including Mmp12 (Table S13). This computational inference was confirmed by ATF3 chromatin immunoprecipitation sequencing (ChIP-seq) results (Krebs et al., 2014), with a 3.4-fold difference (>2 is significant) between WT and Atf3 KO macrophages for the Mmp12 gene region in addition to 29 genes (out of 95) from the IR-ATM signature also targeted by ATF3 (Table S13). Both OV CFS and TLR2 ligands induced the expression of Atf3 in macrophages (Fig. 8 E). Although ATF3 dampens the classic inflammatory response (M1) in macrophages (De Nardo et al., 2014; Gilchrist et al., 2006), by treating Atf3 KO and control cell lines with OV CFS, we demonstrated that ATF3 is required for the complete induction of Mmp12 expression by macrophages (Fig. 8 F, left panel), despite its inhibitory effect on Tnf and Il6 expression (Fig. 8 F, right panel).
Discussion
Using a robust multiorgan network and transkingdom causal inference analysis, we were able to infer, in mice and humans, and experimentally confirm novel pathways linking the deleterious effect of HFHS on glucose metabolism and diabetes via gut microbiome alterations. HFHS expands the gut commensal O. valericigenes and a few other bacterial species that, via the release of TLR2 ligands, may induce Mmp12 expression in adipose tissue macrophages in an ATF3-dependent manner. Mmp12 is part of a meta-signature expressed in a subset of WAT macrophages (IR-ATM) associated with T2D in mice and humans. MMP12 is a negative regulator of glucose metabolism that, through the damage of antioxidative and mitochondrial gene expression, causes OXPHOS dysfunction and IR in adipocytes, leading to systemic dysfunction of glucose metabolism in diabetes.
Dysregulation in the OXPHOS pathway in different organs, including WAT, is an important factor contributing to the pathogenesis of T2D (Lee et al., 2018; Rocha et al., 2020). Our study not only pointed to a central role of WAT mitochondrial OXPHOS damage in T2D, but more importantly showed for the first time the importance of gut microbiota in this process. We characterized a gene expression signature of microbiota-dependent MMP12-expressing macrophages (IR-ATM) that is associated with IR in humans. Notably, among six different clinical cohorts (one single cell and five whole tissue transcriptomes), where IR-ATM enrichment was observed, four cohorts represented patients with a similar level of obesity, differing only in IR. Our results agree with the association studies reporting that expression of MMP12 in WAT positively correlates with IR in humans (Lee et al., 2018) and another study that showed MMP12 plasma levels were elevated in patients with T2D and were associated with atherosclerosis (Goncalves et al., 2015).
Previous studies showed the critical role of diverse adipose macrophages, classified into several subtypes beyond classical M1/M2, in animal models of T2D (Russo and Lumeng, 2018). These macrophages can be identified by individual markers (e.g., LGALS3 [Li et al., 2016; Pejnovic et al., 2013], CD36 [Silva et al., 2019], and CD9 [Hill et al., 2018]) and have a negative impact on metabolism. Our analysis of multiple scRNA-seq datasets established that the previously identified subtypes converge into a single subset of metabolically harmful adipose macrophages (IR-ATM) characterized by a gene signature that includes both previously identified and novel marker genes. Moreover, we found that ATF3 (an inhibitor of proinflammatory cytokines such as TNF and IL-6) is a key TF for IR-ATMs and a positive regulator of Mmp12 expression. This is an additional strong indicator that a special type of macrophage (IR-ATMs), rather than classical inflammatory M1 (Chylikova et al., 2018; Fujisaka, 2021), promotes IR in WAT.
The negative effect of MMP12 on OXPHOS and consequently glucose uptake by adipocytes is a novel mechanism of IR, which could not be predicted based on the existing knowledge about MMP12. Thus, our study adds a new chapter to the variety of roles played by this protein ranging from a pathological factor in COPD (Hunninghake et al., 2009) to an antiviral response molecule (Marchant et al., 2014).
Our observation that TLR2 and MYD88 are involved in the control of Mmp12 is in line with reports demonstrating a role of TLR2 in T2D (Ehses et al., 2010) and of MYD88 in HFHS-induced inflammation in WAT (Tran et al., 2020). Accordingly, our findings suggest that microbial regulators of Mmp12 expression are not limited to one pathobiont (i.e., OV). Importantly, OV’s effect on the IR-ATM signature in adipose tissue in mice shows high similarity with gene expression alterations in humans, suggesting that microbiota-modifying treatments targeting IR-ATMs may offer a therapeutic strategy for T2D. Furthermore, our analysis predicted that at least two other bacteria (Barnesiella viscericola and Hydrogenoanaerobacterium saccharovorans) may impair systemic metabolism by inducing Mmp12 in WAT. Notably, these three bacteria (OV, B. viscericola, and H. saccharovorans) and/or their parent genera were negatively associated with metabolic health in some studies (Anhe et al., 2017; Jung et al., 2016; Wang et al., 2015). Our study, however, is the first to pinpoint a mechanistic role for these bacteria in T2D as well as to propose a molecular mechanism of their potential action. Although our study identified the Mmp12 pathway to be instrumental in mediating effects of HFHS on IR and T2D, alternative pathways are expected to exist, and possibly, to become prevalent in the presence of a different gut microbiota (Leshem et al., 2020). For example, among bacteria associated with a HFHS in the current data, OV acts through the induction of Mmp12 in macrophages, while we previously described that R. ilealis worsens glucose tolerance by inhibiting insulin levels, which may be relevant to more advanced stages of disease (Thévenod, 2008; Rodrigues et al., 2021).
In conclusion, our study developed a multiorgan model corresponding to initial stages of T2D and revealed a new mechanism of interaction between HFHS and microbiota in the pathogenesis of this illness. Specifically, we demonstrated that certain members of the gut microbial community (e.g., pathobionts such as O. valericigenes) exacerbate the detrimental effects of a HFHS on mitochondrial dysregulation in WAT by promoting IR-ATMs and their effector molecule MMP12. Our work sheds light on the one of most enigmatic phenomena in the field of T2D, potentially explaining why some patients with obesity develop diabetes while others do not.
Materials and methods
Animals and diets
7-wk-old SPF C57BL/6 male mice were purchased from The Jackson Laboratory and housed in a controlled environment (12-h daylight cycle) in groups of four to five per cage at the Laboratory Animal Resources Center at Oregon State University, with ad libitum access to food and water. After 1 wk of acclimatization, mice were either switched to a WD designated in this study as HFHS diet D12451 (40 kcal % lard and 17 kcal % sucrose) or to a matched ND D12450K (ND; 4 kcal % lard and 0% sucrose) produced by Research Diets (Table S14). For initial experiments (shown in Fig. 1), diets were γ-irradiated at 50 kGy at the Oregon State University Radiation Center. In follow-up experiments (shown in Figs. 2 and 4), diets irradiated by the manufacturer were used.
Mice were on these diets for 8 wk. C57BL/6 GF were bred at the Oregon State University Germ-Free Mouse Core at the Laboratory Animal Resources Center. At 8 wk of age, male mice were transferred to ventilated racks (Innovive) to fully sealed disposable cages with HEPA-filtered air supply and fed either irradiated ND or HFHS for 8 wk. Each group had five to six mice per experiment and repeated twice.
Mmp12 knockout (B6.129X-Mmp12tm1Sds/J) and C57BL/6J controls initially purchased from The Jackson Laboratory were bred to generate heterozygous Mmp12 KO mice. The heterozygous mice were then interbred to generate littermate KO Mmp12−/− and WT Mmp12+/+ mice. Genotyping was proceeded according to the protocol provided by The Jackson Laboratory with modifications. The following primers were used: mutant forward, 5′-GCTACTTCCATTTGTCACGTCC-3′; WT forward, 5′-ACTGGGCAACTGGACAACTC-3′; common, 5′-CTCCCATGTGCTGGGATTAC-3′. Briefly, a small piece of ear tissue was suspended in 50 μl lysis buffer containing 0.2% SDS, 100 mM Tris-HCl, 100 mM NaCl, 10 mM EDTA (pH = 8), and 1 μl proteinase K (Ambion, 20 mg/ml), and then digested at 55°C for 2 h. After being diluted with 450 μl distilled water, vortexed, and spun down, the supernatant was used for PCR (GoTaq Master Mixes) with the following program: initial denaturation at 94°C for 3 min, 10 cycles of 94°C for 30 s, 65°C (touch-down, 0.5°C/cycle) for 30 s, and 68°C for 30 s, 28 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 2 min, hold at 4°C.
