Abstract
The soluble MMO (sMMO) gene clusters from group I methanotrophs were characterized. An 8.1-kb KpnI fragment from Methylomonas sp. strain KSWIII and a 7.5-kb SalI fragment from Methylomonas sp. strain KSPIII which contained the sMMO gene clusters were cloned and sequenced. The sequences of these two fragments were almost identical. The sMMO gene clusters in the fragment consisted of six open reading frames which were 52 to 79% similar to the corresponding genes of previously described sMMO gene clusters of the group II and group X methanotrophs. The phylogenetic analysis of the predicted amino acid sequences of sMMO demonstrated that the sMMOs from these strains were closer to that from M. capsulatus Bath in the group X methanotrophs than to those from Methylosinus trichosporium OB3b and Methylocystis sp. strain M in the group II methanotrophs. Based on the sequence data of sMMO genes of our strains and other methanotrophs, we designed a new PCR primer to amplify sMMO gene fragments of all the known methanotrophs harboring the mmoX gene. The primer set was successfully used for detecting methanotrophs in the groundwater of trichloroethylene-contaminated sites during in situ-biostimulation treatments.
Trichloroethylene (TCE) is one of the most common contaminants in the soil environment and groundwater in many countries. Several methane-oxidizing bacteria efficiently degrade the contaminant, and a number of investigations of biological removal of TCE from contaminated soil by using methanotrophs have been reported (15, 22). The enzyme responsible for the biodegradation of TCE is methane monooxygenase (MMO), which catalyzes the oxidation of methane to methanol. Two distinct types of MMOs are known: membrane-bound particulate MMO, present in all methanotrophs (43, 54), and soluble MMO (sMMO), which has been found in only several species of methanotrophs (12, 30, 36, 47). Both types of MMO can degrade TCE, but sMMO degrades it at a very high rate compared with particulate MMO (50). Therefore, sMMO has received special attention in the bioremediation of TCE.
Methanotrophs are taxonomically classified into three groups: group I, group II, and group X (22). The extensively characterized sMMO proteins are those purified from group II methanotrophs, Methylosinus trichosporium OB3b (3, 17, 18) and Methylocystis sp. strain M (37), and a group X methanotroph, Methylococcus capsulatus Bath (8, 20, 53). The enzyme complexes of these three strains have very similar properties and consist of three components: a hydroxylase component (MMOH), a reductase component (MMOR), and a regulatory protein B (MMOB) (7). The X-ray crystal structures of the hydroxylase components from M. trichosporium OB3b and M. capsulatus Bath have also been reported (16, 40). The DNA sequence of the gene cluster that codes for the sMMO proteins has been determined for the three methanotrophs (5, 6, 34, 45, 46). In each strain, six genes, mmoX, mmoY, mmoB, mmoZ, orfY, and mmoC, were detected (5, 6, 34, 45, 46). MMOH is encoded by mmoX, mmoY, and mmoZ genes. MMOR and MMOB are encoded by mmoC and mmoB, respectively.
On the other hand, there is little published information on the properties of sMMO in group I methanotrophs. The presence of sMMO protein has been demonstrated in two strains, Methylomonas methanica 68-1 and Methylomonas sp. strain GYJ3 (26, 44). From strain GYJ3, the hydroxylase component and the reductase component of sMMO were purified, and the regulatory protein B was partially purified (44). Fuse et al. reported the partial sequence of mmoX from Methylomicrobium sp., which belongs to group I (19). However, the complete DNA sequence of an sMMO gene cluster from the group I methanotrophs has not been reported.
Recently, two TCE-degrading methanotrophic strains, KSWIII and KSPIII, from a TCE-contaminated field were isolated in our laboratory (21). Phylogenetic analysis based on 16S rRNA sequences suggested that they were affiliated with the genus Methylomonas of group I methanotrophs (21). In this report, we characterize the strains in terms of morphological and chemotaxonomic aspects and analyze the sMMO genes from the strains. This is the first report on characterization of an sMMO gene cluster from the group I methanotrophs. We also designed a gene probe for sMMO genes based on the sequence data and tested its validity for detection of methanotrophs in an aquifer during in situ-biostimulation treatments.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The methanotrophic strains KSWIII and KSPIII were collected from the site at Kururi (Kimitsu, Chiba Prefecture, Japan), which is contaminated with TCE, and isolated in the previous study (21). The reference strains M. trichosporium OB3b (ATCC 35070) and M. capsulatus Bath (ATCC 33009) were obtained from the American Type Culture Collection (Manassas, Va.). Methlocystis sp. strain M was a kind gift from H. Uchiyama, National Institute for Environmental Studies, Tsukuba, Japan. The strains were grown on NMS medium (51) with gentle agitation (100 rpm) at 30°C (strains KSWIII, KSPIII, OB3b, and M) and at 37°C (strain Bath) under a methane-containing air atmosphere (air methane ratio, 8:2).
Taxonomic studies.
Cell morphology was examined by phase-contrast microscopy and transmission electron microscopy. For transmission electron microscopy, a centrifuged cell pellet was fixed with 5% (vol/vol) glutaraldehyde and 1% (vol/vol) osmium tetroxide. Ultrathin sections of the sample embedded in epoxy resin (28) were prepared with a Reinchert ultramicrotome. Samples were stained with uranyl acetate and lead citrate and examined with a Hitachi H-7000 transmission electron microscope. In vivo absorption spectra were measured in cell extracts with a Beckman DU 640 spectrophotometer after disruption by sonication (100 W; 3 min). Quinones were extracted from the cells with chloroform-methanol (2:1 [vol/vol]) and analyzed by reverse-phase high-performance liquid chromatography (Beckman System Gold), as previously described (48), with ubiquinone standards, including 18-methylene-ubiquinone-8 extracted from methanotrophically grown cells of M. capsulatus Bath (9). The G+C content of the total DNA was measured according to the method described previously (24). The extracted total DNAs were digested with P1 nuclease and alkaline phosphatase with a Yamasa GC Kit (Yamasa Shoyu Co., Choshi, Japan) and analyzed by high-performance liquid chromatography (Shimadzu LC-9A with a CLC-ODS column).
Probes for the sMMO gene.
