Abstract
The latent infection by herpes virus type 1 (HSV-1) may be lifelong in trigeminal ganglia and a suspected cause of Alzheimer's Disease (AD) and Amyotrophic lateral sclerosis (ALS). Whether and how N6-methyladenosine (m6A) modification of viral RNAs affects virus infection are poorly understood. Here, we report that HSV-1 infection enhanced the expression of m6A writers (METTL3, METTL14) and readers (YTHDF1/2/3) at the early infection stage and decreased their expression later on, while suppressed the erasers' (FTO, ALBKH5) expression immediately upon infection to facilitate viral replication. Inhibiting m6A modification by 3-deazaadenosine (DAA) significantly decreased viral replication and reduced viral reproduction over 1000 folds. More interestingly, depleting the writers and readers by siRNAs inhibited virus replication and reproduction; whereas depleting the erasers promoted viral replication and reproduction. Silencing YTHDF3 strikingly decreased viral replication by up to 90%, leading to reduction of up to 10-fold viral replication and over 100-fold virus reproduction, respectively. Depletion of m6A initiator METTL3 (by 60%–70%) by siRNA correlatedly decreased viral replication 60%–70%, and reduced virus yield over 30-fold. Consistently, ectopic expression of METTL3 largely increased virus yield. METTL3 knockdown suppressed the HSV-1 intermediate early and early genes (ICP0, ICP8 and UL23) and late genes (VP16, UL44, UL49 and ICP47); while ectopic expression of METTL3 upregulated these gene expression. Results from our study shed the lights on the importance for m6A modification to initiate HSV-1 early replication. The components of m6A modification machinery, particularly m6A initiator METTL3 and reader YTHDF3, would be potential important targets for combating HSV-1 infections.
Keywords: Gene silencing, HSV-1 infection, m6A modification, Virus replication, Virus reproduction
Introduction
N6-methyladenosine (m6A) was firstly discovered in 1970s while a subset of mRNA from Novikoff hepatoma was methylated by DEAE-cellulose (borate) chromatography.1 Later findings revealed that the methylated nucleotides in mRNA played a special role in both physiological and pathological processes.2, 3, 4 M6A usually occurs in two motifs, -G-m6A-C (70%) and -A-m6A-C-(30%), mostly in the region close to 3′ untranslated region (UTR) and the 5′ terminus of mRNA.3,5 Currently, more than 100 kinds of chemical RNA modifications have been observed in types of RNAs, including but not limited to mRNA, transfer RNA (tRNA), ribosomal RNA (rRNA), non-coding RNA (ncRNA, including microRNA and long non-coding RNA). Among them, the m6A modification is the most abundant one that presents in 0.1%–0.4% of all adenosines, and accounts for nearly 50% of all methylated ribonucleotides.2,5,6 It affects RNA splicing, translation and stability,7 as well as unique structural and catalytic functions of RNA, the so-called m6A-derived transcriptome topology8 (Table 1).
Table 1.
The sequences of the primers and siRNAs.
| gene | sequence |
|---|---|
| mettl3 f | TCAGCTCCAGGGGTCATTTT |
| mettl3 r | CTGGGCTGTCACTACGGAAG |
| mettl4 f | GTGGACTGGAAGGGGCATTT |
| mettl4 r | TTGGCCAAAGGGGGTTAAAA |
| ythdf1 f | CACCCAGAGAACAAAAGGACA |
| ythdf1 r | TGCCCAAAAACAGCATCGTG |
| ythdf2 f | CCGAGTGTCAGGGACAAAAGC |
| ythdf2 r | TTTGGTCTCTGCTCCAAGAGG |
| ythdf3 f | GGAGCGGAAGTGAGACTAGG |
| ythdf3 r | TGGCCGAGTGATTGTTCCAG |
| FTO f | GGATGAGCCAGCTTCACTGT |
| FTO r | GATTTCTCCAACCCTGTTGCAC |
| alkbh5 f | CCAGCTATGCTTCAGATCGCCT |
| alkbh5 r | GGTTCTCTTCCTTGTCCATCTCC |
| HSV-1 f | CAACTACCCCGATCATCAGTTA |
| HSV-1 r | ACAGTTGCCTCCCATCCGAAACCAA |
| gapdh f | GATTCCACCCATGGCAAATTCCA |
| gapdh r | TGGTGATGGGATTTCCATTGATGA |
| UL29 f | TGCGAGGGCGTCAGTTTCAG-3 |
| UL29 r | GCGTGTCCGTCCGAAGGC |
| GC f | GTGACGTTTGCCTGGTTCCTGG |
| GC r | GCACGACTCCTGGGCCGTAACG |
| UL23 f | ACCCGCTTAACAGCGTCAACA |
| UL23 r | CCAAAGAGGTGCGGGAGTTT |
| ICP0 f | GGCCCCCTTGTCAACAGA |
| ICP0 r | GGGAGTCGCTGATCACTATGG3′ |
| ICP47 f | GACAGAAACCCACCGGTCCGCCT |
| ICP47 r | CGCATGTTGTCCAGGAAGGTGTC |
| UL48 f | CCGGGTCCGGGATTTACC |
| UL48 r | CTCGAAGTCGGCCATATCCA |
| UL49 f | GGGAGTGCGGCGGATTCTGGCT |
| UL49 r | GGCTCGTCATCCGAAGACGACGA |
| 18S rRNA f | CCAGTAAGTGCGGGTCATAAGC |
| 18S rRNA r | GCCTCACTAAACCATCCAATCGG |
| scramble | AAGAACUGAUGACAGGGAGGCTT |
| simettl3-1 | CAAGCUGCACUUCAGACGAA |
| simettl3-2 | GCUUGGCGUGUGGUCUUU |
| simettl14-1 | AAUGGCCGUUCUGUGCUCAU |
| simettl14-2 | AAGGACCCAUCACAGGCAAG |
| siythdf1-1 | CACGAUGCUGUUUUUGGGCA |
| siythdf1-2 | GCUGACGTCCCCAAUCUUCA |
| siythdf2-1 | CGCGCGUAAAAGCAUAGAGAC |
| siythdf2-2 | UGCAUUAUUGGGCCUUGCCU |
| siythdf3-1 | UAGGGAGUCUGUCCGCCAUU |
| siythdf3-2 | GACAUUCUUCACCGCAACCC |
| siFTO-1 | CGGUAUCUCGCAUCCUCAUUGGUAA |
| siFTO-2 | UCAGCGGUGGCAGUGUACAGUUAUA |
| sialkbh5-1 | CCCAUCGUGUCCGUGUCCUUCUUUA |
| sialkbh5-2 | CGGUUGGAAACAAAGUCCCUGAGCA |
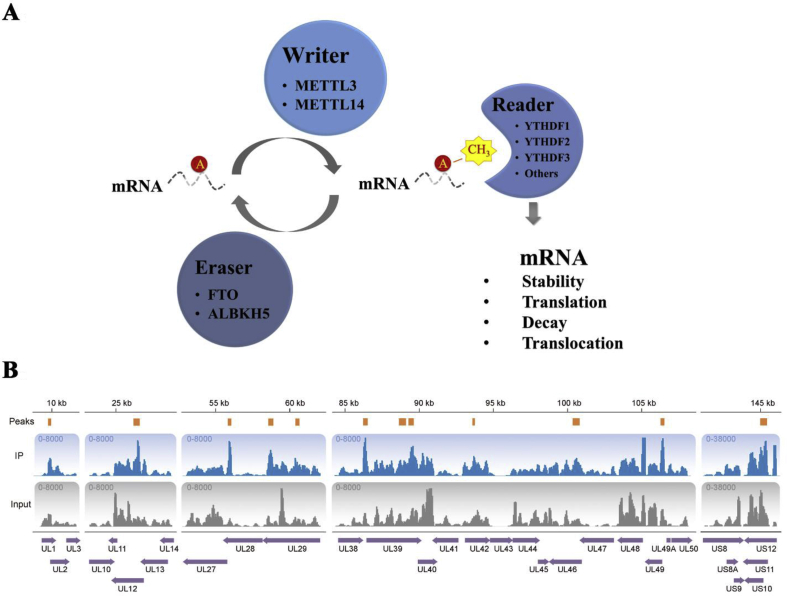
The m6A modification process is reversibly modulated by three parts: the writer, reader and eraser, respectively (Fig. 1A).9 The writer, mainly consisting of adenosine methyltransferase, refers to the METTL3-METTL14 methyltransferase complex in mammalian cells.10 METTL3 functions as a catalytic subunit while METTL14 is a pseudo-methyltransferase that stabilizes METTL3 and contributes to RNA binding. The complex is regulated by the tumor suppressor Wilms' tumor 1 (WT1) associated protein (WTAP) for localization to the nuclear speckle and recruitment of RNAs.11 Besides, the m6A is recognized by the YTH domain-containing proteins, which exert regulatory functions through selective recognition of methylated RNAs.4 YTHDF1 promotes ribosome loading of m6A-modified mRNAs and interacts with initiation factors to facilitate translation initiation.12 YTHDF2 accelerates the decay of m6A-modified transcripts and induces RNA degradation via nuclear processing bodies (P bodies).4,13 YTHDF3 promotes protein synthesis in synergy with YTHDF1, and affects the methylated mRNA decay through YTHDF2.14 What's more, m6A can be reverted to adenosine by the m6A RNA demethylases.13 The “eraser” enzyme controls mRNA export and RNA metabolism as well as the assembly of mRNA processing factors in nuclear speckles.15 Fat mass and obesity-associated protein (FTO) exerts oxidative demethylation activity targeting the m6A residues in RNA,16,17 and ALKB homolog 5 (ALKBH5) demethylates the m6A-containing single-stranded RNA. Above all, m6A modification has been suggested to participate in almost all RNA metabolism processes, thus affecting a variety of cellular normal physiological processes including but not limit to cell fate transition in embryonic stem cells,18 synaptic plasticity,19 signal transduction,20 endocytosis,21 immune response,22 etc. On the other hand, accumulating evidence shows that m6A abnormality is involved in many pathological states, such as Alzheimer's disease,23 diabetes,5 cancers24 and virus infections.25
Figure 1.