Adult male littermates (Mmp12 KO and littermate WT) were fed a HFHS for 5–6 wk before conducting the glucose tolerance test (GTT) and insulin tolerance tests. All mice were housed individually. The experiment was performed twice.
Work on mice was carried out in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the U.S. National Institutes of Health and was approved by Oregon State University’s Animal Care and Use Committee (protocol IACUC-2021-0202).
Mmp12 inhibitor experiments
Adult male C57BL/6J SPF and GF mice were used. Mice were first fed a HFHS for 4 wk to induce glucose intolerance then divided into two groups. Mice were injected intraperitoneally with an MMP12 selective inhibitor MMP408 (3.5 mg/kg, dissolved in PBS containing 10% DMSO) or vehicle every day for 2–3 wk while continuing HFHS feeding. All mice were housed individually. The experiment was performed twice.
Microbe supplementation experiment
8-wk-old male C57BL/6J mice were fed with an ND and gavaged with 109 CFU of O. valericigenes DSM 18026 or R. ilealis DSM 25109 in PBS for 4–5 wk every day.
GTT
Mice were fasted for 6 h during the light phase with free access to water. A concentration of 2 or 1 mg kg−1 glucose (Sigma-Aldrich) was injected intraperitoneally. Blood glucose was measured at 0 (immediately before glucose injection), 15, 30, 60, and 120 min with a Freestyle Lite glucometer (Abbot Diabetes Care).
Insulin tolerance testing
Mice were fasted for 6 h during the light phase with free access to water. Insulin (Novolin R) was injected intraperitoneally using the dose of 1 U/kg body weight. Blood glucose was measured at 0 (immediately before insulin injection), 15, 30, 60, and 120 min with a Freestyle Lite glucometer (Abbot Diabetes Care).
Fasting insulin and fasting glucose
Mice were fasted for 6 h with free access to water. Fasting blood was collected either via submandibular bleed or from the tail vein. Insulin and glucose levels in fasting plasma or serum was measured with Mouse Insulin ELISA Kit (Crystal Chem) and Glucose Colorometric Assay Kit (Cayman Chemical), respectively, according to the manufacturer’s protocol. Homeostatic model assessment for insulin resistance (HOMA-IR) was calculated according to formula: HOMA-IR = glucose (mg/dl) × insulin (μU/ml)/405.
Antibiotic administration experiments
SPF mice were fed ND or HFHS for 4 wk, followed by the administration of antibiotics in drinking water for 4 wk. The following doses of antibiotic were used: ampicillin (Amp), 1 g/liter; neomycin, 1 g/liter; metronidazole (Met), 1 g/liter; vancomycin, 0.5 g/liter; norfloxacin, 1 g/liter; and cefoperazone (Cef) 0.5 g/liter. Body weight and feed intake were measured each week. GTT, insulin, and fasting glucose was measured every 4 wk. At the end of 8 wk of the experiment (i.e., 4 wk on antibiotics), Mmp12 gene expression was measured in WAT using quantitative PCR (qPCR) and stool microbiota was sequenced using 16S rRNA sequencing. Two independent experiments were conducted with n = 5 each.
Fecal transplantation experiments
GF mice were fed either irradiated ND or HFHS for 4 wk and then gavaged with 250 μl of inoculum prepared either from fecal contents from ND- or HFHS-fed mice. Gavaging was done on day 1 and day 3. The change in body weight, feed intake, glucose tolerance, and insulin were measured on day 4, 9, 16, and 30 after first gavage.
For humanized microbiota mice experiments, 7–8 wk old, male, GF mice were gavaged with 100 μl inoculum prepared from stool of a healthy human on day 1 and day 3 and fed with either irradiated ND or HFHS for 8 wk. Systemic measurements were done on week 3 and 8.
Tissue collection
At the end of each experiment, mice were euthanized by cervical dislocation. The right lobe of the liver, terminal ileum, epididymal fat pad, subcutaneous fat, and gastrocnemius muscle were collected and snap frozen at −80°C until future use.
Protein carbonyl assay
Around 80 mg in epididymal adipose tissue was homogenated in 600 μl radioimmunoprecipitation assay buffer containing protease inhibitors using an OMNI Bead Ruptor and 2.8 mm ceramic beads (OMNI International). Protein carbonyl levels were measured with Protein Carbonyl Content Assay Kit (Sigma-Aldrich) according to the manufacturer’s protocol. The final values were normalized by total protein amount measure by the bicinchoninic acid assay.
Preparation of inoculum for fecal transfer
L-cystein at 0.05% solution was prepared in PBS in sterile condition. Stool from ND or HFHS fed mice was mixed with L-cystein solution to final concentration of 10% g/ml and vortexed vigorously for 5 min. This solution was centrifuged at 200 g for 5 min and aliquots from supernatant was prepared and stored at −80°C until gavaging. Inoculum from human sample was prepared in the same way.
Bacterial culture
O. valericigenes DSM 18026 was cultured in peptone yeast glucose (PYG) medium (AS 825; Anaerobe Systems) medium for 2–3 d at 30°C in an anaerobic GasPak jars (BD), then harvested, and stored in PBS containing 15% glycerol at −80°C. The R. ilealis DSM 25109 culture was provided by DSMZ in carboxymethyl cellulose medium after culture for 24 h at 37°C.
To prepare CFS from O. valericigenes, the bacterium was grown in PYG medium for 3 d, culture was centrifuged at 4,000 rpm for 20 min at 4°C, supernatant was filtered through 0.2 μM low protein binding filter and aliquoted in 2 ml tubes, then stored at −80°C. To prepare the HK O. valericigenes, the bacterial suspension was washed twice with broth, heated at 70°C for 10 min, aliquoted, and stored at −20°C. To confirm that there were no viable bacteria, one aliquot was thawed and plated in PYG agar plate, then incubated anaerobically in 30°C for 3 d.
Cell culture
3T3-L1 cells were grown in DMEM (high glucose) media supplemented with 10% FBS in a 5% CO2 incubator at 37°C. For differentiation, cells were seeded in a 96-well plate at a density of 5,000 cells/well. After 24 h, the medium was shifted to differentiation medium (90% DMEM, 10% FBS, 1.0 µM dexamethasone, 0.5 mM methylisobutylxanthine, 1.0 µg/ml insulin). After 48 h, the medium was removed and the cells were cultured in adipocyte maintenance medium (90% DMEM, 10% FBS, 1.0 µg/ml insulin) for another 48 h. Then the medium was changed to regular growth medium (90% DMEM, 10% FBS) and the cells were treated with activated rMmp12 or vehicle for 24 h. Afterward, the cells were incubated in glucose uptake testing medium (low glucose DMEM containing 0.2% BSA) with or without 100 nM insulin for 12 h. Then, 3 μl medium from each well was used for glucose assay using a glucose assay kit (Cayman). Levels of glucose uptake were calculated by subtracting the glucose readout from that of the fresh medium. Cells were frozen in RLT buffer and stored in −80°C for RNA extraction.
RAW cells were grown in DMEM (high glucose) media (Thermo Fisher Scientific) supplemented with 10% FBS in 5% CO2 at 37°C. Cells were than seeded in a 24-well plate with 250,000 cells per well in 500 μl complete media for 24 h. Cells were then treated with either 5% or 10% of CFS or HK for 24 h. For the control, respective concentration of PYG media was added to separate wells. Following 24 h treatment, media was removed, cells were scraped in RLT buffer (Qiagen), and stored at −80°C until RNA extraction.
For the siRNA knock down experiments, Myd88, Ticam-1, Ripk2, Mavs siRNAs, and scrambled controls were purchased from Thermo Fisher Scientific. Then 100,000 RAW cells per well in 500 μl complete medium were seeded in 24-well plate for 24 h. After 24 h, medium was replaced with fresh medium and cells were treated with siRNAs or scrambled control for 24 h. The final concentration of each siRNA was 20 pmol. After 24 h of siRNA transfection, 5% CFS or its control was added to respective wells and incubated for 24 h, followed by collection of cell lysates in RLT buffer.
