Abstract
Background
Using time series and spatiotemporal analyses, this study aimed to establish an Early Warning System (EWS) for COVID-19 in Fars province Iran.
Methods
A EWS was conducted on (i) daily basis city-level time series data including 53 554 cases recorded during 18 February–30 September 2020, which were applied to forecast COVID-19 cases during 1 October–14 November 2020, and (ii) the spatiotemporal analysis, which was conducted on the forecasted cases to predict spatiotemporal outbreaks of COVID-19.
Results
A total of 55 369 cases were forecasted during 1 October–14 November 2020, most of which (26.9%) occurred in Shiraz. In addition, 65.80% and 34.20% of the cases occurred in October and November, respectively. Four significant spatiotemporal outbreaks were predicted, with the Most Likely Cluster (MLC) occurring in ten cities during 2–22 October (P < 0.001 for all). Moreover, subgroup analysis demonstrated that Zarrindasht was the canon of the epidemic on 6 October (P=0.04). As a part of EWS, the epidemic was triggered from Jahrom, involving the MLC districts in the center, west, and south parts of the province. Then, it showed a tendency to move towards Zarrindasht in the south and progress to Lar in the southernmost part. Afterwards, it simultaneously progressed to Fasa and Sepidan in the central and northwestern parts of the province, respectively.
Conclusion
EWS, which was established based on the current protocol, alarmed policymakers and health managers on the progression of the epidemic and on where and when to implement medical facilities. These findings can be used to tailor province-level policies to servile the ongoing epidemic in the area; however, governmental level effort is needed to control the epidemic at a larger scale in the future.
1. Introduction
The surveillance systems for infectious diseases are encouraging well-developed techniques that can warn them by detecting the exact time and place of disease occurrence and, subsequently, targeting interventions and resources to more risky areas [1]. Some infections follow space and time trends for which Early Warning Systems (EWS) can be useful tools to find the potential outbreaks [2]. EWS are alert systems to predict epidemic outbreaks in the region. In the 1910s, Captain S.R Christophers from the British army set a system to predict malaria in India. Since then, it has been used as a useful tool to aid infected nations, make health policies, and manage services to the infected area [1]. The epidemic statue of SARS-CoV-2 disease (COVID-19) was officially declared by the World Health Organization in January 2020 [3, 4]. What makes it important to set a EWS for ongoing COVID-19 epidemic is that it could early notify the policy makers and health managers on the time and location of the outbreaks, so that they could focus on more needy areas and dedicate more medical facilities. Finally, people are helped with less COVID-19 threat, infections, and death.
The question is that, using COVID-19 time series data, if the current study could establish a EWS for future COVID-19 outbreaks, the surveillance system could act timely, prioritize times and locations for targeting interventions, allocate resources more efficiently, and improve decision-making in the area.
Time series data refer to a sequence of successive naturally temporal data, which are equally spaced in points of time. A time series analysis consists of methods for analyzing time series data and forecasting the future values based on the previously observed values [5]. Spatial and Temporal Scan (SaTScan) modeling discovers more about the space and time of an event occurrence. Space-time permutation scan statistic, which is derived from SaTScan, was first introduced by Martin Kulldorff in 2015 [6]. This model aims at determining whether the cases are distributed uniformly over time and space. The statistic utilizes scanning windows moving systematically across the study area to identify the clusters of cases. This modeling helps surveillance systems decide timely on more at risk areas [7]. Using one model output as another mode input, this study could develop a protocol of EWS in the area by (i) forecasting the COVID-19 cases, (ii) discovering the future space-time outbreaks of COVID-19, (iii) predicting the progression of epidemic, and (iv) having more control on the epidemic.
2. Methods
2.1. Study Design, Data Source, and Study Variable
Daily basis city-level time series data including 53 554 cases recorded in 26 cities of Fars province, Iran, during 18 February–30 September 2020 (225 days) were applied to forecast COVID-19 cases during 1 October–14 November 2020 (45 days). The cases were obtained from 44 COVID-19 sampling centers affiliated to Shiraz University of Medical Sciences (SUMS). All data were reported to SUMS either online, https://www.coronalab.sums.ac.ir, or by manual registration. There were no exclusion criteria, meaning that all cases were included in the study. The COVID-19 cases were the positive cases confirmed by real-time Reverse Transcription Polymerase Chain Reaction (RT-PCR). Additionally, the geographical coordinates of the locations were obtained through Google-Earth (US Dept. of State Geographer 2021) based on the latitude-longitude coordinate system.
2.2. Case Study and Area
Fars province, the fourth most populated (4 852 274 dwellers) and widespread (122 608 km2) province amongst 31 provinces in Iran, is located at 27°3′ and 31°40′ northern latitude and 50°36′ and 55°35′ eastern longitude in south of Iran. It includes 26 cities, with Shiraz, the fifth most populated city in Iran, being the capital. Marvdasht, Jahrom, Fasa, Kazerun, Darab, Firoozabad, Lar, and Abade, also, are the most populated areas (more than 100 00 dwellers per capita). 67.6% of the province are urban dwellers (urban areas and suburbs), 31% are villagers (small towns and rural areas), and the rest are nomads (no fixed habitation who regularly move to and from the same areas breeding livestock) [8].
2.3. Statistical Analysis
Mean difference and Std. Error of difference were used to describe quantitative variables, and frequency (relative frequency) was used to describe qualitative variables.
The distribution of COVID-19 cases across cities and times are tested using independent-sample one way ANOVA and pair wise comparisons (LSD test). Additionally, time series analysis including (i) Akaik Information Criterion (AIC)/Bayesian Information Criterion (BIC), which are the estimators of the relative quality of time series models comparative to other models for a given set of data, (ii) Auto Correlation Function (ACF)/Partial Auto Correlation Function (PACF) sample/model, which assess the conformity of the observed and fitted pattern in the data, and (iii) residual analysis, which evaluates the presumptions of white noise parameter, was used to forecast the COVID-19 cases during 1 October–14 November 2020 (45 days). A set of time series models were applied, and the best ones were selected based on lower AIC/BIC scores, ACF/PACF conformity, and residual analysis.
Descriptions on time series models:
- A simple additive decomposed time series model is
(1) mt is the trend, st is the seasonality, and et is the error or random white noise.
To be used to predict well founded values, a time series must be stationary, meaning that it should have a constant mean and variance across the time series. So, in order to allow forecasting, several time series models can be created including Autoregressive Model (AR), Moving Average Model (MA), and Autoregressive Moving Average (ARMA) model, which are used on stationary time series. Seasonal Autoregressive Moving Average (SARMA) model, however, is used for nonstationary time series.
Autoregressive Model: AR (p)
AR model uses observations from previous time steps as input to a regression equation to predict the value at the next step. It takes one argument, p, which determines how many previous time steps will be input. The order, p, can be determined by looking at the PACF. The PACF gives the partial correlation of a time series with its own lagged value, regressed of the time series at all shorter lags.
Moving Average Model: MA (q)
MA model is a time series model that accounts for very short-run autocorrelation. Basically, it states that the next observation is the mean of every past observation. The order of MA model, q, can usually be estimated by looking at ACF plot.
Autoregressive Moving Average: ARMA (p, q)
ARMA and ARIMA models are simply a combination of an AR model and MA model.