Two probes for detecting mmoX and mmoC genes were generated by PCR with the primer sets described by Miguez et al. (35). A 370-bp fragment of the mmoX gene amplified from Methylomonas sp. strain KSWIII with the mmoX-specific primers mmoX1 and mmoX2 (35) was used as the mmoX-specific probe. A 890-bp fragment of the mmoC gene amplified from M. capsulatus Bath with primers mmoC1 and mmoC2 (35) was used as the mmoC-specific probe. Amplification reactions were performed with the reagents supplied with AmpliTaq Gold (Perkin-Elmer Applied Biosystems, Foster City, Calif.) at a magnesium ion concentration of 2 mM, with 500 ng to 1 μg of DNA and 40 pmol of each primer. The reactions were carried out with a TaKaRa PCR Thermal Cycler MP (Takara Shuzo, Kyoto, Japan) with preincubation at 95°C for 9 min and 35 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 2 min. The PCR products were purified with Microspin S-200 HR columns (Amersham Pharmacia Biotech, Uppsala, Sweden).
Cloning and sequencing of sMMO gene clusters.
The total DNAs of methanotrophic strains were isolated with the Qiagen (Hilden, Germany) genomic DNA buffer set and Qiagen Genomic-Tip 20/G. The KpnI-digested total DNA of strain KSWIII was analyzed by Southern hybridization with the digoxigenin (DIG)-labeled mmoX- or mmoC-specific probe. DNA labeling was carried out with a DIG DNA labeling and detection kit (Boehringer Mannheim GmbH, Mannheim, Germany). Hybridizations were carried out by the standard methods (42) with DIG Easy Hyb (Boehringer Mannheim) at 42°C. Washing of membrane filters after hybridization was carried out at 68°C. An 8.1-kb fragment was detected with both the mmoX- and mmoC-specific probes. Restriction fragments were ligated into ZAP Express vector (Stratagene, La Jolla, Calif.). The ligated DNA was packaged in bacteriophage particles with a Gigapack III Gold (Stratagene) and transfected into Escherichia coli XL1-Blue MRF′. The clone library was then probed with the DIG-labeled mmoX-specific probe to identify the 8.1-kb KpnI fragment. The fragment was sequenced by the primer-walking method with an ABI 377 automated DNA sequencer (Perkin-Elmer Applied Biosystems) by cycle sequencing with a dRhodamine terminator cycle sequencing FS ready-reaction kit (Perkin-Elmer Applied Biosystems). The 7.5-kb SalI fragment containing sMMO genes of strain KSPIII was also cloned and sequenced as described above.
Sequence analysis.
DNA and deduced amino acid sequences were analyzed with the GENETYX-WIN (version 3.1.0) software package (Software Development Co. Ltd., Tokyo, Japan). Searching for homologous proteins was done with the BLAST (version 2.0) program (1). Multiple alignments were run under the Clustal W (version 1.6) program (49). Secondary-structure predictions were made with the SIMPA96 program (29) based on the nearest-neighbor method. Phylogenetic trees were constructed by the neighbor-joining method (41) with the MEGA program (27). The reference DNA sequences used in the comparison were retrieved from the DDBJ, EMBL, and GenBank nucleotide sequence databases, and the reference protein sequences were retrieved from the SWISS-PROT database.
Field treatments of the in situ-biostimulation experiment.
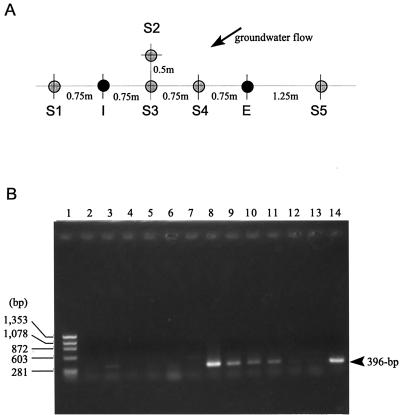
The field treatments for in situ biostimulation were carried out at the Kururi test site. The TCE concentration in groundwater at the test site was approximately 200 μg/liter. The groundwater was continuously pumped from the extraction well at an extraction rate of 1.5 liters/min and injected into the injection well (for the locations of the injection, sampling, and extraction wells at the test site, see Fig. 5A). At first, to survey the movement of the groundwater during the treatments, bromide was continuously added to the extracted water and injected. The concentration of bromide was measured after 10 days of circulation of the groundwater. Then the biostimulation was started on 25 September 1998 by the addition of (i) methane to a dissolved concentration of 10 mg/liter, (ii) oxygen to a dissolved concentration of 30 mg/liter, and (iii) KNO3 (18 mg/liter) and KH2PO4 (15 mg/liter) to serve as nutrient salts. The addition was repeatedly carried out in the following cycle: addition of methane, 190 min; no addition, 50 min; addition of oxygen and nutrient salts, 190 min; and no addition, 50 min.
FIG. 5.
(A) Location of the injection (I), sampling (S1 to S5), and extraction (E) wells at Kururi test site. The direction of groundwater flow is indicated by the arrow. (B) Amplification of mmoX from total DNA extracted from groundwater samples. Lanes: 1, size marker (HaeIII-digested φX174 DNA); 2 to 7, PCR product from the groundwater at the S1, S2, S3, S4, E, and S5 wells, respectively, collected on 24 September 1998; 8 to 13, PCR product from the groundwater at the S1, S2, S3, S4, E, and S5 wells, respectively, collected on 5 October 1998; 14, PCR product from 100 pg of purified total DNA of M. trichosporium OB3b.
PCR amplification of mmoX gene from total DNA extracted from groundwater samples.
Groundwater samples were collected 1 day before the start of the biostimulation treatments (24 September) and 10 days after the start (5 October) at the five sampling wells (S1, S2, S3, S4, and S5) and at the extraction well (E). Groundwater samples (200 ml) were then filtered with a 0.2-μm-pore-size Isopore membrane filter (Millipore Corporation, Bedford, Mass.) to collect bacteria. The DNA from bacteria collected with the filter was extracted with the FastDNA SPIN kit (for soil) (Bio101 Inc., Vista, Calif.). The DNA sample was then dissolved in 100 μl of Tris-EDTA, and 1 μl was used for PCR. The amounts of DNA samples which were used for amplification were 23.4, 12.4, 23.8, 11.7, and 31.7 ng for the DNA samples collected from wells S1, S2, S3, S4, E, and S5, respectively, on 24 September, and 26.2, 14.4, 15.0, 11.1, 12.4, and 14.9 ng for the samples collected from wells S1, S2, S3, S4, E, and S5, respectively on 5 October. The PCR was carried out with a combination of primer mmoXr901 (5′-TGGGTSAARACSTGGAACCGCTGGGT-3′; nucleotide positions 926 to 901 from the beginning of mmoX), which was newly designed based on the sequence data including the mmoX genes from strains KSWIII and KSPIII, and primer mmoX1 (35). Amplification reactions were performed as described above with one modification: the annealing temperature was altered to 60°C.