The status of m6A modification on HSV-1 RNA. (A) Schematic of the m6A modification system. (B) Identification of m6A peaks by MeRIP-seq of HSV-1 transcripts in RD cell. RD cells were infected with HSV-1 at a MOI 5. Total RNAs were extracted with Trizol (Invitrogen) 24 h post infection. Coverage tracks illustrate enrichment signal normalized by counts per million for the RIP and pre-MeRIP input (negative control) samples, respectively. The identified m6A peaks were shown by blocks colored in orange. All genes were shown and overlaid as purple arrows in the bottom track.
Herpes simplex virus type 1 (HSV-1) is a member of human herpesviridae family with a large double-stranded linear DNA genome.26 HSV-1 is mainly transmitted by oral-to-oral contact and cause clinical symptoms including painful blisters and cold sores.27 By estimation, over 67% people under the age of 50 are infected by HSV-1 worldwide. What's more, the infection may be lifelong due to the latent infection in trigeminal ganglia, and is reported as a suspected cause of Alzheimer's Disease (AD) and Amyotrophic lateral sclerosis (ALS).28
The HSV-1 genome contains two unique regions, a unique long region (UL) and a unique short region (US), flanking by terminal and internal repeat sequences (TRL and IRL, TRS and IRS). HSV-1 genome encodes over 70 proteins. Among them, those involved in DNA replication and envelope glycoproteins are first expressed; then followed by proteins for forming virion particles. Outer membrane glycoproteins of HSV-1 play essential roles in the virus entry and spreading. HSPG, 3-OS HS, PILRα, MAG and NMHC-IIA are most well studied host receptors specific for HSV-1 entry.27 Following entry, nucleocapsid covered by inner tegument proteins US3 and UL36, then UL37 are transported to the nucleus, where the genome is deposited and replication starts.29 The role of m6A in regulating viral transcripts has been known for near half a century.30,31 In addition to RNA viruses, the m6A modified transcripts of DNA viruses can also be detected.25,32 Influenza virus, simian virus 40 (SV40), Rous sarcoma virus (RSV), Hepatitis B and C Virus, as well as human immunodeficiency virus (HIV), have been reported internal m6A modifications upon virus reproduction.1,25,30,32 However, the function of m6A modification in HSV-1 infection has not yet been reported.
Materials and methods
Cell culture, HSV-1 infection, viral DNA extraction and TCID50 assay
HeLa cells (American Type Culture Collection, ATCC) and human rhabdomyosarcoma cell (RD, ATCC) were grown in Dulbecco's modified Eagle's medium (Gibco) containing 10% fetal bovine serum (Gibco) and 10 U/ml penicillin–streptomycin (Gibco) in a humidified 5% CO2 incubator at 37 °C.
To test the effect of m6A genes in HSV-1 reproduction, RD and HeLa cells were transfected with the indicated siRNAs or plasmids for 48 h. The HeLa and RD cells were then infected with HSV-1 strain SM44 at a MOI of 5. After 1 h of infection with occasional shaking, the virus was discarded and replaced with the fresh medium. Samples were collected at 24 h after infection. To test the effect of 3-DAA on HSV-1 reproduction, RD and HeLa cells were incubated in DMEM with 2% FBS and various concentrations of DAA (Cayman) for 2 h prior to infection. Then virus was added to cells at a MOI of 0.01. After 1 h of infection with occasional shaking, the virus was discarded and the DAA-containing medium was added back to the infected cells respectively. Samples were collected at 48 h after infection.
Viral DNA was extracted from the cell lysates with the QIAamp DNA blood mini kit (Qiagen) according to the manufacturer's protocol. Virus titers were determined by TCID50 and/or plaque-forming assays. Vero cells (ATCC) were inoculated with 200 μl of serially diluted viral fluid for 1 h. After viral adsorption, the HSV-1-infected cells were overlaid with medium containing 2% FBS for 48 h and stained with 1% crystal violet in 4% formaldehyde overnight to visualize the plaques.
MTT assay
Both RD and HeLa cells were seeded in 96-well plates at 20,000 cells/well in DMEM with 2% FBS. Once cells were attached, the media were replaced with 100 μl/well of media containing various concentrations of DAA. After two days of cell culture in DAA, an MTT assay was performed as described.33 In brief, the culture media were replaced with 100 μL/well containing MTT (5 mg/ml) in serum-free DMEM, and cells were further incubated for an additional 4 h at 37 °C. Next, the medium was carefully removed and subsequently 100 μL of dimethyl sulfoxide were added. The 590 nm absorbance was read with a microplate reader (BioTek SynergyTM).
For proliferation detection, cells were plated at 4 × 103 per well in 96-well plate and transfected with simettl3 siRNAs or pCDNA-METTL3 respectively. Cell viability was determined at different time points accordingly. Each group had six repeat wells and the experiment was repeated three times.
Cell transfection, RNA isolation, reverse transcription and Quantitative real-time PCR
All of the synthetic siRNAs and the negative control siRNA (scramble) were purchased from Shanghai GenePharma Co., Ltd. All the siRNAs were transfected with Hiperfect transfection reagent (Qiagen) according to the manufacturer's protocol. The plasmid pCDNA-METTL3 was a gift from Xin Deng's lab and transfected with FuGENE® HD Transfection Reagent (Promega). The sequences of primers and siRNAs used in this study were listed in Table 1.
Total RNA was isolated with RNAiso Plus (Takara) according to the manufacturer's protocol. For viral mRNA extraction, total mRNA was extracted from RD and HeLa cells at 2 and 8 h after HSV-1 infection and treated with TURBO DNA free DNAse (Ambion, USA). Reversed transcription was performed with PrimeScript RT reagent Kit (Takara), as specified by manufacturers.
Quantitative real-time PCR was performed with SYBR® Premix Ex Taq™ (Tli RNaseH Plus) (Takara). All mRNA levels were measured and normalized to that of 18rs mRNA. The sequences of the siRNAs and primers used in this study are listed in Table S1.34
Western blotting
Cells were lysed in ice-cold RIPA buffer supplemented with complete protease inhibitor cocktail (Roche). The cell extracts were resolved with SDS-PAGE and analyzed with Western blotting. The protein bands were visualized with ECL Blotting Detection Reagents.33,35 The antibodies used for Western blotting included an anti-HSV-1 thymidine kinase (KITH_HHV1antibody, Invitrogen), anti-GAPDH antibody (Santa Cruz).