Seasonal Autoregressive Moving Average model: SARMAs(p, q)
SARMA model is an extension of ARMA model. The only difference is that a seasonal component is added now. s is the number of periods in the season. SARMA can adjust a nonstationary time series by removing trend and seasonality. [9].
Appling time series forecasted cases in spatiotemporal analysis, high-rate clusters were detected considering the minimum and maximum temporal sizes that were equal to one day and 50% of the study period, respectively. The minimum and maximum spatial sizes were also equal to two cases and 50% of at risk population, respectively. Temporal data were checked to ensure that all the cases were within the specified temporal study period. In addition, geographical data check was done to ensure that all the observations were within the specified geographical area. No geographical overlap and subgroup analysis were used.
R v.3.6.3, ITSM 2002, ArcGIS v. 10, and SaTScan software were applied for data analysis. Significance level was considered 0.05 for all tests.
2.4. Validation Study
A validation study using real data from 1 October–14 November 2020 was done to assess the accuracy of the results.
2.5. Ethical Statement
All ethical steps including data collection and analysis as well as reporting the results were in accordance with the standards approved by the Ethic Committee of the Ministry of Health, Treatment, and Medical Education (IR.SUMS.REC.1399.574). The work processes were anonymous, and the results were reported to the study participants.
3. Results
3.1. Distributions of 53 554 COVID-19 Cases in Fars Province by City and Month, February 18–September 30, 2020
From 53 554 COVID-19 cases, the maximum and minimum cases happened in Shiraz (53.49%) and Sarvestan (0.2%), respectively. In addition, the maximum and minimum cases happened in July (24%) and February (0.3%), respectively. The distribution of 53 554 COVID-19 cases by city and month is shown in Tables 1 and 2.
Table 1.
Distribution of 53 554 COVID-19 cases in 26 cities of Fars province, Iran, February 18–September 30, 2020.
| City | Shiraz | Lar | Jahrom | Fasa | Kazerun | Marvdasht | Darab | Abade |
| COVID-19 (%) | 53.49 | 8.64 | 6.25 | 5.95 | 5.24 | 3.32 | 2.07 | 1.63 |
| City | Zarindasht | Firoozabad | Lamerd | Mamasani | Ghirkarzin | Safashahr | Sepidan | Stahban |
| COVID-19 (%) | 1.39 | 1.18 | 1.16 | 1.10 | 1.08 | 1.04 | 0.99 | 0.85 |
| City | Eghlid | Farashband | Pasargad | Neireez | Arsenjan | Rostam | Khonj | Kharame |
| COVID-19 (%) | 0.78 | 0.73 | 0.67 | 0.57 | 0.47 | 0.38 | 0.29 | 0.29 |
| City | Bavanat | Sarvestan | ||||||
| COVID-19 (%) | 0.24 | 0.20 |
Table 2.
Distribution of 53 554 COVID-19 cases in Fars province, Iran, February 18–September 30, 2020.
| Month | February | March | April | May | June | July | August | September |
| COVID-19 (%) | 0.30 | 3.50 | 6.50 | 4.70 | 19.70 | 24 | 18.70 | 22.60 |
COVID-19 cases were not distributed uniformly across cities (test statistics = 170.83, df = 25, P < 0.001) and months (test statistics = 21.3, df = 7, P < 0.001) in Fars province during February 18–September 30, 2020. Pairwise comparisons of 53 554 COVID-19 cases by city and month have been shown in Tables 3 and 4.
Table 3.
Pairwise comparisons of 53 554 COVID-19 cases across 26 cities of Fars province, Iran, during February 18–September 30, 2020.
| City | Mean difference | P | City | Mean difference | P | City | Mean difference | P | City | Mean difference | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Shiraz-Safashahr | 124.28 | <0.001 | Safashahr-Fasa | −11.62 | <0.001 | Neireez-Jahrom | −13.46 | <0.001 | Jahrom-Kharame | 14.13 | <0.001 |
| Shiraz-Fasa | 112.66 | <0.001 | Safashahr-Jahrom | −12.33 | <0.001 | Neireez-Kazerun | −11.07 | <0.001 | Jahrom-Farashband | 13.10 | <0.001 |
| Shiraz-Neireez | 125.41 | <0.001 | Safashahr-Kazerun | −9.95 | <0.001 | Neireez-Marvdasht | −6.52 | 0.01 | Jahrom-Khonj | 14.11 | <0.001 |
| Shiraz-Abade | 122.9 | <0.001 | Safashahr-Marvdasht | −5.39 | 0.04 | Neireez-Lar | −19.13 | <0.001 | Jahrom-Darab | 9.91 | <0.001 |
| Shiraz-Arsenjan | 125.63 | <0.001 | Safashahr-Lar | −18 | <0.001 | Abade-Jahrom | −10.95 | <0.001 | Jahrom-Rostam | 13.90 | <0.001 |
| Shiraz-Stahban | 124.73 | <0.001 | Fasa-Neireez | 12.7 | <0.001 | Abade-Kazerun | −8.56 | 0.001 | Jahrom-Zarindasht | 11.1 | <0.001 |
| Shiraz-Eghlid | 124.92 | <0.001 | Fasa-Abade | 10.24 | <0.001 | Abade-Lar | −16.62 | <0.001 | Jahrom-Sarvestan | 14.32 | <0.001 |
| Shiraz-Bavanat | 126.18 | <0.001 | Fasa-Arsenjan | 12.98 | <0.001 | Arsenjan-Jahrom | −13.69 | <0.001 | Jahrom-Firoozabad | 12 | <0.001 |
| Shiraz-Pasargad | 125.17 | <0.001 | Fasa-Stahban | 12.08 | <0.001 | Arsenjan-Kazerun | −11.3 | <0.001 | Jahrom-Ghirkarzin | 12.26 | <0.001 |
| Shiraz-Jahrom | 111.95 | <0.001 | Fasa-Eghlid | 12.25 | <0.001 | Arsenjan-Marvdasht | −6.7 | 0.01 | Jahrom-Marvdasht | 6.94 | 0.009 |
| Shiraz-Kharame | 126.08 | <0.001 | Fasa-Bavanat | 13.52 | <0.001 | Arsenjan-Lar | −19.36 | <0.001 | Jahrom-Kharame | 1.13 | <0.001 |
| Shiraz-Farashband | 125.03 | <0.001 | Fasa-Pasargad | 12.51 | 0.01 | Stahban-Jahrom | −12.78 | <0.001 | Jahrom-Lamerd | 12.07 | <0.001 |
| Shiraz-khonj | 126.03 | <0.001 | Fasa-Kharame | 13.42 | <0.001 | Stahban-Kazerun | −10.4 | <0.001 | Jahrom-Mamasani | 12.04 | <0.001 |
| Shiraz-Darab | 121.85 | <0.001 | Fasa-Farashband | 12.37 | <0.001 | Stahban-Marvdasht | −5.84 | 0.03 | Jahrom-lar | −5.67 | 0.03 |