WT, Ticam-1ko, and Myd88ko immortalized mouse macrophages (IMM) were originally generated in the Katherine A. Fitzgerald lab and grown in DMEM high glucose media supplemented with 10% FBS and 20 mM HEPES. Then 100,000 cells were seeded in 24-well plate in 500 μl complete medium for 24 h. All three types were seeded in the same plate. After 24 h of seeding, cells were treated with either 5% CFS or control for 24 h, then collected in RLT buffer and stored at −80°C.
TLR agonist experiments
WT IMM were originally generated in the Katherine A. Fitzgerald lab and grown in DMEM media (4.5 g/liter glucose) supplemented with 10% FBS, 1% antibiotics, and 20 mM HEPES. A total of 30,000 cells/well were seeded in triplicate in 96-well plates for 18–24 h in complete medium and incubated at 37°C with 5% CO2. Cells were then stimulated for 6 h with either 1% vol/vol O. valericigenes CFS or PYG media as control, TLR2 agonist (HKLM at 10E+8 cells/ml; InvivoGen), or TLR5 agonist (FLA-ST Ultrapure 1 µg/ml; InvivoGen). Cells were then collected in RLT lysis buffer by scraping before being stored at −80°C until RNA extraction.
Atf3 KO experiments
WT and Atf3ko IMM were originally generated by Dominic de Nardo (De Nardo et al., 2014) and grown in DMEM media (4.5 g/liter glucose) supplemented with 10% FBS, 1% antibiotics, and 20 mM HEPES. Then 30,000 cells/well were seeded in triplicate in 96-well plates for 18–24 h in complete medium and incubated at 37°C with 5% CO2. Cells were then stimulated for 6 h with 1% vol/vol O. valericigenes CFS or PYG media as control (O. valericigenes growth media), then collected in RLT lysis buffer by scraping before being stored at −80°C until RNA extraction.
RNA extraction and qPCR
RNA was extracted using an OMNI Bead Ruptor and 2.8-mm ceramic beads (OMNI International) followed by Qiashredder and an RNeasy kit (the right lobe of liver, terminal ileum, and cells), RNeasy Fibrous Tissue Mini Kit (for gastrocnemius muscle), and an RNeasy Lipid Tissue Mini Kit (for epididymal fat) using Qiacube (Qiagen) automated extraction according to the manufacturer’s specifications with DNase treatment (RNase-Free DNase Set; Qiagen). Total RNA was quantified using Quant-iT RNA Assay Kit (Thermo Fisher Scientific). Complementary DNA was prepared using qScript reverse transcription kit (QuantaBio). qPCR was performed using the Perfecta SYBR Green Fastmix mix (Quantabio) and the StepOne Plus Real Time PCR system and software (Applied Biosystems).
The primers used are listed in Table 1.
Table 1.
Primer sequences used in this study
| Gene name | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| Mmp12 | CAGGTCACACACACATAGTTACACA | AAACCAGTTGGCCTCTGA AC |
| Polr2c | CTCACCGAAGAGAACGTCAAG | TCGATGGCTATTATGGGCACC |
| Myd88 | AGGACAAACGCCGGAACTTTT | GCCGATAGTCTGTCTGTTCTAGT |
| Mavs | TGGGGTCACAGTATCAGCC | ACTGGGACCAATCTGGGAGAA |
| Ticam1 | AACCTCCACATCCCCTGTTTT | GCCCTGGCATGGATAACCA |
| Ripk2 | ATGCCACCTGAGAACTATGAGC | GCAAAGGATTGGTGACCTCTT |
| Atf3 | GAGGATTTTGCTAACCTGACACC | TTGACGGTAACTGACTCCAGC |
| Il6 | TCTATACCACTTCACAAGTCGGA | GAATTGCCATTGCACAACTCTTT |
| Tnf | CCCTCACACTCAGATCATCTTCT | GCTACGACGTGGGCTACAG |
Normalization of qPCR data and statistics
Raw cycle threshold (CT) values of genes of interest from qPCR runs were normalized to CT values of the housekeeping gene (Polr2c) via delta CT method before calculation of relative expression using the 2−∆CT method. Relative expression levels of genes were median-normalized and/or also log2 transformed before departure from normality was assessed with a Shapiro–Wilk test. If data were normally distributed, one- or two-tailed parametric t tests were used, otherwise a Mann–Whitney test was used to compare treatments. Individual plot legends note how data are visualized specifically.
Sequencing of RNA (RNA-seq)
RNA libraries were prepared with the QuantSeq 3′mRNA-Seq Library Prep Kit (Lexogen) for the Apollo 324 NGS Library Prep System and sequenced using Illumina NextSeq. Sequences were processed to remove the adapter, polyA, and low-quality bases by BBTools (https://jgi.doe.gov/data-and-tools/bbtools/) using bbduk parameters of k = 13, ktrim = r, forcetrimleft = 12, useshortkmers = t, mink = 5, qtrim = r, trimq = 15, minlength = 20. Reads were aligned to mouse genome and transcriptome (ENSEMBL NCBIM37) using Tophat (v. 2.1.1) 70 with default parameters. Number of reads per million for mouse genes were counted using HTSeq (v. 0.6.0) 71 and quantile normalized. BRB-ArrayTools was used to identify DEGs.
Western blots
Adipose tissue samples (n = 31) were blocked and randomized to mitigate any potential systematic experimental biases (Oberg and Vitek, 2009). The tissue samples were homogenized in cell lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% vol/vol Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, and 1 mM PMSF [added immediately before use]). Homogenization was achieved using a Bioruptor Plus sonicator and Bioruptor Protein Extraction Beads (Diagenode, Inc.) at 4°C. The samples were microcentrifuged (20 min; 4°C; 20,000 g) and the supernatants were retained. Bicinchoninic acid protein concentration assays were performed using a Micro BCA Protein Assay Kit (Thermo Fisher Scientific).
Multiplexed Western blots were performed using the following primary antibodies from Cell Signaling Technology: total AKT (C67E7; cat# 4691S), AKT phospho-Ser473 (D9E; cat# 4060S), total ERK1/2 (L34F12; cat# 4696S), ERK1/2 phospho-Thr202/Tyr204 (D13.14.4E; cat# 4370S), total p38 MAPK (D13E1; cat# 8690S), p38 MAPK phospho-Thr180/Tyr182 (D3F9; cat# 4511S), total PTEN (D4.3; cat# 9188S), PTEN phospho-Ser380/Thr382/Thr383 (cat# 9554S), total GSK-3β (D5C5Z; cat# 12456S), total JNK (cat# 9252S), and total cofilin (D3F9; cat# 5175S). For each tissue, a sample pool was prepared and probed using each primary antibody individually, and these results were used to ensure that the analytes would not be misidentified due to multiplexing. Between one and three primary antibodies were used per Western blot. Each Western blot was used to analyze either adipose or liver samples (not both). At least two lanes were used for a MW ladder standard and at least two lanes were used for the pooled tissue sample. Imaging was performed using a ChemiDoc MP imager, and densitometry was performed using Image Lab v. 6.0.1 (both Bio-Rad, Inc.).
DNA extraction and 16S rRNA gene libraries preparation
For microbial measurements, stool pellets were collected at T1 (4 wk) and T2 (8 wk). Ileum contents were collected at T2 (8 wk) only. To get microbial DNA, frozen fecal pellets were resuspended in 1.4 ml ASL buffer (Qiagen) and homogenized with 2.8 mm ceramic beads followed by 0.5-mm glass beads using an OMNI Bead Ruptor (OMNI International). DNA was extracted from the entire resulting suspension using a QiaAmp mini stool kit (Qiagen) according to the manufacturer’s protocol. DNA was quantified using a Qubit broad range DNA assay (Life Technologies). The V4 region of the 16S rRNA gene was amplified using universal primers (515f and 806r) as previously described. Individual samples were barcoded, pooled to construct the sequencing library, and then sequenced using an Illumina Miseq (Illumina) to generate pair-ended 250 bp reads.