MeRIP-Seq
For MeRIP seq, protocols were followed as previous described.36, 37, 38 RD cells were infected with HSV-1 in a MOI = 5. Total RNAs were extracted with Trizol (Invitrogen) 24 h after infection. PolyA RNA was purified from total RNA for RD cell by using polyA Spin mRNA isolation kit (New England Biolabs). Samples were fragmented into preferable sizes and then immunoprecipitation with anti-m6A antibody (5 μg, Synaptic Systems) conjugated to Protein G Dynabeads (Thermo Fisher Scientific) overnight at 4 °C. Beads were then washed 5 times with MeRIP buffer, and bound RNA was eluted in proteinase K(Qiagen) digestion. Eluted RNA was purified with the RNeasy mini kit (Qiagen) and concentrated by ethanol precipitation. Libraries were constructed with the NEBNext® Ultra™ DNA Library Prep Kit according to the manufacture's protocol. MeRIP-seq reads were mapped to the Human herpesvirus 1 genome (NC_001806.2) using ‘TopHat 2’ [TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions]. Uniquely mapped reads were retained for the subsequent analyses. Standard mapping BAM files were converted to bigWig coverage tracks by ‘deepTools’ [deepTools 2: a next generation web server for deep-sequencing data analysis]. Binding peaks (Q < 0.05) were identified using MACS2 software [Model-based Analysis of ChIP-Seq (MACS)]. The enriched peaks were annotated to reference genome using the R package ‘ChIPpeakAnno’ [ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP–chip data].
Statistical analysis
Each experiment was repeated three times. The results are presented as the mean ± SD; ∗P < 0.05, ∗∗P < 0.01. Comparisons between two groups were evaluated with a two-sample t test. For three or more groups, standard oneway analysis of variance (ANOVA) followed by Bonferroni's test for multiple comparisons was completed. A two-tailed probability value < 0.05 was considered statistically significant.
Result
The RNA transcripts of HSV-1 are modified by m6A in host cells
To investigate whether HSV-1 RNA was m6A-modified, we isolated total RNA from HSV-1-infected RD cells. We performed RNA immunoprecipitation (RIP) assays using an m6A-specific antibody, followed by MeRIP-seq as previous described.36,39 The results showed that in vitro HSV-1 RNA was pulled down by the anti-m6A antibody.
We identified 12 m6A RNA peaks commonly across the HSV-1 genome to both experimental replicates (Fig. 1B). These data present evidence that mRNA of HSV-1 is modified by m6A during viral infection. The m6A peaks mainly locates in the upstream, overlap start and inside of the transcript region, covering multiple viral transcripts with m6A methylation, including UL1, UL2, UL12, UL28, UL29, UL38, UL39, UL42, UL46, UL49, US10, US11, and US12.
M6A methylation inhibitor suppresses HSV-1 replication and reproduction
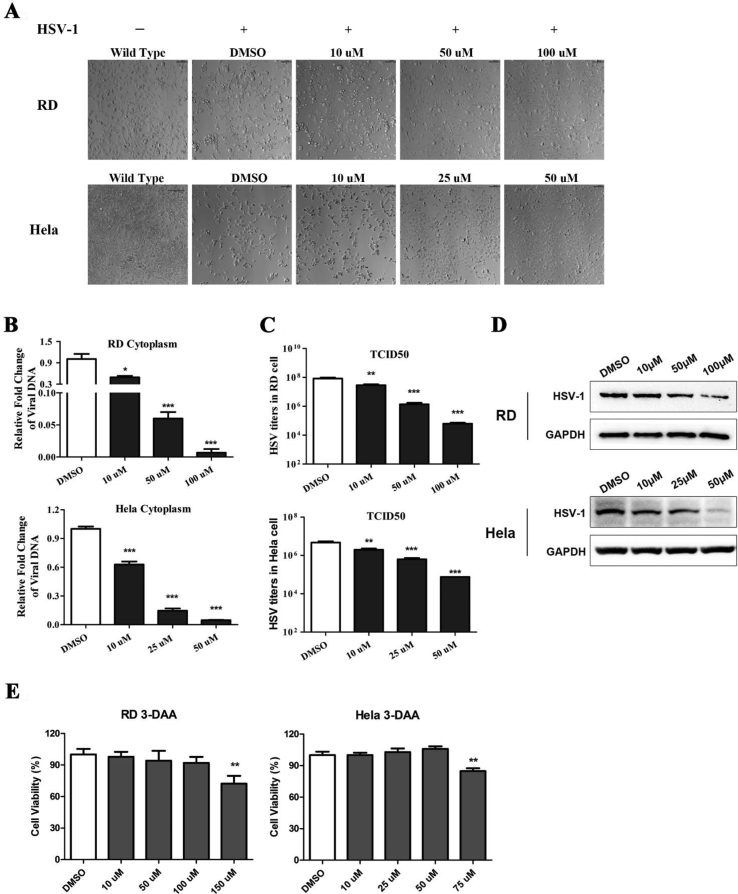
The m6A methylation inhibitor 3-deazaadenosine (DAA) globally suppress the formation of S-adenosyl methionine (SAM) from S-adenosyl homocysteine (SAH).40 3-DAA works well with high tolerance in mammal cells, as well as in mice models.41 RD and HeLa cells were pretreated at different concentrations of DAA for 1 h and then infected with HSV-1 at a MOI 0.01 for 1 h. Then the media was replaced with fresh media to remove uninfected virus, followed by treating the cells with various concentrations of DAA for 48 h. As shown in Figure 2A, DAA protected the cytopathic effects (CPE) from HSV-1 infection in both RD and HeLa cells in a dose dependent manner. Both the intercellular virus DNA levels (in cytoplasm) virus titers (extracellular virions) were markedly decreased up to over 100-fold (Fig. 2B, C), indicating the viral replication and reproduction are inhibited by depletion of m6A modification. As shown by Western blot assay, when cells were treated with the indicated concentrations of DAA for 48 h after HSV-1 infection (MOI 0.01), the viral KITH_HHV1 protein was also remarkably reduced in a does dependent manner (Fig. 2D). No obvious toxicity was observed at the above test concentrations in both RD and HeLa cells as indicated by cell viability (Fig. 2E).
Figure 2.
Inhibition of HSV-1 reproduction by methylation inhibitor 3-deazaadenosine (DAA). Both RD and HeLa cells were pretreated with the indicated concentrations of DAA for 1 h before infection with HSV-1 at MOI 0.01. The cytopathic effects were recorded, and samples were collected 48 h after infection. (A) Image of cytopathic effects was taken in RD and HeLa cells (200×). (B) RD (upper panel) and HeLa (lower panel) cells treated with DAA with indicated concentrations and infected with/without HSV-1 at MOI 0.01. The intracellular viral DNA was isolated and measured by quantitative PCR (qPCR) at 48 h. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (C) Viral titers were quantitated by TCID50 assays. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (D) The levels of HSV-1 KITH_HHV1 protein were detected by Western blot assays 48 h after DAA treatment in the HSV-1 infected (MOI 0.01) RD and HeLa cells. The endogenous GAPDH was used as a loading control. (E) Cell viability was analyzed using the MTT assay after 48 h of DAA treatment.
Depletion of m6A erasers enhances HSV-1 replication and reproduction
To further address how m6A writers, readers and erasers contribute to HSV-1 replication and reproduction, we performed loss of function studies by knockdown of corresponding genes using specific siRNAs (Table 1).