Nucleotide sequence accession numbers.
The complete sequence of the sMMO gene cluster of Methylomonas sp. strains KSWIII and KSPIII will appear in the DDBJ, EMBL, and GenBank databases with accession no. AB025022 and AB025021.
RESULTS AND DISCUSSION
Classification of strains KSWIII and KSPIII.
The phylogenetic analysis based on 16S rRNA sequences revealed that strains KSWIII and KSPIII were almost identical, with a sequence similarity of 99.2%, and that both strains were closely related to M. methanica, with sequence similarities of 97.8 and 98.1%, respectively (21). It suggested that the isolates belonged to the genus Methylomonas.
Several findings in this study support the suggestion. The cells of the strains were short rods that were 0.8 to 1.1 μm wide and 1.3 to 2.0 μm long. The cultures of the strains were pink. Ultrasonically disrupted cells of both strains had three absorption maxima, at 474, 504, and 539 nm, which reflected the presence of carotenoids. The possession of carotenoids is a distinctive feature of species in the genus Methylomonas and is rarely observed in other species of methanotrophs (2). Ultrathin-section electron microscopy showed that the cells grown on NMS medium in the presence of methane and 1 mM CuSO4 possessed bundle-like intracytoplasmic membrane structure, which is typical of the group I methanotrophs (52). Strain KSWIII predominantly contained an 18-methylene-ubiquinone-8, which has been found in several members of the group I methanotrophs, e.g., M. methanica, M. capsulatus, and Methylocaldum gracile (9). Strain KSWIII had 52.0 mol% G+C content, which was within the range of the previously described DNA base composition in M. methanica (51 to 53%) (2). On the basis of these data, we decided to designate the strains KSWIII and KSPIII Methylomonas sp.
Cloning and sequencing of sMMO gene clusters in strains KSWIII and KSPIII.
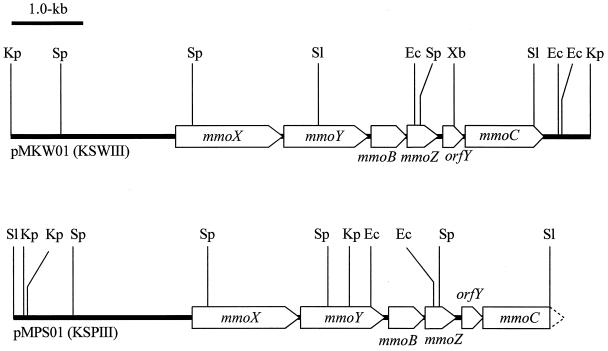
An 8,107-bp KpnI fragment from strain KSWIII and a 7,531-bp SalI fragment from strain KSPIII were cloned and sequenced. Six open reading frames which showed high homology to mmoX, mmoY, mmoB, mmoZ, orfY, and mmoC (partial in the fragment from strain KSPIII) were found in each fragment (Fig. 1). The deduced amino acid sequences of mmo gene products from the two strains were almost identical (only one amino acid mismatch was found in MmoC) (Table 1).
FIG. 1.
Restriction maps of the mmo gene clusters. The upper line shows the cloned 8.1-kb KpnI fragment from strain KSWIII. The lower line shows the cloned 7.5-kb SalI fragment from strain KSPIII. The open boxes represent the mmoX, -Y, -Z, -B, orfY, and mmoC genes. The locations of restriction sites are marked as follows: S1, SalI; Kp, KpnI; Sp, SphI; Ec, EcoRI; Xb, XbaI.
TABLE 1.
Identity matrix comparing DNA sequences and derived amino acid sequences of mmo genesa
| Sequence and source | % Identity
|
||||||
|---|---|---|---|---|---|---|---|
| KSPIII | KSWIII | Bath | OB3b | M | Kb | Peatbog 24b | |
| mmoX/MmoX | |||||||
| KSPIII | 100.0 | 86.7 | 78.7 | 79.5 | 77.0 | 75.3 | |
| KSWIII | 96.6 | 86.7 | 78.7 | 79.5 | 77.0 | 75.3 | |
| M. capsulatus (Bath) | 78.5 | 78.7 | 81.6 | 82.3 | 79.9 | 77.0 | |
| M. trichosporium OB3b | 72.3 | 72.3 | 76.0 | 96.2 | 80.5 | 90.8 | |
| Methylocystis M | 72.0 | 71.9 | 76.8 | 92.1 | 79.9 | 93.7 | |
| Strains K, M131, S6b | 74.0 | 73.7 | 78.1 | 78.4 | 79.0 | 79.3 | |
| Peatbog bacterium 24b | 69.2 | 69.6 | 75.7 | 88.1 | 88.9 | 78.2 | |
| mmoY/MmoY | |||||||
| KSPIII | 100.0 | 56.3 | 57.2 | 61.0 | |||
| KSWIII | 96.5 | 56.3 | 57.2 | 61.0 | |||
| M. capsulatus (Bath) | 61.9 | 61.8 | 54.0 | 56.3 | |||
| M. trichosporium OB3b | 60.8 | 61.1 | 66.6 | 89.1 | |||
| Methylocystis M | 60.5 | 61.3 | 67.0 | 91.4 | |||
| mmoZ/MmoZ | |||||||
| KSPIII | 100.0 | 52.7 | 49.7 | 50.3 | |||
| KSWIII | 98.4 | 52.7 | 49.7 | 50.3 | |||
| M. capsulatus (Bath) | 60.0 | 60.6 | 48.8 | 50.6 | |||
| M. trichosporium OB3b | 57.1 | 56.9 | 62.6 | 87.0 | |||
| Methylocystis M | 58.1 | 58.4 | 63.8 | 86.9 | |||
| mmoB/MmoB | |||||||
| KSPIII | 100.0 | 70.0 | 70.3 | 70.3 | |||
| KSWIII | 99.3 | 70.0 | 70.3 | 70.3 | |||
| M. capsulatus (Bath) | 68.7 | 69.0 | 65.9 | 65.9 | |||
| M. trichosporium OB3b | 66.9 | 67.1 | 67.2 | 96.4 | |||
| Methylocystis M | 67.6 | 67.9 | 68.0 | 94.7 | |||
| orfY/OrfY | |||||||
| KSPIII | 100.0 | 38.1 | 34.7 | 37.7 | |||
| KSWIII | 97.7 | 38.1 | 34.7 | 37.7 | |||
| M. capsulatus (Bath) | 52.7 | 51.5 | 40.0 | 36.9 | |||
| M. trichosporium OB3b | 55.3 | 54.0 | 54.4 | 58.8 | |||
| Methylocystis M | 57.8 | 56.5 | 54.5 | 74.4 | |||
| mmoC/MmoC | |||||||
| KSPIII | 99.7 | 60.6 | 46.2 | 49.0 | |||
| KSWIII | 98.4 | 59.0 | 47.5 | 49.9 | |||
| M. capsulatus (Bath) | 61.8 | 61.2 | 48.7 | 51.3 | |||
| M. trichosporium OB3b | 55.2 | 55.8 | 58.9 | 80.5 | |||
| Methylocystis M | 54.4 | 55.3 | 60.6 | 80.8 | |||
DNA sequence identities (%) and amino acid sequence identities (percentages, in boldface type) were obtained by using the Clustal W (version 1.6) program.