16S rRNA gene sequencing data analysis
The samples were demultiplexed, and forward-end fastq files were analyzed using QIIME v. 1.9.1. The default quality filter parameters from QIIME’s split_libraries_fastq.py were applied to retain high quality reads (Phred quality score ≥20 and minimum read length = 75% of 250 nucleotides). To cluster 16S rRNA gene sequence reads into operational taxonomic units (OTUs) and assign taxonomy, a closed reference OTU picking with 97% sequence similarity was performed using UCLUST and the Greengenes reference database v. 13.8. The reference sequence of candidate OTUs from the Greengenes database was used to obtain species level taxonomic assignment using Megablast (top hit using default parameters). A threshold of 99% cumulative abundance across all samples in an experiment was used to retain abundant microbes, thus removing OTUs with approximately 1% abundance across all samples in that experiment. The read counts were normalized using cumulative sum scaling, accounted for bacterial DNA quantity based on qPCR, followed by quantile normalization. The principal component analysis for the 16S sequencing data was created using Clustvis and the GraphPad Prism software.
Isolation of SVF
Epididymal adipose tissue was minced in HBSS (Invitrogen) containing calcium, magnesium, and 0.5% BSA and 3 mg/ml collagenase type 1 (Rockland). After incubation at 37°C for 1 h, the cell suspension was filtered through 100-μm strainer and then washed with 20 ml wash buffer (DPBS containing 1 mM EDTA and 0.5% BSA). After centrifugation at 700 g for 10 min, the supernatant was removed and the cell pellet was re-suspended in 5 ml of RBC lysis buffer (Tonbo Biosciences) and incubated for 10 min at room temperature. A total of 10 ml wash buffer was added and the cell suspension was filtered through 40 μm and centrifuged again. CD45+ cells from the SVF were isolated using CD45 MicroBeads (Miltenyi Biotec) according to the manufacturer’s instruction.
The isolated cells were re-suspended in 1 ml FBS containing 10% DMSO and stored at liquid nitrogen tank until use.
Analyses
Gene categorization
The genes whose expressions are significantly changed by HFHS (false discovery rate [FDR] < 15%) in the conventional SPF mice were grouped into two main categories: category 1 (microbiota-dependent) and category 2 (microbiota-independent genes). Category 1A are genes whose expressions are significantly changed by the interaction effect of diet and microbiota (two-way ANOVA interaction effect FDR < 15%). Category 1B are genes whose expressions are not affected by the interaction effect (FDR > 15%) but are significantly regulated by microbiota under either ND or HFHS conditions (SPF vs. GF FDR < 15%). Genes whose expressions are not detectable in GF mice are grouped in Category 1 and cannot be categorized further. Category 2 are genes whose expression are independent of microbiota (interaction FDR > 15%, SPF vs. GF FDR > 15%).
Reconstructing the multiorgan network
First, Spearman rank correlations were calculated between all pairs of genes from four tissues (muscle, ileum, liver, and fat) and metabolic parameters (phenotypes) by pooling the samples per diet (HFHS, ND) from both experiments. Meta-analysis was performed to retain edges with the same sign of correlation coefficient in both the diets. Edges were further filtered by the following criteria: individual P value of correlation within each diet from pooled experiments <30%, combined Fisher’s P value over diets from pooled experiments <5%, and FDR cutoff of 5% for edges within tissues and 10% for phenotypes and between tissues, and edges needed to satisfy principles of causality (i.e., satisfied fold change relationship between the two partners in the HFHS vs. ND comparison). Next, correlations were calculated per diet per experiment. Finally, the edges obtained from pooling were retained if they had the same sign of correlation coefficient as in two groups (two experiments × two diets). False positive edges due to pooling experiments were removed in the creation of the network. The proportion of genes from each tissue type that made it to the final network (following statistical cutoffs) was determined by dividing the number of DEGs in each tissue after applying correlation cutoffs by the number of DEGs in each tissue prior to applying correlation cutoffs.
Detecting subnetworks essential for regulation of systemic parameters of metabolic disease
The networks were visualized in the Cytoscape Software 2.6.3. To identify subnetworks of correlated genes in fat and liver, we used the MCODE v. 1.2 (Molecular Complex Detection) plug-in for Cytoscape to identify clusters (subnetworks) of correlated genes. The largest two subnetworks in each tissue were selected for further analyses, as all other subnetworks were representing parts of them.
Finding the shortest paths between subnetworks
Dijkstra’s algorithm was used in the python module NetworkX v. 2.2 to calculate all pairwise shortest paths between the nodes of each subnetwork–subnetwork pair and each subnetwork–phenotype pair. The weighted average (reciprocal) of the shortest paths was calculated to determine the distance of each subnetwork from systemic parameters of metabolic disease. A chi-square test was then conducted on the shortest path distributions for each pair of subnetworks.
Identifying important nodes for the information flow between subnetworks
BiBC has previously been used to find nodes that control the information flow between two parts of a network (Morgun et al., 2015). Node BiBC was calculated between each subnetwork–phenotype pair of the multiorgan network. Per pair, we counted the number of nodes in the top 20% (ranked in decreasing order of BiBC) belonging to each of the identified subnetworks. A chi-square test was used to compare the distributions of the number of top BiBC nodes belonging to each subnetwork for each subnetwork–phenotype pair.
Establishing the information flow within network by incorporating the results of the shortest paths and BiBC analyses
To combine the results from the previous two analyses, we created a model that demonstrates how close each subnetwork is to systemic parameters of metabolic disease as well as how much one of the identified subnetworks relies on another subnetwork to influence those systemic parameters. Both the line weights and distance from systemic parameters indicate the average shortest path between the subnetwork and systemic parameters. Arrow weights indicate the number of nodes in the top 20% BiBC of the network indicated by the arrow direction (i.e., the OXPHOS/mitochondria subnetwork has more nodes in the top 20% BiBC than the inflammation subnetwork when using fatty acid metabolism and systemic parameters as the two groups of the BiBC analysis).
Analysis of association of IS proteins and gene expression
Spearman correlations between the expression levels of DEGs (ND vs. HFHS, FDR < 15%), and p-Akt/Akt protein levels in WAT were calculated using the following cutoffs: (1) correlation sign (±) is consistent between two experiments, (2) when the two experiments are pooled, the correlation P value is <0.05 (two-tailed). Spearman correlations between the expression levels of DEGs and p-p38/p38 protein levels in WAT were also calculated (two-tailed P < 0.05). To compare the relationship between the sign of the correlation (the levels of gene expression vs. the levels of IS protein) with the regulation of HFHS on this gene (up/down), a 2 × 2 contingency table was made and chi-square test was performed. Next, ToppGene was used to perform gene enrichment analysis on the two groups of genes: (1) upregulated by HFHS and negatively correlated with IS proteins; (2) downregulated by HFHS and positively correlated with IS proteins.
Analysis of IR signature in WAT
A gene expression meta-signature of IR in adipose tissue obtained from Jung et al. (2018) was used for this analysis. Dimension reduction using median summary metric was carried out to compare the presence of the signature in different groups using RNA-seq data. For this, we first normalized each gene expression using the median in all groups. For the genes downregulated in T2D, the reciprocal values were used. Then the sum of all genes in the signature was used to indicate the level of presence of the signature in each sample.
Analysis of the relationship between the effects of HFHS and insulin on gene expression
To analyze the relationship between the effects of HFHS and that of insulin, global gene expression was profiled by RNA-seq of the WAT of mice fed with ND or HFHS (n = 10) and 3T3-L1 cells treated with insulin or vehicle (n = 3), respectively. Spearman correlations were conducted between fold change of gene expression in mice (HFHS/ND, FDR < 15%) and fold change of gene expression in 3T3-L1 cells (insulin/vehicle) using GraphPad Prism7.
scRNA-seq analysis
The raw gene expression matrix (UMI counts per gene per cell) was filtered, normalized, and clustered using R (https://www.R-project.org/). Cell and gene filtering were performed as follows: cells with a very small library size (<2,500) and a very high (>0.5) mitochondrial genome transcript ratio were removed. Genes detected (UMI count > 0) in less than three cells were removed. After log normalization, clustering was performed using standard Seurat package procedures. Principal component analysis was used to reduce the number of dimensions representing each cell. The number of components used for analysis was determined based on the elbow of a scree plot. Selection of a biologically relevant number of clusters was based on differential expression between neighboring clusters. Differential expression between clusters was calculated using a likelihood-ratio test for single-cell gene expression implemented in Seurat at a family-wise error rate of 5%. Neighboring clusters in principal component space were identified as the next-nearest cluster to each cell after the cell’s assigned cluster. Clusters were visualized using t-distributed stochastic neighbor embedding of the principal components (spectral t-SNE), as implemented in Seurat. The cell-type identities for each cluster were determined manually using a compiled panel of available known immune cell marker expression.