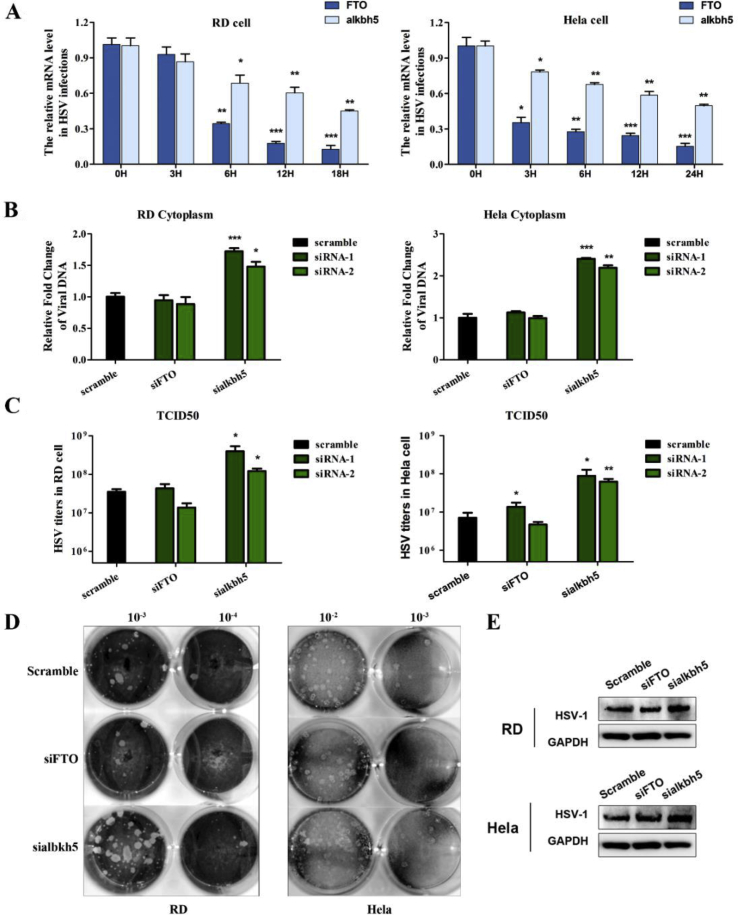
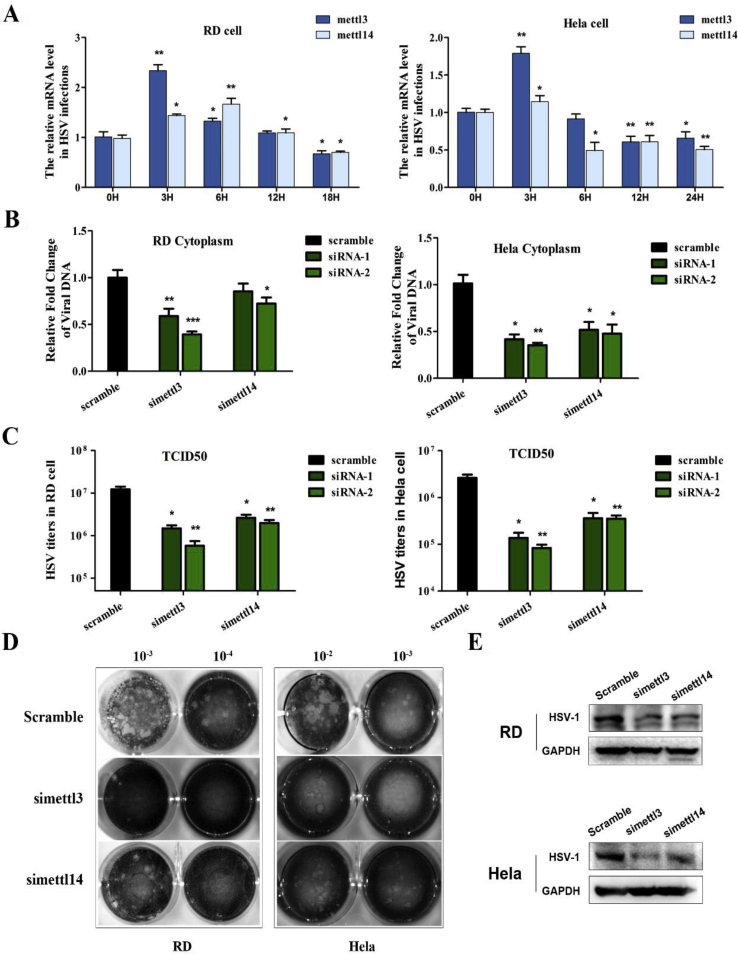
The mRNA level of FTO and ALKBH5 was firstly detected by RT-qPCR after infection with HSV-1 in RD and HeLa cells. As shown in Figure 3A, the expression pattern of FTO and ALKBH5 was quite different from that of the writers and readers (Fig. 4, 5) in the HSV-1 infected cells. Surprisingly, the mRNA levels of FTO and ALKBH5 decreased very early after infection, and FTO decreased even over 80% (in HeLa cells) and 90% (in RD cells) 12 h p.i. (Fig. 3A). These results indicated that HSV-1 infection suppressed m6A eraser immediately upon infection to facilitate fast viral replication.
Figure 3.
Promotion of HSV-1 replication and reproduction after depleting ALKBH5. (A) The expression pattern of the m6A erasers in HSV-1 infected RD (left) and HeLa (right) cells. The cells were infected with HSV-1 at MOI 5. The intracellular RNA was isolated at different time points after infection. The mRNA levels of FTO and ALKBH5 and FTO were determined by RT-qPCR. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (B) The cells were transfected with indicated siRNAs and infected with HSV-1 at MOI 5. The intracellular viral DNA was measured by qPCR at 24 h. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (C) The secreted infectious viral particles were determined by titration of virus titers using TCID50 assay. ∗P < 0.05, ∗∗P < 0.01. (D) Representative photograph of plaques was generated by HSV-1 in FTO and ALKBH5 knockdown cells as compared to the scramble siRNA treated group. (E) HSV-1 KITH_HHV1 protein level in was shown in the HSV-1 infected RD and HeLa cells. Cell were pretreated with siRNAs for 48 h and the cell lysates were prepared 24 h post infection (MOI = 5). GAPDH used as a loading control.
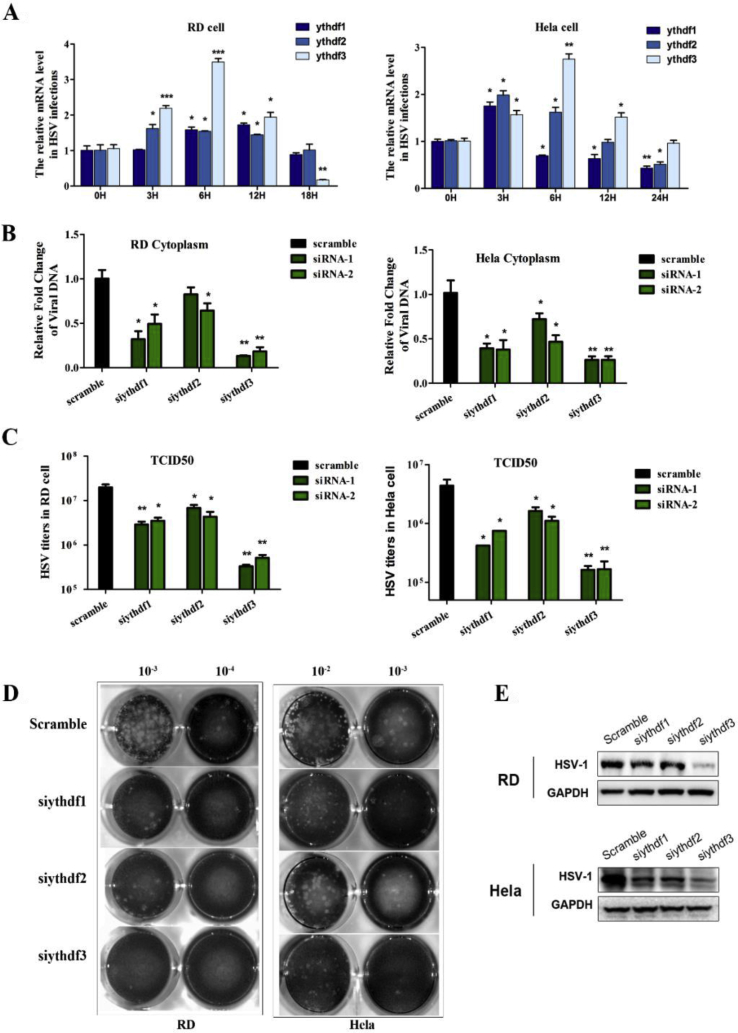
Figure 4.
Inhibition of HSV-1 replication and reproduction by deleting m6A binding protein YTHDFs. (A) The expression pattern of the m6A readers was shown in the HSV-1 infected RD (left) and HeLa (right) cells. The cells were infected with HSV-1 at MOI 5, then the total cellular RNA was isolated at different time points. The mRNA levels of YTHDF1, YTHDF2 and YTHDF3 were determined by RT-qPCR. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (B) RD (left) and HeLa (right) cells were treated with indicated siRNAs and infected with HSV-1 at MOI = 5. To examine the status of virus replication, the intracellular viral DNA level was quantitated by qPCR. ∗P < 0.05, ∗∗P < 0.01. (C) The yield of virus reproduction in RD (left) and HeLa (right) cells was determined by titration of virus titers using TCID50 assay. ∗P < 0.05, ∗∗P < 0.01. (D) Representative photograph of plaques was generated by HSV-1 infections in cells with YTHDF1, YTHDF2 and YTHDF3 knockdown as compared to the control. (E) HSV-1 KITH_HHV1 protein level was detected in HSV-1 infected RD and HeLa cells. Cell were pretreated with siRNAs for 48 h and the cell lysates were prepared 24 h post infection (MOI = 5). GAPDH used as a loading control.
Figure 5.