The deduced amino acid sequences of MmoX, MmoY, MmoZ, and MmoC proteins from the two strains were aligned with those of Mmo proteins from M. capsulatus Bath (45, 46), M. trichosporium OB3b (5, 6), Methylocystis sp. strain M (34), and the partial sequences of MmoX from acidophilic methane-oxidizing strains K, M131, and S6 (14) and an uncultured bacterium (13) (see Fig. 3 and 4).
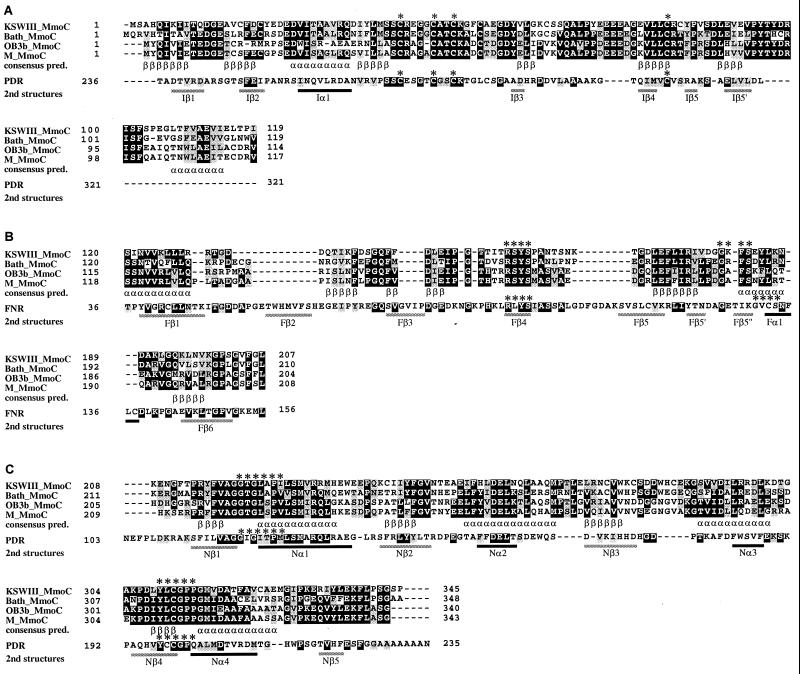
FIG. 3.
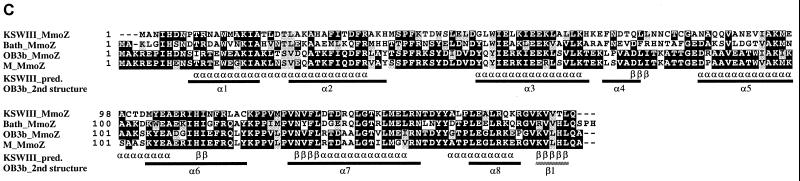
Alignments of the [2Fe-2S] domain (A), the [FAD] domain (B), and the [NAD] domain (C) of the deduced amino acid sequences of the mmoC genes from strain KSWIII, M. capsulatus Bath, M. trichosporium OB3b, and Methylocystis sp. strain M. The primary structures of the [2Fe-2S] domain (A) and the [NAD] domain (C) of PDR from Burkholderia cepacia (accession no. P33164) (10) and the [FAD] domain (B) of FNR from spinach (P00455) (25) are also aligned. Identical residues are in solid boxes, and similar residues are in shaded boxes. Underneath the aligned sequences, the secondary-structural features of PDR (A and C) or FNR (B) obtained in the crystal structure analyses are shown (2nd structures) as solid bars (α-helical regions [α]) and shaded bars (β-strand regions [β]), together with the consensus features of predicted β-strand (β) and α-helical regions (α) for the MmoCs (consensus pred.). The asterisks above the aligned sequences indicate the consensus sequences for FNR family proteins.
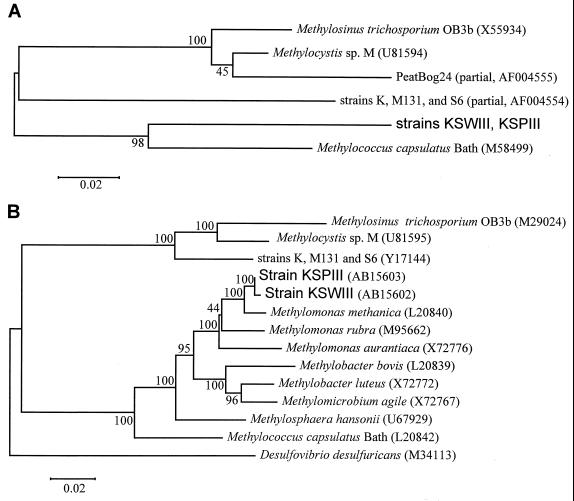
FIG. 4.