Identifying Mmp12-positive cells
The detectable expression of Mmp12 was identified in subset of cells in each of the single cell RNA-seq data set for further analysis.
Meta-correlation
The gene expression was correlated with the expression of Mmp12 in these cells in each dataset using the Pearson correlation coefficient and a t test was performed to determine the significance. The Bonferroni-Hochberg multiple comparisons procedure was used to compute the adjusted P value. Genes with mean Spearman correlation coefficients above 0.2 and a P value of 0.001 or less was selected for meta-analysis.
Meta-differential expression
Mmp12 cluster cells expression was analyzed for differential expression across other clusters. The DEGs whose fold change is >2 in at least three or more datasets with P value <0.001 were selected. Significant genes were further shortlisted based on Bonferroni-Hochberg corrections.
Mmp12 meta-signature
The Mmp12 meta-signature list of genes was selected based on the meta-analysis of overlapped gene sets from both Mmp12-positive cell meta-correlations and Mmp12 cluster meta-differential lists of genes.
Dimension reduction of Mmp12 meta-signature using median summary metric was carried out to compare the metabolic macrophage cell types from different publicly available studies with data from the Gene Expression Omnibus (GEO; GSE126407, GSE128518 and GSE113595). For this, we normalized each gene expression and calculated the median value of all genes for each sample, obtaining a single value for each macrophage cell type in each of the study.
Human diabetic patient adipose tissue SVF scRNA-seq analysis
scRNA-seq profiling of SVF from subcutaneous and visceral adipose tissue of obese individuals with or without T2D available in GEO as Discovery cohort (GSE129363) were analyzed to check the relevance of mouse Mmp12 meta-signature and signature expressing macrophages to human diabetes. The analysis was performed using Seurat package as described previously.
The gene average expression of T2D and non-T2D patients’ samples were calculated separately. The percentage of genes in each condition for Mmp12 meta-signature was calculated and observed to be significantly higher in T2D condition. The single cell gene expression data for cluster specific markers were used to identify and classify the clusters. The cluster enriched in Mmp12 meta-signature identified using the expression of marker genes as part of myeloid cells were then labeled MC1 cluster. Percentage distribution of SVF of MC cluster cells as compared with other clusters in these single cell samples were estimated to show the enrichment of MC1 cells as compared with other clusters in T2D samples. This was also evident in the dot plot of average expression and percentage of cells with Mmp12 meta-signature enrichment in MC1 cluster cells among all clusters of the adipose tissue SVF. Additionally, for the specific cell type identification of MC1 cluster as macrophages, scQuery, a single cell database was queried with highly differential and are also part of Mmp12 meta-signature genes in MC1 cluster.
Analysis of TFs regulating IR-ATMs signature
The gene regulatory network (GRN) was inferred from 1,758 obese adipose tissue macrophages scRNA-seq data (GSE117176/GSM3272967) using standard SCENIC (Aibar et al., 2017) workflow. The normalized enrichment score for the GRN was estimated and 38 TFs were identified in this data. Atf3 was the only TF, with Mmp12 as part of predicted regulon with the normalized enrichment score. There were additionally 13 genes from the Mmp12 meta-gene signature identified to be part of Atf3 regulon. A single gene from the signature was predicted to be part of Maf regulon in this GRN. Expression matrix units: UMI counts.
Meta-analyses of Mmp12 meta-signature in human studies
GEO was searched for studies that compared the expression of one group (either obese or obese with T2D) to another group (either non-obese or obese with T2D, respectively). Studies were further filtered out if (1) either body mass index or fasting glucose were not listed, (2) the T2D group consisted of non-obese individuals, or (3) individuals in the study were not over the age of 18 (adults). After filtering out, five studies remained: GSE133786 (Yang et al., 2019), GSE24883 (Klimcakova et al., 2011), GSE20950 (Hardy et al., 2011), GSE29231 (Jain et al., 2013), and GSE26637 (Soronen et al., 2012).
Using BRB-ArrayTools (https://brb.nci.nih.gov/BRB-ArrayTools/), the gene expression data were then log2-transformed and quantile-normalized, then subset for the 87 genes in the human Mmp12 signature. Since these studies were all microarray data, duplicate arrays were managed by selecting the arrays in the disease group (the group with either obese insulin-resistant individuals or obese noninsulin-resistant individuals) that had the maximum geometric mean. Meta-Mar v2.7.0 (http://www.meta-mar.com/) was used to perform meta-analysis, while GraphPad Prism 8.2.1 was used to create the volcano plots. The online tool Morpheus (https://software.broadinstitute.org/morpheus) was used to create the heatmaps of genes.
Transkingdom causal inference analysis: Prediction of microbes that induce Mmp12 gene expression and worsen glucose metabolism (steps refer to Fig. S4 A)
Step 1. SPF mice were treated with six antibiotics, systemic metabolic parameters were measured, stool microbiota was sequenced, and Mmp12 gene expression in WAT was measured using qPCR at the end of 8 wk. Outliers were identified using Grubbs (α = 0.05). Statistical significance was calculated by comparing the ND and antibiotics groups against HFHS.
Step 2. Microbes were selected if they consistently showed the same (antibiotics vs. HFHS) fold-change direction in the three antibiotics group where Mmp12 was changed (Amp, Met, Cef) in both experiments and were significantly changed (Fisher’s P value <0.1) by at least two of the three antibiotics.
Step 3. OTUs passing the above criteria were correlated (Spearman correlation) with systemic measurements, and Mmp12 across all the samples treated with antibiotics. Meta-analysis was performed to retain correlations with same sign of coefficient in both experiments and accounting for correlation inequalities (Yambartsev et al., 2016; i.e., satisfied fold change relationship between the two partners in the antibiotics vs. HFHS comparison).
Step 4. Next, the OTUs were correlated with genes from Mmp12/IR-ATM meta-signature and direct neighbors of Mmp12 from the multi-organ network by pooling the HFHS samples from both experiments (i.e., samples from mice that did not receive antibiotics), therefore eliminating the common cause (Pearl et al., 2009; Reichenbach et al., 1991) and accepted edges that satisfied correlation inequalities (Yambartsev et al., 2016; i.e., satisfied fold change relationship between the two partners in the HFHS vs. ND comparison). Correlations were calculated in HFHS samples per experiment. Edges obtained from pooling were retained if they had the same sign of correlation coefficient as in HFHS group per experiment. False positive edges due to pooling experiments were removed. The cumulative scores obtained by adding the (OTU-gene) correlation coefficients were used to select top candidate OTUs for experimental validation.
Online supplemental material
Fig. S1 shows additional details of validation of network inferences revealing microbiota-dependent IR in WAT. Fig. S2 shows Mmp12/IR-ATM signature and Mmp12 blockade in mice. Fig. S3 shows Mmp12/IR-ATM signature in patients with T2D. Fig. S4 shows inference and validation of microbes inducing Mmp12/IR-ATMs. Fig. S5 shows testing innate adaptor molecules used by OV to stimulate Mmp12 expression and IR-ATM signature. Table S1 shows analysis of systemic metabolic parameters in SPF and GF mice fed with HFHS and control diets. Table S2 lists DEGs between HFHS and ND in conventional (SPF) mice (FDR < 15%) in four organs and their expression in GF mice. Table S3 shows the total number and percentage of DEGs and percentage of DEGs that were retained in the networks in the WAT, liver, muscle, and ileum. Table S4 shows chi-square values of each pairwise comparison between subnetworks for the shortest paths and BiBC analyses performed in Fig. S2 E. Table S5 shows the intensity values of each individual sample (WAT) from Western blot. Table S6 shows the effects of HFHS on gene expression in WAT of SPF mice and of insulin on gene expression in adipocytes. Table S7 shows Mmp12 and CD45(Ptprc) positive cell counts for eight publicly available scRNA-seq datasets. Table S8 shows Mmp12-IR-ATM meta-signature genes. Table S9 shows gene ontology enrichment analysis of genes regulated by MMP12 inhibitor (ex vivo). Table S10 shows genes significantly regulated in WAT in MMP12 inhibitor-treated mice and their correlation to metabolic parameters (ex vivo). Table S11 shows genes significantly regulated by rMMP12 and insulin in 3T3-L1 cells (in vitro). Table S12 shows Mmp12 meta-signature gene expression in human datasets and in macrophage cell lines stimulated with OV supernatant. Table S13 shows genes from IR-ATM signature targeted by Atf3 as predicted by SCENIC and identified via ChIP-seq. Table S14 shows composition of experimental (HFHS) and control (normal) diets.