Inhibition of HSV-1 replication and reproduction by silencing METTL3/METTL14. (A) The expression pattern of the m6A writers in HSV-1 infected RD (left) and HeLa (right) cells. The cells were infected with HSV-1(MOI = 5) for 1 h, the total cellular RNA was collected at different time points after infection. The mRNA levels of METTL3 and METTL14 were determined by RT-qPCR. ∗P < 0.05, ∗∗P < 0.01. (B) METTL3 and METTL14 were knocked down by siRNAs in RD (left) and HeLa (right) cells, and infected with HSV-1 at MOI 5. To measure viral replication status, the viral DNA in cytoplasm was quantitated by qPCR at 24 h. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (C) The infectious virions in RD (left) and HeLa (right) condition media were measured by titration of viral titers using TCID50 assays. ∗P < 0.05, ∗∗P < 0.01. (D) Representative photographs of plaques were generated by HSV-1 infection in METTL3 (middle) and METTL14 (bottom) knockdown cells as compared to the scramble control (top). (E) HSV-1 KITH_HHV1 protein levels in RD and HeLa cells were detected by Western blot assay 24 h after HSV-1 infection at MOI 5. GAPDH was used as a loading control. (F) METTL3 was overexpressed in RD (left) and HeLa (right) cells, and infected with HSV-1 at MOI 5. The viral DNA in cytoplasm was quantitated by qPCR at 24 h. ∗∗∗P < 0.001. (G) As in F, the infectious virions in RD (left) and HeLa (right) cells were measured by titration of viral titers using TCID50 assays. ∗P < 0.05, ∗∗P < 0.01. (H) HSV-1 KITH_HHV1 protein levels in RD and HeLa cells were detected by Western blot assay 24 h after HSV-1 infection at MOI = 5.
To further confirm our hypothesis, we performed loss-of-function assay by knockdown of both FTO and ALKBH5 by specific siRNAs. The knockdown efficiency was around 70% as shown in the supplement data S1. After depletion of the expression of ALKBH5 protein, the virion production increased as much as 1.8 times in RD cells and 2.5 in HeLa cell, respectively (Fig. 3B). As expected, FTO knockdown played less effective role in the HSV-1 replication as compared to the scramble siRNA (Fig. 3C), because FTO already decreased to very low level upon HSV-1 infection. In consistence, plaque assay further confirmed that knockdown of ALKBH5 increased the yield of live virions (Fig. 3D). The viral KITH_HHV1 protein level significantly increased in the ALBKH5-depeleted RD and HeLa cells as compared to the scramble or the FTO-depleted cells (Fig. 3E).
Depletion of m6A readers inhibits HSV-1 replication and reproduction
We then analyzed the mRNA levels of YTHDF1, YTHDF2 and YTHDF3 during the process of viral infection. YTHDF1 and YTHDF2 were upregulated about 2-fold from 3 to 12 h post infection in RD cells. The expression of these two genes were observed a nearly 2-fold increase at 3 h in HeLa cells; then dropped down to about 50% at 24 h with cell-type specific regulation patterns (Fig. 4A).
To study the function of m6A readers in HSV-1 replication, we applied two different siRNAs to knockdown the specific genes in both RD and HeLa cells. The knockdown efficiency was more than 60% as shown in supplement data S2. Except YTHDF2, knockdown of YTHDF1/3 significantly decreased both the intracellular DNA level (Fig. 4B) and the yield of live virus (Fig. 4C). Among the three m6A readers, depletion of YTHDF3 by siRNAs strikingly decreased the intracellular viral DNA levels between 3- to 10-fold of the control in HeLa and RD cells respectively; and reduced the virion production over 100-fold. The same result was obtained in plaque assay. Knockdown of YTHDF3 almost completely eliminated plaque formation (Fig. 4D). The viral KITH_HHV1 protein expression level was significantly decreased in both RD and HeLa cells after silencing each of the YTHDF proteins; particularly, when knockdown of YTHDF3 (Fig. 4E).
Depletion of m6A writers suppresses HSV-1 replication and reproduction
To reveal the regulatory pattern of m6A writers upon HSV-1 infection, we analyzed the mRNA levels of METTL3 and METTL14 during the process of viral infection. As shown in Figure 5A, the mRNA level of METTL3 notably increased by approximately 2-fold as early as 3 h post infection (p.i.), while the mRNA level of METTL14 only slightly increased at the same time point. The expression of both writer genes then quickly decreased thereafter at 6, 9, 12 and 18 h p.i. in both RD and HeLa cells. These results indicated that the writers, particularly METTL3, must have played very important role in HSV-1 infection, and stringently regulated at different stages of infection. It seems that the fast increase of m6A writers is particularly crucial to facilitate the early replication of HSV-1.
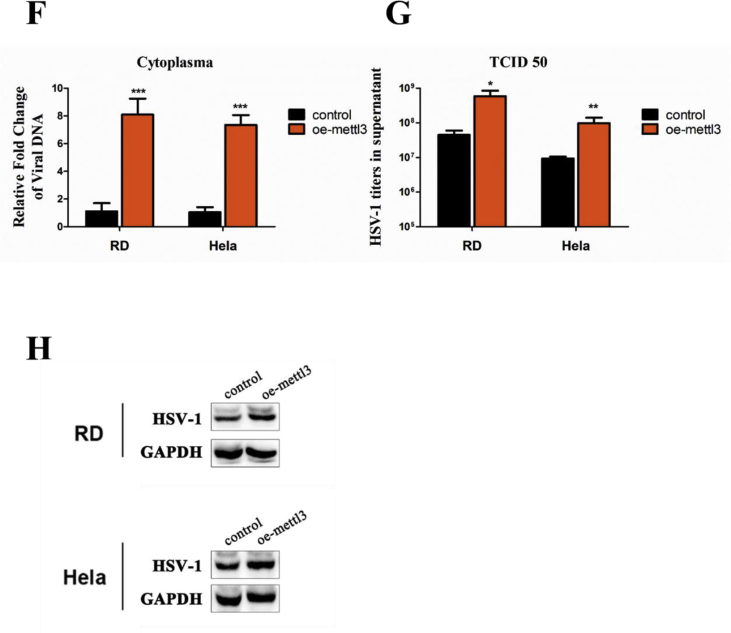
To address our hypothesis, an RNAi approach was employed to knockdown the respective genes. The mRNA level was reduced by more than a half as shown in supplement data S3. RD and HeLa cells were transfected with specific siRNA or scramble siRNA at 40 nM. After transfection for 48 h, the cells were infected with HSV-1 at a MOI of 5. Both the intracellular viral DNA level (Fig. 5B) and the viral titers (Fig. 5C) were respectively decreased by 60–70% and over 30-fold in METTL3-specific siRNA-treated cells when compared with the cells treated with non-targeting sequences (P < 0.01 in RD cells and P < 0.05 in HeLa cells). Obviously, results from TCID50 assay showed that the viral titers decreased near 30-fold after knockdown of METTL3. As compared to METTL3, depleting METTL14 decreased the viral DNA level by 30%–50%, and the viral titers was decreased about 10-fold in RD and HeLa cells. Plaque assay further confirmed our findings and showed that knockdown of METTL3 and METTL14 largely decreased the plaques (Fig. 5D). Strikingly, knockdown of METTL3 almost completely eliminated plaque formation, although the knockdown efficiency was only approximate 50%. Furthermore, the viral KITH_HHV1 protein level also significantly decreased in both RD and HeLa cells after silencing METTL3 or METTL14 in both RD and HeLa cells (Fig. 5E).
Because METTL3 is the initiator of m6A modification and played the fundamental role in initiate HSV-1 replication as shown above, we further investigated the detailed mechanisms of METTL3 in regulating HSV-1 replication. Firstly, we ectopically expressed METTL3 and observe its effect on HSV-1 replication. As shown in Figure 5F and G, both of the intracellular viral DNA level and the and the viral titers largely increased by approximately 10-fold in METTL3-ectopic expressed cells as compared with the control. The viral KITH_HHV1 protein was consistent with the previous result (Fig. 5H). Besides, we have performed MTT assay to detect the cell viability in METTL3 knockdown or overexpression cells. Though some studies revealed that METTL3 might promote cell proliferation in some cancers,42,43 we observed no significant changes in cell proliferation affected by METTL3 knockdown or overexpression (Supplement data S4 and S5).