Phylogenetic relationship of the deduced amino acid sequences of the mmoX genes (A) and of the 16S ribosomal DNA (rDNA) sequences (B) among methanotrophs. The trees were constructed from evolutionary distances obtained by the neighbor-joining method (41). The bars represent 2 amino acid substitutions per 100 amino acids in MmoXs (A) and 2 nucleotide substitutions per 100 nucleotides in 16S rDNA sequences (B). The 16S rDNA sequence of Desulfovibrio desulfuricans that belongs to the δ subclass of Proteobacteria was used to root the tree (B). Bootstrap probabilities (27) are indicated at the branching points. The accession number of each reference sequence is shown in parentheses.
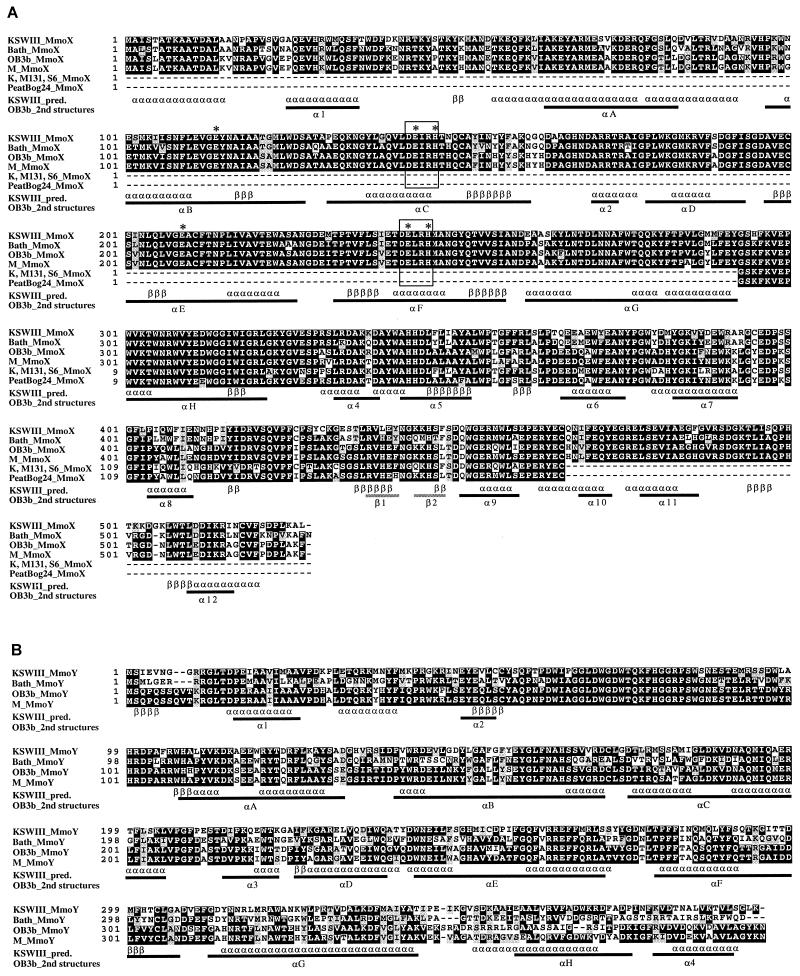
The two consensus regions for putative iron binding sites, which were found in the three known MmoXs (5, 34, 46), and other iron binding proteins (4, 38, 39) were well-conserved in the MmoXs of the strains KSWIII and KSPIII, located at amino acid positions 143 to 147 and 242 to 246 (Fig. 2A). The six amino acids, 114E, 144E, 147H, 209E, 243E, and 246H, which were found by X-ray crystal structure analysis to form the diiron center on the hydroxylase component of sMMOs (MMOH) from M. capsulatus Bath and M. trichosporium OB3b (16, 40), were also conserved in the MmoXs from the two strains (Fig. 2A). The secondary-structure prediction analysis of the MmoXs of the strains KSWIII and KSPIII demonstrated that most of the α-helical and β-strand structure elements from the X-ray structure analysis of strain Bath and strain OB3b were present in the MmoXs of the strains KSWIII and KSPIII, with only the one exception of α-helical element 5 (α5) (Fig. 2A).
FIG. 2.
Alignments of deduced amino acid sequences of the mmoX (A), mmoY (B), and mmoZ (C) genes from strain KSWIII, M. capsulatus Bath (accession no. M90050 and M32314) (45, 46), M. trichosporium OB3b (X55394 and S81887) (5, 6), and Methylocystis sp. strain M (U81594) (34). The partial sequences of the mmoX genes from strains K, M131, and S6 (K, M131, S6_MmoX; AF004554) (14) and an uncultured bacterium (PeatBog24_MmoX; AF004555) (13) are also aligned (A). Identical residues are in solid boxes, and similar residues are in shaded boxes. Underneath the aligned sequences, the secondary structural features of the MMOH from strain OB3b obtained from the crystal structure analysis are marked with bars (2nd structures); the helical (α) and sheet (β) regions are assigned as described previously (16). The predicted β-strand (β) and α-helical (α) regions for MmoX, -Y, and -Z from strain KSWIII (KSWIII_pred.) are also marked. Two conserved regions containing amino acids which are believed to serve as potential iron ligands are enclosed by open boxes. The asterisks above the aligned sequences indicate the amino acids which form the diiron center revealed by X-ray crystal analysis of the MMOHs from strain OB3b and strain Bath.
The multiple alignments of the deduced amino acids sequences of MmoY and MmoZ showed that there is relatively low conservation among the C-terminal portions of MmoYs (amino acid positions between 300 and 393 in strain KSWIII) and the N-terminal portions of MmoZs (amino acid positions between 1 and 100 in strain KSWIII). However, the locations of most putative secondary-structure elements of MmoY and MmoZ from strains KSWIII and KSPIII agreed with those from the X-ray structure analysis of the MMOHs of strains Bath and OB3b, with only one exception for MmoY (α2 was not predicted) and only one exception for MmoZ (α4 was not predicted) (Fig. 2B and C). These findings suggest that the MMOHs from the Methylomonas sp. strains KSWIII and KSPIII have structures similar to those from M. capsulatus Bath and M. trichosporium OB3b.