Supplementary Material
lists analysis of systemic metabolic parameters in SPF and GF mice fed with HFHS and control diets.
lists DEGs between HFHS and ND in conventional (SPF) mice (FDR < 15%) in four organs and their expression in GF mice.
shows the total number and percentage of DEGs and the percentage of DEGs that were retained in the networks in the WAT, liver, muscle, and ileum.
lists chi-square values of each pairwise comparison between subnetworks for the shortest paths and BiBC analyses performed in Fig. 2 E. P values of each comparison <0.0001.
lists intensity values of each individual sample (WAT) from Western blot.
lists effects of HFHS on gene expression in WAT of SPF mice and of insulin on gene expression in adipocytes.
lists Mmp12 and CD45(Ptprc)-positive cell counts for eight publicly available scRNA-seq datasets.
lists Mmp12-IR-ATM meta-signature genes.
shows gene ontology enrichment analysis of genes regulated by MMP12 inhibitor (ex vivo).
shows genes significantly regulated in WAT in MMP12 inhibitor-treated mice and their correlation to metabolic parameters (ex vivo).
shows genes significantly regulated by rMMP12 and insulin in 3T3-L1 cells (in vitro).
shows Mmp12 meta-signature gene expression in human datasets and in macrophage cell lines stimulated with OV.
shows genes from IR-ATM signature targeted by Atf3 as predicted by SCENIC and identified via ChIP-seq.
shows composition experimental (HFHS) and control (normal) diets
contains original blots for Fig. S1.
Acknowledgments
The authors would like to thank Oksana Gavrilova, Mark Rochman, Allison K. Ehrlich, Donald B. Jump, Barbara Gvakharia, and Valerian Dolja for helpful discussions and feedback; Ian Fraser (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD) for kindly providing immortalized macrophage cell lines; Philip Proteau (Oregon State University, Corvallis, OR) for help with reagents to activate recombinant MMP12; and the Oregon State University Center for Quantitative Life Sciences for sequencing and computational support.
This work was supported by the National Institutes of Health R01 grant DK103761 (N. Shulzhenko), an Oregon Medical Research Foundation award (A. Morgun), and intramural National Institutes of Health funding (R.R. Rodrigues, A.K. Dzutsev, A. Nita-Lazar, and G. Trinchieri).
Author contributions: Conceptualization: Z. Li, M. Gurung, R.R. Rodrigues, N.K. Newman, J. Padiadpu, A.K. Dzutsev, A. Nita-Lazar, G. Trinchieri, N. Shulzhenko, A. Morgun; data curation: Z. Li, M. Gurung, R.R. Rodrigues, N.P. Manes, R.L. Greer, S. Vasquez-Perez; formal analysis: Z. Li, M. Gurung, R.R. Rodrigues, N.K. Newman, J. Padiadpu; funding acquisition: N. Shulzhenko, A. Morgun, A. Nita-Lazar, G. Trinchieri; investigation: Z. Li, M. Gurung, R.R. Rodrigues, N.K. Newman, J. Padiadpu, N.P. Manes, R.L. Greer, S. Vasquez-Perez, H. You, K.A. Hioki, A. Fel, A.K. Dzutsev, D. De Nardo, Z. Moulton, J.W. Pederson; methodology: Z. Li, M. Gurung, R.R. Rodrigues, N. Shulzhenko, A. Morgun; project administration: N. Shulzhenko, A. Morgun, G. Trinchieri; resources: A. Nita-Lazar, G. Trinchieri, D. De Nardo, N. Shulzhenko, A. Morgun; software: R.R. Rodrigues, N.K. Newman, J. Padiadpu; supervision: A. Nita-Lazar, G. Trinchieri, N. Shulzhenko, A. Morgun; validation: Z. Li, M. Gurung, R.R. Rodrigues, N.K. Newman, N. Shulzhenko, A. Morgun; visualization: Z. Li, M. Gurung, R.R. Rodrigues, N.K. Newman, J. Padiadpu, J.W. Pederson; N. Shulzhenko, A. Morgun; writing – original draft: Z. Li, M. Gurung, R.R. Rodrigues, N.K. Newman, J. Padiadpu, N. Shulzhenko, A. Morgun; writing – review & editing: Z. Li, M. Gurung, R.R. Rodrigues, N.K. Newman, J. Padiadpu, N.P. Manes, A.K. Dzutsev, A. Nita-Lazar, G. Trinchieri, N. Shulzhenko, A. Morgun.
Data availability
Sequencing data are deposited in under BioProject accession number PRJNA558801 and GEO accession numbers GSE145132, GSE145133, GSE145052, GSE203516, GSE203488, and GSE203489.
References
- Aibar, S., Gonzalez-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., et al. 2017. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods. 14:1083–1086. 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, M., Moreno Viedma V., Sarabi A., Grun N.G., Itariu B., Leitner L., Steiner I., Bilban M., Kodama K., Butte A.J., et al. 2016. Identification of matrix metalloproteinase-12 as a candidate molecule for prevention and treatment of cardiometabolic disease. Mol. Med. 22:487–496. 10.2119/molmed.2016.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anhe, F.F., Nachbar R.T., Varin T.V., Vilela V., Dudonne S., Pilon G., Fournier M., Lecours M.A., Desjardins Y., Roy D., et al. 2017. A polyphenol-rich cranberry extract reverses insulin resistance and hepatic steatosis independently of body weight loss. Mol. Metab. 6:1563–1573. 10.1016/j.molmet.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed, F., Manchester J.K., Semenkovich C.F., and Gordon J.I.. 2007. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA. 104:979–984. 10.1073/pnas.0605374104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauters, D., Van Hul M., and Lijnen H.R.. 2013. Macrophage elastase (MMP-12) in expanding murine adipose tissue. Biochim. Biophys. Acta. 1830:2954–2959. 10.1016/j.bbagen.2012.12.024 [DOI] [PubMed] [Google Scholar]
- Blyth, C.R. 1972. On Simpson’s paradox and the sure-thing principle. J. Am. Stat. Assoc. 67:364–366. 10.1080/01621459.1972.10482387 [DOI] [Google Scholar]
- Chylikova, J., Dvorackova J., Tauber Z., and Kamarad V.. 2018. M1/M2 macrophage polarization in human obese adipose tissue. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech. Repub. 162:79–82. 10.5507/bp.2018.015 [DOI] [PubMed] [Google Scholar]
- De Nardo, D., Labzin L.I., Kono H., Seki R., Schmidt S.V., Beyer M., Xu D., Zimmer S., Lahrmann C., Schildberg F.A., et al. 2014. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat. Immunol. 15:152–160. 10.1038/ni.2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X., Yambartsev A., Ramsey S.A., Thomas L.D., Shulzhenko N., and Morgun A.. 2015. Reverse engeneering of regulatory networks from big data: A roadmap for biologists. Bioinform. Biol. Insights. 9:61–74. 10.4137/BBI.S12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses, J.A., Meier D.T., Wueest S., Rytka J., Boller S., Wielinga P.Y., Schraenen A., Lemaire K., Debray S., Van Lommel L., et al. 2010. Toll-like receptor 2-deficient mice are protected from insulin resistance and beta cell dysfunction induced by a high-fat diet. Diabetologia. 53:1795–1806. 10.1007/s00125-010-1747-3 [DOI] [PubMed] [Google Scholar]
- Fujisaka, S. 2021. The role of adipose tissue M1/M2 macrophages in type 2 diabetes mellitus. Diabetol. Int. 12:74–79. 10.1007/s13340-020-00482-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist, M., Thorsson V., Li B., Rust A.G., Korb M., Roach J.C., Kennedy K., Hai T., Bolouri H., and Aderem A.. 2006. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 441:173–178. 10.1038/nature04768 [DOI] [PubMed] [Google Scholar]
- Goncalves, I., Bengtsson E., Colhoun H.M., Shore A.C., Palombo C., Natali A., Edsfeldt A., Duner P., Fredrikson G.N., Bjorkbacka H., et al. 2015. Elevated plasma levels of MMP-12 are associated with atherosclerotic burden and symptomatic cardiovascular disease in subjects with type 2 diabetes. Arterioscler. Thromb. Vasc. Biol. 35:1723–1731. 10.1161/ATVBAHA.115.305631 [DOI] [PubMed] [Google Scholar]