| Shiraz-Rostam | 125.85 | <0.001 | Fasa-khonj | 13.4 | 0.01 | Stahban-Lar | −18.46 | <0.001 | Jahrom-Sepidan | 12.46 | <0.001 |
| Shiraz-Zarindasht | 123.46 | <0.001 | Fasa-Darab | 9.19 | 0.001 | Eghlid-Jahrom | −12.96 | <0.001 | Kharame-Kazerun | −11.74 | <0.001 |
| Shiraz-Sarvestan | 126.27 | <0.001 | Fasa-Rostam | 13.19 | <0.001 | Eghlid-Kazerun | −10.57 | <0.001 | Kharame-Marvdasht | −7.19 | 0.007 |
| Shiraz-Firoozabad | 123.96 | <0.001 | Fasa-Zarindasht | 10.8 | <0.001 | Eghlid-Marvdasht | −6.02 | 0.02 | Kharame-Lar | −19.8 | <0.001 |
| Shiraz-Ghirkarzin | 124.21 | <0.001 | Fasa-Sarvestan | 13.61 | <0.001 | Eghlid-Lar | −18.63 | <0.001 | Farashband-Kazerun | −10.69 | <0.001 |
| Shiraz-Kazerun | 114.34 | <0.001 | Fasa-Firoozabad | 11.30 | <0.001 | Bavanat-Jahrom | −14.23 | <0.001 | Farashband-Marvdasht | −6.14 | 0.02 |
| Shiraz-Marvdasht | 118.89 | <0.001 | Fasa-Ghirkarzin | 11.55 | <0.001 | Bavanat-Kazerun | −11.8 | <0.001 | Farashband-Lar | −18.75 | <0.001 |
| Shiraz-Lamerd | 124.02 | <0.001 | Fasa-Marvdasht | 6.23 | 0.01 | Bavanat-Marvdasht | −7.29 | 0.006 | Khonj-Kazerun | −11.73 | <0.001 |
| Shiraz-Mamasani | 124.15 | <0.001 | Fasa-Lamerd | 11.36 | <0.001 | Bavanat-Lar | −19.9 | <0.001 | Khonj-Marvdasht | −7.17 | 0.007 |
| Shiraz-Lar | 106.28 | <0.001 | Fasa-Mamasani | 11.50 | <0.001 | Pasargad-Jahrom | −13.22 | <0.001 | Khonj-Lar | −19.78 | <0.001 |
| Shiraz-Sepidan | 124.41 | <0.001 | Fasa-Lar | −6.38 | 0.02 | Pasargad-Kazerun | −10.84 | <0.001 | Marvdasht-Mamasani | 5.26 | 0.04 |
| Darab-Kazerun | −7.52 | 0.005 | Fasa-Sepidan | 11.75 | <0.001 | Pasargad-Marvdasht | −6.28 | 0.02 | Marvdasht-Lar | −12.61 | <0.001 |
| Darab-Lar | −15.57 | <0.001 | Sarvestan-Kazerun | −11.94 | <0.001 | Pasargad-Lar | −19 | <0.001 | Marvdasht-Sepidan | 5.52 | 0.03 |
| Rostam-Kazerun | −11.52 | <0.001 | Sarvestan-Marvdasht | −7.38 | 0.005 | Ghirkarzin-Kazerun | −9.87 | <0.001 | Kazerun-Lamerd | 9.68 | <0.001 |
| Rostam-Marvdasht | −6.95 | 0.009 | Sarvestan-Lar | −19.99 | <0.001 | Ghirkarzin-Marvdasht | −5.32 | 0.04 | Kazerun-Mamasani | 9.82 | <0.001 |
| Rostam-Lar | −19.57 | <0.001 | Firoozabad-Kazerun | −9.62 | <0.001 | Ghirkarzin-Lar | −18 | <0.001 | Kazerun-Lar | −8.06 | 0.002 |
| Zarindasht-Kazerun | −9.13 | 0.001 | Firoozabad-Lar | −17.68 | <0.001 | Lamerd-Lar | −17.74 | <0.001 | Kazerun-Sepidan | 10.08 | <0.001 |
| Zarindasht-Lar | −17.19 | <0.001 | Lar-Sepidan | 18.13 | <0.001 | Mamasani-Lar | −17.88 | <0.001 |
Note: std. error for all difference s is 2.66; just statistically significant differences were reported.
Table 4.
Pairwise comparisons of 53 554 COVID-19 cases by month in Fars province, Iran, during 18 February–30 September 2020.
| Sammonth | Mean difference | Std. error | Adj. P | Sample median | Mean difference | Std. error | Adj. P |
|---|---|---|---|---|---|---|---|
| February–June | −13.02 | 2.45 | <0.001 | April–June | −9.05 | 1.85 | <0.001 |
| February–July | −16 | 2.45 | <0.001 | April–July | −12.04 | 1.85 | <0.001 |
| February–August | −12 | 2.42 | <0.001 | April–August | −7.23 | 1.81 | <0.001 |
| February–September | −15.08 | 2.45 | <0.001 | April–September | −11.2 | 1.85 | <0.001 |
| March–June | −11.15 | 1.84 | <0.001 | May-June | −10.30 | 1.85 | <0.001 |
| March–July | −1.14 | 1.84 | <0.001 | May–July | −13.30 | 1.85 | <0.001 |
| March–August | −9.33 | 1.80 | <0.001 | May–August | −8.48 | 1.81 | <0.001 |
| March–September | −13.21 | 1.84 | <0.001 | May–September | −12.36 | 1.85 | <0.001 |
| July-August | 4.81 | 1.81 | 0.008 | August-September | −3.88 | 1.81 | 0.03 |
Note: just statistically significant differences were reported.
COVID-19 cases were significantly higher in Shiraz than those in 25 other cities (P < 0.05 for all). Jahrom had more cases than Farashband, Khonj, Darab, Rostam, Kharame, Zarindasht, Sarvestan, Firoozabad, Ghirkarzin, Marvdasht, Lamerd, Mamasani, and Sepidan (P < 0.05 for all); however, fewer cases were seen in Jahrom compared with Lar (P < 0.05). In addition, Safashahr has lower cases compared with Fasa, Jahrom, Kazerun, Marvdasht, and Lar (P < 0.05 for all). Also, there were more cases in Fasa than those in Neireez, Abade, Arsenjan, Stahban, Eghlid, Bavanat, Pasargad, Kharame, Farashband, khonj, Darab, Rostam, Zarindasht, Sarvestan, Firoozabad, Ghirkarzin, Marvdasht, Lamerd, Sepidan, and Mamasani (P < 0.05 for all); there, however, were fewer cases in Fasa compared with Lar (P < 0.05). Moreover, fewer cases were seen in Neireez, Arsenjan, Stahban, Eghlid, Pasargad, and Bavanat than those in Jahrom, Kazerun, Marvdasht, and Lar (P < 0.05 for all, respectively). Furthermore, the cases were lower in Kharame, Khonj, Rostam, Ghirkarzin, Sarvestan, and Farashband than those in Kazerun, Marvdasht, and Lar (P < 0.05 for all, respectively). Additionally, the cases were lower in Abade than those in Jahrom, Kazerun, and Lar (P < 0.05 for all). Likewise, the cases were lower in Darab, Firoozabad, and Zarrindasht than those in Kazerun, and Lar (P < 0.05 for all, respectively). Besides, the cases were more in Kazerun than those in Lamerd, Mamasani, and Sepidan (P < 0.05 for all); however, less cases were seen in Kazerun compared with Lar (P < 0.05). Also, the cases were higher in Marvdasht than those in Mamasani, and Sepidan (P < 0.05 for both); however, fewer cases were seen compared with Lar (P < 0.05). Moreover, there were fewer cases in Lamerd and Mamasani than those in Lar (P < 0.05 for both). The cases were higher in Lar than those in Sepidan (P < 0.05).