METTL3 regulates the genes of HSV-1 crucial for viral replication
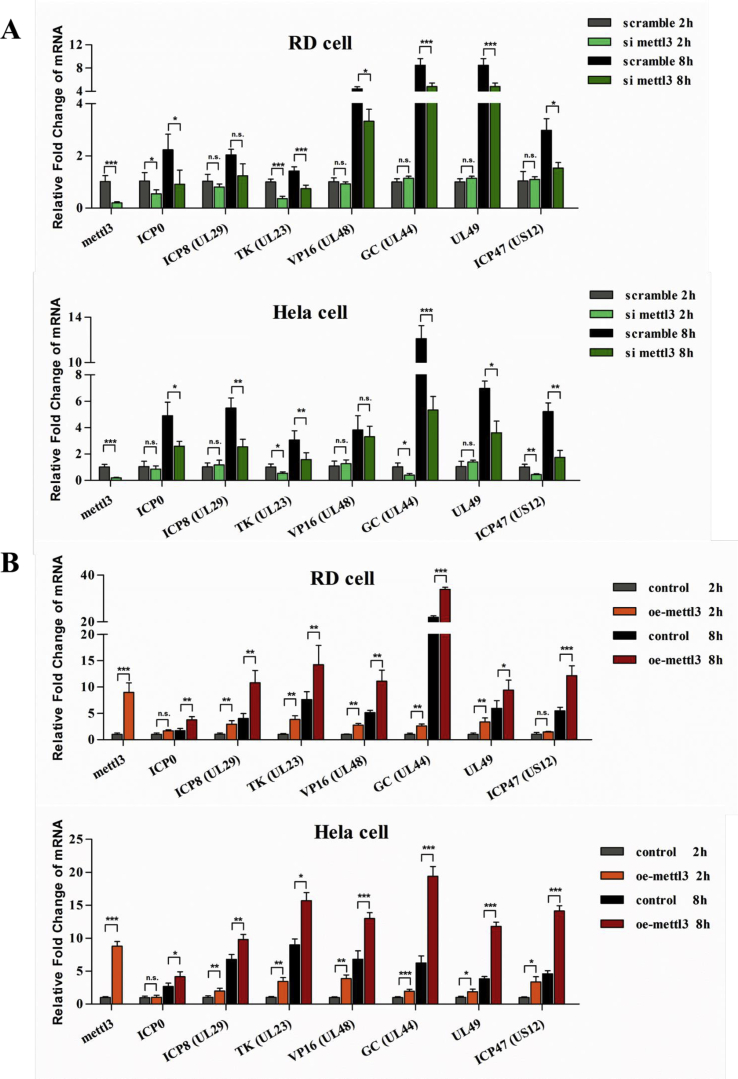
To elucidate the molecular mechanism of METTL3 in promotion of HSV-1 replication, we measured its effects on viral replication-associated genes. ICP0, ICP8 and UL23, UL44, and VP16 are essential HSV genes responsible for viral immediate-early (IE), early (E) and late (L) replication, respectively. We also studied infection-related gene UL49 and ICP47. UL49 is a viral structure protein and play fundamental role in viral protein expression and viral entry.44 ICP47 is a late viral protein responsible for evading host innate immunity.45,46 We silenced or ectopically expressed METTL3 (Fig. 6) in RD and HeLa cells, then collected samples at 2 h and 8 h p.i. and detected mRNA levels of indicated genes were determined by qPCR.
Figure 6.
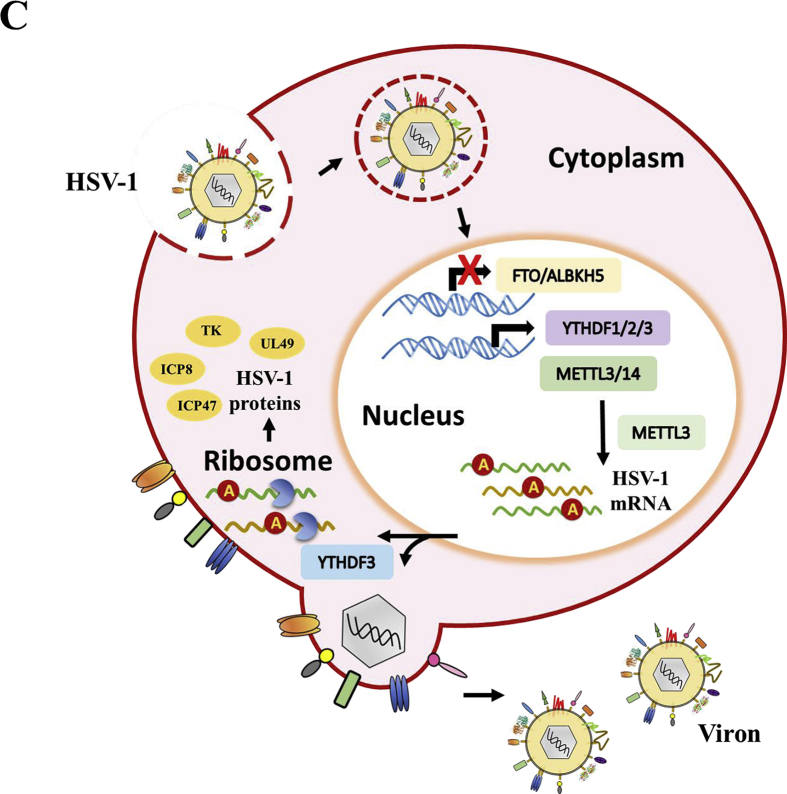
METTL3 regulates genes of HSV-1 important for viral replication. The cells were infected with HSV-1 (MOI = 5) for 1 h, the total cellular RNA was collected at different time points after infection. The mRNA levels were determined by RT-qPCR. The expression pattern of the m6A writers in HSV-1 infected RD (left) and HeLa (right) cells. The cells were infected with HSV-1 (MOI = 5) for 1 h, the total cellular RNA was collected at different time points after infection. The mRNA levels of METTL3 and METTL14 were determined by RT-qPCR. (A, B) Relative fold change of the expression of HSV-1 genes in METTL3 knockdown (A) or METTL3-ectopic exressing cells (B). The level of gene transcription was normalized to 18 S rRNA. n = 4. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (C) The diagram of m6A modification for promotion of HSV-1 replication and reproduction. HSV-1 infection inhibited the transcription of m6A erasers. Meanwhile the mRNA level of m6A writers and readers was increased at first then declined during the infection. In turn, METTL3 induced multiple viral mRNA transcription during virus replication. YTHDF3 normally affected mRNA stabilization and degradation. Both METTL3 and YTHDF3 played an important role in HSV infection.
We showed that the mRNA level of IE genes ICP0 and UL23 increased at least 2-fold from 2 h to 8 h post-infection (Fig. 6A). UL23 encodes a thymidine kinase, which is important to peripheral DNA replication in nerve cells.47,48 Interestingly, the increase levels of UL23 seemed variable. The relative increase of UL23 mRNA levels from 2 h to 8 h in RD cells was much less than that in HeLa cells, possibly reflected cell-type specificity. In METTL3-depleted cells, we observed that the mRNA levels of ICP0 and UL23 significantly decreased approximately 50% and even more at both 2 and 8 at h (p.i.) in both RD and HeLa cell. Although similar reduction of ICP8 mRNA levels was detected at 8 h p.i., there was no such a significant change at 2 h p.i. in METTL3 knockdown cells. The mRNA levels of the late genes VP16, UL44, UL49 and ICP47 greatly increased at least 5- to 8- fold from 2 h to 8 h p.i. in both RD and Hla cells, indicating the importance of these genes for viral activities in the late infection stages. The mRNA level of these genes markedly decreased by 43%–70% at 8 h but not at 2 h p.i. in METTL3 knockdown cells.
Further we compared the indicated genes in the METTL3-ectopic expressed group and the control group. Except ICP0 at 2 h p.i., we showed that all tested viral genes were significantly upregulated in the METTL3-ectopic expressed cells at both 2 and 8 h p.i. (Fig. 6B).
Discussion
The role of m6A modification has attracted great attention in viral infections. A variety of both RNA and DNA viruses, including but not limit to HIV, Zika virus (ZIKV), dengue virus (DENV), West Nile virus (WNV), enterovirus A71(EV A71),49 and some tumor associated virus such as HBV,38 HCV.39 In addition to multiple m6A conserved editing and binding sites on various viral RNA transcripts,27,38,49 virus transcripts as well as other processes of viral life cycle, may be affected by interfering m6A proteins.32,39,49,50 In this study, we revealed the m6A mapping sites of HSV-1 transcripts (Fig. 1), and an inhibitory effect of methylation inhibitor DAA on HSV-1 reproduction (Fig. 2). More importantly, we demonstrated that the early HSV-1 infection suppressed the eraser expression to enhance viral reproduction (Fig. 3); and gradually enhanced the expression of m6A readers and writers to facilitate viral early infection (Figure 4, Figure 5).