- Gurung, M., Li Z., You H., Rodrigues R., Jump D.B., Morgun A., and Shulzhenko N.. 2020. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 51:102590. 10.1016/j.ebiom.2019.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, O.T., Perugini R.A., Nicoloro S.M., Gallagher-Dorval K., Puri V., Straubhaar J., and Czech M.P.. 2011. Body mass index-independent inflammation in omental adipose tissue associated with insulin resistance in morbid obesity. Surg. Obes. Relat. Dis. 7:60–67. 10.1016/j.soard.2010.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartstra, A.V., Bouter K.E.C., Backhed F., and Nieuwdorp M.. 2015. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care. 38:159–165. 10.2337/dc14-0769 [DOI] [PubMed] [Google Scholar]
- Hill, D.A., Lim H.W., Kim Y.H., Ho W.Y., Foong Y.H., Nelson V.L., Nguyen H.C.B., Chegireddy K., Kim J., Habertheuer A., et al. 2018. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc. Natl. Acad. Sci. USA. 115:E5096–E5105. 10.1073/pnas.1802611115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinkhani, S., Aazami H., Hashemi E., Dehghanbanadaki H., Adibi-Motlagh B., and Razi F.J.D.. 2021. The trend in application of omics in type 2 diabetes researches: A bibliometric study. Diabetes Metab. Syndr. 15:102250. 10.1016/j.dsx.2021.102250 [DOI] [PubMed] [Google Scholar]
- Hunninghake, G.M., Cho M.H., Tesfaigzi Y., Soto-Quiros M.E., Avila L., Lasky-Su J., Stidley C., Melén E., Söderhäll C., and Hallberg J., et al. 2009. MMP12, lung function, and COPD in high-risk populations. N. Engl. J. Med. 361:2599–2608, 10.1056/NEJMoa0904006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, P., Vig S., Datta M., Jindel D., Mathur A.K., Mathur S.K., and Sharma A.. 2013. Systems biology approach reveals genome to phenome correlation in type 2 diabetes. PloS One. 8:e53522. 10.1371/journal.pone.0053522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin, D.A., Adlung L., Thaiss C.A., Weiner A., Li B., Descamps H., Lundgren P., Bleriot C., Liu Z., Deczkowska A., et al. 2019. Lipid-associated macrophages control metabolic homeostasis in a trem2-dependent manner. Cell. 178:686–698.e14. 10.1016/j.cell.2019.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, J., Kim G.W., Lee W., Mok C., Chung S.H., and Jang W.. 2018. Meta- and cross-species analyses of insulin resistance based on gene expression datasets in human white adipose tissues. Sci. Rep. 8:3747. 10.1038/s41598-017-18082-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, M.J., Lee J., Shin N.R., Kim M.S., Hyun D.W., Yun J.H., Kim P.S., Whon T.W., and Bae J.W.. 2016. Chronic repression of mTOR Complex 2 induces changes in the gut microbiota of diet-induced obese mice. Sci. Rep. 6:30887. 10.1038/srep30887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, D.J., Kuchibhotla S., Westfall K.M., Silverstein R.L., Morton R.E., and Febbraio M.. 2011. A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signalling. Cardiovasc. Res. 89:604–613. 10.1093/cvr/cvq360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimcakova, E., Roussel B., Marquez-Quinones A., Kovacova Z., Kovacikova M., Combes M., Siklova-Vitkova M., Hejnova J., Sramkova P., Bouloumie A., et al. 2011. Worsening of obesity and metabolic status yields similar molecular adaptations in human subcutaneous and visceral adipose tissue: Decreased metabolism and increased immune response. J. Clin. Endocrinol. Metab. 96:E73–E82. 10.1210/jc.2010-1575 [DOI] [PubMed] [Google Scholar]
- Kraegen, E.W., Clark P.W., Jenkins A.B., Daley E.A., Chisholm D.J., and Storlien L.H.. 1991. Development of muscle insulin resistance after liver insulin resistance in high-fat-fed rats. Diabetes. 40:1397–1403. 10.2337/diab.40.11.1397 [DOI] [PubMed] [Google Scholar]
- Krebs, W., Schmidt S.V., Goren A., De Nardo D., Labzin L., Bovier A., Ulas T., Theis H., Kraut M., Latz E., et al. 2014. Optimization of transcription factor binding map accuracy utilizing knockout-mouse models. Nucleic Acids Res. 42:13051–13060. 10.1093/nar/gku1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusminski, C.M., and Scherer P.E.. 2012. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol. Metab. 23:435–443. 10.1016/j.tem.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.T., Pamir N., Liu N.C., Kirk E.A., Averill M.M., Becker L., Larson I., Hagman D.K., Foster-Schubert K.E., van Yserloo B., et al. 2014. Macrophage metalloelastase (MMP12) regulates adipose tissue expansion, insulin sensitivity, and expression of inducible nitric oxide synthase. Endocrinology. 155:3409–3420. 10.1210/en.2014-1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.S., Wollam J., and Olefsky J.M.. 2018. An integrated view of immunometabolism. Cell. 172:22–40. 10.1016/j.cell.2017.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem, A., Segal E., and Elinav E.. 2020. The gut microbiome and individual-specific responses to diet. mSystems. 5:e00665-20. 10.1128/mSystems.00665-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Wang J.J., Peng Q., Chen C., Humphrey M.B., Heinecke J., and Zhang S.X.. 2012. Macrophage metalloelastase (MMP-12) deficiency mitigates retinal inflammation and pathological angiogenesis in ischemic retinopathy. PloS One. 7:e52699. 10.1371/journal.pone.0052699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., Liu S., Lu M., Bandyopadhyay G., Oh D., Imamura T., Johnson A.M.F., Sears D., Shen Z., Cui B., et al. 2016. Hematopoietic-derived galectin-3 causes cellular and systemic insulin resistance. Cell. 167:973–984.e12. 10.1016/j.cell.2016.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Li J., Wu Y., Wu J., Hotchandani R., Cunningham K., McFadyen I., Bard J., Morgan P., Schlerman F., et al. 2009. A selective matrix metalloprotease 12 inhibitor for potential treatment of chronic obstructive pulmonary disease (COPD): Discovery of (S)-2-(8-(methoxycarbonylamino)dibenzo[b,d]furan-3-sulfonamido)-3-methylbutanoic acid (MMP408). J. Med. Chem. 52:1799–1802. 10.1021/jm900093d [DOI] [PubMed] [Google Scholar]
- Long, X., Li S., Xie J., Li W., Zang N., Ren L., Deng Y., Xie X., Wang L., Fu Z., and Liu E.. 2015. MMP-12-mediated by SARM-TRIF signaling pathway contributes to IFN-gamma-independent airway inflammation and AHR post RSV infection in nude mice. Respir. Res. 16:11. 10.1186/s12931-015-0176-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant, D.J., Bellac C.L., Moraes T.J., Wadsworth S.J., Dufour A., Butler G.S., Bilawchuk L.M., Hendry R.G., Robertson A.G., Cheung C.T., et al. 2014. A new transcriptional role for matrix metalloproteinase-12 in antiviral immunity. Nat. Med. 20:493–502. 10.1038/nm.3508 [DOI] [PubMed] [Google Scholar]
- Martinez-Santibanez, G., Singer K., Cho K.W., DelProposto J.L., Mergian T., and Lumeng C.N.. 2015. Obesity-induced remodeling of the adipose tissue elastin network is independent of the metalloelastase MMP-12. Adipocyte. 4:264–272. 10.1080/21623945.2015.1027848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgun, A., Dzutsev A., Dong X., Greer R.L., Sexton D.J., Ravel J., Schuster M., Hsiao W., Matzinger P., and Shulzhenko N.. 2015. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut. 64:1732–1743. 10.1136/gutjnl-2014-308820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg, A.L., and Vitek O.. 2009. Statistical design of quantitative mass spectrometry-based proteomic experiment. J. Proteome Res. 8:2144–2156. 10.1021/pr8010099 [DOI] [PubMed] [Google Scholar]