COVID-19 cases in February, March, April, and May were significantly lower than those in June, July, August, and September (P < 0.05 for all, respectively); COVID-19 cases in July were significantly higher than those in August (P < 0.05 for all); COVID-19 cases in August were significantly lower than those in September (P < 0.05 for all).
3.2. Forecasting COVID-19 Cases in Fars Province during 1 October–14 November 2020
From the 53 554 observed cases during 18 February–30 September 2020, 55 369 cases were forecasted during 1 October–14 November 2020 using variety of time series models. The frequency of 55 369 forecasted cases alongside the results of time series analysis for the 26 cities has been presented in Table 5.
Table 5.
The frequency 55 369 forecasted cases during 1 October–14 November 2020 accompanied by the results of time series analysis in the 26 cities of Fars province, Iran.
| Time series analysis results | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| City | Forecasted cases | Model | Residual analysis tests (P) | Order of min AICC YW | AIC | BIC | ||||||
| n | % | Ljung-box | Diff sign points | Rank test statistic | McLeod-Li | Turning points | Jarque-bera | |||||
| Shiraz | 14895 | 26.9 | MA (8) | 0.230 | 0.30 | 0.69 | 0.030 | 0.100 | <0.001 | 0 | 8280 | 8303 |
| Lar | 2762 | 4.99 | SARMA12 (2, 2) | 0.400 | 0.9 | 0.49 | 0.040 | 0.560 | <0.001 | 0 | 1098 | 1101 |
| Jahrom | 1716 | 3.10 | SARMA12 (3, 4) | 0.770 | 0.73 | 0.37 | <0.001 | 0.010 | <0.001 | 0 | 1221 | 1225 |
| Fasa | 1772 | 3.20 | ARMA (1, 1) | 0.460 | 0.73 | 0.12 | <0.001 | 0.090 | <0.001 | 0 | 1371 | 1371 |
| Kazerun | 1661 | 3 | AR (1) | 0.440 | 0.30 | 0.04 | 0.020 | 0.220 | <0.001 | 0 | 1267 | 1266 |
| Marvdasht | 2215 | 4 | SARMA12 (1, 1) | 0.610 | 0.9 | 0.04 | 0.010 | 0.560 | <0.001 | 0 | 1247 | 1247 |
| Darab | 1440 | 2.60 | ARMA (1, 1) | 0.960 | 0.02 | 0.80 | 0.040 | 0.180 | <0.001 | 0 | 1360 | 1361 |
| Abade | 1661 | 3 | SARMA12 (1, 1) | 0.960 | 0.42 | 0.60 | 0.040 | 0.180 | <0.001 | 0 | 1360 | 1361 |
| Zarindasht | 1883 | 3.40 | AR (1) | 0.860 | 0.03 | 0.85 | 0.010 | 0.950 | 0.030 | 0 | 1357 | 1356 |
| Firoozabad | 1495 | 2.70 | SARMA12 (1, 2) | 0.060 | 0.08 | 0.21 | 0.030 | 0.010 | <0.001 | 0 | 1391 | 1389 |
| Lamerd | 1440 | 2.60 | SARMA12 (2, 2) | 0.190 | 0.21 | 0.34 | <0.001 | 0.0002 | 0.030 | 0 | 1364 | 1368 |
| Mamasani | 1495 | 2.70 | SARMA12 (1, 2) | 0.580 | 0.04 | 0.32 | 0.040 | 0.490 | <0.001 | 0 | 1381 | 1380 |
| Ghirkarzin | 1440 | 2.60 | ARIMA (3, 1) | 0.980 | 0.73 | 0.51 | 0.020 | 0.450 | 0.010 | 0 | 1364 | 1366 |
| Safashahr | 1440 | 2.60 | ARIMA (1, 1) | 0.500 | 0.45 | 0.76 | 0.0010 | 0.350 | 0.020 | 0 | 1348 | 1348 |
| Sepidan | 1993 | 3.60 | SARMA12 (3, 1) | 0.130 | 0.30 | 0.92 | 0.030 | 0.950 | 0.040 | 0 | 1344 | 1342 |
| Stahban | 1440 | 2.60 | ARMA (1, 1) | 0.980 | 0.2 | 0.81 | <0.001 | 0.180 | <0.001 | 0 | 1391 | 1392 |
| Eghlid | 1440 | 2.60 | MA (1) | 0.140 | 0.73 | 0.17 | 0.020 | 0.670 | <0.001 | 0 | 1391 | 1388 |
| Farashband | 1440 | 2.60 | SARMA12 (2, 2) | 0.920 | 0.73 | 0.40 | <0.001 | 0.040 | 0.001 | 0 | 1369 | 1370 |
| Pasargad | 1661 | 3 | MA (1) | 0.060 | 0.04 | 0.47 | 0.010 | 0.490 | 0.050 | 0 | 1334 | 1332 |
| Neireez | 1440 | 2.60 | AR (1) | 0.230 | 0.45 | 0.34 | 0.002 | 0.450 | 0.010 | 0 | 1364 | 1363 |
| Arsenjan | 1440 | 2.60 | ARMA (2, 1) | 0.080 | 0.42 | 0.14 | 0.030 | 0.830 | <0.001 | 0 | 1372 | 1367 |
| Rostam | 1440 | 2.60 | AR (1) | 0.090 | 0.07 | 0.20 | <0.001 | <0.001 | <0.001 | 0 | 1321 | 1321 |
| Khonj | 1440 | 2.60 | SARMA12 (2, 1) | 0.680 | 0.06 | 0.05 | <0.001 | 0.830 | <0.001 | 0 | 1295 | 1291 |
| Kharame | 1440 | 2.60 | SARMA12 (1, 3) | 0.450 | 0.43 | 0.23 | 0.001 | 0.001 | 0.010 | 0 | 1356 | 1357 |
| Bavanat | 1440 | 2.60 | MA (1) | 0.220 | 0.91 | 0.02 | <0.001 | 0.310 | <0.001 | 0 | 1308 | 1306 |
| Sarvestan | 1440 | 2.60 | SARMA12 (2, 2) | 0.540 | 0.9 | 0.96 | 0.010 | 0.590 | 0.130 | 0 | 1260 | 1258 |
AR, autoregressive; MA, moving average; ARMA, autoregressive moving average; ARIMA, autoregressive integrated moving average; SARMA, seasonal autoregressive moving average. It is noteworthy to mention that if four out of the six residual analysis test statistics are statistically significant, it is enough to say that the model is a good fit; based on residual analysis McLeod- Li test statistic needs to be significant and the other five tests should be greater than or equal to 0.05; in addition, the order of min AICCYW, which assesses the mean of white noise residual, needs to be zero. The lower the AIC/BIC scores, the better the model fits.
From the 55 369 forecasted cases during 1 October–14 November 2020, the highest proportion was related to Shiraz (26.9%). Additionally, 65.80% and 34.20% of the cases were forecasted to occur in October and November, respectively. The time series results for Jahrom and Shiraz have been presented in Figures 1 and 2, respectively. The interpretation of the results was similar for the 24 other cities.