Our mapping result from MeRIP-seq analysis showed 12 peaks across the HSV-1 RNA genome covering multiple transcripts (Fig. 1). Among the m6A residues, UL1 encodes glycoprotein L (gL), UL10 encodes glycoprotein M (gM), UL27 encodes glycoprotein B (gB), UL44 encodes glycoprotein C (gC), US5 encodes glycoprotein J (gJ) and US8 encodes glycoprotein E (gE). These proteins are components of virion surface and membrane, play crucial roles in virus-host cells interaction for entry and uncoating (early infection in virus life cycle). UL2 codes for a uracil-DNA glycosylase and UL3 codes for a polypeptide of unknown function.26,51 Notably, UL11 protein is the main contributor of nucleocapsid envelopment at the inner nuclear membrane for virion exit and secondary envelopment.26,52 UL14, UL41, UL46, UL47, US9 and US10 encoded proteins are viral tegument protein necessary for viral replication, especially viral assembly (intermediate and late infection).53,54 VP16 (alpha TIF, Vmw65), encoded by UL48, is a potent transactivator of immediate-early gene expression in HSV-1 infection. VP16 interacts with the host cell transcription factors and is crucial in the upstream HSV-1 promoter activity.55 ICP47, encoded by gene US12, is a polymorphous protein related to virus immune escape. ICP47 directly binds antigen-dependent transporter (TAP), blocking antigen trafficking, causing empty MHC-I.45 Our mapping result indicates that m6A modifications engage in almost every aspect of HSV-1 life cycle.
Upon entry into host cells, HSV-1 actively undergoes a series of transcription of HSV-1 genes, including IE, E and L genes, which are regulated coordinately and sequentially in a cascade fashion. We applied loss- and gain-of-function experiments to reveal the modulation pattern for METTL3 in HSV-1 infection. Our data showed that downregulation of METTL3 declined mRNA level of ICP0, ICP8 and UL23, and UL44 and VP16, belonging to IE, E and L genes, respectively. The expression of UL49 and ICP47, which were vital for HSV infection, were also decreased in METTL3 knockdown cells. Our data suggested a special pattern for m6A gene in regulation of HSV-1 entry, replication and reproduction. DAA is a universal methylation inhibitor well tolerant in vitro and in vivo and exhibits antivirus activity against a wide range of viruses.40,41 Our results revealed that m6A modification enhanced HSV-1 replication, while inhibition of m6A with DAA repressed the virus reproduction.
How m6A modification system affect virus infection has recently been attracting interests of virologists. It has been reported that the proteins involved in m6A modification, including methyltransferase, demethylase and binding proteins, participate in the life cycle of virus. YTHDF proteins, which colocalized with the HCV core protein around lipid droplets in infected cells, negatively regulate HCV particle production.39 Meanwhile, The m6A writer protein METTL3, as well as the reader protein YTHDF2, induced rapid SV40 replication and larger viral plaques in BSC40 cells, while mutational inactivation of those two genes had the opposite effect.56 METTL3 could conduct sumoylation and ubiquitination on EV-A71 RNA-dependent RNA polymerase 3D and boost viral replication.49 In addition, HCV virion assembly mainly occurs in cytosolic lipid droplets closed to endoplasmic reticulum (ER) membranes. Depletion of the m6A methyltransferases METTL3 and METTL14 promoted infectious HCV viral particle production without affecting viral RNA replication, while depletion of the m6A demethylase FTO, but not ALKBH5, has the opposite effect. In this study, we found that the mRNA level of proteins responsible for m6A modification, including the writers, readers and erasers, are regulated by HSV-1 infections. Particularly, the m6A erasers, FTO and alkbh5, are downregulated during the virus infection. However, METTL3 and YTHDF3 are the most significantly upregulated genes in the HSV-1 early infection and then turn down subsequently. These data indicate that HSV-1 infection alters host proteins expression to enhance RNA m6A modifications, which favors the early replication.
It was reported that knockdown of m6A methyltransferases or m6A readers decreased virion replications while the m6A demethylases worked the opposite in EV-A71 and HIV-1 infection.49,50,56,57 However, some studies reported conflicting results during HCV,39 HIV58 and ZIKA59 infections, because silencing METTL3 or FTO enhanced virus replication, probably due to the special requirement for RNA stability or translation, specific protein expression, special intracellular virus synthesis locations and so on. The controversy findings may be caused by the different host cells used or the status of infections in these studies, or strategies used by different viruses at certain stages. In this study, we showed that knockdown of the m6A writers and readers suppressed HSV-1 reproduction; on the contrary, depleting m6A eraser ALKBH5 boosted the viral replication and reproduction. We noted that the FTO was quickly decreased to very low level upon virus infection while exhibited little effects on virus replication and reproduction after silencing FTO, indicating FTO may be the main player in erasing m6A modification of HSV-1 mRNAs. We speculated that's caused by its much higher mRNA levels than FTO after HSV-1 infections. Overall, the m6A methyltransferases, readers and demethylases regulate HSV-1 replication, which is in agree with the recent reports that the m6A machinery regulates EV-A71, HIV and SV40 infection.49,56,58
In summary, our results demonstrated that m6A modification promoted HSV-1 infection, because (1) inhibition of m6A modification by DAA decreased HSV-1 viral production; (2) HSV-1 infection enhanced the expression of m6A writers and readers at early stage while simultaneously suppressed the eraser expression in all the test time points post infection; (3) knockdown of the m6A erasers promoted viral replication and reproduction; and (4) depleting the m6A writer and reader expression inhibited virus replication and reproduction. METTL3 plays crucial role in regulating HSV-1 immediate early and early genes (ICP0, ICP8 and UL23) as well as late genes (VP16, UL44, UL49 and ICP47) at variable degrees. We have demonstrated that the m6A writer METTL3 and reader YTHDF3 are idea targets for treating HSV-1 infections (Fig. 6C). Our results shed the lights on the importance for m6A modification machinery in the regulation of HSV-1 replication, suggesting m6A modification system would be a good target for combating HSV-1 infections. Nevertheless, since HSV-1 contains a relatively large genome coding for more than 70 proteins, further studies are necessary to elucidate the detailed mechanisms as well as the specific role of the methylated mRNA transcripts in virus reproduction and host responses.
Conflict of interests
No conflict of interest
Funding
This work was supported by grants from National Science Foundation of China (No. 81671995); The Science Technology and Innovation Committee of Shenzhen Municipality (No. JCYJ20180507181627057); and Strategic funds from City University of Hong Kong.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2021.02.004.
Contributor Information
Xin Wang, Email: xin.Wang@cityu.edu.hk.
Xin Deng, Email: xindeng@cityu.edu.hk.