The sequences of MmoCs of strains KSWIII and KSPIII agreed with the previous assignment of the reductase components (MMOR) to the extended flavoprotein reductase (FNR) family (32). The consensus sequences, which were observed in the members of the extended FNR family (10, 11), including the known MMORs (6, 46), conserved cysteine residues for the N-terminal [2Fe-2S] domain, RXYS and GXXS for the central [FAD] domain, and GTGIXP, YXCGP, and EXF for the C-terminal [NAD] domain (10), were observed in the MmoCs of strains KSWIII and KSPIII (Fig. 3). The deduced amino acid sequences of MmoCs from strains KSWIII and KSPIII enabled improved accuracy of the secondary-structure prediction analysis of the multiple alignment of MmoCs, which showed that most of the locations of the putative α-helix and β-strand structures were identical to those of the X-ray crystal structure of FNR (25) and phthalate dioxygenase reductase (PDR) (10), although the sequence similarities between MmoCs and FNR or PDR were not so high (Fig. 3). The three domains of MmoCs, including those from Methylomonas sp. strains KSWIII and KSPIII were, therefore, likely to form structures similar to those of the corresponding domains of other proteins in the extended FNR family.
In the regulatory protein B of sMMO (MMOB), the 29Q-30V cleavage site, which was responsible for forming a truncated inactive form called B′ in all the known MmoBs (31), was found in amino acid sequences from strains KSWIII and KSPIII. However, the 12M-13G site, which was responsible for forming another inactive form called B" found only in MmoB of M. capsulatus Bath (31), was not found in amino acid sequences of strains KSWIII and KSPIII. The open reading frame orfY has been identified in the sequences of all sMMO genes, including our Methylomonas sp. strains. There was significant conservation, especially in their central regions, among these deduced amino acid sequences. However, the function of this orf is not clear, since it differs from any sequences in the database.
A putative promoter sequence upstream of mmoX from strains KSWIII and KSPIII (C2083TGGCACN5TTGCA2099 in strain KSWIII) was similar to the consensus sequence recognized by E. coli RNA polymerase containing ς54 (23). Similar ς54 promoters were also found upstream of the mmoX genes from M. capsulatus Bath (46), M. trichosporium OB3b (5), and Methylocystis sp. strain M (34).
Phylogenetic analysis of sMMO gene clusters.
DNA and amino acid sequence similarities are shown in Table 1. The sMMO genes from the strains KSWIII and KSPIII were more closely related to those from M. capsulatus Bath than to those from M. trichosporium OB3b or those from Methylocystis sp. strain M (Table 1). The partial amino acid sequence of MmoX from Methylomicrobium sp. strain NI (group I) (19) showed high homology with those from our Methylomonas sp. strains, with 96.6% identity (data not shown).
The phylogenetic analysis of the deduced amino acid sequences of MmoX proteins demonstrated that the sequences form two clusters (Fig. 4A). One cluster consisted of three strains in the γ Proteobacteria (M. capsulatus Bath [group X] and Methylomonas sp. strains KSWIII and KSPIII [group I]), and the other cluster consisted of two strains in the α Proteobacteria (M. trichosporium OB3b [group II] and Methylocystis sp. strain M [group II]) and the partial sequence of a PCR product amplified from environmental samples (Peatbog 24). The partial sequence of the acidophilic methane-oxidizing strains K, M131, and S6, which were affiliated with Beijerinckia indica subsp. indica in the α Proteobacteria (14), showed almost equal distances to the two clusters. The phylogenetic relationship determined from MmoXs was significantly correlated with the relationship determined from 16S rRNA sequences (Fig. 4B). The phylogenetic analysis of the deduced amino acid sequences of the other Mmo proteins also showed similar results (data not shown).
PCR primer design for detection of sMMO gene.
There have been several reports of the detection of methanotrophs possessing sMMO genes in environmental samples by amplification with PCR primers for sMMO genes (13, 33, 35). Miguez et al. used a primer set (mmoX1 and mmoX2) to detect mmoX genes in pure cultures and in environmental samples (35). However, the primer mmoX2 had several mismatches with the gene of our Methylomonas sp. strains, so the primer set did not work to amplify the mmoX gene from a small quantity (below 1 ng) of DNA from the strains.
We therefore designed a new PCR primer (mmoXr901) based on the sequence data including strains KSWIII and KSPIII to cover all of the known methanotrophs harboring the mmoX gene. By using a combination of this primer and primer mmoX1, the corresponding DNA fragment was amplified from a minimum of 100 pg of purified DNA from M. capsulatus Bath, M. trichosporium OB3b, Methylocystis sp. strain M, and strains KSWIII and KSPIII (data not shown).
Detection of sMMO gene at in situ-biostimulation site.
The primer set was applied to detect the methanotrophs in the groundwater samples at the in situ-biostimulation site. There were no amplified products from total DNAs from the groundwater samples 1 day before the start of the biostimulation treatment (Fig. 5B, lanes 2 to 7). However, a distinct amplified product of 396 bp was obtained from the groundwater sample extracted at well S1 10 days after the start (Fig. 5B, lane 8). Significant amplified products were observed from the groundwater sample extracted from wells S2, S3, and S4 (Fig. 5B, lanes 9, 10, and 11). No significant amplification was observed in the samples from wells S4 and S5 (Fig. 5B, lanes 12 and 13). Very weak PCR product signals (less than 10 ng per PCR) were detected in the samples shown in Fig. 5B, lanes 3 and 12. Some of the amplified products were cloned and sequenced and were found to consist of mmoX genes similar to those of Methylocystis sp. strain M or our Methylomonas sp. strains (data not shown). This indicated that our new PCR primer can detect not only the mmoX genes of the group II and group X methanotrophs but also those of the group I methanotrophs in environmental samples. In the tracer experiment with bromide carried out prior to the biostimulation treatments, high concentrations of bromide (23 to 27 mg/liter) were detected at wells S1, S2, S3, and S4. However, 7 mg of bromide/liter was detected at the extraction well, and trace levels of bromide were detected at well S5. These results show that the chemical added to the groundwater spread into the aquifer near the injection well at a high concentration but did not reach well S5 and that the extraction well contained an inflow of external groundwater. The results of PCR amplification showed good agreement with the results of the tracer experiment.