Figure 1.
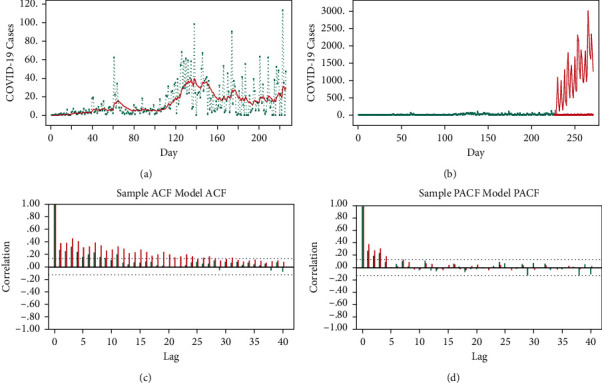
(a) SARMA12 (3, 4) estimating the number of COVID-19 cases in Jahrom during February18th–September30th 2020; the green dashed line represents the number of observed COVID-19 cases during February18th–September30th 2020; the simple red line indicates SARMA12 (3, 4) model estimating COVID-19 cases during February18th–September30th 2020. (b) SARMA12 (3, 4) model predicting COVID-19 cases in Jahrom during October1st-November14th 2020; the green line represents the estimated COVID-19 cases during February18th–September30th 2020; the red line represents the predicted COVID-19 cases during October1st-November14th 2020 with its confidence interval. (c) Sample/model SARMA12 (3, 4) auto correlation function (ACF) and (d) partial auto correlation function (PACF) for Jahrom during October1st-November14th 2020; the green correlations are derived from the sample and the red correlations are taken from the fitted model; two dashed horizontal lines are confidence bands; a model is considered good in case the sample and model correlations out of the bands overlap.
Figure 2.
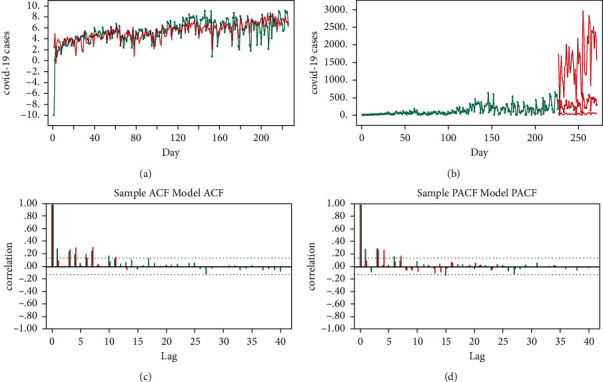
(a) MA (8) estimating the number of COVID-19 cases in Shiraz during February18th–September30th 2020; the green dashed line represents the number of observed COVID-19 cases during February18th–September30th 2020; the simple red line indicates MA (8) model estimating COVID-19 cases during February18th–September30th 2020. (b) MA (8) model predicting COVID-19 cases in Shiraz during October1st-November14th 2020; the green line represents the estimated COVID-19 cases during February18th–September30th 2020; the red line represents the predicted COVID-19 cases during October1st-November14th 2020 with its confidence interval. (c) Sample/model MA (8) auto correlation function (ACF) and (d) partial auto correlation function (PACF) for Shiraz during October1st-November14th 2020; the green correlations are derived from the sample and the red correlations are taken from the fitted model; two dashed horizontal lines are confidence bands; a model is considered good in case the sample and model correlations out of the bands overlap.
3.2.1. COVID-19 Time Series Analysis in Jahrom during 1 October–14 November 2020
To reach the stability of variance, Box-Cox transformation was set zero. Besides, SARMA12 (3, 4) was fitted with the observed quadratic trend and seasonality of 12. The estimated COVID-19 cases during 18 February–30 September 2020, the predicted COVID-19 cases during 1 October–14 November 2020 (45 days), and ACF/PACF sample/model for Jahrom have been presented in Figure 1.
SARMA12 (3, 4) model was shown to be a good fit that can predict COVID-19 cases in Jahrom during October1st-November14th 2020.
3.2.2. COVID-19 Time Series Analysis in Shiraz during 1 October–14 November 2020
To reach the stability of variance, Box-Cox transformation was set zero. Besides, MA(8) was fitted with the observed quadratic trend. The estimated COVID-19 cases during 18 February–30 September 2020, the predicted COVID-19 cases during 1 October–14 November 2020 (45 days), and ACF/PACF sample/model for Shiraz have been presented in Figure 2.
3.3. Spatiotemporal Analysis
The results of the spatiotemporal analysis including 55 369 COVID-19 forecasted cases at the city level on a daily basis together with the results of subgroup analysis have been presented in Table 6.
Table 6.
The spatiotemporal outbreaks of the 55 369 COVID-19 cases in Fars province, Iran during 1 October–14 November 2020.
| Cluster | Location | Radius (km) | Canon | Start date | End date | Test statistic | Critical value† | P ∗ |
|---|---|---|---|---|---|---|---|---|
| Cluster 1 (MLC) | Ghirkarzin, Jahrom, Firoozabad, Khonj, Fasa, Farashband, Sarvestan, Lamerd, Kharame, Stahban | 135.53 | Jahrom | 2020/10/02 | 2020/10/22 | 223.31 | 8.70 | <0.001 |
| Cluster 2 | Sepidan | 0 | Sepidan | 2020/11/03 | 2020/11/13 | 185.98 | 8.70 | <0.001 |
| Cluster 3 | Fasa | 0 | Fasa | 2020/11/03 | 2020/11/13 | 117.80 | 8.70 | <0.001 |
| Cluster 4 | Lar, Zarindasht | 76.03 | Lar | 2020/10/21 | 2020/10/21 | 32.28 | 8.70 | <0.001 |
|
| ||||||||
| Sub-cluster results | ||||||||
| Cluster 4 | Zarrindasht | 0 | Zarrindasht | 2020/10/06 | 2020/10/09 | 3.23 | 3.08 | 0.04 |
| Cluster 1 (MLC) | Fasa | 0 | Fasa | 2020/10/06 | 2020/10/09 | 6.30 | 6.90 | 0.09 |
| Ghirkarzin | 0 | Ghirkarzin | 2020/11/08 | 2020/11/09 | 3.44 | 6.90 | 0.90 | |
∗ Statistical significance was evaluated using Monte Carlo hypothesis testing at 0.05 significance level. †Standard Monte Carlo critical value. MLC, most likely cluster.
Overall, four outbreaks occurred in Fars province during 1 October–14 November 2020, the largest and longest of which occurred in ten cities during 2–22 October, with Jahrom being the canon (MLC). Outbreaks 2 and 3 occurred in Sepidan and Fasa, respectively, and lasted from 3 to 13 November. Besides, outbreak 4 occurred in Lar and Zarrindasht on 21 October. The results of subgroup analysis demonstrated that Zarrindasht was the canon of the epidemic on 6 October.
As a part of EWS, the progression of COVID-19 epidemic resulting from spatiotemporal outbreaks of the 55 369 COVID-19 cases in Fars province, Iran, during 1 October–14 November 2020 has been shown in Figure 3.
Figure 3.