- Passaro, A.P., Marzuillo P., Guarino S., Scaglione F., Del Giudice E.M., and Di Sessa A.. 2021. Omics era in type 2 diabetes: From childhood to adulthood. World J. Diabetes. 12:2027–2035. 10.4239/wjd.v12.i12.2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsouris, D., Li P.P., Thapar D., Chapman J., Olefsky J.M., and Neels J.G.. 2008. Ablation of CD11c-positive cells normalizes insulin sensitivity in obese insulin resistant animals. Cell Metab. 8:301–309. 10.1016/j.cmet.2008.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl, J. 2009. Causality. Cambridge University Press. [Google Scholar]
- Pejnovic, N.N., Pantic J.M., Jovanovic I.P., Radosavljevic G.D., Milovanovic M.Z., Nikolic I.G., Zdravkovic N.S., Djukic A.L., Arsenijevic N.N., and Lukic M.L.. 2013. Galectin-3 deficiency accelerates high-fat diet-induced obesity and amplifies inflammation in adipose tissue and pancreatic islets. Diabetes. 62:1932–1944. 10.2337/db12-0222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R.J., Camporez J.P.G., Kursawe R., Titchenell P.M., Zhang D., Perry C.J., Jurczak M.J., Abudukadier A., Han M.S., Zhang X.M., et al. 2015. Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell. 160:745–758. 10.1016/j.cell.2015.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabot, S., Membrez M., Bruneau A., Gerard P., Harach T., Moser M., Raymond F., Mansourian R., and Chou C.J.. 2010. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 24:4948–4959. 10.1096/fj.10-164921 [DOI] [PubMed] [Google Scholar]
- Rausch, P., Basic M., Batra A., Bischoff S.C., Blaut M., Clavel T., Glasner J., Gopalakrishnan S., Grassl G.A., Gunther C., et al. 2016. Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. Int. J. Med. Microbiol. 306:343–355. 10.1016/j.ijmm.2016.03.004 [DOI] [PubMed] [Google Scholar]
- Reichenbach, H. 1991. The Direction of Time. University of California Press. [Google Scholar]
- Rocha, M., Apostolova N., Diaz-Rua R., Muntane J., and Victor V.M.. 2020. Mitochondria and T2D: Role of autophagy, ER stress, and inflammasome. Trends Endocrinol. Metab. 31:725–741. 10.1016/j.tem.2020.03.004 [DOI] [PubMed] [Google Scholar]
- Rodrigues, R.R., Gurung M., Li Z., Garcia-Jaramillo M., Greer R., Gaulke C., Bauchinger F., You H., Pederson J.W., Vasquez-Perez S., et al. 2021. Transkingdom interactions between Lactobacilli and hepatic mitochondria attenuate western diet-induced diabetes. Nat. Commun. 12:101. 10.1038/s41467-020-20313-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, L., and Lumeng C.N.. 2018. Properties and functions of adipose tissue macrophages in obesity. Immunology. 155:407–417. 10.1111/imm.13002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, C. 1953. The lattice theory of information. 1:105–107. 10.1109/tit.1953.1188572 [DOI] [Google Scholar]
- Silva, H.M., Bafica A., Rodrigues-Luiz G.F., Chi J., Santos P.D.A., Reis B.S., Hoytema van Konijnenburg D.P., Crane A., Arifa R.D.N., Martin P., et al. 2019. Vasculature-associated fat macrophages readily adapt to inflammatory and metabolic challenges. J. Exp. Med. 216:786–806. 10.1084/jem.20181049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, L., Brandon A.E., Turner N., and Cooney G.J.. 2018. Modeling insulin resistance in rodents by alterations in diet: What have high-fat and high-calorie diets revealed? Am. J. Physiol. Endocrinol. Metab. 314:E251–E265. 10.1152/ajpendo.00337.2017 [DOI] [PubMed] [Google Scholar]
- Sonnenburg, J.L., and Backhed F.. 2016. Diet-microbiota interactions as moderators of human metabolism. Nature. 535:56–64. 10.1038/nature18846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soronen, J., Laurila P.P., Naukkarinen J., Surakka I., Ripatti S., Jauhiainen M., Olkkonen V.M., and Yki-Jarvinen H.. 2012. Adipose tissue gene expression analysis reveals changes in inflammatory, mitochondrial respiratory and lipid metabolic pathways in obese insulin-resistant subjects. BMC Med. Genomics. 5:9. 10.1186/1755-8794-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenod, F. 2008. Pathophysiology of diabetes mellitus type 2: Roles of obesity, insulin resistance and β-cell dysfunction. In Diabetes and Cancer. Karger Publishers. 1–18. [Google Scholar]
- Tran, H.Q., Bretin A., Adeshirlarijaney A., Yeoh B.S., Vijay-Kumar M., Zou J., Denning T.L., Chassaing B., and Gewirtz A.T.. 2020. “Western diet”-induced adipose inflammation requires a complex gut microbiota. Cell Mol. Gastroenterol. Hepatol. 9:313–333. 10.1016/j.jcmgh.2019.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uda, S. 2020. Application of information theory in systems biology. Biophys. Rev. 12:377–384. 10.1007/s12551-020-00665-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay, J., Gauthier M.F., Biswell R.L., Louiselle D.A., Johnston J.J., Cheung W.A., Belden B., Pramatarova A., Biertho L., Gibson M., et al. 2020. Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat. Metab. 2:97–109. 10.1038/s42255-019-0152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Tang H., Zhang C., Zhao Y., Derrien M., Rocher E., van-Hylckama Vlieg J.E.T., Strissel K., Zhao L., Obin M., and Shen J.. 2015. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 9:1–15. 10.1038/ismej.2014.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox, C.S. 2010. Effects of tempol and redox-cycling nitroxides in models of oxidative stress. Pharmacol. Ther. 126:119–145. 10.1016/j.pharmthera.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yambartsev, A., Perlin M.A., Kovchegov Y., Shulzhenko N., Mine K.L., Dong X., and Morgun A.. 2016. Unexpected links reflect the noise in networks. Biol. Direct. 11:52. 10.1186/s13062-016-0155-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Wang X., Guo H., Zhang W., Wang W., and Ma H.. 2019. Whole transcriptome analysis of obese adipose tissue suggests u001kfc.1 as a potential regulator to glucose homeostasis. Front. Genet. 10:1133. 10.3389/fgene.2019.01133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists analysis of systemic metabolic parameters in SPF and GF mice fed with HFHS and control diets.
lists DEGs between HFHS and ND in conventional (SPF) mice (FDR < 15%) in four organs and their expression in GF mice.
shows the total number and percentage of DEGs and the percentage of DEGs that were retained in the networks in the WAT, liver, muscle, and ileum.
lists chi-square values of each pairwise comparison between subnetworks for the shortest paths and BiBC analyses performed in Fig. 2 E. P values of each comparison <0.0001.
lists intensity values of each individual sample (WAT) from Western blot.
lists effects of HFHS on gene expression in WAT of SPF mice and of insulin on gene expression in adipocytes.
lists Mmp12 and CD45(Ptprc)-positive cell counts for eight publicly available scRNA-seq datasets.
lists Mmp12-IR-ATM meta-signature genes.
shows gene ontology enrichment analysis of genes regulated by MMP12 inhibitor (ex vivo).
shows genes significantly regulated in WAT in MMP12 inhibitor-treated mice and their correlation to metabolic parameters (ex vivo).
shows genes significantly regulated by rMMP12 and insulin in 3T3-L1 cells (in vitro).
shows Mmp12 meta-signature gene expression in human datasets and in macrophage cell lines stimulated with OV.
shows genes from IR-ATM signature targeted by Atf3 as predicted by SCENIC and identified via ChIP-seq.
shows composition experimental (HFHS) and control (normal) diets
contains original blots for Fig. S1.
Data Availability Statement
Sequencing data are deposited in under BioProject accession number PRJNA558801 and GEO accession numbers GSE145132, GSE145133, GSE145052, GSE203516, GSE203488, and GSE203489.