These results could be valuable for further studies on monitoring the population of methanotrophs during in situ-bioremediation treatments and on examining the diversity of methanotrophs present in environmental samples.
Acknowledgments
We thank Xian-Ying Meng for transmission electron microscopy. We thank Kimitsu City Hall in Chiba Prefecture for their kind cooperation.
This work was conducted as one of the research and development activities for the bioremediation project which is conducted by the Research Institute of Innovative Technology for the Earth (RITE), Japan, and funded by the Ministry of International Trade and Industry (MITI) through the New Energy and Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman J P, Sly L I, Nichols P D, Hayward A C. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int J Syst Bacteriol. 1993;43:735–753. [Google Scholar]
- 3.Burrows K J, Cornish A, Scott D, Higgins I J. Substrate specificities of the soluble and particulate methane monooxygenase of Methylosinus trichosporium OB3b. J Gen Microbiol. 1984;130:3327–3333. [Google Scholar]
- 4.Byrne A M, Kukor J J, Olsen R H. Sequence analysis of the gene cluster encoding toluene-3-monooxygenase from Pseudomonas pickettii PKO1. Gene. 1995;154:65–70. doi: 10.1016/0378-1119(94)00844-i. [DOI] [PubMed] [Google Scholar]
- 5.Cardy D L N, Laidler V, Salmond G P C, Murrell J C. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol Microbiol. 1991;5:335–342. doi: 10.1111/j.1365-2958.1991.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 6.Cardy D L N, Laidler V, Salmond G P C, Murrell J C. The methane monooxygenase gene cluster of Methylosinus trichosporium: cloning and sequencing of the mmoC gene. Arch Microbiol. 1991;156:477–483. doi: 10.1007/BF00245395. [DOI] [PubMed] [Google Scholar]
- 7.Colby J, Dalton H. Resolution of the methane monooxygenase from Methylococcus capsulatus (Bath) into three components: purification and properties of component C, a flavoprotein. Biochem J. 1978;171:461–468. doi: 10.1042/bj1710461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colby J, Dalton H. Characterization of the second prosthetic group of the flacoenzyme NADH-acceptor reductase (Component C) of the methane monooxygenase from Methylococcus capsulatus (Bath) Biochem J. 1979;177:903–908. doi: 10.1042/bj1770903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins M D, Green P N. Isolation and characterization of a novel coenzyme Q from some methane-oxidizing bacteria. Biochem Biophys Res Commun. 1985;133:1125–1131. doi: 10.1016/0006-291x(85)91253-7. [DOI] [PubMed] [Google Scholar]
- 10.Correll C C, Batie C J, Ballou D P, Ludwig M L. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S] Science. 1992;258:1604–1610. doi: 10.1126/science.1280857. [DOI] [PubMed] [Google Scholar]
- 11.Correll C C, Ludwig M L, Bruns C M, Karplus P A. Structural prototypes for an extended family of flavoprotein reductases: comparison of phthalate dioxygenase reductase with ferredoxin reductase and ferredoxin. Protein Sci. 1993;2:2112–2133. doi: 10.1002/pro.5560021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalton H, Smith D D S, Pilkington S J. Towards a unified mechanism of biological methane oxidation. FEMS Microbiol Rev. 1990;87:201–207. [Google Scholar]
- 13.Dedysh S N, Panikov N S, Tiedje J M. Acidophilic methanotrophic communities from Sphagnum peat bogs. Appl Environ Microbiol. 1998;64:922–929. doi: 10.1128/aem.64.3.922-929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dedysh S N, Panikov N S, Liesack W, Großkopf R, Zhou J, Tiedje J M. Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science. 1998;282:281–284. doi: 10.1126/science.282.5387.281. [DOI] [PubMed] [Google Scholar]
- 15.DiSpirito A A, Gulledge J, Shiemke A K, Murrell J C, Lidstrom M E, Krema C L. Trichloroethylene oxidation by the membrane-associated methane monooxygenase in type I, type II and type X methanotrophs. Biodegradation. 1992;2:151–164. [Google Scholar]
- 16.Elango N, Radhakrishnan R, Froland W A, Wallar B J, Earhart C A, Lipscomb J D, Ohlendorf D H. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 1997;6:556–568. doi: 10.1002/pro.5560060305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox B G, Froland W A, Dege J, Lipscomb J D. Methane monooxygenase from Methylosinus trichosporium OB3b: purification and properties of a three component system with high specific activity from a type II methanotroph. J Biol Chem. 1989;264:10023–10033. [PubMed] [Google Scholar]
- 18.Fox B G, Liu Y, Dege J E, Lipscomb J D. Complex formation between the protein components of methane monooxygenase from Methylosinus trichosporium OB3b. J Biol Chem. 1991;266:540–550. [PubMed] [Google Scholar]
- 19.Fuse H, Ohta M, Takimura O, Murakami K, Inoue H, Yamaoka Y, Oclarit J M, Omori T. Oxidation of trichloroethylene and dimethyl sulfide by a marine Methylomicrobium strain containing soluble methane monooxygenase. Biosci Biotechnol Biochem. 1998;62:1925–1931. doi: 10.1271/bbb.62.1925. [DOI] [PubMed] [Google Scholar]
- 20.Green J, Dalton H. Protein B of soluble methane monooxygenase from Methylococcus capsulatus (Bath) J Biol Chem. 1985;260:15795–15801. [PubMed] [Google Scholar]
- 21.Hanada S, Shigematsu T, Shibuya K, Eguchi M, Hasegawa T, Suda F, Kamagata Y, Kanagawa T, Kurane R. Phylogenetic analysis of trichroloethylene-degrading bacteria newly isolated from the polluted soil with the contaminant. J Ferment Bioeng. 1998;86:539–544. [Google Scholar]
- 22.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helmann J D, Chamberlin M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 24.Kamagata Y, Mikami E. Isolation and characterization of a novel thermophilic Methanosaeta strain. Int J Syst Bacteriol. 1991;41:191–196. doi: 10.1099/00207713-42-3-463. [DOI] [PubMed] [Google Scholar]