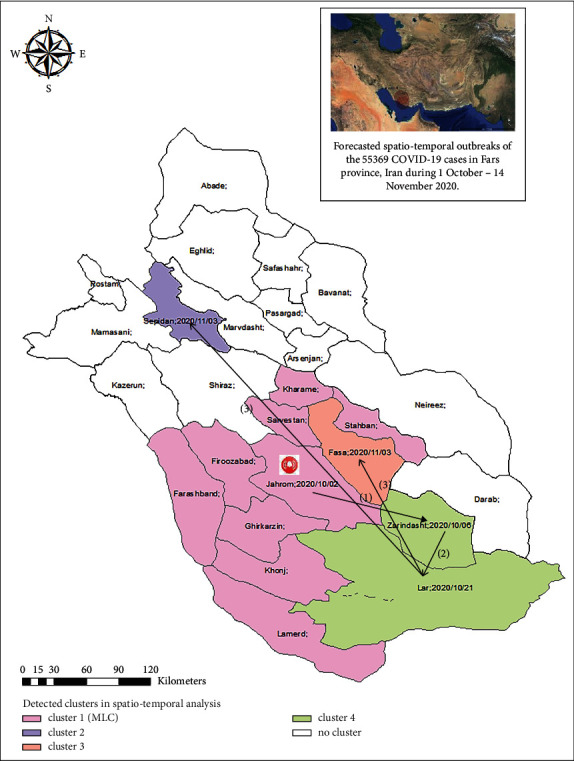
The progression of COVID-19 epidemic in Fars province, Iran during 1 October–14 November 2020.
COVID-19 epidemic inclined to trigger from Jahrom on 2 October, involving the MLC districts in the center, west, and south parts of the province; then, it showed a tendency to move towards the south in Zarrindasht on 6 October. Afterwards, it progressed to the southernmost part in Lar on 21 October. Afterwards, it simultaneously progressed to Fasa and Sepidan in the central and northwestern parts of the province, respectively on 3 November. This progression alarmed the policy makers and health managers on the potential starting point of ongoing epidemic (Jahrom city on 2 October, 2020) and, also, on when and where to prioritize the medical facilities and staffs; moreover, it could intervene with the transmission chain of COVID-19 in the area.
3.4. Validation Study
59 980 cases were recorded during 1 October to 14 November 2020, from which 64.2%, 5.7%, 5.5%, 2.2%, 2.2%, 2.1%, 2%, 2%, 1.8%, 1.4%, 1.2%, 1.2%, 1.1%, 1%, 1%, 0.9%, 0.8%, 0.7%, 0.7%, 0.4%, 0.4%, 0.4%, 0.4%, 0.3%, 0.3%, and 0.2% occurred in Shiraz, Jahrom, Fasa, Kazerun, Lar, Abade, Froozabad, Neireez, Sepidan, Eghlid, Stahban, Mamasani, Darab, Kharame, Pasargad, Safashahr, Bavanat, Arsenjan, Rostam, Sarvestan, Zarindasht, Farashband, Marvdasht, Ghirkazin, Lamerd, and Khonj, respectively. In addition, 51.2% and 48.8% of the cases occurred in October and September, respectively. The space-time outbreaks of the 59 980 COVID-19 incidence cases accompanied by subcluster analysis in Fars province, Iran, during 1 October to 14 November 2020 are shown in Table 7.
Cluster 1:
Real data: an outbreak occurred in Zarindasht (canon), Darab, and Lar during 2020/11/8–2020/11/14. Forecasted in EWS: Zarindasht and Lar (canons) were predicted to get involved in an outbreak on 2020/10/21.
Cluster 2:
Real data: an outbreak occurred in Fasa during 2020/11/4–2020/11/14. Forecasted in EWS: Fasa was predicted to get involved in an outbreak on 2020/11/3.
Cluster 3:
Real data: an outbreak occurred in Sepidan during 2020/11/13–2020/11/14. Forecasted in EWS: Sepidan was predicted to get involved in an outbreak during 2020/11/3–2020/11/13.
Cluster 4:
Real data: an outbreak occurred in Firoozabad (canon), Farashband, Ghirkarzin, and Sarvestan during 2020/11/8–2020/11/14. Forecasted in EWS: Firoozabad, Farashband, Ghirkarzin, and Sarvestan were predicted to get involved in an outbreak during 2020/10/2–2020/10/22.
Table 7.
The space-time outbreaks of the 59 980 COVID-19 incidence cases accompanied by subcluster analysis in Fars province, Iran, during 1 October–14 November 2020.
| Cluster | Location | Radius (km) | Canon | Start date | End date | Test statistic | Critical value† | P value∗ |
|---|---|---|---|---|---|---|---|---|
| Cluster1 | Zarindasht, Darab, Lar | 76.03 | Zarindasht | 2020/11/8 | 2020/11/14 | 189.3 | 5.6 | <0.001 |
| Cluster2 | Fasa | 0 | Fasa | 2020/11/4 | 2020/11/14 | 138.18 | 5.6 | <0.001 |
| Cluster3 | Sepidan | 0 | Sepidan | 2020/11/13 | 2020/11/14 | 55.54 | 5.6 | <0.001 |
| Cluster4 | Firoozabad, Farashband, Ghirkarzin, Sarvestan | 79.70 | Firoozabad | 2020/11/8 | 2020/11/14 | 36.89 | 5.6 | <0.001 |
| Cluster5 | Bavanat | 0 | Bavanat | 2020/11/11 | 2020/11/14 | 21.47 | 5.6 | <0.001 |
| Cluster6 | Rostam | 0 | Rostam | 2020/11/1 | 2020/11/14 | 17.86 | 5.6 | <0.001 |
| Cluster7 | Jahrom | 0 | Jahrom | 2020/10/27 | 2020/11/14 | 6.72 | 5.6 | 0.02 |
|
| ||||||||
| Sub-cluster results | ||||||||
| Cluster1 | Darab, Zarindasht | 46.57 | Darab | 2020/11/14 | 2020/11/14 | 6.08 | 2.4 | <0.001 |
| Zarrindasht | 0 | Zarindasht | 2020/11/5 | 2020/11/14 | 2.1 | 1.53 | 0.01 | |
| Cluster4 | Ghirkarzin | 0 | Ghirkarzin | 2020/11/7 | 2020/11/14 | 18.23 | 2.71 | <0.001 |
| Sarvestan | 0 | Sarvestan | 2020/10/31 | 2020/11/14 | 14.58 | 2.71 | <0.001 | |
| Farashband | 0 | Farashband | 2020/11/8 | 2020/11/14 | 14.25 | 2.71 | <0.001 | |
| Firoozabad | 0 | Firoozabad | 2020/11/14 | 2020/11/14 | 7.61 | 2.71 | <0.001 | |
∗ Statistical significance was evaluated using Monte Carlo hypothesis testing at 0.05 significance level. †Standard Monte Carlo critical value. MLC, most likely cluster.
However, Bavanat (2020/11/11–2020/11/14), Rostam (2020/11/1–2020/11/14), and Darab (2020/11/14), which were detected in validation study, were not predicted in EWS outbreaks.
The prediction of the epidemic consideration: real data showed that the epidemic was started from Jahrom on 2020/10/27 and progressed to Sarvestan, Rostam, Fasa, Zarindasht, Ghirkarzin, Farashband, Bavanat, Sepidan, Firoozabad and Darab; moreover, in EWS, the epidemic was triggered from Jahrom on 2020/10/2 and progressed to MLC districts, Zarindasht, Lar, Fasa, and Sepidan.