Ming-Liang He, Email: mlhe7788@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Nat Acad Sci U S A. 1974;71(10):3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei C.M., Gershowitz A., Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. 1975;4(4):379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 3.Wei C.M., Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977;16(8):1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- 4.Wang X., Lu Z., Gomez A., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei W., Ji X., Guo X., Ji S. Regulatory role of N 6 -methyladenosine (m 6 A) methylation in RNA processing and human diseases. J Cell Biochem. 2017;118(9):2534–2543. doi: 10.1002/jcb.25967. [DOI] [PubMed] [Google Scholar]
- 6.Wei C.M., Gershowitz A., Moss B. 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry. 1976;15(2):397–401. doi: 10.1021/bi00647a024. [DOI] [PubMed] [Google Scholar]
- 7.Batista P.J., Molinie B., Wang J., et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 9.Huang J., Yin P. Structural insights into N6-methyladenosine (m6A) modification in the transcriptome. Genomics Proteomics Bioinformatics. 2018;16(2):85–89. doi: 10.1016/j.gpb.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Huang J., Zou T., Yin P. Human m(6)A writers: two subunits, 2 roles. RNA Biol. 2017;14(3):300–304. doi: 10.1080/15476286.2017.1282025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C., Fu J., Zhou Y. A review in research progress concerning m6A methylation and immunoregulation. Front Immunol. 2019;10:922. doi: 10.3389/fimmu.2019.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Zhao B.S., Roundtree I.A., et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roignant J.Y., Soller M. m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33(6):380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Shi H., Wang X., Lu Z., et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng G., Dahl J.A., Niu Y., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia G., Fu Y., Zhao X., et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y., Jia G., Pang X., et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4:1798. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batista P.J., Molinie B., Wang J., et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widagdo J., Anggono V. The m6A-epitranscriptomic signature in neurobiology: from neurodevelopment to brain plasticity. J Neurochem. 2018;147(2):137–152. doi: 10.1111/jnc.14481. [DOI] [PubMed] [Google Scholar]
- 20.Chen J., Fang X., Zhong P., Song Z., Hu X. N6-methyladenosine modifications: interactions with novel RNA-binding proteins and roles in signal transduction. RNA Biol. 2019;16(8):991–1000. doi: 10.1080/15476286.2019.1620060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu D.F., Koch T., Liang Y.J., et al. Membrane glycoprotein M6a interacts with the micro-opioid receptor and facilitates receptor endocytosis and recycling. J Biol Chem. 2007;282(30):22239–22247. doi: 10.1074/jbc.M700941200. [DOI] [PubMed] [Google Scholar]
- 22.Sekar D., Lakshmanan G. Methylation of N6-adenosine (m6A) modification in miRNAs and its implications in immunity. Epigenomics. 2020;12(13):1083–1085. doi: 10.2217/epi-2020-0131. [DOI] [PubMed] [Google Scholar]
- 23.Han M., Liu Z., Xu Y., et al. Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front Neurosci. 2020;14:98. doi: 10.3389/fnins.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S., Sun C., Li J., et al. Roles of RNA methylation by means of N(6)-methyladenosine (m(6)A) in human cancers. Cancer Lett. 2017;408:112–120. doi: 10.1016/j.canlet.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 25.Brocard M., Ruggieri A., Locker N. m6A RNA methylation, a new hallmark in virus-host interactions. J Gen Virol. 2017;98(9):2207–2214. doi: 10.1099/jgv.0.000910. [DOI] [PubMed] [Google Scholar]
- 26.McGeoch D.J., Rixon F.J., Davison A.J. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117(1):90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Amin I., Vajeeha A., Younas S., et al. HSV-1 infection: role of viral proteins and cellular receptors. Crit Rev Eukaryot Gene Expr. 2019;29(5):461–469. doi: 10.1615/CritRevEukaryotGeneExpr.2019025561. [DOI] [PubMed] [Google Scholar]
- 28.Wan Q., Song D., Li H., He M.L. Stress proteins: the biological functions in virus infection, present and challenges for target-based antiviral drug development. Sign Transduct Target Ther. 2020;5(1):125. doi: 10.1038/s41392-020-00233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danastas K., Miranda-Saksena M., Cunningham A.L. Herpes simplex virus type 1 interactions with the interferon system. Int J Mol Sci. 2020;21(14):5150. doi: 10.3390/ijms21145150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimock K., Stoltzfus C.M. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry. 1977;16(3):471–478. doi: 10.1021/bi00622a021. [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto S.I., Green M. Multiple methylated cap sequences in adenovirus type 2 early mRNA. J Virol. 1976;20(2):425–435. doi: 10.1128/jvi.20.2.425-435.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim G.W., Imam H., Khan M., Siddiqui A. N6-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J Biol Chem. 2020;295(37):13123–13133. doi: 10.1074/jbc.RA120.014260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou F., Wan Q., Lu J., Chen Y., Lu G., He M.L. Pim1 impacts enterovirus A71 replication and represents a potential target in antiviral therapy. iScience. 2019;19:715–727. doi: 10.1016/j.isci.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schremser V., Antoniewicz L., Tschachler E., Geusau A. Polymerase chain reaction for the diagnosis of herpesvirus infections in dermatology : analysis of clinical data. Wien Klin Wochenschr. 2020;132(1-2):35–41. doi: 10.1007/s00508-019-01585-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dan X., Wan Q., Yi L., et al. Hsp27 responds to and facilitates enterovirus A71 replication by enhancing viral internal ribosome entry site-mediated translation. J Virol. 2019;93(9) doi: 10.1128/JVI.02322-18. e02322–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 37.Meng J., Lu Z., Liu H., et al. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69(3):274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imam H., Khan M., Gokhale N.S. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Nat Acad Sci U S A. 2018;115(35):8829–8834. doi: 10.1073/pnas.1808319115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gokhale N.S., Mclntyre A.B.R., McFadden M.J., et al. N6-Methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe. 2016;20(5):654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Clercq E., Montgomery J.A. Broad-spectrum antiviral activity of the carbocyclic analog of 3-deazaadenosine. Antivir Res. 1983;3(1):17–24. doi: 10.1016/0166-3542(83)90011-6. [DOI] [PubMed] [Google Scholar]
- 41.Montgomery J.A., Clayton S.J., Thomas H.J. Carbocyclic analog of 3-deazaadenosine - a novel antiviral agent using S-adenosylhomocysteine hydrolase as a pharmacological target. J Med Chem. 1982;25(6):626–629. doi: 10.1021/jm00348a004. [DOI] [PubMed] [Google Scholar]
- 42.Xiang S., Liang X., Yin S., Liu J., Xiang Z. N6-methyladenosine methyltransferase METTL3 promotes colorectal cancer cell proliferation through enhancing MYC expression. Am J Transl Res. 2020;12(5):1789–1806. [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou L., Yang C., Zhang N., Zhang X., Zhao T., Yu J. Silencing METTL3 inhibits the proliferation and invasion of osteosarcoma by regulating ATAD2. Biomed Pharmacother. 2020;125:109964. doi: 10.1016/j.biopha.2020.109964. [DOI] [PubMed] [Google Scholar]
- 44.Komala Sari T., Gianopulos K.A., Nicola A.V. Glycoprotein C of herpes simplex virus 1 shields glycoprotein B from antibody neutralization. J Virol. 2020;94(5) doi: 10.1128/JVI.01852-19. e01852-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng J.T., Wang Y.Y., Zhu L.Z., et al. Novel transcription regulatory sequences and factors of the immune evasion protein ICP47 (US12) of herpes simplex viruses. Virol J. 2020;17(1):101. doi: 10.1186/s12985-020-01365-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldsmith K., Chen W., Johnson D.C., Hendricks R.L. Infected cell protein (ICP)47 enhances herpes simplex virus neurovirulence by blocking the CD8+ T cell response. J Exp Med. 1998;187(3):341–348. doi: 10.1084/jem.187.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang C.Y., Yao H.W., Wang L.C., Shen F.H., Hsu S.M., Chen S.H. Thymidine kinase-negative herpes simplex virus 1 can efficiently establish persistent infection in neural tissues of nude mice. J Virol. 2017;91(4) doi: 10.1128/JVI.01979-16. e01979–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burrel S., Deback C., Agut H., Boutolleau D. Genotypic characterization of UL23 thymidine kinase and UL30 DNA polymerase of clinical isolates of herpes simplex virus: natural polymorphism and mutations associated with resistance to antivirals. Antimicrob Agents Chemother. 2010;54(11):4833–4842. doi: 10.1128/AAC.00669-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hao H., Hao S., Chen H. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res. 2019;47(1):362–374. doi: 10.1093/nar/gky1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kennedy E.M., Bogerd H.P., Kornepati A.V., et al. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016;19(5):675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klupp B.G., Baumeister J., Karger A., Visser N., Mettenleiter T.C. Identification and characterization of a novel structural glycoprotein in pseudorabies virus, gL. J Virol. 1994;68(6):3868–3878. doi: 10.1128/jvi.68.6.3868-3878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reynolds A.E., Ryckman B.J., Baines J.D., Zhou Y., Liang L., Roller R.J. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol. 2001;75(18):8803–8817. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hernández Durán A., Greco T.M., Vollmer B., Cristea I.M., Grünewald K., Topf M. Protein interactions and consensus clustering analysis uncover insights into herpesvirus virion structure and function relationships. PLoS Biol. 2019;17(6):e3000316. doi: 10.1371/journal.pbio.3000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.You H., Zheng S., Huang Z., Lin Y., Shen Q., Zheng C. Herpes simplex virus 1 tegument protein UL46 inhibits TANK-binding kinase 1-mediated signaling. mBio. 2019;10(3) doi: 10.1128/mBio.00919-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elliott G.D. The extreme carboxyl terminus of the equine herpesvirus 1 homolog of herpes simplex virus VP16 is essential for immediate-early gene activation. J Virol. 1994;68(8):4890–4897. doi: 10.1128/jvi.68.8.4890-4897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai K., Courtney D.G., Cullen B.R. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog. 2018;14(2):e1006919. doi: 10.1371/journal.ppat.1006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kennedy E.M., Bogerd H.P., Kornepati A.V., et al. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016;19(5):675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tirumuru N., Zhao B.S., Lu W., Lu Z., He C., Wu L. Correction: N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife. 2017;6:e31482. doi: 10.7554/eLife.31482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lichinchi G., Zhao B.S., Wu Y., et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. 2016;20(5):666–673. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.