- 25.Karplus P A, Daniels M J, Herriott J R. Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science. 1991;251:60–66. [PubMed] [Google Scholar]
- 26.Koh S-C, Bowman J P, Sayler G S. Soluble methane monooxygenase production and trichloroethylene degradation by a type I methanotroph, Methylomonas methanica 68-1. Appl Environ Microbiol. 1993;59:960–967. doi: 10.1128/aem.59.4.960-967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.0. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- 28.Kushida H. An improved embedding method using ERL 4206 and Quetol 653. J Electron Microsc. 1980;29:193–194. [Google Scholar]
- 29.Levin J M. Exploring the limits of nearest neighbour secondary structure prediction. Protein Eng. 1997;10:771–776. doi: 10.1093/protein/10.7.771. [DOI] [PubMed] [Google Scholar]
- 30.Lipscomb J D. Biochemistry of the soluble methane monooxygenase. Annu Rev Microbiol. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 31.Lloyd J S, Bhambra A, Murrell J C, Dalton H. Inactivation of the regulatory protein B of soluble methane monooxygenase from Methylococcus capsulatus (Bath) by proteolysis can be overcome by a Gly to Gln modification. Eur J Biochem. 1997;248:72–79. doi: 10.1111/j.1432-1033.1997.t01-1-00072.x. [DOI] [PubMed] [Google Scholar]
- 32.Lund J, Dalton H. Further characterisation of the FAD and Fe2S2 redox centres of component C, the NADH:acceptor reductase of the soluble methane monooxygenase of Methylococcus capsulatus (Bath) Eur J Biochem. 1985;147:291–296. doi: 10.1111/j.1432-1033.1985.tb08749.x. [DOI] [PubMed] [Google Scholar]
- 33.McDonald I R, Kenna E M, Murrell J C. Detection of methanotrophic bacteria in environmental samples with the PCR. Appl Environ Microbiol. 1995;61:116–121. doi: 10.1128/aem.61.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald I R, Uchiyama H, Kambe S, Yagi O, Murrell J C. The soluble methane monooxygenase gene cluster of the trichloroethylene-degrading methanotroph Methylocystis sp. strain M. Appl Environ Microbiol. 1997;63:1898–1904. doi: 10.1128/aem.63.5.1898-1904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miguez C B, Bourque D, Sealy J A, Greer C W, Groleau D. Detection and isolation of methanotrophic bacteria possessing soluble methane monooxygenase (sMMO) genes using the polymerase chain reaction (PCR) Microb Ecol. 1997;33:21–31. doi: 10.1007/s002489900004. [DOI] [PubMed] [Google Scholar]
- 36.Murrell J C. Molecular genetics of methane oxidation. Biodegradation. 1994;5:145–159. doi: 10.1007/BF00696456. [DOI] [PubMed] [Google Scholar]
- 37.Nakajima T, Uchiyama H, Yagi O, Nakahara T. Purification and properties of a soluble methane monooxygenase from Methylocystis sp. M. Biosci Biotech Biochem. 1992;56:736–740. doi: 10.1271/bbb.56.736. [DOI] [PubMed] [Google Scholar]
- 38.Nordlund I, Powlowski J, Shingler V. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol. 1990;172:6826–6833. doi: 10.1128/jb.172.12.6826-6833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordlund P, Dalton H, Eklund H. The active site structure of methane monooxygenase is closely related to the binuclear iron center of ribonucleotide reductase. FEBS Lett. 1992;307:257–262. doi: 10.1016/0014-5793(92)80690-i. [DOI] [PubMed] [Google Scholar]
- 40.Rosenzweig A C, Frederick C A, Lippard S J, Nordlund P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 41.Saitou N, Nei M. The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 43.Semrau J D, Chistoserdov A, Lebron J, Costello A, Davagnino J, Kenna E, Holmes A J, Finch R, Murrell J C, Lidstrom M E. Particulate methane monooxygenase genes in methanotrophs. J Bacteriol. 1995;177:3071–3079. doi: 10.1128/jb.177.11.3071-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen R-N, Yu C-L, Ma Q-Q, Li S-B. Direct evidence for a soluble methane monooxygenase from type I methanotrophic bacteria: purification and properties of a soluble methane monooxygenase from Methylomonas sp. GYJ3. Arch Biochem Biophys. 1997;345:223–229. doi: 10.1006/abbi.1997.0239. [DOI] [PubMed] [Google Scholar]
- 45.Stainthorpe A C, Murrell J C, Salmond G P C, Dalton H, Lees V. Molecular analysis of methane monooxygenase from Methylococcus capsulatus (Bath) Arch Microbiol. 1989;152:154–159. doi: 10.1007/BF00456094. [DOI] [PubMed] [Google Scholar]
- 46.Stainthorpe A C, Lees V, Salmond G P C, Dalton H, Murrell J C. The methane monooxygenase gene cluster of Methylococcus capsulatus (Bath) Gene. 1990;91:27–34. doi: 10.1016/0378-1119(90)90158-n. [DOI] [PubMed] [Google Scholar]
- 47.Stainthorpe A C, Salmond G P C, Dalton H, Murrell J C. Screening of obligate methanotrophs for soluble methane monooxygenase genes. FEMS Microbiol Lett. 1990;70:211–216. [Google Scholar]
- 48.Tamaoka K, Katayama-Fujimura Y, Kuraishi H. Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. J Appl Bacteriol. 1983;54:31–36. [Google Scholar]
- 49.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsien H-C, Hanson R S. Soluble methane monooxygenase component B gene probe for identification of methanotrophs that rapidly degrade trichloroethylene. Appl Environ Microbiol. 1992;58:953–960. doi: 10.1128/aem.58.3.953-960.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whittenbury R, Philips K, Willkinson J F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 52.Whittenbury R, Krieg N R. Family IV. Methylococcaceae fam. nov. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1989. pp. 256–261. [Google Scholar]
- 53.Woodland M P, Dalton H. Purification and characterization of component A of the methane monooxygenase from Methylococcus capsulatus (Bath) J Biol Chem. 1984;259:53–60. [PubMed] [Google Scholar]
- 54.Zahn J A, DiSpirito A A. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]