A total of 12 cities got involved in the outbreaks during 1 October–14 November 2020 using real data, nine of which were predicted in the EWS. The EWS predicted time periods were either the same as the observed periods or occurred earlier. Based on the validation study findings, the results of EWS were shown to be valid enough for our purpose.
4. Discussion
Covid-19 cases did not distributed uniformly across cities and times during 18 February–30 September 2020 in Fars province, Iran. In this protocol presented as EWS, 55 369 COVID-19 cases were forecasted from the 53 554 daily basis city-level time series data recorded during 18 February–30 September 2020. The forecasted cases resulted in the prediction of four significant spatiotemporal outbreaks of COVID-19 in the area including the MLC and three other clusters. The MLC, which was also the largest and the longest cluster, included ten cities during 2–22 October, with Jahrom being the canon. This finding, schematically presented as the progression of the epidemic during 1 October–14 November 2020, revealed that the epidemic inclined to trigger from Jahrom on 2 October, involving the MLC districts in the center, west, and south parts of the province; then, it showed a tendency to move towards the south in Zarrindasht on 6 October. Afterwards, it progressed to the southernmost part in Lar on 21 October. Later, it simultaneously progressed to Fasa and Sepidan in the central and northwestern parts of the province, respectively on 3 November. To the best of our knowledge, this protocol alarmed the surveillance system to focus on the progression of the epidemic and especially on the starting point, Jahrom, so that proper interventions could be done on the transmission chain of SARS-CoV-2 resulting in lower prevalence of the disease in the area. Moreover, the surveillance system can dedicate more test sites, medical resources, community quarantine, vaccination coverage, and travel bans on more at risk locations at specified detected times.
Despite being the most important transmission line of the disease, it should be explained why the most populated city, Shiraz, with the maximum number of COVID-19 cases was not included in the predicted outbreaks. Shiraz was ignored in permutation scan statistics analysis, in which clusters are ranked by their likelihood ratio scores rather than by the population density or the area size. This can be attributed to the fact that, with the capital city of Fars province being with advanced medical services, many patients from nearby cities travel to Shiraz to receive health services, thereby transmitting the infection to other cities of the province.
Time series models based on statistical methods were revealed to fit well for forecasting time dependent data. They presented well-fit results in case of the COVID-19 epidemic as well. For instance, the AR model could forecast the confirmed COVID-19 cases in Iran [10]. Additionally, ARIMA-based time series analysis forecasting near future could assist the government to be prepared for the upcoming emergencies of the current epidemic in India [11–14]. In another study, ARIMA models were fitted to forecast short-term COVID-19 cases in the five most affected countries of the world including Italy, Spain, France, the United Kingdom, and the United States of America [15].
Space-time permutation scan statistics could well manage space-time dependent variables through informing the policymakers on the progression of the epidemic in China [16–18]. In the United States, the space-time outbreaks of COVID-19 were detected during two periods of time (22 January to 9 March 2020 and 18 January to 27 March 2020) using space-time statistics [19]. Space-time scan statistics also resulted in the detection of space-time outbreaks in Hong Kong during 23 January–14 April 2020 [20]. Furthermore, space-time permutation scan statistics worked better for predicting highly frequent infectious diseases with high transmission rates [8, 21, 22]. Another study employed almost the same protocol as the one used to establish the EWS in the current study in order to detect the vulnerable districts for COVID-19 in India. Using ARIMA time series model for forecasting short-term COVID-19 cases and performing spatial analysis, they indicated that the western and southern parts of the country were highly vulnerable for COVID-19 infection [23, 24].
EWS establishment could effectively warn the surveillance system about the progression of the epidemic in Fars province, Iran. However, it had several limitations, which might have affected the results. Firstly, the surveillance system of infectious diseases in Iran is a passive system, and many SARS-CoV-2 infected people with mild to moderate symptoms did not refer to health centers and were not diagnosed. Therefore, the forecasted cases of COVID-19 might have been underestimated. This could affect estimating the required health services and medical facilities. Secondly, permutation scan statistics discover more than one cluster in a given area and time. Hence, interpretation of the results could be somewhat difficult, especially when incorporating main-cluster and subcluster results. Regarding clinical and statistical significance, prioritizing the clusters can also be difficult. Another limitation of scan statistics is that it uses a circular scan window. Thus, the precise limits of the detected clusters remain uncertain. In other words, either the site within the detected clusters may not have a high risk, or a high risk site may be out of the cluster. Yet, using subgroup analysis for detecting significant clusters within large clusters could solve the problem. Further investigations are suggested to assess the genetic variation and meteorological risk factors of COVID-19 in the long run.
5. Conclusion
The EWS presented here would alarm the policy makers and health managers on the potential starting point of ongoing epidemic and, also, on when and where to prioritize the medical facilities and staffs. The MLC, which was also the largest and the longest cluster, included ten cities during 2–22 October, with Jahrom being the canon. This finding revealed that the epidemic inclined to trigger from Jahrom on 2 October, involving the MLC districts in the center, west, and south parts of the province; then, it showed a tendency to move towards the south in Zarrindasht on 6 October. Afterwards, it progressed to the southernmost part in Lar on 21 October. Afterwards, it simultaneously progressed to Fasa and Sepidan in the central and northwestern parts of the province, respectively, on 3 November. To the best of our knowledge, this protocol alarmed the surveillance system to focus on the starting point and progression of the epidemic as well as to dedicate more test sites, medical resources, stricter quarantine, vaccination coverage, and travel bans on more at risk locations at specified times. These findings can be used to tailor province-level policies to servile the ongoing epidemic in the area; however, governmental level effort is needed to control the epidemic at a larger scale in the future.
Acknowledgments
The authors would like to thank Ms. A. Keivanshekouh at the Research Consultation Center (RCC) of the Shiraz University of Medical Sciences for improving the use of English in the manuscript. As a purely academic work, this study did not receive any funds and is scientifically supported by research deputy of Shiraz University of Medical Sciences, Shiraz, Iran.
Data Availability
The health service data used to support the findings of this study are restricted by the Ethics Committee of the Ministry of Health, Treatment, and Medical Education affiliated to the Shiraz University of Medical Sciences, Shiraz, Iran, in order to protect patient privacy. Data are available from Marjan Zare, Central Building of Shiraz University of Medical Sciences, Vice Chancellor for Health, fourth floor, Zand St, Shiraz, Iran, telephone number: 07132122368, E-mail: marjan.zare@gmail.com, for researchers who meet the criteria for access to confidential data.
Ethical Approval
All ethical steps including data collection and analysis as well as reporting the results were in accordance with the standards approved by the Ethic Committee of the Ministry of Health, Treatment, and Medical Education (IR.SUMS.REC.1399.574). The work processes were anonymous, and the results were reported to the study participants.
Conflicts of Interest
No conflicts of interest were declared by the authors.
References
- 1.Zare M., Rezaianzadeh A., Tabatabaee H., Faramarzi H., Aliakbarpour M., Ebrahimi M. Establishment of an early warning system for cutaneous leishmaniasis in Fars province, Iran. Asian Pacific Journal of Tropical Biomedicine . 2019;9(6):232–239. [Google Scholar]
- 2.Chaves L. F., Pascual M. Comparing models for early warning systems of neglected tropical diseases. PLoS Neglected Tropical Diseases . 2007;1(1):p. e33. doi: 10.1371/journal.pntd.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasraeian M., Zare M., Vafaei H., et al. COVID-19 pneumonia and pregnancy; a systematic review and meta-analysis. Journal of Maternal-Fetal and Neonatal Medicine . 2020;35:1–8. doi: 10.1080/14767058.2020.1763952. [DOI] [PubMed] [Google Scholar]
- 4.Semati A., Zare M., Mirahmadizadeh A., Hemmati A., Ebrahimi M. Epidemiological study of infection and death due to COVID-19 in Fars province, Iran, from February to September 2020. Iranian Journal of Medical Sciences . 2021;47 doi: 10.30476/IJMS.2021.90768.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laine M. Geodetic Time Series Analysis in Earth Sciences . Berlin, Germany: Springer; 2020. Introduction to dynamic linear models for time series analysis; pp. 139–156. [Google Scholar]
- 6.Kulldorff M. SaTScan—Software for the Spatial, Temporal, and Space-Time Scan Statistics . Boston, MA, USA: Harvard Medical School and Harvard Pilgrim Health Care; 2015. [Google Scholar]
- 7.Zare M., Rezaianzadeh A., Tabatabaee H., Faramarzi H., Aliakbarpour M., Ebrahimi M. Determining endemic values of cutaneous leishmaniasis in Iranian Fars province by retrospectively detected clusters and receiver operating characteristic curve analysis. Asian Pacific Journal of Tropical Biomedicine . 2019;9(9):359–364. doi: 10.4103/2221-1691.267636. [DOI] [Google Scholar]
- 8.Zare M., Rezaianzadeh A., Tabatabaee H., Aliakbarpoor M., Faramarzi H., Ebrahimi M. Spatiotemporal clustering of cutaneous leishmaniasis in Fars province, Iran. Asian Pacific Journal of Tropical Biomedicine . 2017;7(10):862–869. doi: 10.1016/j.apjtb.2017.09.011. [DOI] [Google Scholar]
- 9.Jake Esprabens A. A., Kim J. A Short Guide to Time Series Analysis . 2020. https://bookdown.org/JakeEsprabens/431-Time-Series/introduction-to-time-series.html . [Google Scholar]
- 10.Maleki M., Mahmoudi M. R., Wraith D., Pho K.-H. Time series modelling to forecast the confirmed and recovered cases of COVID-19. Travel Medicine and Infectious Disease . 2020;37 doi: 10.1016/j.tmaid.2020.101742.101742 [DOI] [PubMed] [Google Scholar]
- 11.Tandon H., Ranjan P., Chakraborty T., Suhag V. Coronavirus (COVID-19): ARIMA based time-series analysis to forecast near future. 2020. https://arxiv.org/abs/200407859 .
- 12.Khan F. M., Gupta R. ARIMA and NAR based prediction model for time series analysis of COVID-19 cases in India. Journal of Safety Science and Resilience . 2020;1(1):12–18. doi: 10.1016/j.jnlssr.2020.06.007. [DOI] [Google Scholar]
- 13.Mousavi M., Salgotra R., Holloway D., Gandomi A. H. COVID-19 time series forecast using transmission rate and meteorological parameters as features. IEEE Computational Intelligence Magazine . 2020;15(4):34–50. doi: 10.1109/mci.2020.3019895. [DOI] [Google Scholar]
- 14.Fang L., Wang D., Pan G. Analysis and estimation of COVID-19 spreading in Russia based on ARIMA model. SN Comprehensive Clinical Medicine . 2020;2(12):2521–2527. doi: 10.1007/s42399-020-00555-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh S., Parmar K. S., Makkhan S. J. S., Kaur J., Peshoria S., Kumar J. Study of ARIMA and least square support vector machine (LS-SVM) models for the prediction of SARS-CoV-2 confirmed cases in the most affected countries. Chaos, Solitons & Fractals . 2020;139 doi: 10.1016/j.chaos.2020.110086.110086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mo C., Tan D., Mai T., et al. An analysis of spatiotemporal pattern for COIVD‐19 in China based on space‐time cube. Journal of Medical Virology . 2020;92(9):1587–1595. doi: 10.1002/jmv.25834. [DOI] [PubMed] [Google Scholar]
- 17.Yang W., Deng M., Li C., Huang J. Spatio-temporal patterns of the 2019-nCoV epidemic at the county level in Hubei province, China. International Journal of Environmental Research and Public Health . 2020;17(7):p. 2563. doi: 10.3390/ijerph17072563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye L., Hu L. Spatiotemporal distribution and trend of COVID-19 in the Yangtze river delta region of the people’s Republic of China. Geospatial Health . 2020;15(1) doi: 10.4081/gh.2020.889. [DOI] [PubMed] [Google Scholar]
- 19.Desjardins M., Hohl A., Delmelle E. Rapid surveillance of COVID-19 in the United States using a prospective space-time scan statistic: detecting and evaluating emerging clusters. Applied Geography . 2020;118 doi: 10.1016/j.apgeog.2020.102202.102202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kan Z., Kwan M.-P., Wong M. S., Huang J., Liu D. Identifying the space-time patterns of COVID-19 risk and their associations with different built environment features in Hong Kong. The Science of the Total Environment . 2021;772 doi: 10.1016/j.scitotenv.2021.145379.145379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rezaianzadeh A., Zare M., Aliakbarpoor M., Faramarzi H., Ebrahimi M. Space-time cluster analysis of malaria in Fars province-Iran. International Journal of Infection . 2020;7(3) doi: 10.5812/iji.107238.e107238 [DOI] [Google Scholar]
- 22.Rezaianzadeh A., Zare M., Tabatabaee H., Ali-Akbarpour M., Faramarzi H., Ebrahimi M. Does prospective permutation scan statistics work well with cutaneous leishmaniais as a high-frequency or malaria as a low-frequency infection in Fars province, Iran? Asian Pacific Journal of Tropical Biomedicine . 2018;8(10):478–484. doi: 10.4103/2221-1691.244138. [DOI] [Google Scholar]
- 23.Roy S., Bhunia G. S., Shit P. K. Spatial prediction of COVID-19 epidemic using ARIMA techniques in India. Modeling Earth Systems and Environment . 2021;7(2):1385–1391. doi: 10.1007/s40808-020-00890-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman M. R., Islam A. H. M. H., Islam M. N. Geospatial modelling on the spread and dynamics of 154 day outbreak of the novel coronavirus (COVID-19) pandemic in Bangladesh towards vulnerability zoning and management approaches. Modeling Earth Systems and Environment . 2020;7(3):2059–2087. doi: 10.1007/s40808-020-00962-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The health service data used to support the findings of this study are restricted by the Ethics Committee of the Ministry of Health, Treatment, and Medical Education affiliated to the Shiraz University of Medical Sciences, Shiraz, Iran, in order to protect patient privacy. Data are available from Marjan Zare, Central Building of Shiraz University of Medical Sciences, Vice Chancellor for Health, fourth floor, Zand St, Shiraz, Iran, telephone number: 07132122368, E-mail: marjan.zare@gmail.com, for researchers who meet the criteria for access to confidential data.


